Abstract
Circadian rhythms refer to biologic processes that oscillate with a period of approximately 24 hours. These rhythms are sustained by a molecular clock and provide a temporal matrix that ensures the coordination of homeostatic processes with the periodicity of environmental challenges. We demonstrate the circadian molecular clock controls the expression and function of toll like receptor 9 (TLR9). In a vaccination model using TLR9 ligand as adjuvant, mice immunized at the time of enhanced TLR9 responsiveness presented weeks later with an improved adaptive immune response. In a TLR9-dependent mouse model of sepsis, we found that disease severity was dependent on the timing of sepsis induction, coinciding with the daily changes in TLR9 expression and function. These findings unveil a direct molecular link between the circadian and innate immune systems with important implications for immunoprophylaxis and immunotherapy.
Nearly all organisms have developed mechanisms for anticipating environmental changes in order to optimize their survival. Circadian rhythms are autonomous, self-sustained, ~24 h oscillations in biologic processes entrained by environmental cues, most notably the daily changes in light intensity. Blue light activates a specific group of photoreceptors in the retina connected to the suprachiasmatic nuclei in the hypothalamus, which entrains peripheral circadian clocks via neuroendocrine signals (Bell-Pedersen et al., 2005). At the molecular level, this 24-h rhythmicity is perpetuated by a set of transcription-translation feedback loops controlling the expression of so-called clock genes that comprise the molecular circadian clock (Dibner et al., 2010).
Light and daily rhythms are thought to have a strong influence on immune function (Roberts, 2000). Several studies have described circadian variations in different immune parameters such as lymphocyte proliferation, antigen presentation, and cytokine gene expression (Arjona and Sarkar, 2005; Esquifino et al., 1996). Many of these parameters, including circadian oscillations in cytokine levels, are recapitulated in both mice and humans (Arjona and Sarkar, 2006; Holzheimer et al., 2002; Petrovsky and Harrison, 1998; Petrovsky et al., 1998). It is becoming increasingly evident that disruption of daily rhythms (e.g., sleep deprivation, jet lag) affects the immune response (Bovbjerg, 2003; Majde and Krueger, 2005; Redwine et al., 2004). In addition, the fact that both the autonomic nervous system and the neuroendocrine system (both displaying a strong daily rhythmicity) have been shown to modulate cytokine production, leukocyte trafficking, proliferation, and apoptosis, further supports the notion that the light-dark cycle may influence some immune responses (Petrovsky, 2001). Many components of inflammation (such as pro-inflammatory cytokines) exhibit circadian patterns in humans, typically peaking during the night (Scheff et al., 2010), and several inflammatory diseases such as sepsis (Hrushesky and Wood, 1997) and rheumatoid arthritis (Kowanko et al., 1982) show diurnal variations in their severity.
The recognition of microbial infection by innate immune cells is critical for the overall host immune response (Iwasaki and Medzhitov, 2004). The success of the host immune defense against infections is dependent on the ability of the cells to detect the presence of the invading pathogen (Medzhitov and Janeway, 2000). One the most important advances in immunology has been the identification of an evolutionary conserved family of proteins that recognize conserved pathogen-associated molecular patterns (PAMPs). Microbial molecules can be recognized by these pattern-recognition receptors (PRRs) that are expressed in innate immune cells such as macrophages and dendritic cells (DCs) (Medzhitov, 2001). Upon recognition of microbial components, PRR signaling results in the upregulation of co-stimulatory molecules as well as in the production of type I interferon and inflammatory cytokines, leading to DC maturation, efficient Ag presentation and establishment of adaptive immunity (Akira et al., 2001). Among all the different PRRs described so far, Toll-like receptor 9 (TLR9) is unique in the sense that it recognizes bacterial and viral DNA (Lund et al., 2003) as well as synthetic oligonucleotides. The CpG-motif dependency of TLR9 activation is restricted to synthetic ligands based on phosphorothioate-modified DNA. Natural DNA (phosphodiester sugar backbone) drives TLR9 activation sequence-independently as s the case for RNA recognition mediated by other PRRs (Haas et al., 2008).
The aim of this study was to determine whether innate immune pathogen recognition mechanisms are under circadian control and to assess the tentative consequences in innate and adaptive immune responses. Here, we provide evidence showing that TLR9 expression and function was modulated by core circadian molecular clock components. The circadian regulation of this innate immune pathway shaped subsequent long-term adaptive immune mechanisms. Thus, the magnitude of anamnestic responses in the context of an immunization adjuvanted with a TLR9 ligand correlated with daily variations in TLR9 responsiveness. In a TLR9-dependent model of sepsis, we found that disease severity was dependent on the timing of sepsis induction, which correlated with daily changes in TLR9 expression and function. These findings unveil a direct molecular link between the circadian and innate immune systems with important implications for immunoprophylaxis and immunotherapy.
RESULTS
Defective TLR9-dependent cytokine response in Per2Brdm1 macrophages
We initially hypothesized that the circadian system regulates innate immune responses in order to maximize antimicrobial responses at the time of the day when they are needed most. In order to evaluate whether this would be a common finding among multiple innate immune recognition mechanisms or to the contrary, would be exclusive for a particular innate immune pathway, we isolated peritoneal macrophages from circadian-deficient Per2-mutant mice and challenged them with different pathogen-associated molecular patterns (PAMPs) targeting multiple TLRs. Per2Brdm1 mice carry a mutant Per2 gene with a deletion in the Per-Arnt-Sim (PAS) dimerization domain, which is critical for the interaction with other clock proteins, thus rendering a non-functional PER2 protein and a defective circadian molecular clock (Zheng et al., 1999). Upon challenge with CpG ODNs (TLR9 ligand), Per2Brdm1 macrophages produced significantly less TNF-α and IL-12 than wild type (WT) macrophages (Figure 1A). This finding was restricted to TLR9-mediated responses, as cytokine production in Per2Brdm1 macrophages was comparable to that seen in WT macrophages when PAMPs other than CpG ODNs were used (Figure 1A).
