Abstract
We have shown that the DNA demethylation complex isolated from chicken embryos has a G⋅T mismatch DNA glycosylase that also possesses 5-methylcytosine DNA glycosylase (5-MCDG) activity. Herein we show that human embryonic kidney cells stably transfected with 5-MCDG cDNA linked to a cytomegalovirus promoter overexpress 5-MCDG. A 15- to 20-fold overexpression of 5-MCDG results in the specific demethylation of a stably integrated ecdysone-retinoic acid responsive enhancer-promoter linked to a β-galactosidase reporter gene. Demethylation occurs in the absence of the ligand ponasterone A (an analogue of ecdysone). The state of methylation of the transgene was investigated by Southern blot analysis and by the bisulfite genomic sequencing reaction. Demethylation occurs downstream of the hormone response elements. No genome-wide demethylation was observed. The expression of an inactive mutant of 5-MCDG or the empty vector does not elicit any demethylation of the promoter-enhancer of the reporter gene. An increase in 5-MCDG activity does not influence the activity of DNA methyltransferase(s) when tested in vitro with a hemimethylated substrate. There is no change in the transgene copy number during selection of the clones with antibiotics. Immunoprecipitation combined with Western blot analysis showed that an antibody directed against 5-MCDG precipitates a complex containing the retinoid X receptor α. The association between retinoid receptor and 5-MCDG is not ligand dependent. These results suggest that a complex of the hormone receptor with 5-MCDG may target demethylation of the transgene in this system.
DNA demethylation and methylation of specific genes occurs at specific stages of embryonic development. Methylation or demethylation of DNA can occur as a global genome-wide event or in a developmentally regulated fashion for specific genes of a given cell lineage. Genome-wide demethylation occurs in the early mouse blastocytes where the paternal genome is demethylated (1–3). Specific gene demethylation occurs throughout the embryonic development (4–9) and, in specific cases, in terminally differentiated cells (10–13). The avian vitellogenin gene in the liver of immature chickens treated with estrogen is such an example (13).
Mechanistically, DNA demethylation can occur via different pathways: passive or active demethylation or a combination of both processes. In passive demethylation, an inhibition of the maintenance DNA methyltransferase during cycles of replication leads to the progressive loss of methylation (14–16). Active DNA demethylation occurs enzymatically by the hypothetical removal of the methyl group from 5-methylcytosine (17–19) or by a base excision mechanism (20–25). In the latter case, 5-methylcytosine DNA glycosylase (5-MCDG) initially removes the methylated base resulting in the cleavage of the abasic sugar, which is then replaced by cytosine (23–26). So far we have identified two enzymes with 5-MCDG activities. One was identified in the DNA demethylation complex purified from developing chicken embryos (25). Its human homologue has been identified as a G⋅T mismatch-specific thymine-DNA glycosylase (27). The other was MBD4, another G⋅T mismatch DNA glycosylase (26).
In the present work, we show that overexpression of human 5-MCDG demethylates the modified ecdysone responsive enhancer-promoter of a lacZ reporter gene. This leads to the inducibility of the reporter gene by the steroid hormone ponasterone A (a stable analogue of ecdysone). The presence of the hormone receptor–5-MCDG complex could possibly explain the targeting of DNA demethylation.
Materials and Methods
Construction of Recombinant Plasmids.
A 1.4-kb cDNA encoding human 5-MCDG was generated by reverse transcription–PCR with total RNA from human embryonal kidney 293 cells as template. The following oligonucleotides were used as primers: forward, 5′-TTCAGGATCCTGAAGTCGGGAGCTTGAGTC-3′; reverse, 5′-CTTAGAGCTCGCACCATTCTTAAGCATGGC-3′. After SacI digestion, the cDNA was blunt-ended with T4 DNA polymerase, then digested with BamHI, and inserted into the BamHI–HpaI sites of the mammalian expression vector pIREShyg2 (CLONTECH). The sequence of one clone corresponded exactly to the published human 5-MCDG/G⋅T mismatch DNA glycosylase sequence (GenBank accession no. U51166) and was designated pHGC2. A mutant clone that carries five amino acid transitions in the coding region (R61 → K, F102 → L, S166 → P, S359 → P, and D390 → G), caused by PCR errors, was designated pHGC2M.
