Abstract
Motivation: Polychromatic flow cytometry (PFC), has enormous power as a tool to dissect complex immune responses (such as those observed in HIV disease) at a single cell level. However, analysis tools are severely lacking. Although high-throughput systems allow rapid data collection from large cohorts, manual data analysis can take months. Moreover, identification of cell populations can be subjective and analysts rarely examine the entirety of the multidimensional dataset (focusing instead on a limited number of subsets, the biology of which has usually already been well-described). Thus, the value of PFC as a discovery tool is largely wasted.
Results: To address this problem, we developed a computational approach that automatically reveals all possible cell subsets. From tens of thousands of subsets, those that correlate strongly with clinical outcome are selected and grouped. Within each group, markers that have minimal relevance to the biological outcome are removed, thereby distilling the complex dataset into the simplest, most clinically relevant subsets. This allows complex information from PFC studies to be translated into clinical or resource-poor settings, where multiparametric analysis is less feasible. We demonstrate the utility of this approach in a large (n=466), retrospective, 14-parameter PFC study of early HIV infection, where we identify three T-cell subsets that strongly predict progression to AIDS (only one of which was identified by an initial manual analysis).
Availability: The ‘flowType: Phenotyping Multivariate PFC Assays’ package is available through Bioconductor. Additional documentation and examples are available at: www.terryfoxlab.ca/flowsite/flowType/
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: rbrinkman@bccrc.ca
1 INTRODUCTION
The immune response to infection, vaccination or malignancy can be characterized by examining changes in the expression of a wide array of proteins expressed on leukocytes (either generally or on antigen-specific B- or T-cells). These proteins identify an enormous variety of cell types, and it is often not known which subsets of cells are clinically relevant. In some settings, the immunologically-relevant cell subset represents a small minority of the bulk cell population. Therefore, gross measurements taken from heterogeneous samples (as generally done with microarrays) may mask immunologically or clinically significant signals. This limitation can be overcome with polychromatic (>5 color) flow cytometry (PFC), where protein expression can be assessed among a large number of cell subsets, at the single cell level (Chattopadhyay et al., 2008b; Perfetto et al., 2006).
The need for PFC is particularly apparent in studies of HIV, were the strongest cellular correlate of clinical outcome (CD4+ T-cell count) provides little help in identifying those individuals who would benefit from early initiation of highly active anti-retroviral therapy (HAART) (Burgoyne and Tan, 2008; Conway and Coombs, 2011; Kitahata et al., 2009; Sax and Baden, 2009). Recent studies of simian immunodeficiency virus (SIV) infection of non-human primates provide some guidance, demonstrating that the level of central memory T-cells may be a relevant predictor of the need for early therapy (Kuhrt, 2010; Mattapallil et al., 2005; Veazey et al., 1998). Similarly, our recent study of early HIV infection suggests the presence of long-lived T-cells during early infection correlates with long-term progression, as does the absence of proliferating cells (Ganesan et al., 2010). Likewise, measurements of polyfunctional T-cells (simultaneously producing at least three of the following: IFNγ, IL2, CD107a, MIP1β and TNFα) are relevant in individuals whose disease progresses slowly (Klausner et al., 2003; Voronin et al., 2010). Importantly, enumeration of central memory, long-lived, proliferating or polyfunctional cells requires PFC technology, since many markers are needed to discriminate each of these cell types from other populations of leukocytes.
Thus, it is evident that highly multiplexed approaches (such as PFC (Bendall et al., 2011; Ornatsky et al., 2010)) are critical, at least as exploratory tools to identify potential correlates of pathogenesis; however, despite recent developments in hardware and reagents, the technology has not yet reached its full potential. In particular, data analysis methodologies have not kept up with instrumentation advances (Chattopadhyay and Roederer, 2010), because of the challenges presented by the highly multidimensional data. First, the identification of cell populations using manual approaches requires a time-consuming and complex process of successively applying polygon filters (i.e. gates) to 2D scatter plots. This can also be an important source of experimental variation in large, longitudinal or multicenter studies, and limits down-stream analysis (De Rosa et al., 2001; Lugli et al., 2010; Maecker et al., 2005, 2010). Second, even though a PFC experiment collects data describing tens of thousands of cell subsets, only a small proportion of those can be reasonably queried against a clinical outcome. The choice of these subsets depends heavily on the investigator; therefore, important immunophenotypes that were not initially hypothesized to be important may be ignored (Chattopadhyay et al., 2008a). A third challenge emerges when assessing the statistical rigor of findings from manual data analysis. Since the number of exploratory attempts at the analysis is rarely reported, adjustment for multiple comparisons is not usually performed. Multiple testing correction is complicated further when the choice of candidate cell populations for exploratory analysis is biased by the results of previous similar studies. A fourth challenge is the identification of the minimal set of markers that describe a clinically relevant cell type. Although thousands of immunophenotypes can be identified in a PFC experiment, it is not clear how many of these subsets represent functionally distinct cell populations. Moreover, for those cells that are clinically relevant, the exact set of markers needed to identify that cell subset is rarely known. This is a particularly important problem, because it prevents the translation of results from PFC studies to more widespread use in clinical or resource-poor settings where complex instrumentation is often not available.