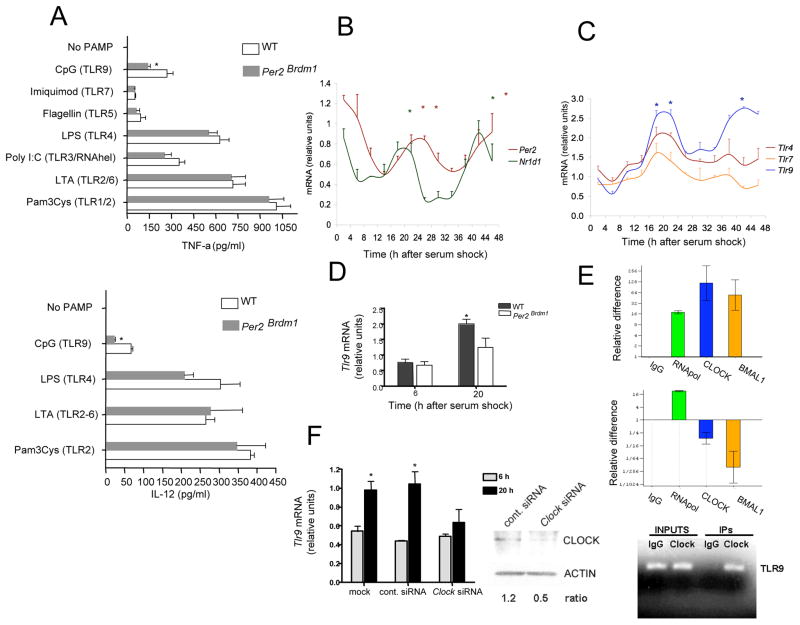
Figure 1. Circadian control of TLR9 expression and function.
(A) WT and Per2Brdm1 thioglycolate-elicited peritoneal macrophages were challenged with PAMPs targeting different TLRs. TNF-α and IL-12 levels in culture supernatants were determined by ELISA 16 h after challenge from 4 independent experiments; *, significantly different from WT. (B) Circadian oscillations of Nr1d1 and Per2 mRNA expression in serum-entrained peritoneal macrophages. *, significantly different from time points showing the lowest mRNA expression. (C) Circadian oscillations of Tlr mRNA expression in serum-entrained peritoneal macrophages, significantly different from T10 and T30 time points. (D) Impaired serum-induced Tlr9 expression in Per2Brdm1 peritoneal macrophages, significantly different from T6. (E) ChiP analysis of CLOCK and BMAL1 binding to the Tlr9 promoter. Tlr9 (top) and Gapdh (bottom) promoter specific sequences were amplified by qPCR from DNA-protein complexes pulled-down with specific antibodies. (F) Impaired Tlr9 expression after siRNA-mediated knockdown of Clock, significantly different from T6. (A–D, F) Bars represent means ± s.e.m. *P < 0.05; significantly different as per two-sided t test. See also Figure S1.
Circadian regulation of Tlr9 expression
We then considered that the circadian clock could be modulating TLR9-dependent cytokine production by controlling Tlr9 expression. The circadian molecular clock can be triggered in vitro by exposing immortalized or primary cells to a high serum concentration (serum shock) (Balsalobre et al., 1998). Triggering of the circadian clock is evidenced by the observation of circadian (near 24-h period) oscillations in the expression of canonical clock genes (Balsalobre et al., 1998). We exploited this system to entrain the circadian clock in murine peritoneal macrophages in vitro and assessed the subsequent temporal variations in gene expression. We detected robust circadian oscillations in the mRNA expression of the canonical clock genes Per2 and Nr1d1 (Figure 1B) after serum shock, indicating that macrophages are equipped with a functional circadian molecular clock. This finding is consistent with other reports using resident peritoneal macrophages and other innate immune cells (Arjona and Sarkar, 2005; Keller et al., 2009). In line with our hypothesis, we found that along with these two clock genes, Tlr9 mRNA expression also fluctuated in a circadian fashion after serum shock treatment, for two significant expression peaks ~24-h apart were detected (Figure 1C). This pattern was not seen when the mRNA expression of Tlr4 or Tlr7 were analyzed. Canonical clock gene expression has previously been analyzed in the spleen of Per2Brdm1 mice over the daily cycle. In these mice, Per1 and Per2 expression rhythms are phase advanced, overall transcript levels of Per1, Per2, and Bmal1 are increased, and Bmal1 expression does not undergo significant daily oscillations (Arjona and Sarkar, 2006). When comparing Tlr9 expression between WT and Per2Brdm1 peritoneal macrophages ~20 h after SS (peak of Tlr9 expression), Tlr9 mRNA expression in Per2Brdm1 macrophages was significantly lower than those in WT macrophages (Figure 1D). Regardless of whether this finding reflects an overall reduction of Tlr9 expression or to a change in its rhythmic parameters, it shows that disruption of the molecular clock affects the Tlr9 mRNA expression profile after SS-induced circadian entrainment.
Circadian transcription factors bind the Tlr9 promoter
The core molecular clock machinery generates circadian outputs by coordinating the expression of the clock-controlled genes (CCGs). Direct regulation of target genes is accomplished by the core circadian transcription factors CLOCK:BMAL1 binding to E-boxes in the promoter region and activating transcription (Ueda et al., 2005). We examined the murine Tlr9 proximal promoter region as described by Schroder et al. (Schroder et al., 2007) and found two putative non-canonical E-boxes (Leclerc and Boockfor, 2005) near the transcription initiation site (Figure S1). The binding of CLOCK:BMAL1 heterodimer to the Tlr9 promoter was examined by conducting ChIP assays in serum-entrained peritoneal macrophages (Figure 1E), for the Tlr9 promoter was amplified by PCR when pulling down with BMAL1 or CLOCK antibodies. In addition, we found that under serum shock conditions, peak Tlr9 mRNA expression in Clock siRNA-transfected macrophages were significantly lower than those found in control siRNA-transfected or mock-transfected macrophages (Clock siRNAs did not have an affect on Tlr9 expression 6 h after serum shock) (Figure 1F), which further supports the notion that CLOCK:BMAL1 regulates Tlr9 expression.
In vivo daily and circadian variations in Tlr9 expression and function
We then evaluated the existence of in vivo daily variations in Tlr9 expression in immune tissue and cells obtained every 4 h from mice entrained to a 12 h light and12 h dark cycle (LD). Splenic Tlr9 mRNA expression showed a daily rhythm that peaked at Zeitgeber time (ZT) 19, coinciding with the mouse active phase (lights off at ZT12; Figure 2A). Of note, the splenic mRNA expression of Tlr4 and Tlr7 did not show substantial daily fluctuations, which is in line with the circadian specificity for TLR9 shown in the in vitro experiments. Tlr9 is highly expressed in antigen-presenting cells such as macrophages, B cells and DCs (Hornung et al., 2002). Figure 2B shows that in macrophages, Tlr9 mRNA expression showed a daily rhythm that peaked at ZT11, whereas in B cells Tlr9 mRNA reached maximum levels from ZT15 through ZT19 (Figure 2C). In contrast, no substantial changes in the expression of Tlr9 mRNA over the daily cycle were seen in DCs (Figure 2D), which indicates that the circadian control of TLR9-mediated responses may be restricted to specific immune cell types. A defining feature of circadian rhythms is that the rhythm persists in the absence of an environmental cue (e.g., light-dark cycle), therefore, we assessed the Tlr9 in vivo expression rhythm in the spleen, macrophages, B cells and DCs under constant darkness (DD). Despite detecting some changes in phase, amplitude or acrophase, the Tlr9 rhythm is maintained in the spleen, macrophages, and B cells (Figure 2E–2G) of mice kept in DD. We verified the clock was functional in the spleen of these animals by assessing canonical clock gene expression (Figure S2A). We did not observe marked daily fluctuations in Tlr9 mRNA expression in DCs from LD mice, (Figure 2D) however, under DD conditions a significant expression peak was detected from circadian time (CT) 15 to CT23 (Figure 2H). The absence of marked Tlr9 mRNA fluctuations in DCs under LD conditions is likely due to light masking. The persistence of 24-h rhythms in Tlr9 mRNA expression in splenic macrophages and B cells obtained from mice kept in DD further demonstrates the circadian nature of these rhythms.
