Abstract
Motivation: Scholarly biomedical publications report on the findings of a research investigation. Scientists use a well-established discourse structure to relate their work to the state of the art, express their own motivation and hypotheses and report on their methods, results and conclusions. In previous work, we have proposed ways to explicitly annotate the structure of scientific investigations in scholarly publications. Here we present the means to facilitate automatic access to the scientific discourse of articles by automating the recognition of 11 categories at the sentence level, which we call Core Scientific Concepts (CoreSCs). These include: Hypothesis, Motivation, Goal, Object, Background, Method, Experiment, Model, Observation, Result and Conclusion. CoreSCs provide the structure and context to all statements and relations within an article and their automatic recognition can greatly facilitate biomedical information extraction by characterizing the different types of facts, hypotheses and evidence available in a scientific publication.
Results: We have trained and compared machine learning classifiers (support vector machines and conditional random fields) on a corpus of 265 full articles in biochemistry and chemistry to automatically recognize CoreSCs. We have evaluated our automatic classifications against a manually annotated gold standard, and have achieved promising accuracies with ‘Experiment’, ‘Background’ and ‘Model’ being the categories with the highest F1-scores (76%, 62% and 53%, respectively). We have analysed the task of CoreSC annotation both from a sentence classification as well as sequence labelling perspective and we present a detailed feature evaluation. The most discriminative features are local sentence features such as unigrams, bigrams and grammatical dependencies while features encoding the document structure, such as section headings, also play an important role for some of the categories. We discuss the usefulness of automatically generated CoreSCs in two biomedical applications as well as work in progress.
Availability: A web-based tool for the automatic annotation of articles with CoreSCs and corresponding documentation is available online at http://www.sapientaproject.com/software http://www.sapientaproject.com also contains detailed information pertaining to CoreSC annotation and links to annotation guidelines as well as a corpus of manually annotated articles, which served as our training data.
Contact: liakata@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Since the launch of the first scientific journal in 1665, Philosophical transactions of the Royal Society, the scientific literature has developed into the core medium for the exchange of ideas and findings across all scientific communities. In recent years, numerous initiatives have emerged to automatically process electronic documents in the life sciences, add semantic markup to them and facilitate access to scientific facts. Most work in biological text mining (Ananiadou et al., 2010; Cohen and Hersh, 2005) has concentrated on identifying biological entities and extracting the relations between these entities as facts or events appearing in article abstracts while recently, the focus has shifted towards full text articles (Kim et al., 2011). While system performance on biomolecular event extraction is improving (Kim et al., 2011), there is little progress in the analysis of the context of extracted events and relations which help to characterize the knowledge conveyed within the text and build the argumentation within the article discourse.
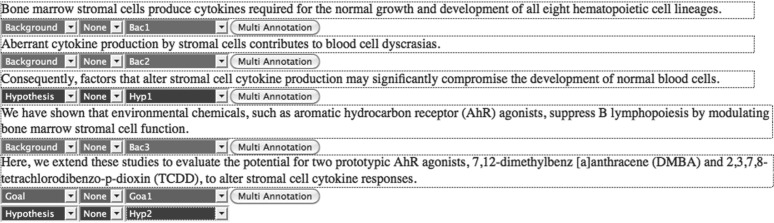
The analysis of the scientific discourse plays a key role in differentiating between the nature of the knowledge encoded in relations and events, e.g. ‘AhR agonists suppress B lymphopoiesis’ in the fourth sentence of Figure 1 is a known fact whereas ‘the potential of two AhR agonists to alter stromal cell cytokine responses’ in sentence 5 is a hypothesis to be investigated. Such a distinction between events or relations is currently ignored in standard biomedical information extraction. Discourse analysis of this type would improve the distinction between facts, speculative statements, pre-existing and new work. In Figure 1, factual sentences (denoted as ‘Background’, sentences 1, 2 and 4) are distinguished from a sentence containing information inferred from the ‘Background’, a hypothesis driving and justifying the work presented in the article (‘Hypothesis’, sentence 3). Sentence 5 which conveys the aim of the work as being that of evaluating a certain hypothesis, is annotated as both Goal and Hypothesis.
Fig. 1.
Example of discourse labelling using CoreSC.
The categorization of sentences within scientific discourse has been studied in previous work and from a number of different angles. Simone Teufel (Teufel et al., 1999; Teufel, 2010) created argumentative zoning (AZ), an annotation scheme which models rhetorical and argumentational aspects of scientific writing and concentrates on author claims. AZ has been modified for the annotation of biology articles (Mizuta et al., 2006) and chemistry articles (Teufel et al., 2009). Other work has looked at the annotation of information structure in abstracts, based on abstract sections (Hirohata et al., 2008; Lin et al., 2006; McKnight and Srinivasan, 2003; Ruch et al., 2007). A separate line of work has looked at the characterization of scientific discourse in terms of modality and speculation (Kilicoglu and Bergler, 2008; Light et al., 2004; Medlock and Briscoe, 2007) while Shatkay et al. (2008) and Wilbur et al. (2006) annotate sentences according to various dimensions such as focus, polarity and certainty. There is as yet no general consensus among researchers in scientific discourse regarding the optimal unit of annotation. Most of the previous research considers sentences as their basic unit while de Waard et al. (2009) has proposed the annotation at the clause level and Nawaz et al. (2010) and Thompson et al. (2011) consider a multi-dimensional scheme for the annotation of biological events in texts (bio-events).
Existing schemes vary in their scope and granularity, with ones designed for abstracts considering only four categories and schemes for full articles generally consisting of at most seven content-related categories. However, especially for the case of full articles, it is becoming apparent that more information is required to characterize statements and claims. Researchers are interested in identifying hypotheses and different types of evidence to support claims (Ciccarese et al., 2008), which are not readily identifiable by current schemes.
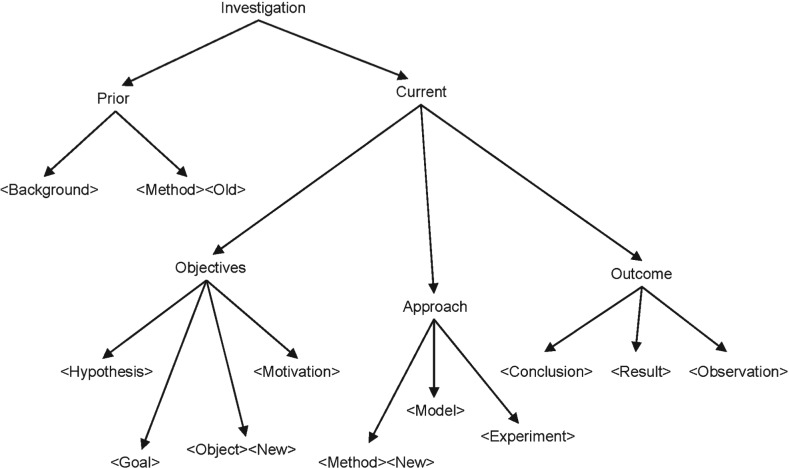
Our work fills the need for finer-grained annotation to capture the content and conceptual structure of a scientific article. Inspired by the definitions in the EXPO ontology for scientific experiments (Soldatova and King, 2006) and the CISP meta-data (Soldatova and Liakata, 2007), in Liakata and Soldatova (2008) and Liakata et al. (2010) we introduced a sentence-based, three layer scheme which recognizes the main components of scientific investigations as represented in articles (see Fig. 2 and Supplementary Material). The first layer consists of 11 categories which describe the main components of a scientific investigation, the second layer is properties of those categories (e.g. Novelty, Advantage), and the third layer provides identifiers that link together instances of the same concept.
Fig. 2.
Hierarchical representation of concepts and properties in the CoreSC scheme.
