Abstract
Chx10
and Vsx1 are the only Paired-like CVC (Prd-L:CVC) homeobox genes in the mouse genome. Both are expressed in the retina and have important but distinct roles in retinal development. Mutations in Chx10 cause reduced retinal progenitor cell (RPC) proliferation and an absence of bipolar cells, while mutations in Vsx1 impair differentiation of cone bipolar cells. Given their structural similarities and importance in retinal development, we sought to determine if a regulatory interaction exists between these genes and whether inactivation of both genes blocks initiation of retinal development. We found that Chx10 binds to a specific sequence in the Vsx1 5′-intergenic region and represses the activity of a luciferase reporter under the control of the Vsx1 promoter. This is consistent with our observation that there is an inverse relationship between the levels of Chx10 and Vsx1 immunostaining within the bipolar cell class. Furthermore, Vsx1 mRNA is upregulated in the RPCs of Chx10 deficient mice and zebrafish embryos injected with a chx10 morpholino. In mice deficient for both Chx10 and Vsx1 and zebrafish embryos co-injected with chx10 and vsx1 morpholinos, the changes in embryonic retinal development and marker expression are similar in magnitude to embryos with Chx10 loss of function only. From these studies, we propose that Vsx1 is a direct target of Chx10-mediated transcriptional repression. Although Vsx1 mRNA is upregulated in Chx10 deficient RPCs, Vsx1 does not genetically compensate for loss of Chx10, demonstrating that Prd-L:CVC genes, although important, are not absolutely required to initiate retinal development.
Keywords: homeobox, homeodomain, development, eye, microphthalmia, bipolar cell
1. Introduction1
A multitude of homeobox genes is required for retinal development, from the initial patterning events of the optic vesicle to the completion of terminal differentiation (reviewed in Chow and Lang, 2001; Dyer, 2003; Fuhrmann et al., 2000a; Levine and Green, 2004; Lupo et al., 2000; see also de Melo et al., 2003). In many cases, paralogous homeobox genes retain close relationships as indicated by similarities in expression patterns and mutant phenotypes. It is not uncommon, however, for these genes to develop cross-regulatory networks and diverge in function (for examples, see Czerny et al., 1999; Plouhinec et al., 2005). Identifying these relationships is essential for sorting out the complex transcription factor networks driving retinal development.
Prd-L:CVC proteins comprise a subgroup of homeodomain (HD) proteins based on two distinguishing structural characteristics: the presence of a Paired-like HD with a glutamine at position 50 (Prd-L, Q50; Galliot et al., 1999), and a region of approximately 60 amino acids of unknown function positioned immediately C-terminal to the HD. This region of extended conservation is referred to as the CVC domain after the three genes from which it was originally identified, ceh-10 (C.elegans; Svendsen and McGhee, 1995), vsx1 (goldfish; Levine et al., 1994), and Chx10 (mouse; Liu et al., 1994). Whereas ceh-10 is the only identified Prd-L:CVC gene in C. elegans, essentially all vertebrate genomes examined to date have two Prd-L:CVC genes that parse into two ortholog clusters, Chx10-like and Vsx1-like, named after their founding members. It is likely that duplication of a single Prd-L:CVC gene occurred prior to the vertebrate radiation to give rise to the Chx10-like and Vsx1-like paralogs (Chow et al., 2001; Passini et al., 1998).
Two conserved features of Prd-L:CVC genes during development are their expression in interneurons and importance in differentiation. ceh-10 is expressed in a restricted set of sensory interneurons and has a role in their fate specification (Altun-Gultekin et al., 2001; Svendsen and McGhee, 1995). In vertebrates, Chx10 and Vsx1 are expressed in interneuron populations in the spinal cord, hindbrain, and in retinal bipolar cells. Both genes are required for bipolar cell differentiation, although in different ways (see below). Biochemical studies also suggest a high degree of functional overlap between Prd-L:CVC proteins. Chx10 and Vsx1 proteins bind with high affinity to the same DNA sequence (Ferda Percin et al., 2000; Hayashi et al., 2000; Heon et al., 2002) and overexpression of human CHX10 or VSX1 protein represses transcription from the same heterologous reporter construct (Dorval et al., 2005). All Prd-L:CVC proteins contain an octapeptide motif that functions as a nuclear export signal (Knauer et al., 2005) and a nuclear localization signal(Kurtzman and Schechter, 2001). These motifs may work together to regulate the sub-cellular localization of Prd-L:CVC proteins.
Despite their close structural and biochemical relationships, Chx10 and Vsx1 differ in several ways. Sequences N-terminal of the HD and C-terminal of the CVC domain vary considerably and contain motifs not shared across the paralog groups such as the OAR motif in Chx10 orthologs (named for othopedia/aristaless/Rax; Furukawa et al., 1997) and the RV motif in Vsx1 orthologs (named for Rinx/Vsx1; Hayashi et al., 2000). Chx10 and Vsx1 also differ in their expression patterns. Chx10 expression (mRNA and protein) initiates during optic cup formation in presumptive retinal progenitor cells (RPCs) and remains expressed until RPCs exit the cell cycle and in the adult, Chx10 is expressed in bipolar cells and a subset of Muller glia (reviewed in Levine and Green, 2004; see also Rowan and Cepko, 2004). Vsx1 mRNA is detected during embryonic retinal development in several species (Chen and Cepko, 2000; D’Autilia et al., 2006; Passini et al., 1998; Passini et al., 1997). In contrast, Vsx1 protein is first detected late in retinal development and is expressed in differentiating bipolar cells (Chow et al., 2001; Decembrini et al., 2006; Ohtoshi et al., 2001; this study). In the adult, Vsx1 is expressed in a subset of cone bipolar cells.
Evidence for distinct contributions of Chx10 and Vsx1 to retinal biology is illustrated by their mutant phenotypes. Mutations in Chx10 cause microphthalmia in humans (Bar-Yosef et al., 2004; Ferda Percin et al., 2000) and mice (Burmeister et al., 1996), and antisense-chx10 RNA injected into zebrafish embryos causes a small eye phenotype (Barabino et al., 1997). Studies in ocular retardation J (orJ) mice, which carry a spontaneously-derived nonsense mutation in the HD (Y176stop) of Chx10 (Chx10orJ; Burmeister et al., 1996; Theiler et al., 1976), show that in addition to a lack of bipolar cells, the Chx10orJ homozygote (Chx10orJ/orJ) retina exhibits a profound decrease in RPC proliferation, a propensity to transdifferentiate along a pigmentation pathway, delays in embryonic neurogenesis, persistent neurogenesis in the adult retina, and an enrichment of adult ciliary epithelium derived retinal stem cells (Bone-Larson et al., 2000; Burmeister et al., 1996; Coles et al., 2006; Dhomen et al., 2006; Green et al., 2003; Horsford et al., 2005; Livne-Bar et al., 2006; Rowan et al., 2004; Rutherford et al., 2004).
On the other hand, retinal abnormalities associated with Vsx1 mutations are considerably less severe and more restricted. Microphthalmia is not observed and retinal histology appears normal in Vsx1 knockout mice (Chow et al., 2004; Ohtoshi et al., 2004). However, humans and mice with Vsx1 mutations have abnormal photopic electroretinogram (ERG) profiles associated with cone bipolar cell dysfunction (Heon et al., 2002; Mintz-Hittner et al., 2004; Valleix et al., 2006). Consistent with this, a restricted set of cone bipolar cells fails to complete their differentiation even though the full cohort of bipolar cells appear to be specified in Vsx1 knockout mice (Chow et al., 2004; Ohtoshi et al., 2004). Additionally, Vsx1 mutations in humans are also associated with corneal dystrophies such as keratoconus, possibly because of a role in corneal wound repair that is independent from its retinal function (Barbaro et al., 2006).
Since Chx10 and Vsx1 have several features in common but also mediate distinct aspects of retinal development, we set out to determine if a regulatory interaction exists between Chx10 and Vsx1 to control their expression. We also wanted to determine if Vsx1 promotes what remains of histogenesis in the Chx10 deficient retina. Data presented here provide evidence that Chx10 negatively regulates Vsx1 expression by direct transcriptional control. However, once relieved from this regulation, Vsx1 does not fill in for Chx10 during embryonic retinal development.