To address these problems, we developed a computational approach for identifying biomarkers in PFC data with clinical outcomes. Briefly, this approach first defines all possible immunophenotypes within a dataset and assesses the relationship between each and the clinical outcome. Importantly, the approach combines completely automated analysis of markers with some level of expert guidance to facilitate identification of rare subsets. Next, it reveals the minimal set of markers needed to identify the cell populations of interest. We demonstrate the utility of this approach by applying it to a dataset derived from a large retrospective study of individuals at the early stage of HIV infection. The dataset included a well-defined clinical outcome (time to AIDS diagnosis or death), against which the frequency of each immunophenotype was correlated. We identified three groups of related T-cell subsets whose frequency during early infection had a statistically and clinically significant relationship with progression to AIDS. One of these groups was closely related to a cell population identified previously using standard manual approaches (Ganesan et al., 2010).
2 MATERIALS AND METHODS
2.1 The cohort
The United States Military HIV Natural History Study has collected clinical data on HIV-infected military personnel since 1985. Basic demographic characteristics of this dataset are described elsewhere (Weintrob et al., 2008). We studied a subset of these subjects (n=466) with cryopreserved peripheral blood mononuclear cells (PBMCs) stored within 18 months of their date of seroconversion. The seroconversion date was estimated as the midpoint between the documented HIV− and HIV+ dates. The cohort included 135 death/AIDS events as defined by 1993 guidelines (Castro et al., 1992). The date of the last follow-up or initiation of HAART was considered a censoring event. The immunologic and virologic characteristics of this subset were previously published (Ganesan et al., 2010).
2.2 Flow cytometry assays
Antibodies, staining procedures and instrumentation were described previously (Ganesan et al., 2010). Briefly, the staining panel enumerated various subsets of naïve and memory T-cells defined by CD3, CD4, CD8, CD45RO, CD27, CD28, CD57, CCR5, CCR7, CD127 and KI-67. CD14 and V-amine dye were used to exclude monocytes and dead cells, respectively. All study samples were treated the same way using methods common to the field (i.e. gradient centrifugation of whole blood, isolation of PBMC, cryopreservation and thawing). Therefore, the results presented are not confounded by sample manipulation, and are applicable to most of the settings in which HIV pathogenesis/vaccination studies are performed. On average ≃400 000 cells including ≃120 000 T-cells were measured (Supplementary Figure S6).
2.3 Population identification
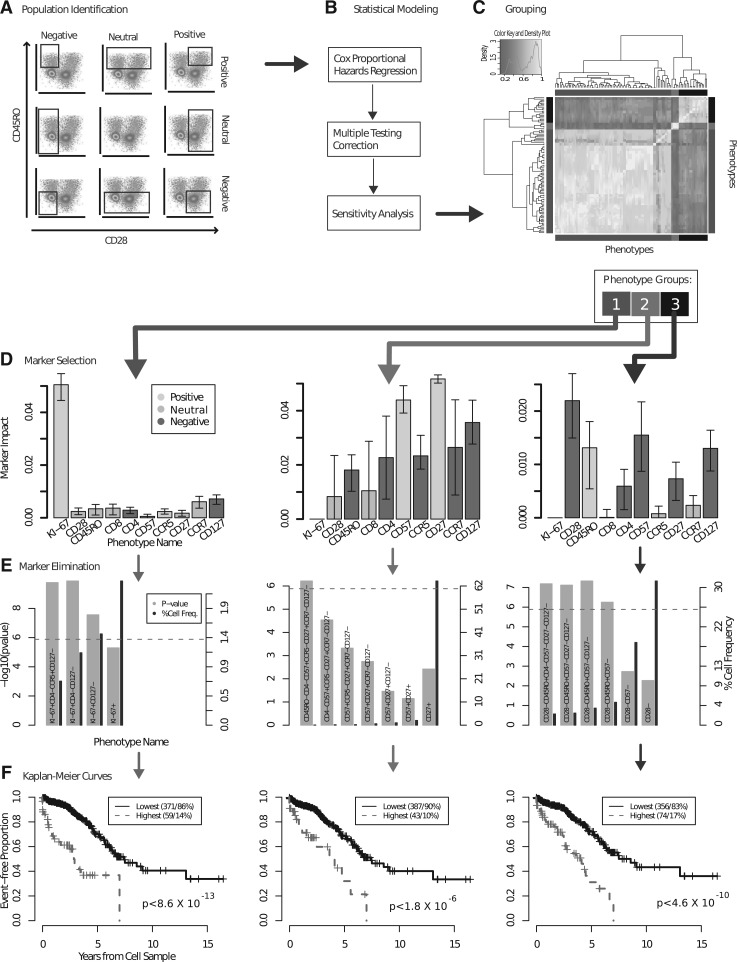
Dead cells, doublets and cellular debris were removed and live T-cells were selected by manual gating as previously described (Ganesan et al., 2010). The flowMeans algorithm was used for cell population identification within the T-cell compartment (Aghaeepour et al., 2011). The software package, as well as the infrastructure for PFC data analysis (Hahne et al., 2009) are available through Bioconductor (Gentleman et al., 2004). More specifically, flowMeans identified many clusters in the data and repeatedly merged adjacent ones based on the Mahalanobis distance between them until the desired number of clusters was reached. For each of the 10 markers in our data, flowMeans was used to identify a partition that divided the cells into a positive and a negative population (a movie demonstrating this partitioning is available in the Supplementary Material). This was based on the assumption that the expression was either on or off (i.e. there are two distinct cell populations). These 10 partitions could be combined in 210 possible ways, resulting in 1024 cell populations. To allow exclusion of markers from subset identification (which later enabled us to identify the most clinically meaningful markers), each marker could be assigned a ‘neutral’ value (i.e. that marker was excluded from the clustering—see Discussion); thus, for any single subset, each marker could be negative, positive or neutral (ignored). This increased the number of possible cell populations to 310 (59 049). An example of all possible combinations of gates (partitions) for two markers is shown in Figure 1A. Notably, the Ki-67+ population was rare (<5% of the total number of cells), and could not be identified by flowMeans. Therefore, for this marker, historical negative controls provided a static gate to partition the cells. The appropriateness of gate was confirmed manually, by visual inspection of each participant's data.