In comparison to closely related schemes (de Waard, 2007; Nawaz et al., 2010; Teufel et al., 2009), none of which have been automated yet, the Core Scientific Concept (CoreSC) scheme makes finer grained distinctions between the different types of objective (Hypothesis–Goal–Motivation–Object), approach (Method–Model–Experiment) and outcome (Observation–Result–Conclusion) and constitutes the most fine grained analysis of knowledge types of any such scheme. The distinction between the above types of objective, approach and outcome are important to expert needs (For more details, see the definitions and explanations in the Supplementary Material.).
The CoreSC scheme has been applied to articles in biochemistry and chemistry to create a corpus of 265 annotated articles (ART/CoreSC corpus, 39 915 sentences + 265 titles, over 1 million words) (Liakata and Soldatova, 2009; Liakata et al., 2010). Guo et al. (2011) showed that a finer level of annotation of cancer risk assessment (CRA) abstracts using CoreSC categories, increased experts' efficiency in extracting information from the text while White et al. (2011) argue that the CoreSC scheme is ‘uniquely suited to recovering common types of scientific arguments about hypotheses, explanations, and evidence’.
In this article, we automate the annotation of full scientific articles with categories from the first layer of the CoreSC scheme, provide intrinsic evaluation of the results and discuss existing and future applications of this work. The article is structured as follows: In Section 2, we describe how we trained and tested machine learning classifiers on automatic recognition of CoreSCs in full articles. In Section 3, we analyse the classifier performance and discuss the features used for building the classifiers and their contributions to each category. Finally in Section 4, we discuss existing and future applications of the work. Our system for the classification of CoreSCs, our guidelines and annotated articles are all available online for researchers in biology to use.
To our knowledge this is the first time a discourse annotation scheme is being used to automatically annotate full articles in the biosciences on this scale. It is also the first such scheme for which machine learning classifiers have been trained and tested on chemistry articles. Both the resources and the tools for automatic annotation are available online.
2 METHODS
The data
the training and test data used as input to the machine learning classifiers consist of 265 articles from biochemistry and chemistry annotated at the sentence level by experts using the CoreSC annotation scheme. These articles constitute the ART/CoreSC corpus (Liakata and Soldatova, 2009; Liakata et al., 2010), which was developed in three phases (training, evaluation and expansion). During the first-phase 20 annotators, all chemistry experts at postdoc or PhD level, recruited from UK Universities, were trained on four full papers with the first version of the guidelines and detailed explanations resulting from error analysis. This data and individual comments from annotators were used to improve the annotation guidelines. The second phase was designed to evaluate both the guidelines and expert performance in terms of κ-inter-annotator agreement (κ-IAA). Our goal was to obtain IAA for a reasonable amount of papers, while ensuring at least three annotators per paper, so as to minimize the chance of random agreements. Thus, 16 annotators from the first phase were split into 5 groups of 3 annotators each, where each group annotated 8 different papers and 1 additional paper was common across all 5 groups. The 16th annotator annotated across groups to provide a normalizing factor. The κ-IAA for the 41 papers obtained in this manner, measured according to Cohen's κ (Cohen, 1960), was κ=0.55 (median average for the 9 best annotators across all groups and the paper common to all annotators). The third and final phase of corpus development aimed at expanding the size of the corpus by selecting the nine best performing annotators (according to IAA) from the second phase to annotate 25 papers each. While no IAA could be obtained for the 225 papers1 annotated in this way, the assumption is that it would be the same as the average of the agreement achieved by each of the nine annotators in the second phase of development. The 265 journal articles were chosen by a chemistry expert with extensive experience in publishing, so as to cover a wide range of topics and journals. The 265 articles cover 16 different chemistry journals and 25 topics, with the majority involving spectroscopy, biochemistry, kinetics and theoretical work. Article length ranges between 32 and 379 sentences and numbers of authors range between 1 and 11, with the majority attributed to 2–3 authors and being 150 sentences long. More details about the papers can be found in the Supplementary Material. The corpus has therefore good coverage of the field and was designed in three phases with the contribution of multiple experts so as to minimize classifier bias.
Statistics on the corpus are available in Table 1. The corpus consists of 39 915 sentences (>1 million words) with the majority categories being Result (21%) and Background (19%). The next most populous category is Observation (14%), followed by Method (11%), Experiment (10%), Conclusion (9%) and Model (9%). Finally, the categories designating the Objectives (Hypothesis, Object, Motivation and Goal) altogether amount to 7 with Object and Hypothesis the most prominent at 3% and 2%, respectively.
Table 1.
Statistics on the training data (ART/CoreSC corpus)
| Measure | Bac | Con | Exp | Goa | Met | Mot | Obs | Res | Mod | Obj | Hyp | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of sentences | 7606 | 3636 | 3858 | 582 | 4281 | 541 | 5410 | 8404 | 3656 | 1161 | 780 | 39 915 |
| Number of words | 193 930 | 102 173 | 93 882 | 16 564 | 107 309 | 13 737 | 123 394 | 224 353 | 99 313 | 29 215 | 21 315 | 1 025 185 |
| Percentage of sentences | 19 | 9 | 10 | 1 | 11 | 1 | 14 | 21 | 9 | 3 | 2 | |
| Number of words p/s (mean) | 25.5 | 28.1 | 24.33 | 28.46 | 25.07 | 25.39 | 22.81 | 26.7 | 27.16 | 25.16 | 27.33 | |
| Number of words p/s (SD) | 12.32 | 12.49 | 20.6 | 12.69 | 11.4 | 10.34 | 11.44 | 12.65 | 14.76 | 11.16 | 12.01 | |
| κ-IAA | 0.87 | 0.89 | 0.65 | 0.60 | 0.74 | 0.46 | 0.79 | 0.78 | 0.43 | 0.81 | 0.46 |
To segment sentences we used the XML aware sentence splitter SSSplit, described in Liakata et al., 2009. The choice of the sentence as our unit of annotation stems mainly from the fact that sentences are the most common unit of text selection for summaries (Brandow et al., 1995; Kupiec et al., 1995). We also regard the sentence as the most meaningful minimal unit for the analysis of scientific discourse, in agreement with earlier work (Teufel, 2000, Chapter 3).
The methods
we have used state of the art supervised machine learning algorithms to train classifiers on the automatic annotation of papers with the first layer of the CoreSC scheme, that is, the following 11 categories: Background (BAC), Hypothesis (HYP), Motivation (MOT), Goal (GOA), Object (OBJ), Method (MET), Model (MOD), Experiment (EXP), Observation (OBS), Result (RES) and Conclusion (CON) (Liakata et al., 2010). From a machine learning perspective we treat the recognition of CoreSCs as: (i) text classification and (ii) sequence labelling. In text classification sentences are classified independently of each other and any dependencies between sentences need to be added explicitly. On the other hand, in sequence labelling the assignment of labels is such as to satisfy dependencies between sentences. The latter is a more natural approach when considering discourse annotation since the flow of the narrative is influenced by what has already been mentioned. For classification, we employed support vector machines (SVMs) and for sequence labelling conditional random fields (CRFs). Previous work on discovering information structure from papers and abstracts has made successful use of both of these methods (Guo et al., 2010; Hirohata et al., 2008; Mullen et al., 2005). While experimental settings vary in each of the above cases, most notably in the number and type of classification categories, the amount of training data available and whether abstracts of full papers are used, the best performing algorithms were SVMs and CRFs.
SVM and LibLinear
we used the LibSVM (LibS) implementation of SVMs (Chang and Lin, 2011) coded in C++. Our experiments were conducted using a linear kernel, known to perform well in document classification. We used the default values for the C, γ and ϵ parameters and concentrated on the input features. When we experimented with different types of cross-validation and feature configuration we used LibLinear (LibL) (Fan et al., 2008) instead of LibS as the latter is costly timewise both in training and testing. LibL is a classifier for large scale data, which uses linear SVMs, splits data into blocks and considers one block at a time. To give an indication about the gain in speed using LibL as opposed to LibS, it takes 29 h 41 min to train one of our models with LibS and 8 h 15 min for testing a single fold versus 10 min and 4 h 36 min,2 respectively, for LibL.