2. Results
2.1 Vsx1 is a candidate direct target of transcriptional repression by Chx10
We generated a polyclonal antiserum against a peptide corresponding to a unique sequence in the C-terminal variable region of mouse Vsx1. The Vsx1 antiserum detects nuclei positioned in the INL in a pattern consistent with previous reports of Vsx1 expression in bipolar cells of the adult wild type retina, whereas retinal sections from adult Vsx1 null mice (Vsx1τlacz/τlacz) are devoid of any staining (Fig. 1A,B). Western blots from retinal lysates P14 and older show that the antiserum detects a band of approximately 39 kDa in the wild type retina, which is consistent with the predicted size of the endogenous Vsx1 protein, and this band is absent in lysates from the Vsx1τlacz/τlacz retina (Fig. 1C). A second band at 64 kDa is detected in both lysates and is therefore assumed to not originate from the Vsx1 locus. In addition, embryonic retinal extracts contain a band of the same size as Vsx1 that persists in the Vsx1τlacz/τlacz lysates (data not shown). Importantly, immunohistochemical detection of these ‘non-Vsx1’ proteins in cryosections is not apparent. These observations demonstrate the utilities and limitations of our antiserum for analyzing Vsx1 expression in the mouse retina.
Figure 1. Endogenous expression pattern of Vsx1 and Chx10 in postnatal retina.
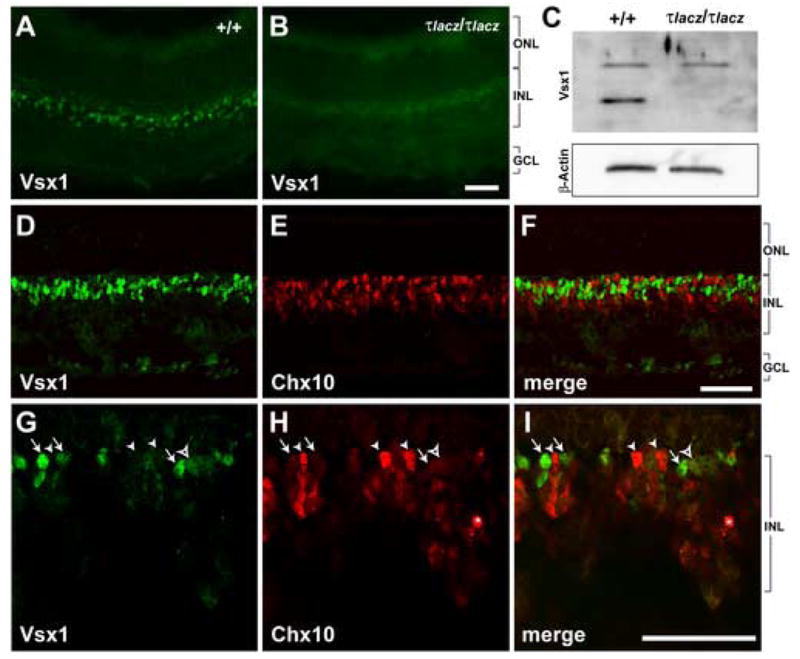
(A-C) The specificity of Vsx1 antibody was determined by immunohistochemistry (A, B) and by western blot analysis (C) on retinal samples from wild type (+/+) and Vsx1τlacz/τlacz (τlacz/τlacz) mice at P14 or older. The nuclear pattern of Vsx1 staining in the INL is observed in wild type (A) and absent in Vsx1τlacz/τlacz retina (B). (C) A band of the expected size (approximately 39 kDa) is present in wild type and absent in Vsx1τlacz/τlacz P14 retinal lysates. Another band of approximately 64 kDa is observed in both lysates. β-Actin was used as a loading control. (D-I) Confocal images showing Vsx1 (D, G) and Chx10 (E, H) expression patterns in P8 wild type retina. Merged images are shown in F and I. Arrows point to examples of cells with high Vsx1 and low Chx10 expression. Arrowheads point to examples of cells with low Vsx1 and high Chx10 expression. The open arrowhead points to a cell that co-expresses both proteins at low levels. Asterisks in H and I indicate a staining artifact. Scale bars: 40 μm. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Using double-label indirect-immunofluorescence, we directly compared the expression patterns of Chx10 and Vsx1 in the postnatal mouse retina (Fig. 1D–I). As expected, both proteins were localized to cells in the outer half of the inner nuclear layer (INL; Fig. 1D–F). However, cells showing the brightest staining with each antibody segregated into distinct populations as indicated by the lack of yellow nuclei in the merged images (Fig. 1F, I). Closer examination (Fig. 1G–I) revealed that most cells with Chx10 staining had little or no Vsx1 staining (closed arrowheads); some cells with low Chx10 staining had high Vsx1 staining (arrows); and occasional cells with low Chx10 staining also had low Vsx1 staining (open arrowheads). These data show that Chx10 and Vsx1 can be co-expressed in the same cells but not in a manner in which both proteins are expressed at their highest levels.
Prior to bipolar cell differentiation, Chx10 expression predominates. Chx10 expression is activated to relatively high levels in RPCs at the earliest stages of optic cup formation whereas Vsx1 is expressed at low levels, if at all. Two exceptions are the Vsx1 orthologues Chx10-1 in chick and vsx1 in Xenopus, whose transcripts are robustly detected in RPCs (Chen and Cepko, 2000; D’Autilia et al., 2006). Even so, the sum of the expression data suggests a regulatory mechanism exists to keep Vsx1 expression at low levels in Chx10 expressing cells. Consistent with this, Vsx1 mRNA is upregulated in the newborn Chx10orJ/orJ retina (Fig. 2A).
Figure 2. Vsx1 mRNA levels are increased in the Chx10orJ retina and Chx10 binds to a site upstream of the Vsx1 gene in vivo and in vitro.

(A) RT-PCR amplification products using wild type and Chx10orJ/orJ P0 retinal RNA and primers for Vsx1 and Gapdh. (B) Chromatin immunoprecipitation (ChIP) assay. Native chromatin-protein lysates were isolated from E16, P0, and P1 wild type retinas and immunoprecipitated with a sheep polyclonal Chx10 antibody (Chx10 ab). The precipitated chromatin was amplified by PCR using two non-overlapping primer sets that flank sequences conforming to the Chx10 consensus at positions −576 and −1275 relative to the Vsx1 transcription start site. The input lane shows amplification from chromatin not subjected to immunoprecipitation. Sheep IgG or H2O were used as negative controls. (C) Electrophoretic mobility shift assays (EMSA). Lane 1 shows the pattern of [32]P-labeled oligonucleotide probe containing the Chx10 consensus site at -576 (Vsx1wt) without addition of Chx10 protein. Lane 2 shows the resulting band pattern when in vitro translated Chx10 protein is mixed with the [32]P-labeled Vsx1wt probe in a binding reaction. Lane 3 and 4 show results of competition assays when unlabeled Vsx1 oligonucleotides are added in excess to the binding reaction (Vsx1wt comp (Lane 3) and Vsx1mut comp (Lane 4)). Addition of Chx10 ab to the binding reaction (lane 5) results in a supershifted band. The line over the sequences shown indicates the position of the Chx10 binding site sequence and asterisks indicate the nucleotide changes in the Vsx1mut oligonucleotide.