Fig. 1.
The computational pipeline for discovering correlates of HIV protection using PFC. (A) 59 069 cell populations were identified for 466 patients; a CPHR model was used to select the immunophenotypes with significant predictive power; (C) the correlation between the immunophenotypes suggested 3 internally correlated groups, shown in the side-bar colors and circumscribed by the bright yellow squares on the diagonal; (D) each group was represented by a specific combination of markers. The markers that were consistently positive or negative across all immunophenotypes are colored yellow and red, respectively, the markers with a mix of positive and negative values are grey; (E) the redundant markers were removed without affecting the predictive power; (F) the resulting immunophenotypes were used to partition the patients to two groups with different survival patterns.
2.4 Predictive analysis
To measure the predictive power of each immunophenotype, a Cox proportional hazards model (CPHR) was used to calculate the correlation between the measured phenotypes' cell frequencies (the number of cells in that immunophenotype divided by the total number of T-cells) and the clinical outcome (survival time) (Breslow, 1975). Next, the immunophenotypes with a statistically significant correlation to the survival time were identified by the log-rank test, after multiple testing correction using the Bonferroni method.
The sensitivity of the predictive power (measured by coefficient of determination (R2) as the effect size of the log-rank test) was determined using a bootstrapping procedure that tested the phenotypes of different subsets of the cohort (Hesterberg et al., 2005). Specifically, for a given vector S of subjects, a 95% confidence interval (CI) for the effect size can be calculated using the following procedure:
Repeat for 104 times: from S, draw a uniform random sample of size |S| with replacement, fit the CPHR model and record the R2.
Report the 2.5′th and 97.5′th percentiles of the distribution of R2 values from Step (1) as the lower and upper bounds of the CI, respectively.
Thus, if an immunophenotype was measured over 104 subsets of the cohort and every subject's probability of selection [as defined in Equation (1)] Pselection=0.63, then in 95% of the trials the R2 (and therefore the P-value) would have been within the range of the CI.
| (1) |
2.5 Phenotype extraction
Many of the cell populations identified were subsets of others (e.g. CD28+CD45RO− cells are also CD28+), and therefore could be redundant. We used an approach known as complete linkage hierarchical clustering to find homogeneous groups of immunophenotypes that are similar to each other (Everitt et al., 2001). Let fi be the vector of cell frequencies across all subjects for immunophenotypes i∈{1, 2,…, 59 049}. For the hierarchical clustering, we used the distance function disti,j=cor(fi, fj) where i and j are immunophenotype numbers and cor is the Pearson's correlation coefficient. The output of this procedure consists of several groups of immunophenotypes; however, the immunophenotypes in each group were highly correlated and likely to be subsets of the same parent cell type. Therefore, two additional steps were employed to identify the cell populations underlying these overlapping immunophenotypes.
2.5.1 Marker selection:
This step was designed to identify the markers that had a positive impact on the predictive power of a group of immunophenotypes. To investigate this, we let the impact of a marker be the absolute difference between (i) the means of CPHR R2 goodness-of-fit scores for the given groups of immunophenotypes and (ii) the scores after forcing that marker to be neutral. The impact value reflected the increase in the error of the CPHR model when that specific marker was excluded (see Supplementary Material for example). To identify the markers with impacts significantly higher than zero, the same bootstrapping procedure described in the predictive analysis section was applied to given groups of immunophenotypes. Combining these markers identified the candidate cell population representative of the immunophenotypes in the respective group.