Conditional random fields
we chose CRFs because they do not assume independent features but do not suffer from the label bias problem, where preference is given to states with fewer transition possibilities. For our purposes we used CRFSuite (Okazaki, 2007) an algorithm for linear-chain, first-order CRFs, optimized for speed and implemented in C. Stochastic Gradient Descent was employed for parameter estimation.
Features for classification
features are extracted from each sentence and are represented in a sparse binary matrix format. In selecting features our aim was to take into account different aspects of a sentence, ranging from its location within the paper and the document structure (global features), to its length and sentence-internal features such as the citations, verbs, n-grams and grammatical triples (GRs) it may contain (local features). Below we describe all our features in detail. The following are all implemented as binary features:
Absolute location (absloc): we divide the document into 10 unequal segments (as in Loc of (Teufel, 2000)) and assign 1 of the 10 locations, A–J, to the sentences. Larger segments, containing more sentences, are designated to be in the middle of the paper.
SectionId: a sequentially incremented section number (up to 10) is assigned to each section and inherited at sentence level. SectionId is assigned independently of the section heading, which is addressed by feature Struct-3 below.
Struct-1: the location of a sentence within seven unequal segments of a section.3 Each section is first divided into three equally sized slices; the first and the last sentence of the section are considered separate segments (1 and 7) whereas the second and the third sentence of the section also form a segment (2). The rest of the first slice is segment 3 and the second slice is segment 4. Segment 6 consists of the second and third sentence from the end of the section and the rest of the third slice is segment 5 (Teufel, 2000).
Struct-2: location within a paragraph split in five equal segments. (Teufel, 2000)
Struct-3: one of 16 heading types assigned to a sentence by matching its section heading against a set of regular expressions (a variant on Struct-3 of Teufel, 2000). SectionId and Struct-3 are complementary features since the first pertains to the absolute location of a section and is dependent on the length of the paper, while the other follows section structure irrespective of paper length. Details on header matching are available in the Supplementary Material.
Location in section (sectionloc): like Struct-2 but at section level.
Length: sentences are assigned to one of nine bins, representing a word count range. More details are available in the Supplementary Material.
Citation: we distinguish three cases: no citations, one citation, and two or more citations present.
History: the CoreSC category of the previous sentence. Only used in LibS and LibL, implicit in first-order CRF.
N-grams: binary values for significant unigrams (Uni), bigrams (Bi) and trigrams. N-grams are lemmatized using morpha (Minnen et al., 2001). Significant unigrams have frequency >3. Bigrams and trigrams are filtered according to the measure of Fair Symmetrical Conditional Probability and the LocalMaxs algorithm, defined in Silva et al. (1999). We considered filtering our n-grams by adapting an online stop word list.4 However, classifier performance was better when we did not filter stop words. In this latter case, no trigrams exceeded the threshold. Examples of significant n-grams are available in the Supplementary Material.
Verb POS (VPOS): for each verb within the sentence we determine which of the six binary POS tags (VBD, VBN, VBG, VBZ, VBP and VB) representing the tense, aspect and person of a verb are present.
Verbs: all verbs in our training data with frequency >1.
Verb Class: ten verb classes, obtained by clustering together all verbs with a frequency >150 as in Guo et al. (2010). The verb classes can be found in the Supplementary Material.
Grammatical triples (GRs): dependency–head-dependent triples (Briscoe and Carroll format) generated using C&C tools (Curran et al., 2007). We used the model of the supertagger trained on biomedical abstracts (Rimell and Clark, 2009) and applied self-training on our papers according to Kummerfeld et al. (2010). We considered dependencies subj, dobj, iobj and obj2 with frequency >3. Examples of significant GRs can be found in the Supplementary Material.
Other GR: subjects (Subj), direct objects (Dobj), indirect objects (Iobj) and second objects of ditransitive verbs (Obj2) with frequency >1.
Passive (P): whether any verbs are in passive voice.
3 RESULTS AND DISCUSSION
To test classification accuracy and establish feature contributions to CoreSC recognition we performed a number of runs, including multi-class (CRF, LibL and LibS) and binary classification using 9-fold cross-validation and a variety of feature configurations (All features, Leave-out-one-feature (LOOF) and Single feature with and without stop words). Our results (Table 2) show we can achieve accuracy of >50 in classifying the 11 CoreSCs in full papers. This is a promising result given the difficulty of the task. It is the first time the automatic recognition of such a fine grained set of categories is being attempted for full papers. F-score for the categories ranges from 76% for EXP (Experiment) to 18% for the low frequency category MOT (Motivation). The distribution of categories in papers is shown in Table 1, with RES the most frequent category and MOT and GOA the least frequent. Our feature analysis shows that the most important role is played by n-grams (primarily bigrams), GRs and verbs as well as global features such as history (sequence of labels) and section headings. It is important to note that particular features do not affect all categories in the same way. In the following, we present our results in detail. Section 4 discusses various CoreSC-based applications already implemented on the basis of current results.