Since Chx10 can act as a transcription repressor, we asked whether Vsx1 is a target of Chx10. Based on a collection of known Chx10 binding sequences, Dorval and colleagues defined the following sequence as a consensus for Chx10 binding: PyTAATTPuPu (Py, pyrimidine; Pu, purine; Dorval et al., 2006). A scan of genomic DNA associated with the Vsx1 locus revealed two potential sites: one at −576 nt (TTAATTAG) and another at −1275 nt (CTAATTGG) relative to the Vsx1 transcriptional start site as predicted by ensembl (www.ensembl.org). Chx10 preferentially binds at or near the site positioned at −576 as determined by chromatin immunoprecipitation (ChIP) using native neonatal retinal lysates (Fig. 2B). Consistent with this, electrophoretic mobility shift assays (EMSA) show that in vitro translated Chx10 binds to the 32P-labeled Vsx1 probe containing the site at −576 (Vsx1wt; Fig. 2C). The binding of Chx10 to this probe is diminished with unlabeled Vsx1wt oligonucleotide in excess, but not by a variant containing a mutated Chx10 binding site (Vsx1mut). These data show that the Chx10 binding is dependent on the sequence conforming to the consensus. Addition of Chx10 antibody to the binding reaction resulted in a supershifted band, which indicates that the band observed in these assays is due to the association of Chx10 protein with the Vsx1wt probe.
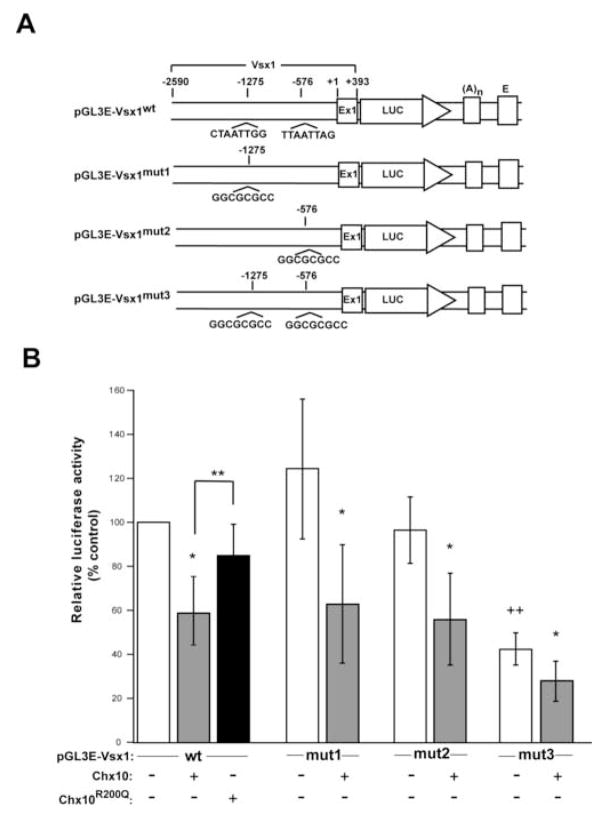
To further explore a potential transcriptional regulation of Vsx1 by Chx10, a Chx10 expression construct was co-transfected with the pGL3E luciferase reporter construct into HEK293 cells (Fig. 3). Approximately 2.6kb of Vsx1 genomic DNA containing the two putative Chx10 binding sites (−1275 and −576), the Vsx1 promoter, and 0.4 kb of exon 1 was cloned directly upstream of the luciferase cDNA. Consistent with its role as a transcriptional repressor, we found that Chx10 significantly inhibits luciferase activity (p-value <0.0001). We also tested a variant of Chx10 containing an arginine to glutamine replacement at residue 200 (Chx10R200Q). This mutation resides in the DNA binding helix of the HD (residue 53) and eliminates high affinity binding to Chx10 consensus sites and causes microphthalmia when inherited in a homozygous manner in humans (Ferda Percin et al., 2000). Chx10R200Q does not repress expression from the construct containing both Chx10 binding sites (pGL3E-Vsx1wt; p-value 0.076). Additionally, the level of reporter activity is significantly higher in the presence of Chx10R200Q compared to Chx10 (p-value < 0.0001).
Figure 3. Chx10 expression inhibits luciferase activity in a DNA-binding dependent manner and Chx10 binding sites near the Vsx1 promoter are required for high luciferase activity in HEK293 cells.
(A) Schematic diagrams of reporter constructs with Vsx1 promoters that contain both Chx10 binding sites (pGL3E-Vsx1wt) or with site specific mutations in these sites (pGL3E-Vsx1mut1, mutated at -1275; pGL3E-Vsx1mut2, mutated at −576; pGL3E-Vsx1mut3, mutated at both sites). Region bracketed corresponds to Vsx1 genomic DNA and numbering is relative to predicted Vsx1 transcription start site. (B) HEK293 cells were transfected with each reporter construct, pRL-TK to monitor transfection efficiency, and either an empty expression vector (pCMV; white bars), pCMV-Chx10 (gray bars), or pCMV-Chx10R200Q (black bar). Relative luciferase activity is normalized to the pGL3E-Vsx1wt co-transfected with empty expression vector (the first white bar) and is arbitrarily assigned as 100% activity. The standard deviation for each test condition is shown. Statistical significances (p-values) were tested by two-way repeated measures ANOVA and Tukey tests for multiple comparisons with values transformed by log2. *p<0.001 (p-value of each gray bar compared to each respective white bar); **p =0.0001; ++ p <0.001 (p-value of pGL3E-Vsx1mut3 (last white bar) compared to other reporter constructs (white bars)). Abbreviations: Ex1, exon 1 of Vsx1; LUC, luciferase cDNA; (A)n, SV40 late poly(A) signal; E, SV40 enhancer.
To determine the importance of the Chx10 consensus sites for transcription from the Vsx1 promoter, we tested reporter constructs with mutations in the sites at −576 and −1275, either alone or in combination. Chx10 still inhibits luciferase activity in the context of the single site mutations (mut1 and mut2; p-value < 0.001) and inhibition is not completely eliminated when both sites are mutated (mut3). Interestingly, the basal level of luciferase activity is significantly lower with the mut3 reporter than observed for the other promoters (wt, mut1, and mut2; p-value < 0.001, the same value for all three comparisons). Although Chx10 continues to inhibit luciferase activity in the absence of both Chx10 binding sites (see section 3.1), our results indicate that both of these sites are necessary for high levels of transcription from the Vsx1 promoter and adjacent regions, and the sum of our observations support the model that Chx10 is a direct negative regulator of Vsx1 transcription.
2.2 Vsx1 fails to compensate for the loss of Chx10 function during embryonic retinal development
Although histogenesis is severely affected by the absence of Chx10, several aspects of retinal development are still evident. This suggests Chx10 is not absolutely required and Vsx1 may partially compensate for the Chx10 deficiency. The upregulation of Vsx1 mRNAs in the Chx10orJ retina (Fig. 2A) supports this idea. Therefore, we crossed Chx10orJ mice with Vsx1τlacz mice to produce compound mutants and compared the resultant phenotypes.
During normal retinal development in mice, histogenesis is well underway by E14.5. RPC proliferation and differentiation into postmitotic neurons are robust as indicated by the expression patterns of PCNA and CycD1 in proliferating RPCs found in the neuroblast layer (NBL; Fig 4A, C) and Tuj1 in differentiating neurons that are scattered through the NBL and accumulating in the nascent ganglion cell layer and presumptive INL (collectively defined here as differentiated cell layer, DCL; Fig 4G). Pax6, which is important for optic vesicle development, RPC proliferation, and maintenance of the multipotential-state in RPCs (reviewed in Ashery-Padan and Gruss, 2001), is expressed initially in RPCs and subsequently shifts to differentiating neurons. At E14.5, Pax6 is detected in most if not all cells, and appears to be detected at higher levels in differentiated cells compared to RPCs (Fig. 4E). Compared to the wild type retina, overall ocular morphology and expression of PCNA, CycD1, Pax6, and Tuj1 are unaffected in the Vsx1τlacz/τlacz retina (Fig. 4B, D, F, H). Consistent with these observations, Chx10 expression appears normal in the Vsx1τlacz/τlacz retina (data not shown). In the Chx10orJ/orJ mouse, overall eye size is reduced, but retinal expression of PCNA is still abundant (Fig. 5A). CycD1 is also expressed but in a more dispersed pattern compared to wild type and Vsx1τlacz/τlacz retinas (Fig. 5C). Tuj1 expression is more centrally restricted (Fig. 5G) consistent with a delay in neurogenesis (Bone-Larson et al., 2000; Rutherford et al., 2004; Green et al., submitted). Pax6 is widely expressed in a pattern consistent with RPCs, and this could be due to developmental delay (Fig. 5E). Compared to the Chx10orJ/orJ mouse, ocular morphology and the expression patterns of the markers analyzed are similar in the Chx10orJ/orJ, Vsx1τlacz/τlacz compound mutant (Fig. 5B, D, F, H). Although retinal development appears enhanced in the compound mutant, the phenotype is within the range of variation observed for Chx10orJ/orJ single mutants. From these observations, we conclude that Vsx1 is dispensable in early retinal development and does not compensate for the loss of Chx10.