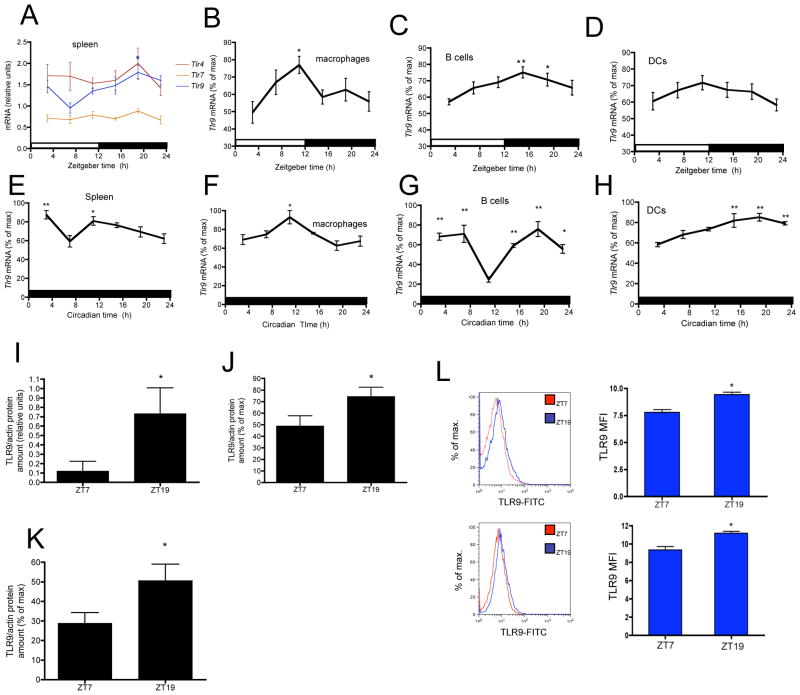
Figure 2. In vivo daily and circadian oscillations of Tlr9 mRNA and protein levels.
Daily oscillations of Tlr mRNA levels in (A) spleen, (B) macrophages, (C) B cells, (D) DCs. Relative mRNA levels at each time point were calculated as the percentage of the maximum value over the 24-h period. Data are mean ± s.e.m. (A) n = 5 mice per time point (B) n = 10 animals per time point, compiled from 2 independent experiments (C) n = 15 animals per time point, compiled from 3 independent experiments (D) n = 15 animals per time point, compiled from 3 independent experiments. Oscillations of Tlr9 mRNA levels under DD in (E) spleen, (F) macrophages (G) B cells (H) DCs. Data are mean ± s.e.m. (E–H) n = 5 mice per time point. (A–H) * P < 0.05, ** P < 0.01, significantly different from lowest level of mRNA (nadir) by one-way ANOVA with the Dunnett posttest. Increased TLR9 protein levels at ZT19 in (I) spleen (J) splenic CD11b+ cells (K) splenic CD19+ cells. TLR9 protein amounts were determined by immunoblot and actin levels were used as normalizing factor. Relative protein amounts were calculated as the percentage of the maximum value. (I) n = 3 animals per time point (J) n = 10 animals per time point, compiled from 2 independent experiments (K) n = 15 animals per time point, compiled from 3 independent experiments. (L) TLR9 levels determined by flow cytometry in MHC-II+ cells (top) and CD19+ cells (bottom). n = 5 animals per time point. (I–L) Data are mean ± s.e.m. * P < 0.05, significantly different from ZT7 as per two-sided t test. See also Figure S2.
Daily variations in TLR9 were also detected at the protein level. TLR9 protein amounts measured by immunoblot were significantly higher at ZT19 in whole spleen lysates, splenic CD11b+ and CD19+ cells isolated by magnetic separation (Figure 2I–2K and S2B) when compared to ZT7. We also observed a significant increase in TLR9 amounts by flow cytometry in MHC-II+ and CD19+ cells isolated from the spleen at ZT19 when compared to those isolated at ZT7 (Figure 2L). Variations in TLR9 protein amounts mimicked those of Tlr9 mRNA expression. Nevertheless, TLR9 protein degradation, not only mRNA synthesis, may also determine the daily variations in TLR9 overall protein amounts, as it has been shown for circadian SUMOylation of BMAL1 (Cardone et al., 2005).