Table 2.
Micro precision, recall and F-measure for different system configurations, with highest value for each measure per category in bold
| Acc | BAC |
CON |
EXP |
GOA |
MET |
MOT |
OBS |
RES |
MOD |
OBJ |
HYP |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features | Classifier | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | |
| Base | Multinomial | 14 | 19 | 19 | 19 | 9 | 9 | 9 | 10 | 10 | 10 | 1 | 1 | 1 | 11 | 11 | 11 | 1 | 1 | 1 | 13 | 14 | 14 | 21 | 22 | 21 | 9 | 9 | 9 | 3 | 3 | 3 | 2 | 2 | 2 |
| ngrams | CRFSuite | 45.3 | 45 | 60 | 51 | 35 | 28 | 31 | 72 | 74 | 73 | 42 | 17 | 24 | 29 | 28 | 28 | 24 | 11 | 15 | 49 | 49 | 49 | 43 | 43 | 43 | 49 | 47 | 48 | 42 | 26 | 32 | 24 | 12 | 16 |
| LibLinear | 39.9 | 41 | 47 | 44 | 26 | 23 | 25 | 61 | 67 | 64 | 27 | 18 | 22 | 24 | 24 | 24 | 15 | 12 | 14 | 43 | 45 | 44 | 38 | 37 | 38 | 39 | 39 | 39 | 31 | 25 | 27 | 17 | 13 | 15 | |
| LibSVM | 41.2 | 41 | 50 | 45 | 30 | 22 | 25 | 66 | 66 | 66 | 30 | 23 | 26 | 26 | 25 | 26 | 22 | 15 | 18 | 48 | 44 | 46 | 39 | 44 | 41 | 45 | 38 | 41 | 33 | 28 | 30 | 21 | 13 | 16 | |
| Hist+ngram | LibLinear | 41.2 | 44 | 52 | 47 | 29 | 27 | 28 | 68 | 70 | 69 | 32 | 19 | 24 | 26 | 26 | 26 | 15 | 12 | 13 | 44 | 45 | 45 | 40 | 38 | 39 | 43 | 42 | 42 | 31 | 23 | 26 | 17 | 12 | 14 |
| LibSVM | 44.9 | 45 | 62 | 52 | 36 | 27 | 30 | 74 | 66 | 70 | 39 | 12 | 19 | 28 | 31 | 29 | 26 | 05 | 08 | 49 | 43 | 46 | 42 | 49 | 45 | 52 | 42 | 46 | 39 | 19 | 26 | 23 | 09 | 13 | |
| All Binary | CRFSuite | 34.7 | 60 | 51 | 55 | 51 | 32 | 39 | 78 | 72 | 75 | 39 | 13 | 19 | 33 | 17 | 22 | 28 | 10 | 15 | 53 | 40 | 46 | 46 | 31 | 37 | 58 | 37 | 45 | 42 | 18 | 25 | 28 | 06 | 10 |
| LibLinear | 34.6 | 53 | 60 | 56 | 41 | 39 | 40 | 69 | 73 | 71 | 32 | 21 | 25 | 27 | 25 | 26 | 23 | 18 | 20 | 45 | 47 | 46 | 44 | 43 | 43 | 45 | 45 | 45 | 35 | 26 | 30 | 18 | 12 | 14 | |
| All Multi | CRFSuite | 50.4 | 56 | 65 | 60 | 46 | 42 | 44 | 74 | 78 | 76 | 41 | 21 | 28 | 31 | 29 | 30 | 29 | 13 | 18 | 50 | 52 | 51 | 46 | 49 | 47 | 53 | 52 | 52 | 42 | 28 | 34 | 26 | 14 | 18 |
| LibLinear | 47.7 | 54 | 60 | 57 | 43 | 40 | 41 | 69 | 73 | 71 | 35 | 20 | 25 | 29 | 28 | 28 | 22 | 16 | 18 | 47 | 49 | 48 | 45 | 44 | 45 | 49 | 49 | 49 | 38 | 28 | 32 | 21 | 15 | 18 | |
| Libsvm | 51.6 | 56 | 68 | 62 | 50 | 41 | 45 | 72 | 78 | 75 | 37 | 20 | 26 | 33 | 25 | 29 | 25 | 06 | 10 | 53 | 47 | 50 | 46 | 57 | 51 | 54 | 52 | 53 | 43 | 29 | 34 | 32 | 13 | 19 | |
Classifiers and categories
Table 2 shows that LibS has the highest accuracy at 51.6%, closely followed by CRF at 50.4% with LibL at 47.7%. All three classifiers outperform the simple baseline (Base) by a large margin. The latter consists of multinomial trials, which randomly label new instances according to the percentage of each CoreSC in the training data. We have also considered an n-gram baseline for both CRF and SVM and a history+n-gram baseline for SVM (history is implicit for CRF), which are discussed in the section on Feature Contribution. The best results overall are obtained from multi-class classification using all the features we considered. Interestingly the combination of binary classifiers (one for each CoreSC category) gave the highest precision in most cases but recall was significantly lower than in the multi-class scenario.
There is not a significant difference in performance between LibS+all features and CRF: five categories seem to be predicted better by LibS and for the other six CRF performs better. When the history feature is absent, LibS and LiBL perform much lower than CRF but hist+n-gram for LibS is comparable to the n-gram performance of CRF. This highlights the importance of category sequence information for the task. The performance of LibL lags slightly behind both LiBS and CRF but this is to be expected since it is an approximation for linear SVMs.
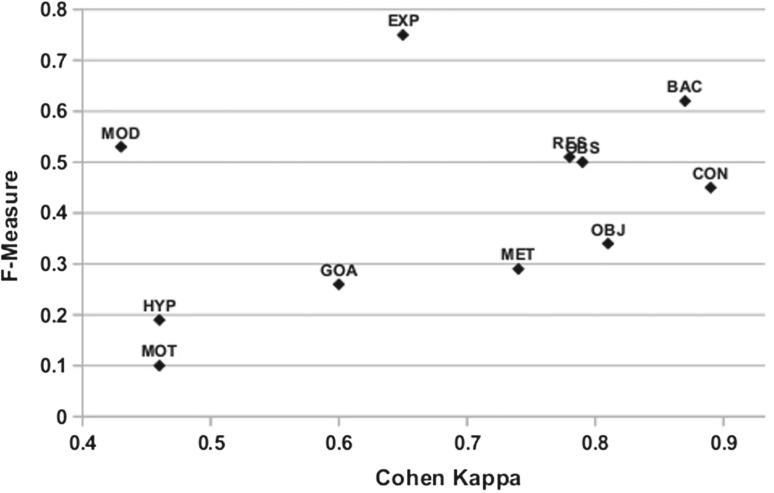
The highest performing categories for all three classifiers are EXP, BAC and MOD with an F-score of 76%, 62% and 53%, respectively. BAC is the second most frequent category (19%) in the corpus after RES, so high recall is not surprising. EXP and MOD (experimental and theoretical methods) are more interesting, as they are moderately frequent (10 and 9%), respectively. Furthermore, EXP and MOD are the only categories which have a higher F-score in automatic recognition compared with κ-IAA (Liakata et al., 2010) as shown in Figure 3. On the other hand categories with high κ such as CON, MET and OBJ were more difficult to classify than expected. While κ was measured on only 41 papers (5022 sentences) (Liakata et al., 2010), which may not be representative of the entire corpus, these results suggest that there is not necessarily a direct correlation between annotator agreement and classifier performance. This is in support of Beigman Klebanov and Beigman, 2009, which argues that IAA is neither sufficient nor necessary for obtaining reliable data from annotated material but rather it is important to focus on non-noisy, ‘easy’ instances.
Fig. 3.
F-score versus κ for CoreSCs.
Beigman Klebanov and Beigman, 2009 suggest researchers should report the level of noise in a corpus and only use non-noisy (easy) instances for testing. They emphasize the importance of requiring the agreement between more than two annotators, which reduces both the chance of random agreements as well as hard case bias, whereby a classifier tends to model the pattern of bias of a particular annotator for instances which are hard to predict. By having different phases of corpus development, with a varied number of annotators for each phase and subset of the corpus as well as a large number of classification categories, we believe that we have minimized the chance of random agreements and hard case bias.