2.5.2 Backward marker elimination
In the previous step, we selected the markers that, on average, had a positive impact on the predictions of the respective groups of immunophenotypes. The next step was to identify the markers that were redundant (i.e. were uninformative in presence of others). For each immunophenotype, we sequentially removed markers starting with the one with lowest impact. At every step, the P-value of the log-rank test was calculated and evaluated (false discovery date =0.05 after adjustment). The last statistically significant cell population was selected. This cell population could define the immunophenotypes in the respective group with a minimum number of markers.
2.6 Sensitivity analysis
The pipeline is an exploratory analysis tool that outputs a list of immunophenotypes (and not a multivariate predictive model). Therefore, cross-validation or holdout-validation (i.e. keeping a test-set) are not meaningfully applicable. Instead, we used the following bootstrapping procedure to assess the generalizability of the selected immunophenotypes to previously unseen data:
Repeat for K times: from the given set of subjects, S, draw a uniform random sample of size |S| with replacement, run the pipeline and record the selected immunophenotypes;
Report the proportion of iterations in Step (1) in which each immunophenotype was selected,
where K is the number of iterations, set manually by considering the amount of variation in the data and the computing resources available. To measure the sensitivity of the pipeline to different subsets of the cohort, this procedure measures the proportion of trials on subsets of the subjects in which a given immunophenotype was selected by the pipeline. Like the previous bootstrapping step, it can be shown that the probability of every sample being included in the subset is 0.63. Therefore, phenotypes that are selected in a high proportion of trials (with different subject compositions of 37% on average) are not sensitive to variations within the cohort of subjects.
3 RESULTS
3.1 Identification of cell subsets related to clinical outcome
Cell populations were identified (as described in Methods) and the frequencies of the 59 049 immunophenotypes were calculated (Fig. 1A). Next, these immunophenotypes were related to each patient's time to AIDS/death by CPHR analysis (Fig. 1B). In total, 101 of these immunophenotypes were revealed as candidate correlates of HIV disease progression by the predictive model; these were analyzed in two ways. First, we examined the correlations between cell frequencies using a clustered heat map, shown in Figure 1C and in more detail in Supplementary Figure S1. The ‘correct’ number of clusters (as in any other clustering algorithm) is subjective; our choice to use three groups is justified later in this section. Second, all 101 immunophenotypes were listed, using the order determined by the heatmap clustering (see Supplementary Table S1). To make it easier to observe patterns among the immunophenotypes represented, the immunophenotype names are illustrated with a heat map in Supplementary Figure S1. The dendrogram and the side-bar are identical to Figure 1C. The immunophenotype names in Supplementary Figure S1 are consistent with the clusters of immunophenotypes identified in Figure 1C based on correlation between cell frequencies. These figures show that closely correlated immunophenotypes have similar combinations of markers. This process allowed us to define the immunophenotypes that exhibited high correlation (i.e. describe almost identical cell types).
Next, we identified the minimum set of markers necessary to describe each of the three groups of immunophenotypes. This helped define the clinically relevant cells using the simplest possible immunophenotype, which described the most general cell population of those measured. As described in the previous section, this process was carried out in two steps: (i) selection of the markers with a positive impact on the predictive power and (ii) elimination of the redundant markers.
3.2 Impact of individual markers
For each immunophenotype group, we selected the markers that had a positive impact on the immunophenotype, as measured by the changes in mean effect size (Fig. 1D). The 95% CIs were calculated using bootstrapping (over the patient cohort). Thus, for the three groups of immunophenotypes, the predictive power depended on the combination of different markers included in the measurements (Fig. 1D). It is important to note that the impact value depends on the effect-size (R2) of the original immunophenotypes in a given group. Different immunophenotype groups had different mean R2 (and P-values); therefore, impact values cannot be compared across multiple groups.
We used the impact value to confirm that the heat map clustered by frequency described three groups (and not two or four; Supplementary Figures S3 and S4). With only two groups, a mix of positive and negative labels was observed, suggesting that the groups consisted of heterogeneous subpopulations. When the impact values for four groups were analyzed, two had very similar marker impacts, suggesting that we had bisected a single homogeneous cell population into two populations artificially. Finally, those markers with impacts significantly higher than zero, as indicated by the CIs (Supplementary Table S3), were selected as representatives of each phenotypic group, in order to define the most clinically relevant immunophenotype. By selecting markers that, on average, had a positive impact on the predictions of the respective groups of immunophenotypes, we narrowed down the list of potential immunophenotypes to three (Supplementary Table S3).