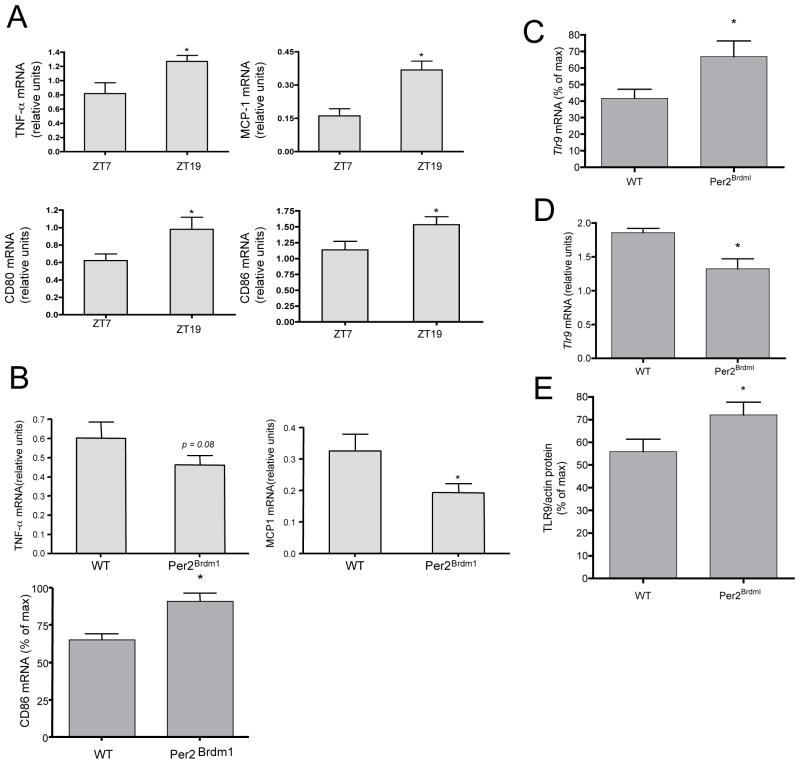
When mice were challenged with CpG ODNs at ZT19 (increased Tlr9 expression; Figure 2A and 2I), they presented with higher splenic mRNA expression of TNF-α, MCP-1, CD80 and CD86 at 2 h after challenge vs. mice that were challenged at ZT7 (Figure 3A). To verify the differences seen in challenged animals were not simply due to an oscillation in baseline levels, we determined that there was no significant difference in mRNA levels of these cytokines and co-stimulatory molecules in unchallenged mice over the daily LD cycle (Figure S3). To evaluate whether these temporal differences in TLR9-mediated innate responses were under circadian control, we also compared the in vivo CpG-induced cytokine response of WT to that of Per2Brdm1 mice. We found that when challenged at ZT19, circadian deficient mice exhibited a reduced CpG-induced cytokine response (TNF-α, MCP-1) but had significantly higher splenic mRNA expression of CD86 (Figure 3B), vs. WT mice. These seemingly discordant results may be due to immune cell specific differences in the circadian regulation of TLR9, as related to both overall expression and rhythmic parameters (e.g., rhythm phase). We also found that Tlr9 mRNA expression was higher in the spleen but lower in splenic macrophages of unchallenged Per2Brdm1 mice vs. WT mice at ZT19 (Figure 3C and 3D). When examining protein amounts between unchallenged WT and Per2Brdm1 mice at ZT19, we found that TLR9 amounts are higher in Per2Brdm1 splenic B cells (Figure 3E). Collectively, these data demonstrate that there are in vivo daily and circadian variations in Tlr9 expression that correlate with TLR9 responsiveness. Importantly, normal TLR9 expression and function is affected by a loss-of-function mutation in Per2, a key component of the circadian molecular clock.
Figure 3. Functional repercussions of the in vivo Tlr9 daily rhythm.
(A) Increased TLR9-mediated cytokine response at ZT19. Cytokine and co-stimulatory molecule mRNA amounts were determined 2 h after CpG challenge, n = 10 mice per time point, compiled from 2 independent experiments. (B) TLR9-mediated cytokine and co-stimulatory response in WT and Per2Brdm1 mice. Cytokine and co-stimulatory molecule mRNA amounts in the spleen were determined 2 h after CpG. CD86 mRNA amount plotted as the percentage of the maximum value, n = 10 animals per time point, compiled from 2 independent experiments. (C) Increased splenic Tlr9 mRNA expression in Per2Brdm1 mice at ZT19. Relative mRNA amounts were calculated as the percentage of the maximum value, n = 8 animals per group compiled from 2 independent experiments. (D) Decreased Tlr9 mRNA expression in splenic macrophages isolated from Per2Brdm1 mice at ZT19, n = 5. (E) Increased TLR9 protein amounts in splenic CD19+ cells in Per2Brdm1 at ZT19, n = 5. (C–D) TLR9 protein amounts were determined by immunoblot. Actin amounts were used as normalizing factor. (A–E) Data are mean ± s.e.m. * P < 0.05, significantly different as per two-sided t test. See also Figure S3.
Circadian control of TLR9 ligand-adjuvanted vaccine responsiveness
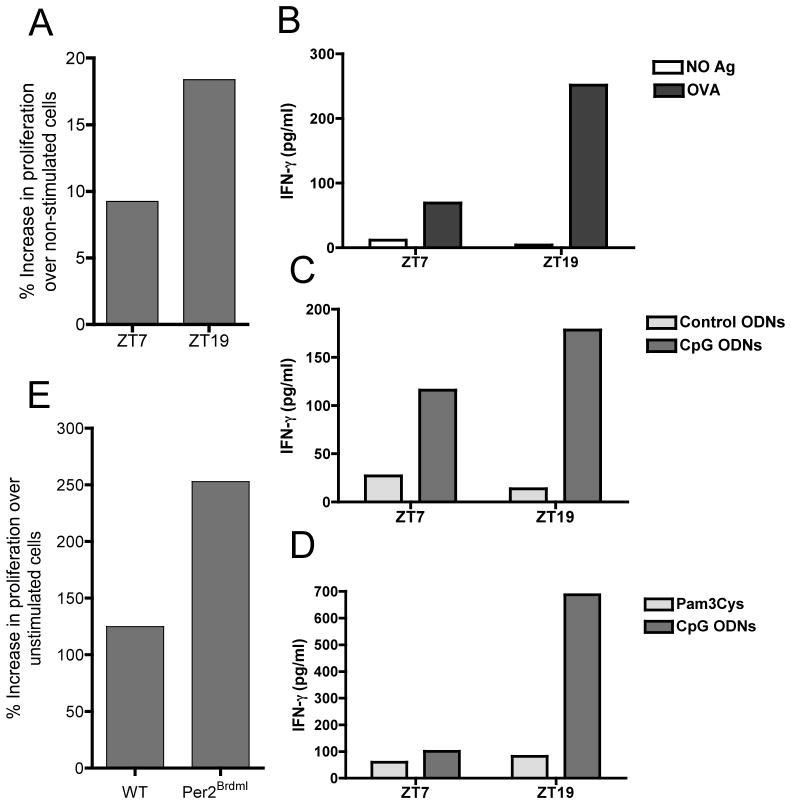
Innate immune activation is critical for mounting an effective adaptive immune response (Medzhitov and Janeway, 1999), and CpG ODNs are being evaluated in the clinic for their adjuvant potential (Huang et al., 2008; Kline, 2007; Vollmer and Krieg, 2009). We examined whether vaccine responsiveness could be enhanced by conducting a CpG ODN-adjuvanted immunization at the time of increased TLR9 expression and function. Mice were injected intraperitoneally with a mixture of OVA and CpG-ODNs at either ZT7 or ZT19, and received a boost 14 days later at their corresponding time point. Four weeks after the first immunization, lymph nodes were harvested and total lymph node cells were cultured to measure OVA-induced proliferation and IFN-γ production. We found that lymphocyte cultures obtained from mice that were immunized 4 weeks earlier with OVA-CpG ODNs at ZT19 (increased TLR9 responsiveness), showed a substantially increased Ag-induced proliferation and IFN-γ production compared to mice that were immunized at ZT7 (Figure 4A and 4B). This phenomenon was reproduced in multiple independent experiments. Of note, this difference was not seen when control ODNs or Pam3Cys (a PAMP that is not detected by TLR9) were used as adjuvants (Figure 4C and 4D). We also immunized WT and Per2Brdm1 mice with OVA-CpG ODNs at ZT19 to determine if their adaptive immune responses differed. Lymphocyte cultures obtained from OVA-CpG immunized Per2Brdm1 mice showed an increased Ag-induced proliferation when compared to WT mice (Figure 4E), which correlated with increased TLR9 protein amounts in Per2Brdm1 mice B cells (Figure 3E). These results show that the daily variations in TLR9 responsiveness have long-term repercussions in the magnitude of the adaptive immune response, which is also modulated by Per2, a canonical component of the circadian molecular clock.