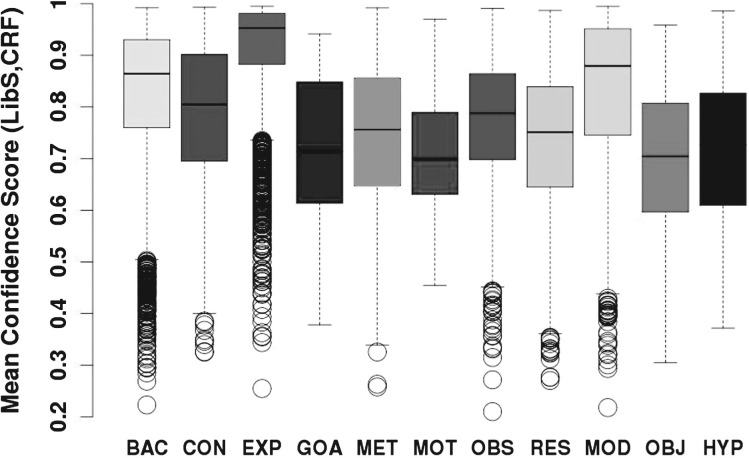
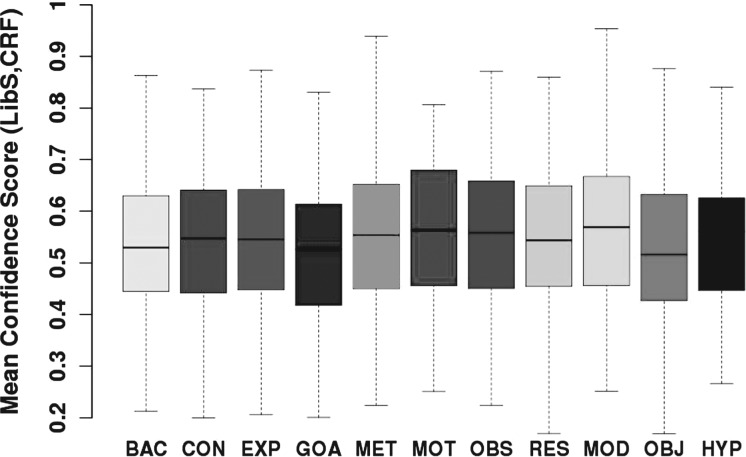
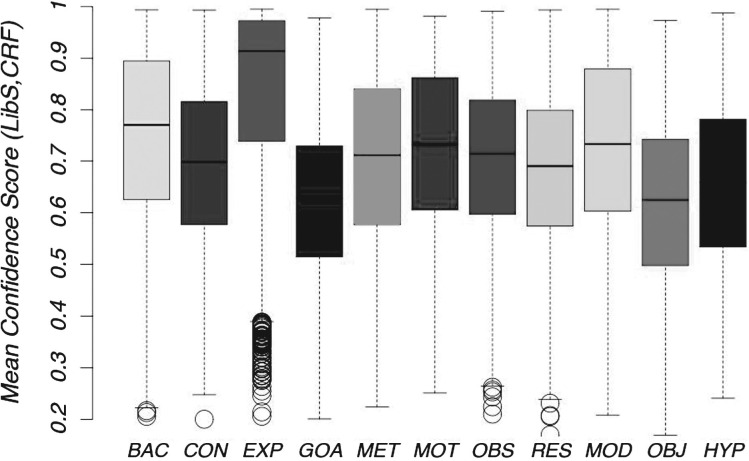
Therefore, we can infer that when our machine learning annotations agree with manual annotations, noise levels will be usually low, instances will be easier to predict and thus classifier confidence will be higher. Indeed this is confirmed both by a Pearson moment correlation test between agreement and classifier confidence and a Welch T-test for classifier confidence values in cases of agreement and disagreement, both of which gave a p<2.2e-16 at 99%. They showed a direct correlation between classifier confidence and agreement between manual annotation and classifiers. Details are in the Supplementary Material. Classifier confidence for an instance is a probability, where a high value indicates high classifier confidence for the particular prediction. As an indication of the noise for different categories in the corpus, we show the confidence of the machine learning classifiers when both classifiers agree with the manual annotation and when there is no agreement between either the classifiers or the manual annotation (see Figs 4 and 5). For the cases where LibSVM agrees with CRF and the manual annotation, confidence scores are high, with over 75% of the data having a confidence value of >0.6, and over 50% of the data having a confidence score of over 0.7. This can be compared against the situation of disagreement where only 25% of the data have a confidence score of 0.6. For EXP, BAC and MOD the confidence scores are especially high in cases of agreement, with 50% of the data having a confidence score of over 0.87. Therefore, agreements for EXP and MOD consist mostly of non-noisy (easy) instances. Classifier confidence for the entire corpus is depicted in Figure 6. Assuming that lack of noise correlates with high classifier confidence, we can say that >50% of data in each category (and in most well beyond 75%) is non-noisy.
Fig. 4.
Confidence value when LibS, CRFSuite and manual annotation agree.
Fig. 5.
Confidence value when there is no agreement on annotation.
Fig. 6.
Confidence value scores per category for the entire corpus.
Classifier performance for the CoreSC categories can be ranked from highest to lowest F-score as follows: EXP > BAC > MOD > RES > OBS > CON > OBJ > MET > GOA > HYP > MOT. OBJ performs well given its low frequency, suggesting that OBJ sentences contain distinct features. The low scores for MET may be due to noise introduced by our neglect of the distinction between MET-Old and MET-New (Liakata et al., 2010). The low F-score for MOT and HYP are due to their low frequency as the levels of noise are similar to those of OBJ. We intend to boost performance for the low frequency categories by using active learning.
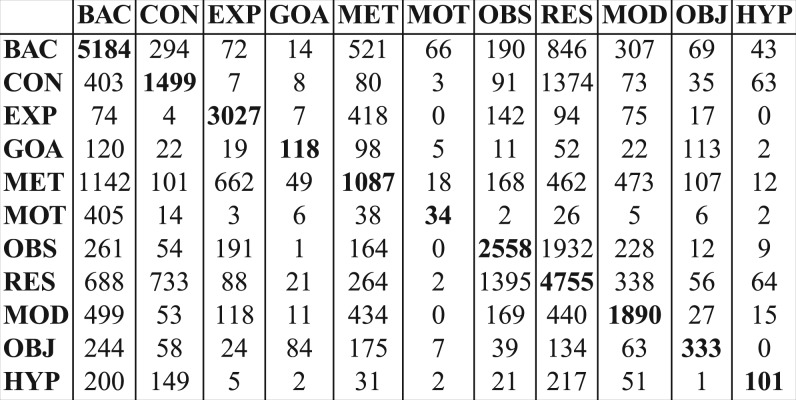
These are promising results given the complexity of the task, the number of the categories and their distribution in the corpus. A confusion matrix (Fig. 7) gives an indication of which categories have consistent overlaps. There is bias in favour of the BAC category due to its high frequency and broad definition, which we will need to counterbalance in the future. CON is often taken as RES whereas RES is often confused with OBS and vice versa. GOA is often assigned to OBJ and MET, the latter presumably because goals and method are often expressed in the same sentence. MET is confused with EXP and BAC, the latter because we have not yet considered the second layer of CoreSC annotation at this stage, which caters for MET-Old, methods mentioned in previous work. OBJ is often confused with MET, since a method can be the object of an investigation.5 Finally, HYP is often assigned to RES, CON and BAC. This can be explained by the fact that a weak result or conclusion is often expressed in the same language as a hypothesis, while a hypothesis may also be expressed as an assumption arising from background knowledge. For examples see the Supplementary Material.
Fig. 7.
Confusion matrix for CoreSC categories according to LibS.
If we merge CoreSC categories so that we consider a coarser grain layer of four categories, namely Prior (BAC), Approach (MET+MOD+EXP), Outcome (OBS+RES+CON) and Objective (MOT+GOA+HYP+OBJT) then our F-measures respectively become: BAC: 59%, Approach: 72%, Outcome: 81%, Objective: 38%. A variant merge with seven categories, roughly corresponding to the scheme proposed by de Waard et al., 2009, which considers BAC, HYP, Problem(=MOT), GOA=(GOA+OBJT), MET= (MET+EXP+MOD), RES=(OBS+RES), Implication(=CON), gives us F1: BAC: 60%, CON: 44%, MET: 72%, GOA: 47%, MOT: 19%, HYP: 18% and RES: 72% This shows the flexibility of our scheme for different applications, which may require different levels of granularity.
Feature contribution
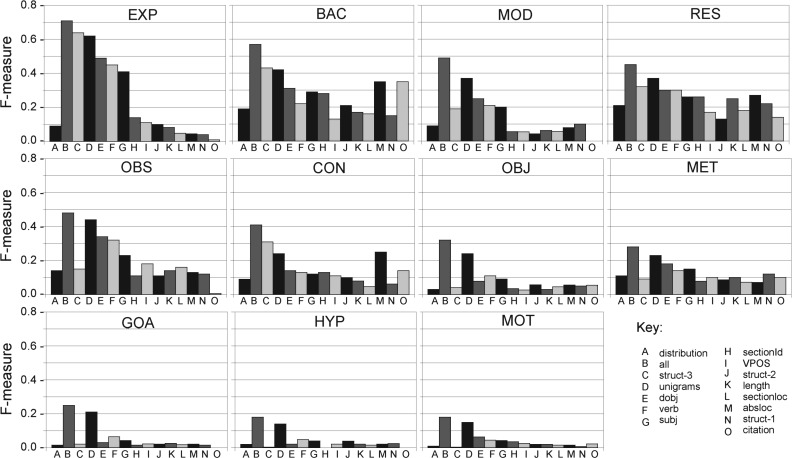
we examine feature contribution in LOOF cross-validation and single feature runs, using CRF and LibL. Tables 3 and 4 show how F-score is affected when each type of feature is omitted. For each CoreSC category we have highlighted the lowest scores (bold), corresponding to the most important features being left out, and the highest scores (italic), corresponding to features whose omission has less impact on classification. Performance for all categories drops when all n-grams are removed. Since features are not independent, many of the important features of other categories are covered in n-grams but this does not necessarily work in both directions. Primarily, bigrams are more important than unigrams, since many of the former contain the latter. Categories affected most by the omission of unigrams are the low frequency categories GOA, MOT and HYP for CRF and MOT, HYP for LibL. Bigrams are not as important for these categories and removing them improves performance in the case of MOT and HYP. This is probably because they are not frequent enough for association with bigrams. Removing the verb feature has a negative effect on MOT, HYP and GOA in CRF and GOA and HYP in LibL. This agrees with our observation of the importance of verbs in single feature classification (Fig. 8). The high frequency categories are more robust to omission of features, whereas the lower frequency categories are dependent on all features.