3.2.1 Marker elimination:
Next, we identified the markers that were uninformative in the presence of others. For each of the immunophenotype groups, we removed the markers one at a time, starting with the one with lowest impact, until only the marker with the highest impact remained. Figure 1E lists the P-values after every removal step. The first phenotypic group was originally described as Ki-67+CD4−CCR5+CD127− (panel A). However, the iterative removal of markers only affected the P-value when CD4 and CCR5 were removed from the analysis, indicating that the relationship to disease progression in this immunophenotype is driven by Ki-67 and CD127. For the second phenotypic group, the P-value remains significant for a combination of eight markers (CD45RO−CD8+CD4−CD57+ CCR5−CD27+CCR7−CD127−). Finally, the representative immunophenotype of the third group was simplified from CD28−CD45RO+CD4−CD57− CD27−CD127− to CD28−CD45RO+ CD57−. The most frequent cell population with a P-value higher than the threshold determined by multiple comparisons adjustment (i.e. the statistically significant immunophenotype with minimum number of markers) was reported as the representative immunophenotype of the respective group (Table 1). Representative examples are illustrated in Supplementary Figures S7–S12.
Table 1.
The final immunophenotypes
| Immunophenotype | P-value | P-value CI | Adjusted P-value | CPHR coefficient | R2 | Cell frequency |
| Ki-67+CD127− | 2.7×10−08 | 2.9×10−15, 2.1×10−6 | 1×10−3 | 19 | 0.069 | 0.01 |
| CD45RO−CD8+CD57+CCR5− CD27+CCR7−CD127− | 3.1×10−07 | 1.5×10−11, 1.6×10−2 | 1×10−2 | 633 | 0.059 | 6×10−4 |
| CD28−CD45RO+CD57− | 5.6×10−7 | 1.1×10−11, 2.6×10−4 | 2e-02 | 12 | 0.056 | 5×10−2 |
The largest cell population (calculated as cell population with the highest mean proportion calculated respective to the total number of T-cells per respective sample) with a P-value higher than the Bonferroni cut-off was selected as the representative of the respective group.
3.3 Confirmatory analysis
We performed several experiments to confirm the results obtained by the pipeline. We manually identified CD28−CD45RO+CD57− cells using conventional methods (polygon gates on two scatter plots as demonstrated in Supplementary Figure S5) and confirmed the relationship between frequencies of these cells and survival time (P=7×10−6). This result is similar to that obtained with the automated pipeline (P=5×10−7); any difference is likely due to minor variations in the data that cannot be captured using the manual analysis. A second confirmatory analysis was performed by using the three identified immunophenotypes to partition the patients into two groups by thresholding the cell frequencies; these groups had different survival patterns (Fig. 1F), confirming the ability of the automated pipeline to identify clinically meaningful cell populations. Finally, the sensitivity of the automated pipeline was determined after 100 bootstrap iterations, which required nearly 2000 CPU days. The immunophenotypes selected in the first and third groups were clearly dominant as demonstrated in Supplementary Figure S5 panels D, E and F. However, the second phenotypic group could be labelled CD4− or CD8+, according to this analysis. Importantly, these populations likely overlap significantly, as expression of CD4 and CD8 are usually mutually exclusive on T-cells in the peripheral blood. Thus, the CD4− label includes primarily CD8+ T-cells (Korber et al., 2006).
4 DISCUSSION
We described a computational approach to analyze a high-dimensional clinical flow cytometry dataset that was previously investigated through laborious manual inspection. The findings from our analysis both replicate and extend the original analysis by human experts, revealing the T cell subsets and markers most highly correlated with HIV progression. The pipeline consists of five steps: (i) automated identification of positive and negative populations for each marker; (ii) quantification of subsets defined by every combination of markers; (iii) identification of those cell subsets whose frequency is most highly associated with clinical outcome; (iv) calculation of the impact of each individual marker; and (v) identification of the minimal set of markers needed to describe significant cell populations.
The first step in the pipeline delineates positive and negative populations for every channel. This step uses a clustering tool that was developed exclusively for PFC data (Aghaeepour et al., 2011). Many such tools have been developed for identifying cell populations in a multidimensional setting, but several limitations have kept these algorithms from replacing manual analysis. Firstly, the use of these algorithms (as any other clustering tool) is highly subjective and complicated – often, the concept of what comprises a cluster/cell population is not well-defined. Clustering tools are also limited in their ability to find rare cell populations. Furthermore, meta-clustering of candidate clusters must be performed to identify clinically relevant immunophenotypes; however, for this, clusters must be linked to subjectively-defined categories of cells. It is also difficult to visualize and interpret results because clusters cannot be described using marker names. Lastly, biological information is rarely incorporated into the clustering process. The algorithm presented here overcomes these limitations by partitioning cells one marker at a time and by using combinations of the partitions to extract immunophenotypes/features for predictive analysis.