Both coding regions of the wild-type and mutant cDNAs were also inserted in-frame into a prokaryotic expression vector and expressed in Escherichia coli. The wild-type 5-MCDG shows both 5-MCDG and G⋅T mismatch DNA glycosylase activities and also binds to a hemimethylated substrate. The 5-MCDG mutant retains binding activity but has no detectable 5-MCDG or G⋅T mismatch DNA glycosylase activity (data not shown).
Establishing Cell Lines.
The ecdysone-inducible mammalian expression system (Invitrogen) was used for construction of all cell lines tested (28). EcR 293 cells (Invitrogen), derived from human embryonal kidney 293 cells, were the parental cell line. This cell line constitutively expresses the ecdysone receptor and retinoid X receptor α (RXRα). Both receptors are required to induce genes controlled by modified ecdysone responsive elements. pIND/lacZ (Invitrogen) or its in vitro methylated form were transfected into EcR293 cells, and the daughter cell lines were designated 293M− or 293M+, respectively. The methylation status of the promoter-enhancer region of the lacZ gene of 293M− and 293M+ cells was confirmed by Southern blot analysis and bisulfite sequencing. The steroid response element of the reporter gene binds heterodimers containing ecdysone and RXRs. However, in 293 M+ cells, the promoter-enhancer region has been fully methylated and is, therefore, silent even in the presence of the ecdysone analogue ponasterone A. To generate cell lines M+vector, M+5-MCDG, and M+mutant 5-MCDG, 293M+ cells were stably transfected (SuperFect transfection reagent, Qiagen, Chatsworth, CA) with pIREShyg2, pHGC2, and pHGC2 M, respectively. The cells were selected and maintained in Zeocin (400 μg/ml), Geneticin (400 μg/ml), and hygromycin B (200 μg/ml), respectively.
Enzyme Activity Assays.
Cells were collected in the growing phase and fractionated by use of nuclear and cytoplasmic extraction reagents from Pierce. 5-MCDG activity assays were performed as described (24, 25). The hemimethylated end-labeled oligonucleotide (only the lower strand is shown) 5′-ATATATATATATATATATATATATATCmCGGATATATATATATATATAT3-′ was used as the substrate in all experiments. Methyltransferase activity assay was performed as described (29). β-Galactosidase activity was assayed by using a β-gal assay kit (Invitrogen). LacZ-expressing cells were stained in situ by using a β-gal staining kit (Invitrogen).
The SssI DNA methylase accepting assay was carried out according to Schmitt et al. (30).
Determination of the Methylation Status of the Promoter-Enhancer Region of the Reporter Gene.
Southern blot analysis.
Genomic DNA was digested to completion with BglII and Af1II to release the entire enhancer-promoter fragment (500 bp). This fragment contains two closely spaced internal MspI–HpaII sites suitable for methylation analysis. Digestion with HpaII releases fragments of around 300 and 180 bp, respectively, when either one or both HpaII sites are demethylated. A radiolabeled probe spanning the entire region was used in detection of the restriction pattern.
Bisulfite sequencing.
Bisulfite sequencing was performed as described (31). The promoter-enhancer region of the reporter gene was amplified by PCR with the following primers: forward, 5′-GGATGTATTTAGAAAAATAAATAAATAGGG-3′; reverse, 5′-ATAATAAAAACCCCCCATAATAAACTTAAA-3′.
Immunoblots and Immunoprecipitations.
Immunoblots were as described (32). As primary antibody, we used the monoclonal antibody YOHGL2/99 (Serotec) directed against human G⋅T mismatch DNA glycosylase (1:500 dilution).