Table 3.
F-measures for CRFSuite LOOF, 9-fold cross-validation
| Feat | BAC | CON | EXP | GOA | MET | MOT | OBS | RES | MOD | OBJ | HYP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| all | 60 | 44 | 76 | 28 | 30 | 18 | 51 | 47 | 52 | 34 | 18 |
| length | 60 | 44 | 76 | 27 | 30 | 18 | 51 | 47 | 53 | 34 | 19 |
| ref | 58 | 44 | 76 | 26 | 30 | 18 | 51 | 47 | 52 | 34 | 17 |
| absloc | 60 | 44 | 76 | 28 | 29 | 18 | 51 | 47 | 52 | 34 | 18 |
| struct1 | 60 | 43 | 76 | 27 | 30 | 18 | 51 | 47 | 52 | 33 | 17 |
| secid | 60 | 44 | 76 | 27 | 30 | 18 | 51 | 48 | 51 | 35 | 17 |
| Struct-2 | 60 | 44 | 76 | 27 | 30 | 17 | 51 | 47 | 52 | 34 | 18 |
| SecLoc | 60 | 44 | 76 | 27 | 30 | 19 | 51 | 47 | 52 | 34 | 18 |
| Struct-3 | 60 | 43 | 75 | 26 | 30 | 18 | 51 | 47 | 52 | 34 | 19 |
| uni | 60 | 44 | 76 | 24 | 29 | 17 | 50 | 47 | 51 | 33 | 15 |
| bi | 59 | 43 | 75 | 27 | 30 | 22 | 50 | 46 | 50 | 32 | 19 |
| ngrams | 58 | 42 | 74 | 25 | 29 | 17 | 47 | 45 | 48 | 29 | 14 |
| gr | 60 | 44 | 75 | 27 | 30 | 18 | 51 | 47 | 52 | 35 | 17 |
| pos | 60 | 44 | 76 | 26 | 29 | 18 | 51 | 47 | 53 | 34 | 16 |
| subj | 60 | 45 | 76 | 27 | 30 | 17 | 51 | 48 | 52 | 34 | 18 |
| dobj | 60 | 45 | 76 | 28 | 30 | 18 | 51 | 47 | 53 | 34 | 19 |
| iobj | 60 | 44 | 76 | 27 | 30 | 18 | 51 | 47 | 52 | 34 | 18 |
| obj2 | 60 | 44 | 76 | 28 | 30 | 18 | 51 | 47 | 52 | 34 | 18 |
| vclass | 60 | 44 | 76 | 27 | 30 | 18 | 51 | 47 | 52 | 34 | 18 |
| verb | 60 | 44 | 76 | 27 | 30 | 17 | 51 | 47 | 52 | 34 | 17 |
Table 4.
F-measures for LibLinear LOOF, 9-fold cross-validation
| Feat | BAC | CON | EXP | GOA | MET | MOT | OBS | RES | MOD | OBJ | HYP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| all | 57 | 41 | 71 | 25 | 28 | 18 | 48 | 45 | 49 | 32 | 18 |
| history | 55 | 41 | 71 | 28 | 27 | 20 | 48 | 43 | 46 | 32 | 17 |
| length | 57 | 42 | 71 | 24 | 28 | 19 | 48 | 45 | 48 | 31 | 17 |
| ref | 55 | 41 | 71 | 25 | 28 | 19 | 48 | 45 | 48 | 31 | 15 |
| absloc | 57 | 40 | 72 | 25 | 28 | 20 | 48 | 45 | 49 | 33 | 18 |
| Struct-1 | 57 | 41 | 71 | 26 | 28 | 21 | 48 | 45 | 49 | 31 | 17 |
| secid | 57 | 41 | 71 | 26 | 28 | 19 | 48 | 44 | 48 | 32 | 17 |
| Struct-2 | 57 | 41 | 71 | 25 | 28 | 19 | 48 | 45 | 49 | 32 | 17 |
| SecLoc | 57 | 41 | 71 | 26 | 28 | 18 | 48 | 45 | 48 | 32 | 18 |
| Struct-3 | 56 | 40 | 70 | 25 | 27 | 19 | 48 | 44 | 47 | 30 | 19 |
| uni | 56 | 41 | 72 | 26 | 27 | 17 | 47 | 44 | 46 | 31 | 16 |
| bi | 54 | 40 | 70 | 25 | 27 | 20 | 46 | 43 | 45 | 27 | 17 |
| ngrams | 53 | 37 | 69 | 23 | 26 | 17 | 44 | 42 | 41 | 26 | 12 |
| gr | 56 | 40 | 71 | 23 | 29 | 19 | 47 | 44 | 49 | 31 | 17 |
| pos | 57 | 42 | 71 | 25 | 28 | 20 | 48 | 45 | 49 | 32 | 16 |
| subj | 57 | 41 | 71 | 25 | 29 | 21 | 47 | 45 | 48 | 31 | 17 |
| dobj | 57 | 42 | 71 | 25 | 28 | 20 | 48 | 45 | 49 | 32 | 19 |
| iobj | 57 | 41 | 71 | 26 | 28 | 19 | 48 | 45 | 48 | 32 | 17 |
| obj2 | 57 | 41 | 71 | 25 | 28 | 18 | 48 | 45 | 49 | 32 | 18 |
| vclass | 57 | 41 | 71 | 26 | 28 | 19 | 48 | 45 | 49 | 33 | 17 |
| verb | 57 | 41 | 71 | 24 | 28 | 20 | 48 | 45 | 49 | 32 | 16 |
Fig. 8.
Single feature classification with LibL, illustrating the contribution of 15 individual features.
Single feature classification is more meaningful with respect to individual feature contributions and Figure 8 paints a clear picture of which features are most important for which category. We believe this to be the most interesting finding of our analysis. While Figure 8 shows the general trend whereby n-grams (D) (bigrams and GRs are not actually shown in Figure 8, but they strongly correlate with unigrams) followed by direct object (E) and verb (F) as accounting for the overall F-measure of a category, this is not true for all categories. For EXP, BAC and CON section headings (C) matter more than n-grams and for BAC, CON and RES absolute location (M) also plays a prominent role, meaning that the location of these three categories tends to be fixed in a paper (presumably in the beginning and the end). Citations (O) play an important role in discriminating BAC and are also prominent for CON, RES and MET to some extent. Verbs (F) are usually more important than subjects (G) but slightly less important than direct objects (E), however verbs (F) feature more prominently for categories such as RES, GOA, HYP and OBJ suggesting that particular verbs are used in the context of these categories. Perhaps more feature engineering involving semantic categories of verbs would benefit the low frequency categories. Verb tense (expressed by VPOS (I)) does not seem to play a major role, though its contribution is higher for OBS and RES. Looking at the feature profile of different categories, RES and MET show the least variation between individual feature contribution but it is clear that RES is more location specific than MET.