A potential shortcoming of this approach is the underlying assumption that every channel has only two well-separated cell populations (i.e. expression is either on or off). However, some cellular proteins exhibit a continuum of expression across a cell population, with cells that lack expression, others with low levels of expression, and some with very high levels of expression. Furthermore, for some markers these differences are known to be biologically meaningful; CCR7 expression is high on naive T-cells, but low on more differentiated central memory T-cells (Ganesan et al., 2010). Thus, a potential limitation of our approach is that CCR7^bright and CCR7^dim cells would be classified as a single cell population, or conceivably, that the CCR7^dim would be grouped with the CCR7−. To address this limitation, the pipeline could be modified to support automatic gating of more than two cell populations. This will become particularly important for bar-coded samples (where dozens of different populations are represented by the bar-code (Krutzik and Nolan, 2006)), although in this case the problem is lessened by having prior knowledge of the number of populations present. Nevertheless, because these cells differ in expression of other markers, the populations may be resolved when the complete phenotypic combinations using the rest of markers are created (Sallusto et al., 1999).
The second step lists all possible combinations of markers, and assesses the frequency of each immunophenotype within patient samples. By designating positive and negative populations for each of the 10 markers studied, 210 (1024) terminal immunophenotypes were identified. Thus, every subset, defined by any combination of markers, was examined. However, this assumes that every marker is relevant to clinical outcome, which is unlikely. To examine immunophenotypes defined both by combinations of all markers, and by combinations of all subsets of markers, our algorithm allowed markers to be neutral. It is thus possible to measure the frequency of each of the parent populations as well as the terminal ones. For example, our algorithm identified and quantified not only CD4+CD45RA−CCR7+Ki−67+CD57−CD27+ cells, but also cells in the CD4+CD45RA−CCR7+ parent population (i.e. CD4+CD45RA−CCR7+Ki−67NCD57NCD27N, where N marks the neutral state). This ability to allow neutral markers is important to discovery efforts, since it enables researchers to include markers in their experimental design without knowing ahead of time whether they are clinically relevant. This process resulted in the identification of 310 (59 049) immunophenotypes, defined by all combinations of positive and negative populations over all combinations of the 10 markers.
The third step determines whether the frequency of each of these immunophenotypes is associated with the clinical outcome by CPHR and the log-rank test. Because of the high number of candidate immunophenotypes, adjustment for multiple comparisons is critical. We chose the conservative approach of using Bonferroni's method, knowing that the level of false positives would be low, at the cost of some statistical power. Alternatively, less conservative approaches used in other high-dimensional biological assays (Noble, 2009) could be employed. At this step, the pipeline identified 101 phenotypes with a statistically significant relationship with the clinical outcome (time to AIDS/death).
However, since the second element of the algorithm allows for inclusion of parent populations, some of the phenotypes identified are overlapping and highly correlated. To unravel relationships that are driven by parent populations from uniquely important cell subsets, the fourth step of our pipeline calculates the impact of each individual marker. This is determined by clustering the immunophenotypes based on the Pearson's correlation between them, and then assuming that each cluster of immunophenotypes represents a single cell type, uniquely related to the clinical outcome. In the dataset presented here, we find three distinct populations of cells that predicted time to AIDS/death.
Finally, the fifth step of the pipeline simplifies the cell populations with the strongest relationship to clinical outcome by identifying the minimal set of markers that can be used to define them. Unlike subjective methods that are based on a researcher's assessment of which markers are important, this step is based on ‘impact’ values calculated by the algorithm. One disadvantage of this method is that it is a greedy approach, capable of finding the subtractively minimal marker set, but potentially not the globally optimal markers. In future, graph theory (Needham et al., 2007) or graphical modeling tools could be developed both to visualize connections between the cell populations that affect clinical outcome, and to find globally optimized marker sets defining them. Nevertheless, even in its current form, the algorithm can distill the complexity of a multivariate data set into immunophenotypes that can be assessed in resource-poor or clinical settings.
The three cell populations defined by the algorithm included one closely related to the CD8+ Ki−67+ (proliferating) cells identified in the previous analysis (Ganesan et al., 2010). However, our computational pipeline showed that the presence of these cells in both the CD4+ and CD8+ T-cell compartment had predictive value. Moreover, the pipeline refined the definition of these cells to include only those that were lacking a receptor involved in homoeostatic proliferation (CD127−). These cells may represent antigen-experienced memory and effector cells, proliferating in response to the immune activation that occurs during HIV infection. A second population identified by the algorithm was CD45RO−CD8+CD57+ CCR5−CD27+CCR7−CD127−. Interestingly, this cell type could not be defined by fewer markers (i.e. it was not flagged as redundant by the backward elimination algorithm in step five, thus demonstrating the importance of multiparametric measurements. The immunophenotype of these cells is consistent with highly differentiated (terminal) effector T-cells (which have re-expressed CD45RA, not measured, and CD27). Notably, these cells represent the polar opposite of naive cells, which were found to have slight predictive power in the manual analysis. The number of markers necessary to define these cells likely reflects the expression of markers of terminal effector cells (like CD57) within other memory cell populations. Thus, the automated algorithm has honed in on the best possible definition of this cell type. Finally, the algorithm identified CD28−CD45RO+CD57− cells as clinically relevant. This population likely includes cells capable of strong effector function, which have not yet lost the ability to proliferate or differentiate. The biological function of these cells is not well understood, but the predictive value of this immunophenotype suggests that studies to further characterize these cells is necessary. In the future, cell ontology approaches may be developed to define a consistent nomenclature for the subsets identified in PFC analysis, particularly those that have unique clinical importance. Such efforts would facilitate our understanding of the underlying biology and would allow simpler meta analysis of data across studies (Bard et al., 2005; Smith et al., 2007). Following this direction, it will be possible to connect PFC studies to the existing efforts of system biologists (Nurse, 2003).