Figure 4. Immunohistochemical analysis of E14.5 eyes from wild type and Vsx1τlacz/τlacz mice.
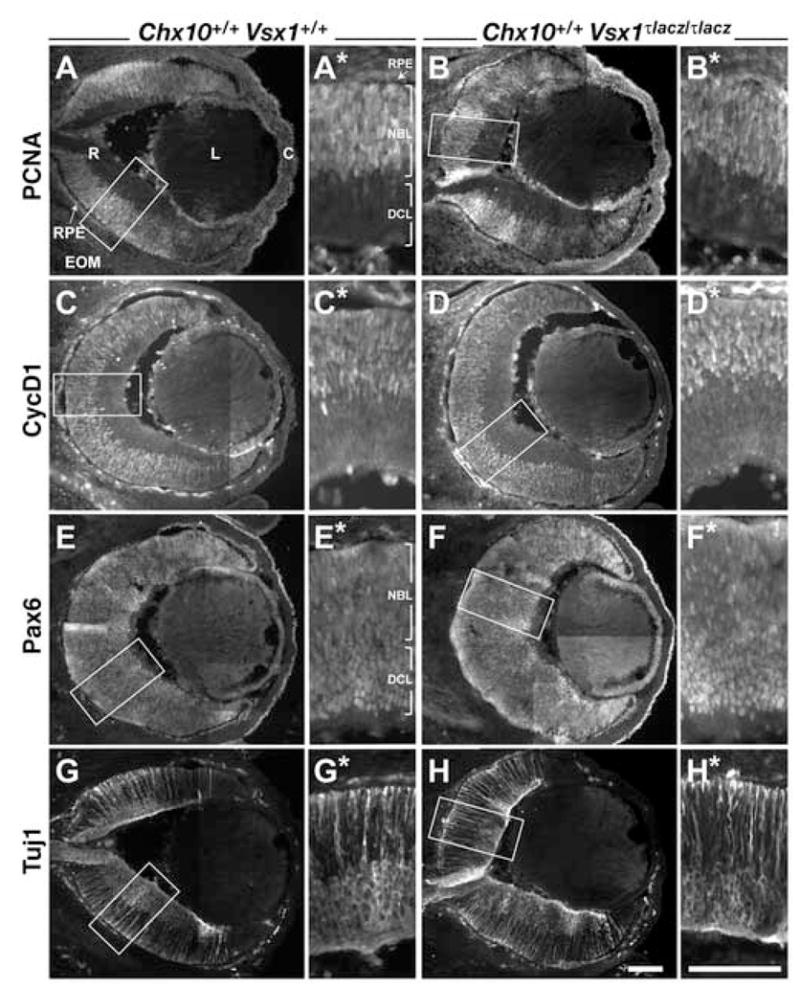
Expression patterns of PCNA (A, B), CycD1 (C, D), Pax6 (E, F), and Tuj1 (G, H) proteins in wild type (A, C, E, G) and Vsx1τlacz/τlacz (B, D, F, H) eyes. All panels with an * show the images in the boxes from each corresponding panel. These images were rotated such that the apical surface of the retina is pointed down. Scale bars: 100 μm. Abbreviations: RPE, retinal pigmented epithelium; R, retina; EOM, extraocular mesenchyme; L, lens; C, cornea; NBL, neuroblast layer; DCL: differentiated cell layer.
Figure 5. Immunohistochemical analysis of E14.5 eyes from Chx10orJ/orJ and Chx10orJ/orJ, Vsx1τlacz/τlacz mice.
Expression patterns of PCNA (A, B), CycD1 (C, D), Pax6 (E, F), and Tuj1 (G, H) proteins in Chx10orJ/orJ (A, C, E, G) and Chx10orJ/orJ, Vsx1τlacz/τlacz (B, D, F, H) eyes. All panels with an * show the images in the boxes from each corresponding panel. These image were rotated such that the apical surface of the retina is pointed down. Scale bars: 100 μm.
Marker expression was examined at P0 to determine whether Vsx1 compensates for the loss of Chx10 in neonatal RPCs (Fig. 6). The expression patterns of PCNA, CycD1, and Tuj1 in the Vsx1τlacz/τlacz retina are similar to wild type (Fig. 6A, B, E, F, I, J), and the expression of these markers in the Chx10orJ/orJ, Vsx1τlacz/τlacz compound mutant retina is similar to the Chx10orJ/orJ single mutant (Fig. 6C, D, G, H, K, L). We also examined the expression of Brn3b, which marks a major fraction of the retinal ganglion cell population (RGCs; Xiang et al., 1993). Whereas Brn3b-positive cells are observed in all genotypes (Fig. 6M-P), the Chx10orJ/orJ and Chx10orJ/orJ, Vsx1τlacz/τlacz mutants showed obvious dispersion of these cells into the NBL. The cause of this dispersion is not known, but possibilities include the delay in neurogenesis, migration defects, or lamination defects. Importantly, these observations extend our finding that Vsx1 is largely dispensable for retinal development through birth, either in the presence or absence of Chx10.
Figure 6. Immunohistochemical analysis of P0 eyes.

Expression patterns of PCNA (A-D), CycD1 (E-H), Tuj1 (I-L), and Brn3b (M-P) in wild type (A, E, I, M), Vsx1τlacz/τlacz (B, F, J, N), Chx10orJ/orJ (C, G, K, O), and Chx10orJ/orJ, Vsx1τlacz/τlacz (D, H, L, P) eyes. Scale bar: 40 μm.
To determine whether this is a common feature of vertebrate retinal development, we performed similar experiments in zebrafish (Fig. 7). Translation and splice blocking morpholino oligonucleotides (MO) targeted against chx10 and vsx1, respectively, were injected into one-cell stage zebrafish and embryos were examined at 24 hpf, a stage of development when RPC proliferation is robust, but neurogenesis has not yet begun. Figure 7A shows that the vsx1 MO blocks splicing of vsx1 mRNA in a dose dependent manner. To assess the effects of the MOs on retinal development, the expression patterns of vsx1, cycD1, and pax6a were examined by in situ hybridization (Fig. 7B–M) and the sum of our observations on vsx1 and cycD1 expression are shown in Fig. 7N and 7O. Chx10 knockdown led to an increase in vsx1 expression specifically in the eye, as well as to a decrease in eye size (Fig. 7B, C). An eye-specific decrease in cycD1 expression was also observed in chx10 morphant embryos (Fig. 7F, G). Vsx1 knockdown had little or no effect on the expression of vsx1 (Fig. 7D), cycD1 (Fig. 7H) or chx10 (not shown). pax6a expression was unaffected relative to eye size in all morphant embryos (Fig. 7J–M), indicating that eye identity was not perturbed. If Vsx1 compensates for loss of Chx10, then it would be expected that cycD1 expression and eye size would be more severely affected in chx10, vsx1 double morphant embryos compared to chx10 single morphants. We did not observe such phenotypes (Fig. 7G, I), suggesting that, as in the mouse, Vsx1 does not compensate for loss of Chx10 in zebrafish eye development.
Figure 7. Vsx1 does not compensate for loss of Chx10 in zebrafish.