Table 5 shows the number of individual features considered for each feature type. The vast majority of features are bigrams (42 438), unigrams (10 515) and GR triples (11 854), which also explains their importance for the classification. This makes the prominence of citations and global structural features such as section headings all the more important whenever we encounter them.
Table 5.
Numbers for each type of feature
| Feat | Uni | Bi | GR | VPOS | Subj | Dobj | Iobj | Obj2 | Verb | VC | P | H | Gl | L | C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 10 515 | 42 438 | 11 854 | 6 | 3843 | 7414 | 45 | 59 | 1543 | 10 | 1 | 12 | 53 | 9 | 3 |
Numbers for each type of feature were: L, length; H, history; C, citation; Gl, global features, including absloc, sectionid, struct1-3, sectionloc
Variants of some of the above features have been used by Teufel and Moens (2002), Mullen et al. (2005) and Merity et al. (2009) to automate AZ. Merity et al., 2009 found that n-grams (unigrams and bigrams) in combination with knowledge of the label of previous sentences (history) constituted a very strong baseline for AZ. This agrees with our findings in general, where n-grams are roughly responsible for 40% of the system accuracy, the history category contributes another 5% and a further 5–6% is due to all other features.
In the future, we intend to consider more elaborate semantic classes for features and also consider training individual classifiers for each category which we would then combine using stacking or ensemble techniques.
Comparison with related work
a direct comparison between our results and earlier work is not possible, as the scope, schemes and experimental settings differ significantly. Earlier work on automating discourse schemes with four categories (Hirohata et al., 2008; Lin et al., 2006; McKnight and Srinivasan, 2003) has reported F-measures in the 80s or 90s. However, in addition to having a third of the number of categories, these schemes only concentrate on abstracts, which are shown to have a very different structure from full articles. Shatkay et al. (2008) annotate sentences from full articles but they evaluate on a small scale and do not attempt to classify an entire article. Their scheme has only three to four categories per dimension, where each dimension is evaluated separately from the rest. Our results are more comparable to Mullen et al. (2005) and Teufel (2000), who have automated AZ for articles with six and seven categories, respectively, reporting respective F-measures 0.44–0.87 and 0.26–0.86. Merity et al. (2009) replicated and significantly improved on the results of Teufel (2000), reporting an F-measure in the 90s for the same categories. However, Teufel et al. (2009) introduced a new scheme, designed specifically for chemistry papers (ChemAZ), containing 15 categories, which has not been yet automated.
It has been shown that a small number of categories annotated by a small number of experts will result in a less challenging annotation task, leading to a higher F-measure. However, a more expressive and thus more complex annotation scheme allows for better representation of the discourse structure of the articles so as to identify hypotheses and relevant evidence (see Section 1). This will contribute to more advanced information extraction solutions in the future.
4 APPLICATIONS AND FUTURE WORK
One of the applications stemming from our work is the use of automatically generated CoreSC annotations for the production of extractive summaries of full papers in chemistry and biochemistry. Such summaries are different from abstracts as they are longer (20 sentences) and represent the entire content of the paper, from Background and Hypotheses to Experiments performed, main Observations and Results obtained. The idea is that such summaries could be read much faster than the paper but convey a lot more of the key information than the abstract, which often acts as a selling point of the paper.
We created summaries so that each contained 1–2 sentences from each CoreSC category (Hypothesis, Background, etc.), extracted from the original paper, following the distribution of categories in the paper. These summaries were given to 12 experts divided into 4 groups, along with summaries created using Microsoft Autosummarize and summaries written by humans. The automatically generated summaries performed significantly better than Miscrosoft autosummarize and achieved a 66% and 75% precision in answering complex content based questions. In some cases they outperformed human summaries.6 Question-based extractive summaries created using CoreSCs could be used to help speed up curation and we plan to explore this in the future.
A different user based study, involved collaboration with experts in CRA, who were presented with abstracts that contained CoreSC annotations and abstracts with no annotations, or annotations originating from simpler schemes (abstract sections or an AZ variant) (Guo et al., 2011). Three experts were timed as they answered questions about the main objectives and methods described in abstracts and it was shown that experts responded consistently faster when given abstracts annotated with CoreSCs than in the rest of the cases, while no significant difference was observed pertaining to the quality of the responses. In the future, we plan to perform more question based user studies with CRA experts, using full papers.
We also plan to use CoreSC annotated papers in biology to guide information extraction and retrieval, characterize extracted events and relations and also facilitate inference from hypotheses to conclusions in scientific papers. Our web-based tool for the automatic annotation of CoreSC categories in full biomedical papers from Pubmed Central is available for biologists to download and use.
The ability to automatically identify and qualify discourse structure from the scientific literature has far-reaching implications. The original facts and results from a scientific publication form the key information to be extracted in order to curate biological resources and validate against resources such as UnitProtKb, EntrezGene, Reactome and others. The different types of conceptualization zones defined by CoreSCs (Background, Hypothesis, Method, etc.) so far have been used to create extractive summaries and more use cases of filtering text during information extraction are in progress. Work in progress also involves the application of CoreSC annotations to full papers involving CRA and drug–drug interactions and preliminary results show that the annotation scheme and categorization methods generalize well to these new domains.
Supplementary Material
ACKNOWLEDGEMENT
We are very grateful to Dr Stephen Clark, Prof. Bonnie Webber, Dr Naoaki Okazaki, Dr Nigel Collier, Dr Anna Korhonen, Yufan Guo, Dr Ian Lewin, Jee-Hyub Kim, Dr Simone Teufel, Dr Amanda Clare and Dr Andrew Sparkes for their useful feedback. We also thank our domain experts (28 in total) who helped us with corpus annotation and summary evaluation and Dr Colin Sauze for his help with the code interfacing the machine learning model and the online tool. Last but not least we would like to thank the anonymous reviewers for their comments.
Funding: This work was funded by JISC (UK) (SAPIENT Automation project), The Leverhulme Trust (Early Career Fellowship for Dr Liakata) and the EMBL-EBI.
Conflict of Interest: none declared.
Footnotes
1one of the 225 papers had been annotated already in phase II, giving a total of 265 unique papers
2Testing is done sentence by sentence and so takes longer than training.
3A section is a block of sentences between two headings.
4www.lextek.com/manuals/onix/stopwords1.html reduced to 186 words
5See definitions in Supplementary Material.
6The details of this experiment is the focus of a separate publication under submission.