Figure 4. Enhanced adaptive immunity in mice vaccinated at ZT19 when using a TLR9 ligand as adjuvant.
(A,E) Enhanced lymphocyte proliferation in total lymph node cell cultures obtained from mice immunized with CpG-OVA at ZT19 compared to ZT7 or WT and Per2Brdm1 mice at ZT19. Bars represent the average of 4 replicates from lymphocyte cultures pooled from 5 mice per treatment and time point. Results were reproduced in multiple independent experiments. (B–D) Enhanced OVA-induced IFN-γ production in lymphocyte cultures obtained from mice challenged with CpG-OVA at ZT19 compared to ZT7. Mice were immunized with OVA-CpG (B) OVA-control ODNs or OVA-CpG ODNs (C) and OVA-Pam3cys or OVA-CpG (D) at the indicated time points. Bars represent the average of 4 replicates from lymphocyte cultures pooled from 5 mice per treatment and time point. Results were reproduced in 3 independent experiments.
Diurnal variations in disease severity in a TLR9-dependent mouse model of sepsis
Next, we investigated if the circadian regulation of TLR9 expression could mediate inflammatory disease in vivo. During sepsis, an infection leads to a systemic inflammatory response syndrome (SIRS), which is harmful to the host. Septic patients have a heightened risk of mortality between the hours of 2 and 6 am (Hrushesky and Wood, 1997). However, the reasons for this are unknown. We examined whether daily rhythms affect disease pathogenesis in the mouse cecal ligation and puncture (CLP) model, wherein the mouse cecum is punctured allowing commensal bacteria to escape into the peritoneum, leading to SIRS and polymicrobial sepsis. This model was previously shown to be TLR9-dependent (Plitas et al., 2008; Yasuda et al., 2008); hence, we reasoned that the daily variations in Tlr9 expression and function might affect sepsis pathogenesis following CLP.
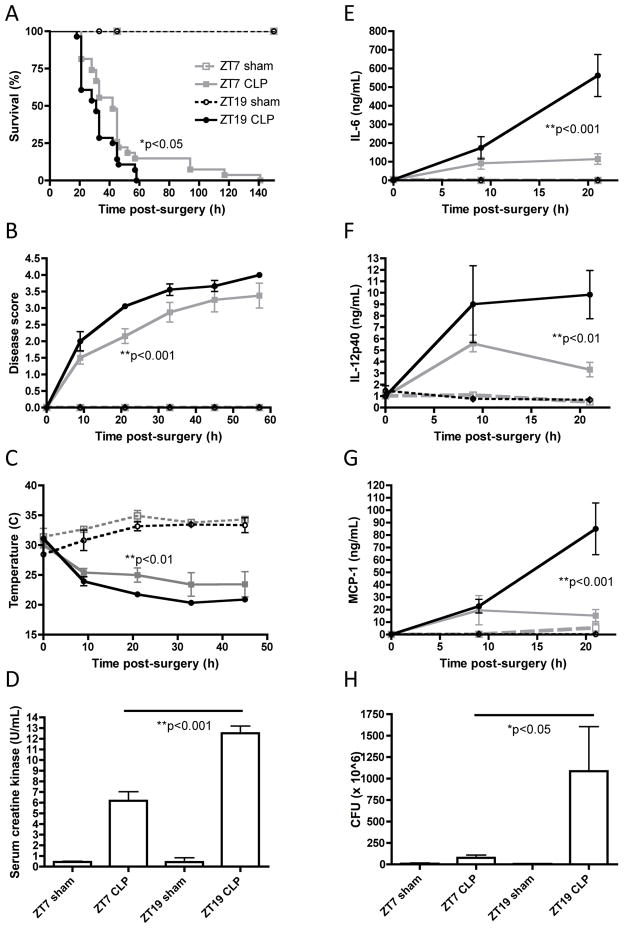
We performed CLP surgeries and sham controls at the nadir and peak of splenic Tlr9 expression (ZT7 and ZT19, respectively). Mice that underwent sham surgeries at either ZT7 or ZT19 survived and exhibited no signs of sepsis (Figure 5A–5H). In line with our hypothesis, mice that underwent CLP at ZT19 exhibited a worse sepsis phenotype vs. mice that underwent CLP at ZT7, including: earlier mortality (Figure 5A, ZT-19 CLP MST=31h, n=28; ZT-7 CLP MST=45h, n=27), worse disease score (Figure 5B), more severe hypothermia (Figure 5C), higher concentrations of serum creatine kinase (Figure 5D, indicative of heart or skeletal muscle damage), higher concentrations of inflammatory mediators in the serum, including cytokines IL-6 and IL-12p40, and chemokine MCP-1 (Figure 5E–5G), and greater numbers of bacterial CFU in the peritoneum (Figure 5H).
Figure 5. Daily variations in TLR9 responsiveness influence disease severity in the TLR9-dependent CLP mouse model of sepsis.
A more severe sepsis pathophysiology ensued in mice that underwent CLP at ZT19 vs. ZT7 including: (A) earlier mortality; (B) worse disease score (disease was scored as follows: 0 = bright, alert, responsive; 1 = slightly lethargic; 2 = lethargic and hunched; 3 = very lethargic, hunched and shaky; 4 = dead) (C) more severe hypothermia; (D) higher concentration of serum creatine kinase; higher concentration of serum inflammatory mediators including (E) IL-6; (F) Il-12/23p40 and (G) MCP-1 and (H) higher amounts of bacterial CFU in the peritoneum.