Importantly, all three cell subtypes are rare after removing the redundant markers (Table 1); this highlights another major advantage of this pipeline over standard methods: manual or computational identification of rare cell subtypes is challenging (Altschuler and Wu, 2010; De Rosa et al., 2001). However, a large number of rare cell subtypes exist in the human immune system, and it is well established that rare cells play an important role in the immune system (e.g. HIV (Franz et al., 2011), stem cell research (Notta et al., 2011) and cancer (Zimmerlin et al., 2011)).
We allowed the automated pipeline to search for clinically relevant subsets from the entire T-cells, rather than within only CD4+ or CD8+ T-cell compartments (as is typically done with standard methods). This approach has two advantages. First, it limits the preliminary gates that are needed to prepare the data, making the analysis easier and less susceptible to error or subjectivity. Second, some of the immunophenotypes identified may be relevant to both CD4+ and CD8+ T-cell biology, as is the case for immunophenotypes where the algorithm identified that the CD4 and CD8 markers are irrelevant. Given the stark differences between CD4+ and CD8+ T-cell biology in HIV (one cell type is infected and depleted, while the other expands), immunophenotypes that are clinically relevant and shared between the two compartments may be particularly interesting for future study. Supplementary Table S2 demonstrates the projection of these populations into the cytotoxic and helper populations. The table shows that the cytotoxic compartment has a stronger predictive power than the helper compartment, which confirms the findings of previous manual analysis (Ganesan et al., 2010). In addition, similar results were reported in a recent comparison of these cells against other components of the immune system (i.e. natural killer (NK) cells and B-cells) in SIV infection (Elemans et al., 2011).
Although much of our effort was geared toward development of an automated pipeline, we embedded a number of opportunities for users to integrate their biological knowledge into the analysis, with the aim of producing a more robust system. For example, biological knowledge could be used to exclude irrelevant cells (e.g. B-cells, dead cells and debris cells, and doublets); therefore, we allowed manual identification of live, CD3+ T-cells. In addition, for low-frequency populations (e.g. Ki-67+ cells), we offered the ability to set a threshold gate based on a negative control. Finally, the number of phenotype groups reported by the algorithm could be limited, based on the investigator's biological knowledge.
In summary, our pipeline allowed the identification of a large number of rare populations associated with clinical outcome and then described these cell types using only the most impactful markers. Although it was applied to an HIV dataset in this work, it can be applied in its current form to any PFC study, across a wide variety of disciplines (including but not limited to studying malaria, tuberculosis, autoimmune diseases and various blood cancer subtypes). In particular, this computational approach holds significant potential for: (i) detailed exploratory analysis of the immune system (using a high number of markers to parse the cell populations); (ii) analysis of large cohorts of subjects (e.g. clinical studies and vaccine/drug trials); and (iii) screening studies to identify appropriate marker panels for further clinical investigation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Wayne Moore for contributing preprocessing analysis tools, Greg Finak for his comments on an earlier version of this work, and Nishant Gopalakrishnan, Jamie Scott, Celsa A. Spina and Armstrong Murira for their contribution to the Bioconductor package. Also, the authors would like to thank the patients enrolled in the US military Natural History Study without whom none of this work would have been possible. Finally, the authors would like to thank the research coordinators and support staff who diligently work on the HIV Natural History Study, as well as the members of the Infectious Disease Clinical Research Program HIV Working Group.
Funding: This work was supported by NIAID Intramural Research Program; NIH/NIBIB grant EB008400; an NSERC discovery grant held by HHH; NIH (contract HSN261200800001E); NCI; Infectious Disease Clinical Research Program; Uniformed Services University of the Health Sciences, The Terry Foundation and The Terry Fox Research Institute. NA was supported by a UBC4YF scholarship and a CIHR/MSFHR scholarship. RRB was supported in part by a Michael Smith Foundation for Health Research Scholar Award. This research was enabled by the use of computing resources provided by the Western Canada Research Grid (WestGrid) and Compute/Calcul Canada.
Conflict of Interest: none declared.