For the immunoprecipitation, nuclear extracts (50 μg) were first incubated with or without 10−8 M 9-cis-[20-methyl-3H]retinoic acid in 50 μl of PBS for 30 min at room temperature. The first antibody (1 μl of anti-G⋅T DNA glycosylase monoclonal antibody or 1 μl of anti-RXRα polyclonal antibody) was then added and incubated for 10 min at 37°C and then for 3 h at 4°C. The second antibody (2 μl of goat anti-mouse or goat anti-rabbit IgG) was added and incubation continued for 3 h at 4°C. The incubation mixture was then overlaid on 300 μl of 1 M sucrose in PBS and centrifuged for 30 min at 10,000 × g in a Sorvall HB rotor at 4°C. The discontinuous gradient was then frozen in liquid nitrogen, the tip of the tubes containing the immunoprecipitates was cut off, and the tip was processed for measuring radioactivity. Tests were done in triplicate, and the control values were subtracted from test values.
Alternatively, immunoprecipitates were separated by SDS/PAGE on a 10% gel, and proteins were electrotransferred to Immobilon membranes (Millipore, Bedford, MA). Western blot analysis was then carried out.
Chemicals and Antibodies.
Ponasterone A was purchased from Invitrogen and 5-azacytidine was from Fluka. Antibodies directed against the estrogen receptor and G⋅T mismatch DNA glycosylase were from Serotec. Polyclonal antibodies directed against the RXRα were obtained from P. Chambon (CNRS, INSERM, Université Louis Pasteur, Strasbourg, France). The hybridoma producing anti-ecdysone receptor antibody, developed by L. M. Riddiford, was obtained from the Developmental Studies Hybridoma Bank developed by the National Institute of Child Health and Human Development and maintained by the University of Iowa (Iowa City). The 9-cis-[20-methyl-3H]retinoic acid (67 Ci/mmol; 1 Ci = 37 GBq) was from NEN Life Science Products and [α-32P]dCTP (3000 Ci/mmol) was purchased from Amersham Pharmacia. Sodium disulfite for bisulfite sequencing reactions was from Merck.
Results
The Methylated Transgene Is Silent and Is Reactivated by 5-Azacytidine.
In the following experiments, it was important to assess the biological relevance of 5-MCDG for the active demethylation of DNA. Human embryonal kidney cells were stably transfected with the hormone-inducible β-galactosidase reporter gene, with the reporter gene under the regulation of the Drosophila minimal heat shock promoter coupled to a hormone inducible enhancer. The enhancer consists of tandem repeats of a hybrid retinoid–ecdysone response element. As a functional requirement, these cells express the RXR and ecdysone receptor that can form heterodimers in the presence of ponasterone A (an ecdysone analogue). Binding of the heterodimer to the retinoid–ecdysone response element of the enhancer triggers expression of the lacZ reporter gene. This system provides a very tight control of the reporter gene. This enhancer-promoter was stably introduced in methylated and nonmethylated forms to create the cell lines 293 M+ (methylated enhancer-promoter) and 293 M− (nonmethylated enhancer-promoter).
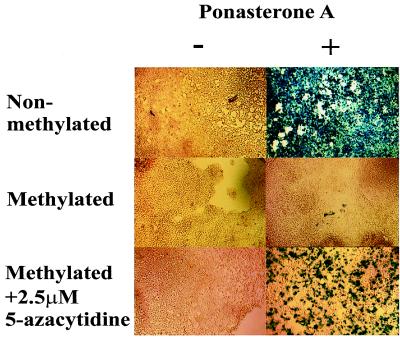
A stably transfected nonmethylated reporter gene was expressed to high levels in the presence of 10 μM ponasterone A (Fig. 1 Top and see Fig. 3). However, the methylated enhancer-promoter of the reporter gene was not induced by ponasterone A even after 48 h (Fig. 1 Middle and see Fig. 3). To show that the silencing of the gene is not caused by a position effect of the transgene but by the methylation of the enhancer-promoter, cells were treated with 2.5 μM 5-azacytidine for 5 days. Treatment with 5-azacytidine restored the ability of 10 μM ponasterone A to induce the reporter gene (Fig. 1 Bottom), thereby confirming that the transgene was silenced by methylating its promoter.