DISCUSSION
Our findings unveil a direct molecular link between the circadian system and the innate immune system. A nonfunctional Per2 gene affects TLR9 expression, responsiveness and absolute levels of the adaptive response when a TLR9 ligand is employed as an adjuvant. We showed that the circadian clock controlled the expression of the pattern recognition receptor TLR9, and this regulation translated to daily variations in TLR9 responsiveness as measured by cytokine response and expression of co-stimulatory molecules. TLR9 is highly expressed in various types of antigen-presenting cells (Hornung et al., 2002), and our data indicate that the circadian regulation and TLR9 rhythm phase is unique to each immune cell type and different from the overall rhythm found in the spleen. Of note, DCs did not exhibit significant changes in Tlr9 mRNA expression throughout the daily cycle. After challenge with CpG ODNs, WT mice had an increased cytokine response but lower CD86 mRNA expression than Per2Brdm1 mice, and Per2 mutation affected Tlr9 mRNA and protein expression differently depending on the tissue or cell type analyzed, reflecting the intricate web woven between the circadian clock and TLR9 expression and function in each of these specific immune cells. Further contributing to this in vivo complexity, some immune cells may not possess a functional circadian clock yet it remains possible that PER2 could have a clock-independent influence on the TLR9 pathway. It is likely that Per2 and other canonical components of the molecular clock may modulate the expression and function of TLR9 in different ways depending on the immune cell type.
The circadian system may target other innate immune pathways depending on the immune cell type or on the activation status. For example, transcriptional analyses in resident peritoneal macrophages have revealed circadian fluctuations in several aspects of the TLR4-LPS response pathway (Keller et al., 2009). Although the latter does not provide direct evidence for downstream signaling effectors of the LPS-TLR4 response pathway being directly controlled by canonical molecular clock components, it clearly shows that circadian mechanisms act at multiple levels in innate response pathways, including cytokine expression. While Keller et al. assessed immune response rhythms after challenge with a PAMP (LPS), in this manuscript we have focused on the basal rhythms in the expression and function of the sensing molecule TLR9 and provide direct evidence for its circadian control. Thus, circadian mechanisms seem to regulate immune response pathways at multiple levels and both in basal and post-challenge conditions. Perhaps an important role for this circadian control is to ensure the temporal coincidence of all the necessary molecular components of the response, from those needed to sense the pathogen to those required for providing the effector molecules. In 1960, Halberg and colleagues reported that the mortality upon LPS-induced endotoxic shock in mice depends highly on the time of day when the mice are challenged, which further suggests that other immune response pathways triggered by pathogen recognition may be also subject to circadian control (Halberg et al., 1960).
Exposure to microbial pathogens may be higher at certain times of the day due to the intrinsic 24-hour rhythms of the host (activity, feeding and behavior). Moreover, many pathogens that cause disease throughout the world such as flaviviruses and Plasmodium falciparum, are transmitted by mosquitoes, which show a marked daily rhythm in their biting activity (Githeko et al., 1996). It is therefore plausible that TLR9 and possibly other components of the innate immune system have evolved a circadian regulation in order to maximize its efficiency at the time of the day when the encounter with the pathogen is more likely to occur. In contrast, arthropod-borne pathogens may take advantage of the vector’s activity rhythms in order to infect the host when innate response mechanisms are at the lowest efficiency.
In support of the translational potential of our findings into the clinic, we have identified up to 3 non-canonical E-boxes in the human Tlr9 promoter at positions −513 (CAGGTG), −386 (CAGCTG), and −148 (CATTTG). Studies assessing the possible circadian control of Tlr9 expression in humans are warranted. The results described herein also demonstrate that the daily variations in TLR9 responsiveness modulate the efficacy of CpG ODN-adjuvanted immunization. Several TLR9 agonists are undergoing extensive clinical evaluation as adjuvants and immunomodulators that could be useful for treatment of important conditions such as multiple sclerosis, asthma and cancer(Vollmer and Krieg, 2009). Considering not only the dosing and immunization timing but also the degree of proper circadian entrainment in patients may be critical for assessing and maximizing the prophylactic and therapeutic efficacy of these agents.
Our findings with the TLR9-dependent CLP model indicate that the daily variations in TLR9 expression and function are correlated with the progression and severity of sepsis. Given the observation that septic patients are at increased risk of mortality between the hours of 2 and 6 am (Hrushesky and Wood, 1997), future studies are warranted to determine if human patients also exhibit circadian regulation of TLR9 and other aspects of the innate immune response (several PRRs other than TLR9 would be concurrently activated during sepsis). If this proves to be the case, septic patients might benefit from receiving immunomodulatory therapies during the hours when they are most vulnerable. Furthermore, patients in the ICU often have disturbed sleep patterns, due to noise, nocturnal light exposure and medications; it will be important to investigate how these factors influence TLR9 expression levels and innate immune responses.
EXPERIMENTAL PROCEDURES
Animals
2–3 month old C57BL/6J and Per2 mutant (mPer2Brdm1) male mice (Jackson ImmunoResearch Laboratories) were fed rodent chow ad libitum, maintained under constant environmental conditions and entrained to a 12 h light/12 h dark cycle (light period from 7:00 a.m. to 7:00 p.m.) for at least 2 weeks before experiments. The Per2Brdm1 mutant strain was originally developed by Dr. Allan Bradley and has a genetic background of C57BL/6 (Zheng et al., 1999). For most daily-cycle experiments, animals were euthanized at 6:00 a.m., 10:00 a.m., 2:00 p.m., 6:00 p.m., 10:00 p.m., and 2:00 a.m. These time points correspond with Zeitgeber times (ZT) 3, 7, 11, 15, 19, and 23, respectively. For experiments under constant conditions, dark/dark (DD), animals were held in constant darkness for 3 days prior to sampling at circadian times (CT) 3, 7, 11, 15, 19 and 23. Tissues were immediately collected for further processing as described below. During the study, animal care and treatment complied with National Institutes of Health policy, were in accordance with institutional guidelines, and were approved by the Yale Institutional Animal Care and Use Committee.
Cells
Peritoneal macrophages from C57BL/6J (Jackson ImmunoResearch Laboratories), or Per2Brdm1 of the same genetic background were obtained 3–4 days after i.p. injection with 3% thioglycollate broth (REMEL) by peritoneal lavage. Macrophages were purified by adherence following standard procedures and cultured overnight in RPMI 1640 containing 10% FBS plus penicillin-streptomycin (Invitrogen Life Technologies).
In vitro entrainment
Cells were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% FBS and transferred to a 6-well cell culture plate (BD Biosciences) were they were incubated overnight. The following day the medium was aspirated, cells were washed with 1 ml of warm PBS, and incubated for 2 h in 2 ml of serum-free medium. At the conclusion of 2 h the cells were serum shocked with 1 ml of FBS (50% final concentration). The medium containing 50% serum was replaced with serum-free medium after 2 h. Cells were harvested every 4 h, lysed in the appropriate buffer and stored at −80° C until their later use for RNA extraction or Western blot analysis.