REFERENCES
- Ananiadou S., et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Beigman Klebanov,B., Beigman E. From annotator agreement to noise models. Comput. Linguist. 2009;35:495–503. [Google Scholar]
- Brandow R., et al. Automatic condensation of electronic publications by sentence selection. Inform. Process. Manag. 1995;31:675–685. [Google Scholar]
- Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:27:1–27:27. [Google Scholar]
- Ciccarese P., et al. The swan biomedical discourse ontology. J. Biomed. Inform. 2008;41:739–751. doi: 10.1016/j.jbi.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.M., Hersh W.R. A survey of current work in biomedical text a survey of current work in biomedical text mining. Brief. Bioinform. 2005;6:57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. [Google Scholar]
- Curran J., et al. Proceedings of the 45th Annual Meeting of the Association for Computational Linguistics Companion Volume Proceedings of the Demo and Poster Sessions. Prague, Czech Republic: Association for Computational Linguistics; 2007. Linguistically motivated large-scale nlp with c&c and boxer; pp. 33–36. [Google Scholar]
- de Waard A., et al. Proceedings of the Eighth International Conference on Computational Semantics, IWCS-8 '09. Stroudsburg, PA, USA: Association for Computational Linguistics; 2009. Identifying the epistemic value of discourse segments in biology texts; pp. 351–354. [Google Scholar]
- de Waard A. Proceedings of the 2nd International Conference on Pragmatic Web, ICPW '07. New York, NY, USA: Association for Computing Machinery; 2007. A pragmatic structure for research articles; pp. 83–89. [Google Scholar]
- Fan R.-E., et al. LIBLINEAR: a library for large linear classification. J. Mach. Learn. Res. 2008;9:1871–1874. [Google Scholar]
- Guo Y., et al. Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. Uppsala, Sweden: Association for Computational Linguistics; 2010. Identifying the information structure of scientific abstracts: an investigation of three different schemes; pp. 99–107. [Google Scholar]
- Guo Y., et al. A comparison and user-based evaluation of models of textual information structure in the context of cancer risk assessment. BMC Bioinform. 2011;12:69 doi: 10.1186/1471-2105-12-69. doi:10.1186/1471-2105-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohata K., et al. Identifying sections in scientific abstracts using conditional random fields. Proceedings of the IJCNLP 2008. 2008 [Google Scholar]
- Kilicoglu H., Bergler S. Recognizing speculative language in biomedical research articles: a linguistically motivated perspective. BMC Bioinform. 2008;9(Suppl. 11):S10. doi: 10.1186/1471-2105-9-S11-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-D., et al. Proceedings of BioNLP Shared Task 2011 Workshop. Portland, Oregon, USA: Association for Computational Linguistics; 2011. Overview of BioNLP shared task 2011; pp. 1–6. [Google Scholar]
- Kummerfeld J.K., et al. Faster parsing by supertagger adaptation. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, ACL '10. 2010:345–355. [Google Scholar]
- Kupiec J., et al. Proceedings of the 18th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR '95. New York, NY, USA: Association for Computing Machinery; 1995. A trainable document summarizer; pp. 68–73. [Google Scholar]
- Liakata M., Soldatova L. Guidelines for the annotation of general scientific concepts. Technical report, JISC Project Report 88, Aberystwyth University; 2008. http://ie-repository.jisc.ac.uk/88/ [Google Scholar]
- Liakata M., Soldatova L. The ART Corpus. Technical report, Aberystwyth University; 2009. http://hdl.handle.net/2160/1979. [Google Scholar]
- Liakata M., et al. Proceedings of BioNLP-09. Boulder, Colorado: 2009. Semantic annotation of papers: interface & enrichment tool (SAPIENT) pp. 193–200. [Google Scholar]
- Liakata M., et al. Proceedings of the Seventh International Conference on Language Resources and Evaluation (LREC'10) Valletta, Maltapp: European Language Resources Association; 2010. Corpora for the conceptualization and zoning of scientific papers; pp. 2054–2061. [Google Scholar]
- Light M., et al. The language of bioscience: facts, speculations, and statements in between. In: Hirschman L., Pustejovsky J., editors. HLT-NAACL 2004 Workshop: BioLINK 2004, Linking Biological Literature, Ontologies and Databases. Boston, Massachusetts, USA: Association for Computational Linguistics; 2004. pp. 17–24. [Google Scholar]
- Lin J., et al. Proceedings of the Workshop on Linking Natural Language Processing and Biology: Towards Deeper Biological Literature Analysis, BioNLP '06. Morristown, NJ, USA: Association for Computational Linguistics; 2006. Generative content models for structural analysis of medical abstracts; pp. 65–72. [Google Scholar]
- McKnight L., Srinivasan P. Categorization of sentence types in medical abstracts. AMIA Annu Symp Proc. 2003:440–444. [PMC free article] [PubMed] [Google Scholar]
- Medlock B., Briscoe T. 45th Annual Meeting of the ACL. Prague, Czech Republic: 2007. Weakly supervised learning for hedge classification in scientific literature; pp. 23–30. [Google Scholar]
- Merity S., et al. Proceedings of the 2009 Workshop on Text and Citation Analysis for Scholarly Digital Libraries, NLPIR4DL '09. Morristown, NJ, USA: Association for Computational Linguistics; 2009. Accurate argumentative zoning with maximum entropy models; pp. 19–26. [Google Scholar]
- Minnen G., et al. Applied morphological processing of english. Nat. Lang. Eng. 2001;7:207–223. [Google Scholar]
- Mizuta Y., et al. Zone analysis in biology articles as a basis for information extraction. Int. J. Med. Inform. 2006;75:468–487. doi: 10.1016/j.ijmedinf.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Mullen T., et al. A baseline feature set for learning rhetorical zones using full articles in the biomedical domain. SIGKDD Explor. 2005;7:52–58. [Google Scholar]
- Nawaz R., et al. Meta-knowledge annotation of bio-events. LREC. 2010:2498–2507. [Google Scholar]
- Okazaki N. CRFsuite: a fast implementation of Conditional Random Fields (CRFs) 2007 http://www.chokkan.org/software/crfsuite/(20 February 2012, date last accessed) [Google Scholar]
- Rimell L., Clark S. Porting a lexicalized-grammar parser to the biomedical domain. J. Biomed. Inform. 2009;42:852–865. doi: 10.1016/j.jbi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Ruch P., et al. Using argumentation to extract key sentences from biomedical abstracts. Int. J. Med. Inform. 2007;76:195–200. doi: 10.1016/j.ijmedinf.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Shatkay H., et al. Multi-dimensional classification of biomedical text: toward automated, practical provision of high-utility text to diverse users. J. Bioinform. 2008;24:18:2086–2093. doi: 10.1093/bioinformatics/btn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.F.d., et al. Proceedings of the 9th Portuguese Conference on Artificial Intelligence: Progress in Artificial Intelligence, EPIA '99. London, UK: Springer; 1999. Using localmaxs algorithm for the extraction of contiguous and non-contiguous multiword lexical units; pp. 113–132. [Google Scholar]
- Soldatova L., King R. An ontology of scientific experiments. J. Roy. Soc. Interf. 2006;3:795–803. doi: 10.1098/rsif.2006.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova L., Liakata M. An ontology methodology and cisp-the proposed core information about scientific papers. Technical Report. JISC Project Report 137, Aberystwyth University; 2007. http://ie-repository.jisc.ac.uk/137/ [Google Scholar]
- Teufel S., Moens M. Summarizing scientific articles: experiments with relevance and rhetorical status. Comput. Linguist. 2002;28:409–445. [Google Scholar]
- Teufel S., et al. An annotation scheme for discourse-level argumentation in research articles. Proceedings of EACL. 1999:110–117. [Google Scholar]
- Teufel S., et al. Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing: Volume 3 - Volume 3, EMNLP '09. Stroudsburg, PA, USA: Association for Computational Linguistics; 2009. Towards discipline-independent argumentative zoning: evidence from chemistry and computational linguistics; pp. 1493–1502. [Google Scholar]
- Teufel S. PhD Thesis. Edinburgh: School of Cognitive Science, University of Edinburgh; 2000. Argumentative Zoning: Information Extraction from Scientific Text. [Google Scholar]
- Teufel S. The Structure of Scientific Articles: Applications to Citation Indexing and Summarization. Stanford, California, United States: Stanford: CSLI Publications, CSLI Studies in Computational Linguistics; 2010. [Google Scholar]
- Thompson P., et al. Enriching a biomedical event corpus with meta-knowledge annotation. BMC Bioinform. 2011;12:393. doi: 10.1186/1471-2105-12-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., et al. Proceedings of BioNLP 2011 Workshop. Portland, Oregon, USA: Association for Computational Linguistics; 2011. Hypothesis and evidence extraction from full-text scientific journal articles; pp. 134–135. [Google Scholar]
- Wilbur W.J., et al. New directions in biomedical text annotations: definitions, guidelines and corpus construction. BMC Bioinform. 2006;7:356. doi: 10.1186/1471-2105-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.