(A) RT-PCR showing accumulation of unspliced vsx1 mRNA in a dose-dependent manner following injection of a splice-blocking morpholino (MO) against vsx1. Lane 1 shows molecular weight marker. Lanes 2, 4, and 6 show spliced and unspliced Vsx1 RNA derived from zebrafish embryos injected with the indicated morpholinos. Lanes 3, 5, and 7 represent no template controls (reverse transcriptase omitted). (B-M) in situ hybridizations for vsx1 (B-E), cycD1 (F-I), and pax6a (J-M) in 24h embryos that were uninjected (B, F, J), injected with 1.5ng chx10 MO (C, G, K), 15ng vsx1 MO (D, H, L), or co-injected with1.5ng chx10 and 15ng vsx1 Mos (E, I, M). Eyes in F-I are within the dotted lines. (M, O) Quantification of relative changes in expression levels for vsx1 and cycD1 mRNAs among morphant embryos (n, number of embryos analyzed).
3. Discussion
3.1 Regulation of Vsx1 by Chx10
In this study, we present evidence supporting a model in which Chx10 directly regulates expression of Vsx1 mRNA through a mechanism of transcription repression. Chx10 preferentially binds at or in close proximity to the Chx10 consensus sequence that is positioned close to the transcription start site (−576) in vivo and requires this site for binding in vitro. Chx10 inhibits luciferase activity when transcription is under the control of the Vsx1 promoter, which is consistent with the proposed role of Chx10 as a transcription repressor. We found that Chx10R200Q does not significantly repress reporter activity, suggesting that the ability of Chx10 to repress transcription is largely dependent on high affinity binding to sequences conforming to the consensus. These findings are in agreement with a previous report showing that mutation of another residue in the DNA binding helix of Chx10 (N51A) also reduces the efficiency of transcriptional repression by Chx10 (Dorval et al., 2005; Dorval et al., 2006).
Interestingly, mutation of both consensus sites in the Vsx1 genomic sequence (mut3) causes a significant drop in basal reporter activity. Since Chx10 is a more effective repressor when it has high affinity for its consensus site, it is possible that Chx10 inhibits transcription by competing with activators for the same sites.
We found that Chx10 still inhibits luciferase activity in the absence of the two consensus sites (mut3). Although it is tempting to propose a mechanism of Chx10 repression that is independent of its binding to sequences fitting the consensus, two additional Chx10 consensus sites are present in the SV40 enhancer region of the reporter construct and it is likely that the repressive effect of Chx10 on the mut3 reporter is mediated through these sites. Currently, the simplest model is that Chx10 inhibits Vsx1 transcription by a mechanism that depends on binding to its consensus sequence, and the site at −567 is sufficient for Chx10 binding and negative regulation of Vsx1 transcription in vivo.
The expression patterns of Chx10 and Vsx1 during retinal development provide further evidence of a regulatory interaction. When Chx10 expression is high in RPCs, Vsx1 is low or nonexistent. Additionally, Vsx1 mRNA levels are increased in the Chx10 deficient retina of mice and zebrafish. Interestingly, the chick Vsx1 ortholog, Chx10-1, is expressed at high levels, and Chx10 appears to be expressed at relatively lower levels in RPCs (Chen and Cepko, 2000). A simple scenario to explain this apparent reversal in expression profile is that Chx10 regulation changed in the developing chick retina such that Chx10 levels decreased, thereby allowing Vsx1 levels to increase. Xenopus vsx1 is also expressed at high levels in RPCs, but the status of Chx10 expression is not known (D’Autilia et al., 2006).
In the postnatal and mature retina of all vertebrates examined, Chx10 and Vsx1 mRNAs are expressed in bipolar cells, but their patterns are not in perfect correspondence. Where examined, Chx10 is expressed at the earliest stages of bipolar cell differentiation and remains expressed in rod bipolar cells and a large subset of cone bipolar cells. Vsx1 expression is subsequently activated and is restricted to a subset of cone bipolar cells (off-cone). We found that the Chx10 and Vsx1 proteins can be co-expressed in the same cells of the postnatal retina, but in a complementary fashion: cells expressing high levels of Chx10 express Vsx1 at low levels, and vice versa, and these relationships are not time dependent (R.L. Chow, ms in prep). While these observations support the model that Chx10 antagonizes Vsx1 expression, they also leave open the possibility that Vsx1 could antagonize Chx10 expression as well.
Further insight into the transcriptional control of Chx10 and Vsx1 is gained from expression studies in Chx10 and Vsx1 single mutant mice. Chx10 mRNA expression in Chx10orJ mice is not altered, suggesting that Chx10 does not regulate its own expression (Rowan and Cepko, 2004; Rowan et al., 2004; Rutherford et al., 2004; Green et al., submitted). Furthermore, the upregulation of Vsx1 mRNA in the developing Chx10orJ retina does not appear to have a significant effect on Chx10 expression as well, either because Vsx1 does not regulate Chx10 levels or its expression is not at sufficient levels to exert an effect. In contrast, Vsx1 may negatively regulate its own expression in bipolar cells (Chow et al., 2004; Ohtoshi et al., 2004).
The sum of these findings suggests a hierarchical transcriptional network shown in Figure 8. In this model, Chx10 expression is not regulated either by Chx10 or Vsx1 in RPCs. However, RPCs are competent to express Vsx1 but are inhibited from doing so by cross-regulation from Chx10. Activation and/or maintenance of Chx10 expression may be mediated by interaction with the surface ectoderm (Hyer et al., 1998; Nguyen and Arnheiter, 2000) and/or exclusion of the extraocular mesenchyme (Fuhrmann et al., 2000b). Transcriptional regulation of Chx10 appears to be complex and involve multiple enhancers (Rowan and Cepko, 2004; Rowan and Cepko, 2005) and candidate pathways and factors for promoting Chx10 expression include Fgf signaling, Bmp signaling, Hes activity, Mab21l2, and Rx3 (Gotoh et al., 2004; Hatakeyama et al., 2004; Horsford et al., 2005; Loosli et al., 2001; Murali et al., 2005; Nguyen and Arnheiter, 2000; Winkler et al., 2000; Yamada et al., 2004). However, the mechanisms that account for their regulation of Chx10 are not known nor is it known whether any of these candidates act in a direct manner. Pou3f2 (Brn2) was recently identified as a candidate direct regulator of Chx10 expression in late RPCs and early, differentiating bipolar cells (Rowan and Cepko, 2005). In this case, however, it is not known if Pou3f2 is sufficient or required for Chx10 expression. As bipolar cells mature, Chx10 is downregulated in a subset of cone bipolar cells and Vsx1 is upregulated, albeit in a controlled manner because of Vsx1-mediated autoregulation (Chow et al., 2001; Chow et al., 2004; Ohtoshi et al., 2004). The factors that maintain Chx10 expression in mature bipolar cells are not known, nor is it known whether negative regulators of Chx10 or positive regulators of Vsx1 feed into the proposed network.
Figure 8. Proposed regulatory network for Chx10 and Vsx1 in RPCs and bipolar cells.
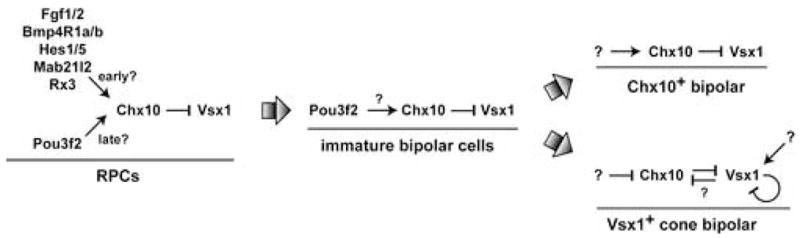
Large arrows represent the indicated developmental transitions. See section 3.2 for details.
3.2 Prd-L:CVC genes in embryonic retinal development
The increase in Vsx1 mRNA in the developing Chx10orJ/orJ mouse retina and zebrafish chx10-morphant retina prompted us to test whether Vsx1 has an effect on retinal development prior to bipolar cell differentiation. Since the HD and CVC domain of Chx10 and Vsx1 are similar and both proteins act as transcription repressors with overlapping DNA binding specificities, we wanted to know if the residual histogenesis occurring in the Chx10orJ/orJ retina is dependent on Vsx1. In mice and zebrafish deficient for both Chx10 and Vsx1, we did not observe any significant changes in retinal development compared to Chx10 deficient animals. Additionally, Vsx1 deficiency did not have any obvious effect on retinal development compared to wild type animals. From these observations, we conclude that Vsx1 does not contribute to embryonic retinal development and that the residual histogenesis occurring in the Chx10 deficient retina is not dependent on Prd-L:CVC genes in general since Chx10 and Vsx1 are the only known Prd-L:CVC genes in mouse and zebrafish.