PAMP challenge and cytokine measurements
Thioglycollate-elicited peritoneal macrophages from WT and Per2Brdm1 mice were plated and challenged the following day with pam3Cys (500 ng/ml), lipoteichoic acid (LTA, 10 μg/ml), poly(I:C) (100 μg/ml), LPS (100 ng/ml), flagellin (5 μg/ml), imiquimod (10 μg/ml) or unmethylated CpG-DNA (10 μg/ml) - synthetic oligonucleotides containing unmethylated CpG dinucleotides in particular sequence contexts. We employed CpG ODNs from Invivogen (ODN 1826), which are type B specific for mouse TLR9. The 20 mer sequence is 5′-tccatgacgttcctgacgtt-3′. TNF-α and IL-12 levels in culture supernatants were determined by ELISA 16 h after challenge using commercially available ELISA kits (R&D Systems).
Cell isolation and purification
Spleens were collected in RPMI 1640 (Invitrogen Life Technologies) supplemented with 10% FBS and placed on ice. To obtain homogeneous splenocyte suspensions, each spleen was homogenized in 1 ml of RPMI, filtered through a 70 μM nylon cell strainer (BD Biosciences) and spun down for 5 min at 1,200 rpm. The supernatant was removed and cells were resuspended in 3 ml red cell lysing buffer (Sigma) and incubated for 10 min at room temperature. 7 ml of PBS were added to the cell suspension and cells were spun down for 5 min at 1,200 rpm. The supernatant was removed, cells were resuspended in 3 ml PBS and aliquoted into separate tubes for labeling with microbeads and subsequent cell separation. CD11b, CD11c and CD19 mouse microbeads (Miltenyi Biotec) were used to label splenic macrophages, dentritic cells and B cells, respectively. The labeling was followed according to manufacturers instructions and the autoMACS™ Pro Separator (Miltenyi Biotec) was utilized for the magnetic cell separation. The purity of the enriched fractions was assessed by flow cytometry using FITC-conjugated CD11b, CD11c and CD19 antibodies (Miltenyi Biotech). The enrichment method consistently yielded a purity of approximately 90, 60 and 80% for CD11b, CD11c and CD19 positive cells, respectively. Immediately after separation, enriched cells were lysed in appropriate buffers for later RNA extraction and protein analysis. For flow cytometry, cells were fixed and permeabilized before staining with purified fluorescent monoclonal anti-TLR9 antibody from Imgenex (clone 26C593.2).
RNA extraction and quantitative PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen). cDNA was synthesized using the high capacity cDNA reverse transcription kit (Applied Biosystems). Relative quantitation of mRNA levels was performed by quantitative PCR via TaqMan Gene Expression Assays (Applied Biosystems) using a real-time PCR 7500 Fast System (Applied Biosystems). Analyses were performed using the standard curve method with β-actin as the normalizing endogenous control. RNA from enriched cells was isolated using the RNeasy Plus-Micro kit (Qiagen). Due to low expression levels and a limited amount of cDNA, the TaqMan PreAmp Master Mix Kit (Applied Biosystems) was used in conjunction with TaqMan Gene Expression Assays (Applied Biosystems) to examine mRNA levels of selected genes from individual cell types. The procedure was followed according to manufacturers instructions.
Protein isolation and Western blot analysis
For protein analysis, spleen tissue and cells were lysed in T-PER and M-PER Protein Extraction Reagent (Thermo Scientific), respectively. Protein samples were subjected to standard SDS-PAGE electrophoresis, transferred to a nitrocellulose membrane, and probed with TLR9 and beta Actin polyclonal Ab (Imgenex). After chemiluminescence detection using ECL Western blot detection kit (Amersham), densitometry analyses were performed using ImageJ64 software.
ChIP, EMSA, and si-RNA-mediated knockdown of Clock
ChIP was performed using EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore) and EMSA was performed using the LightShift® Chemiluminescent EMSA Kit (Thermo Scientific). The procedures were followed according to manufacturers instructions. Clock expression was knocked down using Clock siRNA (Santa Cruz Biotechnology) and the transfection protocol was followed according to manufacturers instructions (Santa Cruz Biotechnology). Clock antibody (Santa Cruz Biotechnology) was used to verify knockdown in CLOCK protein levels.
Immunization experiments
Mice entrained to a 12 h light/12 h dark cycle were injected intraperitoneally with 10 μg OVA and 30 μg CpG ODNs (InvivoGen) at either ZT7 or ZT19. Two weeks later, mice received a boost (10 μg OVA and 30 μg CpG ODNs) at their corresponding time. Twenty-eight days after the initial OVA CpG ODN challenge, mice were sacrificed and lymph nodes were pooled, according to time of immunization or mouse strain, in cell culture medium (RPMI supplemented with 10 % FBS). Lymph nodes were homogenized and passed through a 70 μM nylon cell strainer (BD Biosciences). Lymphocytes were spun down, resuspended in 10 ml of medium and counted using a hemocytometer. 96 well U-bottom tissue culture plates (BD Biosciences) were seeded with 105 or 106 cells per well for lymphocyte proliferation or IFN-γ production, respectively, in 200 μl of medium. Cells were allowed to incubate for 1 h in a 37° C CO2 incubator and then cells were either challenged with 20 μg of OVA, 1 μg of Concanavalin A (Sigma), or were left unstimulated. Lymphocyte proliferation was determined using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega). IFN-γ was determined using the Mouse IFN-γ High Sensitivity ELISA (eBioscience).
CLP and related measurements
CLP was performed as previously described(Rittirsch et al., 2009) (75% of the cecum was ligated and pierced with a 21G needle). Mice received 1 ml warmed saline s.c. post-operatively and buprenorphine analgesia at 12h intervals. Surface temperature was determined with an infrared thermometer. Blood was collected by retro-orbital bleed and serum was prepared in microtainer tubes (Becton Dickinson, San Jose, CA). ELISA kits for IL-6, IL-12/23p40, and MCP-1 (eBiosciences, San Diego, CA) and the CK reagent set (Pointe Scientific, Canton, MI) were used per manufacturer’s instructions. Peritoneal lavage was plated on plated on BBL agar plates to determine bacterial CFU.
Statistical analysis
Data were analyzed using GraphPad Prism 4.0b (GraphPad, San Diego, CA). For parametric comparisons, we employed a two-way T test for pair-wise comparisons and the analysis of variance test for repeated measures. For nonparametric comparisons, we employed Mann-Whitney for pairwise comparisons and Kruskal-Wallis for comparison of three or more groups. Survival curves were compared by the Logrank test. *P < 0.05 was considered to be statistically significant, **P < 0.01 highly significant. Unless otherwise stated, P values result from the statistical comparison of mean values obtained from independent experiments.