REFERENCES
- Aghaeepour N., et al. Rapid cell population identification in flow cytometry data. Cytometry A. 2011;79:6–13. doi: 10.1002/cyto.a.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler S., Wu L. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J., et al. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall S., et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N. Analysis of survival data under the proportional hazards model. Int. Stat. Rev./Revue Internationale de Statistique. 1975;43:45–57. [Google Scholar]
- Burgoyne R., Tan D. Prolongation and quality of life for HIV-infected adults treated with highly active antiretroviral therapy (HAART): a balancing act. J. Antimicrob. Chemother. 2008;61:469–474. doi: 10.1093/jac/dkm499. [DOI] [PubMed] [Google Scholar]
- Castro K., et al. Revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm. Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- Chattopadhyay P., Roederer M. Good cell, bad cell: flow cytometry reveals T-cell subsets important in HIV disease. Cytometry A. 2010;77:614–622. doi: 10.1002/cyto.a.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P., et al. A chromatic explosion: the development and future of multiparameter flow cytometry. Immunology. 2008a;125:441–449. doi: 10.1111/j.1365-2567.2008.02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay P., et al. Techniques to improve the direct ex vivo detection of low frequency antigen-specific CD8+ T cells with peptide-major histocompatibility complex class I tetramers. Cytometry A. 2008b;73:1001–1009. doi: 10.1002/cyto.a.20642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J., Coombs D. A stochastic model of latently infected cell reactivation and viral blip generation in treated HIV patients. PLoS Comput. Biol. 2011;7:e1002033. doi: 10.1371/journal.pcbi.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa S., et al. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- Elemans M., et al. Quantification of the relative importance of CTL, B Cell, NK Cell, and target cell limitation in the control of primary SIV-infection. PLoS Comput. Biol. 2011;7:e1001103. doi: 10.1371/journal.pcbi.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt B., et al. Cluster Analysis. Vol. 4. Arnold, London: 2001. [Google Scholar]
- Franz B., et al. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood. 2011;118:348–357. doi: 10.1182/blood-2011-03-341917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A., et al. Immunologic and virologic events in early HIV infection predict subsequent rate of progression. J. Infect. Dis. 2010;201:272. doi: 10.1086/649430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F., et al. flowCore: a Bioconductor package for high-throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterberg T., et al. Bootstrap methods and permutation tests. Intro. Prac. Stat. 2005;47:1–70. [Google Scholar]
- Kitahata M., et al. Effect of early versus deferred antiretroviral therapy for HIV on survival. N. Engl. J. Med. 2009;360:1815–1826. doi: 10.1056/NEJMoa0807252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R., et al. Enhanced: the need for a global HIV vaccine enterprise. Science. 2003;300:2036. doi: 10.1126/science.1086916. [DOI] [PubMed] [Google Scholar]
- Korber B., et al. Immunoinformatics comes of age. PLoS Comput. Biol. 2006;2:e71. doi: 10.1371/journal.pcbi.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik P., Nolan G. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Meth. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Kuhrt D. PhD Thesis, School of Medicine. University of Pittsburgh; 2010. SIV infection results in detrimental phenotypic and functional alterations of the naive and memory B cell compartments that are initiated during acute infection. [Google Scholar]
- Lugli E., et al. Data analysis in flow cytometry: the future just started. Cytometry A. 2010;77:705–713. doi: 10.1002/cyto.a.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H., et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H., et al. A model for harmonizing flow cytometry in clinical trials. Nat. Immunol. 2010;11:975–978. doi: 10.1038/ni1110-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil J., et al. Massive infection and loss of memory CD4 T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- Needham C., et al. A primer on learning in Bayesian networks for computational biology. PLoS Comput. Biol. 2007;3:e129. doi: 10.1371/journal.pcbi.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W. How does multiple testing correction work? Nat. Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Nurse P. Systems biology: understanding cells. Nature. 2003;424:883. doi: 10.1038/424883a. [DOI] [PubMed] [Google Scholar]
- Ornatsky O., et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Meth. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Perfetto S., et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J. Immunol. Meth. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Sallusto F., et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;402:34–38. [PubMed] [Google Scholar]
- Sax P., Baden L. When to start antiretroviral therapy—⣔ready when you are? N. Engl. J. Med. 2009;360:1897–1899. doi: 10.1056/NEJMe0902713. [DOI] [PubMed] [Google Scholar]
- Smith B., et al. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey R., et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Voronin Y., et al. The future of HIV vaccine research and the role of the global HIV vaccine enterprise. Curr. Opin. HIV AIDS. 2010;5:414. doi: 10.1097/COH.0b013e32833cfe32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintrob A., et al. Increasing age at HIV seroconversion from 18 to 40 years is associated with favorable virologic and immunologic responses to HAART. JAIDS J. Acquir. Immune Defic. Syndr. 2008;49:40. doi: 10.1097/QAI.0b013e31817bec05. [DOI] [PubMed] [Google Scholar]
- Zimmerlin L., et al. Rare event detection and analysis in flow cytometry: bone marrow mesenchymal stem cells, breast cancer stem/progenitor cells in malignant effusions, and pericytes in disaggregated adipose tissue. Methods Mol. Biol. 2011;699:251–273. doi: 10.1007/978-1-61737-950-5_12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.