These findings begin to provide insight into how Chx10 fits into the hierarchy of transcription factors important for early retinal development. Homeobox genes such as Six3, Rx, Pax6, and Lhx2 are expressed in the developing eye field and are necessary for optic cup formation in mice or humans. In each case, genetic inactivation results in anophthalmia, which is a complete failure of eye development (reviewed in Fitzpatrick and van Heyningen, 2005; Graw, 2003). In addition, these genes induce ectopic eyes in Xenopus when expressed together and in combination with six6/optx2, tll, and ET (Zuber et al., 2003). As a result of these features, these genes have been termed the eye field transcription factors (EFTFs) and they are thought to form a network analogous to the retinal determination gene network (RDGN) in Drosophila (reviewed in Hanson, 2001). How Chx10 fits into this paradigm is not known. At present, it is the earliest expressed and most specific marker of the neural retina domain and its inactivation produces a severe and highly penetrant microphthalmia. Since genetic inactivation of both Chx10 and Vsx1 does not push the eye phenotype towards anophthalmia, it is now clear that Vsx1 is not compensating for loss of Chx10 in early eye development and Chx10 has a role distinct from, but possibly downstream of EFTFs such as Six3, Rx, Pax6, and Lhx2.
3.3 Functional overlap between Chx10 and Vsx1
We did not observe a change of phenotype in Chx10orJ/orJ, Vsx1τlacz/τlacz compound mutants and chx10, vsx1 double morphants compared to the Chx10orJ/orJ single mutants and chx10 single morphants, respectively. While this suggests that Chx10 and Vsx1 do not have overlapping functions such as shared transcriptional targets, it is premature to conclude this since we do not know if Vsx1 protein is upregulated to sufficient levels in RPCs lacking Chx10. Even in Xenopus, where vsx1 mRNA is highly expressed in RPCs (D’Autilia et al., 2006), post-transcriptional regulation is thought to keep Vsx1 protein levels at a minimum until bipolar cell formation (Decembrini et al., 2006). Thus, it is still not known if the functional overlap between Chx10 and Vsx1 shown in biochemical studies has significance in retinal development.
Chx10 overexpression inhibits photoreceptor differentiation and several phototransduction genes are candidate direct transcriptional targets of negative regulation by Chx10 (Dorval et al., 2006; Livne-Bar et al., 2006; Toy et al., 2002). These findings combined with the shared DNA binding characteristics and repressive activities of Chx10 and Vsx1 led to the proposal that these genes could function in bipolar cells to prevent inappropriate expression of photoreceptor genes (Dorval et al., 2006). While possible, this scenario is not likely to fully explain how Chx10 and Vsx1 function in bipolar cells. In the Vsx1 deficient mouse retina, several markers of bipolar cells are downregulated in cells that normally express Vsx1 (Chow et al., 2004; Ohtoshi et al., 2004). However, Chx10 remains expressed in the same cells. This suggests that Chx10 is not able to compensate for Vsx1 in cone bipolar cells and that Chx10 and Vsx1 have distinct functions, possibly by targeting different genes for regulation.
Although more work is needed to understand more fully the functional relationships between Chx10 and Vsx1, our work shows that a regulatory relationship has evolved between these paralogs that could allow for overlap of some functions, but at the same time, also allow for functional divergence that contributes to the complex networks driving retinal development.
4. Experimental Procedure
4.1. Mouse strains
Chx10 null mice (Chx10orJ) were purchased from Jackson Labs. Vsx1 null mice (Vsx1τlacz; Chow et al., 2004) were mated to Chx10orJ mice to produce compound mutants. PCR based genotyping was done as previously described (Burmeister et al., 1996; Chow et al., 2004). The Chx10orJ and Vsx1τlacz alleles originated from 129 genetic backgrounds and the mice analyzed for this study are mixed hybrids. For all animals used in this study, efforts were made to minimize pain and discomfort during procedures and in preparing for euthanasia.
4.2. Vsx1 polyclonal antibody production
15-amino acid peptide, HLKKGANKDEDGPER (position 301 to 315), was synthesized and conjugated to KLH at the University of Utah peptide synthesis core facility. Polyclonal antibodies were prepared commercially (Harlan Bioproducts, Inc. Indianapolis IN). IgG fraction was purified using SulfoLink Kit according to manufacturer’s instructions (Pierce, Rockford, IL).
4.3. Immunohistology
Mouse embryonic heads, postnatal whole eyes or isolated retinas were dissected in Hanks buffered saline solution (HBSS) and immediately fixed in 4% neutral-buffered formalin (PFA) for various lengths of time varying from 20 minutes at room temperature to overnight at 4°C. Following fixation, tissue was washed twice with PBS, then replaced with sucrose gradient and finally embedded in OCT (Sakura Finetek, Torrance, CA). 12 μm thick cryosections were used for immunostaining. The primary antibodies used were: rabbit anti-Vsx1 (1:200; this study); sheep anti-Chx10 (1:300; Exalpha Biologicals, Boston, MA); rabbit anti-CycD1 (1:400; Lab Vision, Fremont, CA); rabbit anti-Pax6 (1:300; Mastick et al., 1997); mouse monoclonal anti-PCNA (clone PC10; 1:500; Dako, Denmark); goat anti-Brn3b (1:50; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-neuronal class III tubulin (Tuj1; 1:4000; Covance, Richmond, CA). Primary antibodies were followed with species-specific secondary antibodies conjugated to either Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen/Molecular Probes, Eugene, OR).
4.4. Western Blots
Mouse retinas were dissected in HBSS and stored at −80°C. Frozen retinas were resuspended in a lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS, 1% Triton X-100) complemented with protease inhibitors (Complete mini tablets, Roche, Indianapolis, IN) and sonicated on ice. Protein concentration was determined by a BCA protein assay (Pierce Biotechnology, Rockford, IL). 20 μg of protein lysates were subsequently run on 12% SDS PAGE, then transferred to nitrocellulose membranes. Blots were incubated with antibodies diluted in blocking buffer (5% dry milk, 0.04% Tween-20, TBS pH 7.6). Primary antibodies included rabbit anti-vsx1, mouse anti-β-Actin (Chemicon, Temecula, CA). Detection was performed using SuperSignal West Dura (Pierce Biotechnology, Rockford, IL) and documented by ChemiDoc XRS imaging system (Bio-Rad, Hercules, CA).
4.5. RT-PCR of mouse RNA
Total RNA was extracted from P0 Black Swiss (BS) wild type and Chx10orJ retinas by using Trizol (Invitrogen, Carlsbad, CA). The concentration of total RNA was assessed by spectrophotometry and adjusted to the same level among samples. First –strand cDNA was synthesized by using M-MuLV Reverse Transcriptase and Oligo(dT)18 (Fermentas, Hanover, MD). RT-PCR was carried out in a 30-μl reaction mixture. RT-PCR for a housekeeping gene (Gapdh) was used as a control (22 cycles) to normalize the concentration of the cDNA samples (primer sequences available on request). RT-PCR for Vsx1 was performed with different amounts of cDNA (4, 2 or 1μl) to estimate an appropriate number of cycles to amplify in the linear range (data not shown). The primers for Vsx1 were: 5′-GGATGAGGATGGACCTGAGA-3′ and 5′-AGGTGTTTGTCCAGCTTTGG-3′. PCR conditions were: 94°C 30 seconds, 55°C 30 seconds and 72°C 30 seconds for 30–33 cycles. The size of the product is 208 bp.