Supplementary Material
Acknowledgments
We thank Manchuan Chen for technical assistance with the qPCR and Western blot analysis, Kathleen DePonte and Deborah Beck for assistance with the animal work, Aaron Bozzi for help with the sepsis model. This work was supported by National Institutes of Health Grants AI079360 and AI50031 (to EF), and NIH Training Grant, T32 AI07019 (to AS) and in part by N01 HHSN272201100019C. EF is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nature immunology. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- Arjona A, Sarkar DK. The circadian gene mPer2 regulates the daily rhythm of IFN-gamma. J Interferon Cytokine Res. 2006;26:645–649. doi: 10.1089/jir.2006.26.645. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovbjerg DH. Circadian disruption and cancer: sleep and immune regulation. Brain, behavior, and immunity. 2003;17(Suppl 1):S48–50. doi: 10.1016/s0889-1591(02)00066-1. [DOI] [PubMed] [Google Scholar]
- Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science (New York, NY) 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Esquifino AI, Selgas L, Arce A, Maggiore VD, Cardinali DP. Twenty-four-hour rhythms in immune responses in rat submaxillary lymph nodes and spleen: effect of cyclosporine. Brain, behavior, and immunity. 1996;10:92–102. doi: 10.1006/brbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Githeko AK, Adungo NI, Karanja DM, Hawley WA, Vulule JM, Seroney IK, Ofulla AV, Atieli FK, Ondijo SO, Genga IO, et al. Some observations on the biting behavior of Anopheles gambiae s.s., Anopheles arabiensis, and Anopheles funestus and their implications for malaria control. Experimental parasitology. 1996;82:306–315. doi: 10.1006/expr.1996.0038. [DOI] [PubMed] [Google Scholar]
- Haas T, Metzger J, Schmitz F, Heit A, Muller T, Latz E, Wagner H. The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity. 2008;28:315–323. doi: 10.1016/j.immuni.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine (New York, NY) 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- Holzheimer RG, Curley P, Saporoschetz IB, Doherty JM, Mannick JA, Rodrick ML. Circadian rhythm of cytokine secretion following thermal injury in mice: implications for burn and trauma research. Shock. 2002;17:527–529. doi: 10.1097/00024382-200206000-00015. [DOI] [PubMed] [Google Scholar]
- Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, Endres S, Hartmann G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- Hrushesky WJ, Wood PA. Circadian time structure of septic shock: timing is everything. The Journal of infectious diseases. 1997;175:1283–1284. [PubMed] [Google Scholar]
- Huang CF, Wu TC, Chu YH, Hwang KS, Wang CC, Peng HJ. Effect of neonatal sublingual vaccination with native or denatured ovalbumin and adjuvant CpG or cholera toxin on systemic and mucosal immunity in mice. Scandinavian journal of immunology. 2008;68:502–510. doi: 10.1111/j.1365-3083.2008.02172.x. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nature immunology. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline JN. Immunotherapy of asthma using CpG oligodeoxynucleotides. Immunologic research. 2007;39:279–286. doi: 10.1007/s12026-007-0083-2. [DOI] [PubMed] [Google Scholar]
- Kowanko IC, Knapp MS, Pownall R, Swannell AJ. Domiciliary self-measurement in the rheumatoid arthritis and the demonstration of circadian rhythmicity. Annals of the rheumatic diseases. 1982;41:453–455. doi: 10.1136/ard.41.5.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc GM, Boockfor FR. Pulses of prolactin promoter activity depend on a noncanonical E-box that can bind the circadian proteins CLOCK and BMAL1. Endocrinology. 2005;146:2782–2790. doi: 10.1210/en.2005-0100. [DOI] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. The Journal of experimental medicine. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majde JA, Krueger JM. Links between the innate immune system and sleep. J Allergy Clin Immunol. 2005;116:1188–1198. doi: 10.1016/j.jaci.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nature reviews. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C., Jr Innate immunity. The New England journal of medicine. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Innate immune induction of the adaptive immune response. Cold Spring Harb Symp Quant Biol. 1999;64:429–435. doi: 10.1101/sqb.1999.64.429. [DOI] [PubMed] [Google Scholar]
- Petrovsky N. Towards a unified model of neuroendocrine-immune interaction. Immunology and cell biology. 2001;79:350–357. doi: 10.1046/j.1440-1711.2001.01029.x. [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Harrison LC. The chronobiology of human cytokine production. Int Rev Immunol. 1998;16:635–649. doi: 10.3109/08830189809043012. [DOI] [PubMed] [Google Scholar]
- Petrovsky N, McNair P, Harrison LC. Diurnal rhythms of pro-inflammatory cytokines: regulation by plasma cortisol and therapeutic implications. Cytokine. 1998;10:307–312. doi: 10.1006/cyto.1997.0289. [DOI] [PubMed] [Google Scholar]
- Plitas G, Burt BM, Nguyen HM, Bamboat ZM, DeMatteo RP. Toll-like receptor 9 inhibition reduces mortality in polymicrobial sepsis. The Journal of experimental medicine. 2008;205:1277–1283. doi: 10.1084/jem.20080162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine L, Dang J, Irwin M. Cellular adhesion molecule expression, nocturnal sleep, and partial night sleep deprivation. Brain, behavior, and immunity. 2004;18:333–340. doi: 10.1016/j.bbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JE. Light and immunomodulation. Ann N Y Acad Sci. 2000;917:435–445. doi: 10.1111/j.1749-6632.2000.tb05408.x. [DOI] [PubMed] [Google Scholar]
- Scheff JD, Calvano SE, Lowry SF, Androulakis IP. Modeling the influence of circadian rhythms on the acute inflammatory response. J Theor Biol. 2010;264:1068–1076. doi: 10.1016/j.jtbi.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Schroder K, Lichtinger M, Irvine KM, Brion K, Trieu A, Ross IL, Ravasi T, Stacey KJ, Rehli M, Hume DA, Sweet MJ. PU.1 and ICSBP control constitutive and IFN-gamma-regulated Tlr9 gene expression in mouse macrophages. Journal of leukocyte biology. 2007;81:1577–1590. doi: 10.1189/jlb.0107036. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature genetics. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Advanced drug delivery reviews. 2009;61:195–204. doi: 10.1016/j.addr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Leelahavanichkul A, Tsunoda S, Dear JW, Takahashi Y, Ito S, Hu X, Zhou H, Doi K, Childs R, et al. Chloroquine and inhibition of Toll-like receptor 9 protect from sepsis-induced acute kidney injury. American journal of physiology. 2008;294:F1050–1058. doi: 10.1152/ajprenal.00461.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.