4.6. Chromatin immunoprecipitation (ChIP)
Retinas from E16.0, P0 or P1 BS wild-type animals were dissected and cut into small pieces and then fixed in 1% formaldehyde for 15min at room temperature. Following crosslinking, tissues were washed and sonicated to shear the DNA to lengths between 200 and 1000 bp. Sonicated supernatants were precleared with salmon sperm DNA (Invitrogen)/Protein G agarose beads (Roche, Indianapolis, IN), and incubated overnight at 4°C with 5 μl sheep anti-Chx10 antibody (Exalpha Biologicals, Boston, MA) or sheep IgG (Sigma, St. Louis, MO). The chromatin-antibody complex was collected with salmon sperm DNA/Protein G agarose. Crosslinking was reversed with 5M NaCl at 65°C for four hours. DNA was purified by QIAquick PCR purification kit (QIAGEN, Valencia, CA). 4μl IP DNA was employed for PCR amplification for potential Chx10 binding sites in Vsx1 promoter (−576 and −1275). The following primers were used: Vsx1 (−567): 5′-AGTTGTAAGCTGCCCTGTGG-3′ and 5′-CCTGACTGGCACGTAGGAAT-3′. Vsx1 (-1275): 5′-GCCGAAATTTGGATTTACGA-3′ and 5′-TGGATGAGTGGGGAGAAATC-3′.
4.7. Electrophoretic mobility shift assays (EMSA)
pET26b-chx10 plasmids were in vitro translated using rabbit reticulocyte lysates (Promega, Madison, WI). 2 pmol single-strand probes were end-labeled by T4 kinase (Invitrogen) with [γ-32P]ATP (MP Biologicals, Solon, OH) and were purified using BioSpin6 column (Bio-Rad, Hercules, CA). The probes were boiled for 5 min at 95 °C and cooled to room temperature for 3 hrs to form double-stranded probes. Probe sequences were: Vsx1 wild type (Vsx1wt), top strand: 5′-GCGTTTTAATTAGCTCCAGTTTCA; Vsx1 mutant (Vsx1mut), top strand: 5′-GCGTTTTCCTTAGCTCCAGTTTCA. EMSA assays were performed as described by (Dorval et al., 2005). Gels were dried and visualized with phosphoimager (Bio-Rad, Hercules, CA)
4.8. Luciferase assays
HEK293 cells were transfected with 0.04μg reporter construct, 0.02 μg pRL-TK and either 0.2μg pCMV-Chx10 or pCMV-Chx10R200Q using the lipofectamine method according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Reporter constructs were designed as follows: pGL3E-Vsx1wt (wild type Vsx1 promoter), pGL3E-Vsx1mut1 (Vsx1 promoter mutated in −1275 Chx10 binding site), pGL3E-Vsx1mut2 (Vsx1 promoter mutated in −576 Chx10 binding site) and pGL3E-Vsx1mut3 (Vsx1 promoter mutated in both −1275 and −576 Chx10 binding sites). Cell lysates were prepared 24hr after transfection. The activities of firefly and Renilla luciferase were assayed using a Dynex Technologies MRX Revelation microplate reader (Dynex Technologies, Denkendorf, Germany) using 100 μl D-luciferin reagent and 100 μl coelenterazine (Biotium, Hayward, CA). To standardize for transfection efficiency, the luciferase activities of all transfected cells were divided by the Renilla luciferase activities. Data are presented as mean ±SD from four separate experiments. Statistical significances were tested by two-way repeated measures ANOVA and Tukey tests for multiple comparisons.
4.9. Zebrafish strains and staging
Embryos were obtained from natural spawning of wild-type (AB-1) zebrafish lines. All developmental stages in this study are reported in hours post-fertilization (hpf) at 28.5°C (Kimmel et al., 1995)
4.10. MO injections
chx10 translation blocking, and vsx1 splice-blocking MO antisense oligonucleotides were obtained from Gene Tools (vsx1 MO: 5′-AGCAAAGTGATTCGTACCGGAGTAA -3′ and chx10 MO: 5′-AAACAGCCCCATCCTTTCCTGTCAT -3′). Both MOs were injected into one-cell stage wild-type embryos at doses of 1.5 ng and 15 ng, respectively.
4.11. RT-PCR of zebrafish RNA
Fifty wild-type embryos and vsx1 morphants were used for preparing RNA. Total RNA was isolated using Trizol reagent and standard protocols. Total RNA (1–5 μg/μl) was reverse transcribed by either random hexamers or a gene-specific primer using the Superscript first strand synthesis kit (Invitrogen) following the manufacturer’s protocol. PCR was performed using an exon 1 forward primer (CGC AAT CAC AGA TCT CCT GG) and an exon 2 reverse primer (TCC ATC ATT GCG ATC ACC GG) for 30–35 cycles using an annealing temperature of 55°C, and reactions were visualized on 1% agarose gels in TAE.
4.12. In situ Hybridization
Probe synthesis and in situ hybridization were performed as described elsewhere (Oxtoby and Jowett, 1993), and visualized using BM Purple (Roche, Indianapolis, IN). The following three RNA probes were used: vsx1 (amplified from published cDNA sequence (Passini et al., 1997); cycD1 (869 bp PCR fragment); and pax6a (Krauss et al., 1991)
Acknowledgments
We thank Dr. Sabine Fuhrmann and members of the Levine and Fuhrmann laboratories for their suggestions and insights during the course of this work. We also thank Amy Kircher for her excellent mouse colony management. Funding for this project was generously provided by Research to Prevent Blindness, Inc. (Career Development Award to E.M.L.), Knights Templar Eye Foundation (E.M.L.), NEI R01-EY0013760 (E.M.L.), NEI Vision Core Grant EY0014800, Pew Scholars Program (R.I.D.), NINDS R01-NS0053897 (R.I.D), and Foundation Fighting Blindness, Canada (New Investigator Award to R.L.C.). This work is dedicated to the memory of our friend and colleague, Dr. Kimberly Howes.
Footnotes
Paired-Like:CVC, Prd-L:CVC; homeodomain, HD: retinal progenitor cells, RPCs; neuroblast layer, NBL; morpholino, MO; ocular retardation J, orJ; outer nuclear layer, ONL; inner nuclear layer, INL; ganglion cell layer; GCL; differentiated cell layer, DCL; retinal pigmented epithelium, RPE; extraocular mesenchyme, EOM; orthopedia/aristaless/rax motif, OAR motif; rinx/vsx1 motif, RV motif; Black Swiss, BS
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature References
- Altun-Gultekin Z, et al. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–69. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Gruss P. Pax6 lights-up the way for eye development. Curr Opin Cell Biol. 2001;13:706–14. doi: 10.1016/s0955-0674(00)00274-x. [DOI] [PubMed] [Google Scholar]
- Bar-Yosef U, et al. CHX10 mutations cause non-syndromic microphthalmia/anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–9. doi: 10.1007/s00439-004-1154-2. [DOI] [PubMed] [Google Scholar]
- Barabino SM, et al. Inactivation of the zebrafish homologue of Chx10 by antisense oligonucleotides causes eye malformations similar to the ocular retardation phenotype. Mech Dev. 1997;63:133–43. doi: 10.1016/s0925-4773(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Barbaro V, et al. Expression of VSX1 in Human Corneal Keratocytes during Differentiation into Myofibroblasts in Response to Wound Healing. Invest Ophthalmol Vis Sci. 2006;47:5243–50. doi: 10.1167/iovs.06-0185. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, et al. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Ophthalmol Vis Sci. 1997;38:1293–303. [PubMed] [Google Scholar]
- Bone-Larson C, et al. Partial rescue of the ocular retardation phenotype by genetic modifiers. J Neurobiol. 2000;42:232–47. doi: 10.1002/(sici)1097-4695(20000205)42:2<232::aid-neu7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Burmeister M, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–84. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Chen CM, Cepko CL. Expression of Chx10 and Chx10-1 in the developing chicken retina. Mech Dev. 2000;90:293–7. doi: 10.1016/s0925-4773(99)00251-8. [DOI] [PubMed] [Google Scholar]
- Chow RL, Lang RA. Early Eye Development in Vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Chow RL, et al. Vsx1, a rapidly evolving paired-like homeobox gene expressed in cone bipolar cells. Mech Dev. 2001;109:315–22. doi: 10.1016/s0925-4773(01)00585-8. [DOI] [PubMed] [Google Scholar]
- Chow RL, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci U S A. 2004;101:1754–9. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles BL, et al. Loss of retinal progenitor cells leads to an increase in the retinal stem cell population in vivo. Eur J Neurosci. 2006;23:75–82. doi: 10.1111/j.1460-9568.2005.04537.x. [DOI] [PubMed] [Google Scholar]
- Czerny T, et al. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- D’Autilia S, et al. Cloning and developmental expression of the Xenopus homeobox gene Xvsx1. Dev Genes Evol. 2006;216:829–834. doi: 10.1007/s00427-006-0109-0. [DOI] [PubMed] [Google Scholar]
- de Melo J, et al. Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol. 2003;461:187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- Decembrini S, et al. Timing the generation of distinct retinal cells by homeobox proteins. PLoS Biol. 2006;4:e272. doi: 10.1371/journal.pbio.0040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen NS, et al. Absence of chx10 causes neural progenitors to persist in the adult retina. Invest Ophthalmol Vis Sci. 2006;47:386–96. doi: 10.1167/iovs.05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorval KM, et al. Transcriptional activity of the paired-like homeodomain proteins CHX10 and VSX1. J Biol Chem. 2005;280:10100–8. doi: 10.1074/jbc.M412676200. [DOI] [PubMed] [Google Scholar]
- Dorval KM, et al. CHX10 targets a subset of photoreceptor genes. J Biol Chem. 2006;281:744–51. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- Dyer MA. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle. 2003;2:350–7. [PubMed] [Google Scholar]
- Ferda Percin E, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr Opin Genet Dev. 2005;15:348–53. doi: 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Fuhrmann S, et al. Molecular control of cell diversification in the vertebrate retina. In: Fini E, editor. Vertebrate Eye Development. Vol. 31. Springer Verlag; New York: 2000a. pp. 69–84. [DOI] [PubMed] [Google Scholar]
- Fuhrmann S, et al. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development. 2000b;127:4599–609. doi: 10.1242/dev.127.21.4599. [DOI] [PubMed] [Google Scholar]
- Furukawa T, et al. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A. 1997;94:3088–93. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, et al. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–97. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Gotoh N, et al. Tyrosine phosphorylation sites on FRS2alpha responsible for Shp2 recruitment are critical for induction of lens and retina. Proc Natl Acad Sci U S A. 2004;101:17144–9. doi: 10.1073/pnas.0407577101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw J. The genetic and molecular basis of congenital eye defects. Nat Rev Genet. 2003;4:876–88. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- Green ES, et al. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003;130:539–52. doi: 10.1242/dev.00275. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–84. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–50. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hayashi T, et al. RINX(VSX1), a novel homeobox gene expressed in the inner nuclear layer of the adult retina. Genomics. 2000;67:128–39. doi: 10.1006/geno.2000.6248. [DOI] [PubMed] [Google Scholar]
- Heon E, et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11:1029–36. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- Horsford DJ, et al. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–87. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- Hyer J, et al. FGF1 patterns the optic vesicle by directing the placement of the neural retina domain. Development. 1998;125:869–77. doi: 10.1242/dev.125.5.869. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knauer SK, et al. Nuclear export is evolutionarily conserved in CVC paired-like homeobox proteins and influences protein stability, transcriptional activation, and extracellular secretion. Mol Cell Biol. 2005;25:2573–82. doi: 10.1128/MCB.25.7.2573-2582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, et al. Zebrafish pax[zf-a]: a paired box-containing gene expressed in the neural tube. Embo J. 1991;10:3609–19. doi: 10.1002/j.1460-2075.1991.tb04927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman AL, Schechter N. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc Natl Acad Sci U S A. 2001;98:5602–7. doi: 10.1073/pnas.101129698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine EM, Green ES. Cell-intrinsic regulators of proliferation in vertebrate retinal progenitors. Semin Cell Dev Biol. 2004;15:63–74. doi: 10.1016/j.semcdb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Levine EM, et al. Restricted expression of a new paired-class homeobox gene in normal and regenerating adult goldfish retina. J Comp Neurol. 1994;348:596–606. doi: 10.1002/cne.903480409. [DOI] [PubMed] [Google Scholar]
- Liu IS, et al. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–93. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Livne-Bar I, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006;103:4988–93. doi: 10.1073/pnas.0600083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, et al. Medaka eyeless is the key factor linking retinal determination and eye growth. Development. 2001;128:4035–44. doi: 10.1242/dev.128.20.4035. [DOI] [PubMed] [Google Scholar]
- Lupo G, et al. Homeobox genes in the genetic control of eye development. Int J Dev Biol. 2000;44:627–36. [PubMed] [Google Scholar]
- Mastick GS, et al. Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–97. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- Mintz-Hittner HA, et al. VSX1 (RINX) mutation with craniofacial anomalies, empty sella, corneal endothelial changes, and abnormal retinal and auditory bipolar cells. Ophthalmology. 2004;111:828–36. doi: 10.1016/j.ophtha.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Murali D, et al. Distinct developmental programs require different levels of Bmp signaling during mouse retinal development. Development. 2005;132:913–23. doi: 10.1242/dev.01673. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development. 2000;127:3581–91. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, et al. Isolation and characterization of Vsx1, a novel mouse CVC paired-like homeobox gene expressed during embryogenesis and in the retina. Biochem Biophys Res Commun. 2001;286:133–40. doi: 10.1006/bbrc.2001.5372. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–6. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–95. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, et al. Cloning of zebrafish vsx1: expression of a paired-like homeobox gene during CNS development. Dev Genet. 1998;23:128–41. doi: 10.1002/(SICI)1520-6408(1998)23:2<128::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Passini MA, et al. Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J Comp Neurol. 1997;388:495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Plouhinec JL, et al. Comparative analysis of gnathostome Otx gene expression patterns in the developing eye: implications for the functional evolution of the multigene family. Dev Biol. 2005;278:560–75. doi: 10.1016/j.ydbio.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. A POU factor binding site upstream of the Chx10 homeobox gene is required for Chx10 expression in subsets of retinal progenitor cells and bipolar cells. Dev Biol. 2005;281:240–55. doi: 10.1016/j.ydbio.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Rowan S, et al. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–52. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Rutherford AD, et al. Delayed expression of the Crx gene and photoreceptor development in the Chx10-deficient retina. Invest Ophthalmol Vis Sci. 2004;45:375–84. doi: 10.1167/iovs.03-0332. [DOI] [PubMed] [Google Scholar]
- Svendsen PC, McGhee JD. The C. elegans neuronally expressed homeobox gene ceh-10 is closely related to genes expressed in the vertebrate eye. Development. 1995;121:1253–62. doi: 10.1242/dev.121.5.1253. [DOI] [PubMed] [Google Scholar]
- Theiler K, et al. A new allele of ocular retardation: early development and morphogenetic cell death. Anat Embryol (Berl) 1976;150:85–97. doi: 10.1007/BF00346288. [DOI] [PubMed] [Google Scholar]
- Toy J, et al. Effects of homeobox genes on the differentiation of photoreceptor and nonphotoreceptor neurons. Invest Ophthalmol Vis Sci. 2002;43:3522–9. [PubMed] [Google Scholar]
- Valleix S, et al. H244R VSX1 is associated with selective cone ON bipolar cell dysfunction and macular degeneration in a PPCD family. Invest Ophthalmol Vis Sci. 2006;47:48–54. doi: 10.1167/iovs.05-0479. [DOI] [PubMed] [Google Scholar]
- Winkler S, et al. The conditional medaka mutation eyeless uncouples patterning and morphogenesis of the eye. Development. 2000;127:1911–9. doi: 10.1242/dev.127.9.1911. [DOI] [PubMed] [Google Scholar]
- Xiang M, et al. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11:689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Yamada R, et al. Requirement for Mab21l2 during development of murine retina and ventral body wall. Dev Biol. 2004;274:295–307. doi: 10.1016/j.ydbio.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Zuber ME, et al. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–67. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]