Figure 1.
Silenced methylated reporter genes can be reactivated with 5-azacytidine. (Top) β-Galactosidase activity from cells stably transfected with the β-galactosidase gene under the control of an nonmethylated promoter-enhancer. Ponasterone A (10 μM) was added to the cultures and induction of β-galactosidase was tested 48 h later. (Middle) Same cell type stably transfected with a β-galactosidase construct containing the methylated promoter-enhancer. (Bottom) Pretreatment of above cells (silent reporter gene) with 2.5 μM 5-azacytidine for 5 days and then further a treatment with 10 μM ponasterone A for 48 h.
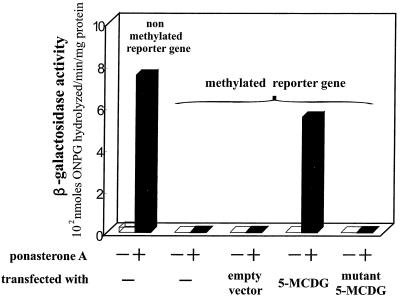
Figure 3.
Relative specific activity of β-galactosidase from cells not treated or treated with ponasterone A. These cell lines were permanently transfected with the nonmethylated or the demethylated reporter gene. In the latter, the cell line was further stably transfected with an empty vector or a vector containing 5-MCDG or a mutant of 5-MCDG. Cells treated with 10 μM ponasterone A were tested for β-galactosidase activity 48 h later. ONPG, ortho-nitrophenyl-β-D-galactopyranoside.
Overexpression of 5-MCDG Overcomes the Repression Caused by DNA Methylation.
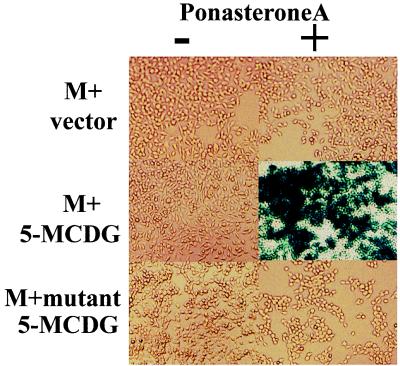
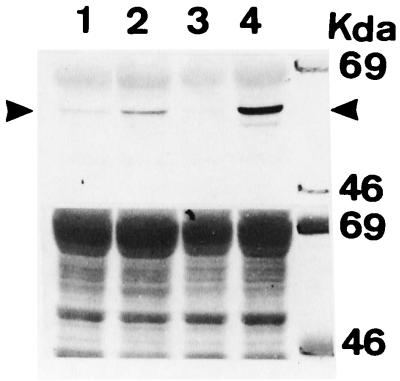
Introduction of the empty vector into 293M+ cells (15 clones tested) did not elicit a response to ponasterone A (Figs. 2 Top and 3). However, if the human 5-MCDG cDNA in the same vector is stably introduced into these cells, ponasterone A (6 positive clones of 10) induced considerable β-galactosidase activity (Figs. 2 Middle and 3). In contrast, no ponasterone A-induced expression of β-galactosidase was detected when an inactive mutant of 5-MCDG was introduced (18 clones tested; Figs. 2 Bottom and 3). We then tested whether the stable introduction of 5-MCDG had any effect on the total levels of enzyme protein and activity. Fig. 4 shows an immunoblot of nuclear extracts from cells overexpressing or not overexpressing 5-MCDG. When compared with the parent cell line (lane 1) or the cell line transfected with the empty vector (lane 3), overexpression of 5-MCDG resulted in a large increase in immunoreacting proteins (lane 4). A more modest increase was observed for the mutant of 5-MCDG (lane 2). Moreover, Fig. 5 shows that the cells overexpressing 5-MCDG had about 19-fold more 5-MCDG activity than the parent cell line M or cells transfected with the empty vector or the inactive mutant of 5-MCDG. In the cells overproducing 5-MCDG, there was no apparent change in DNA methyltransferase(s) activity compared with the cells transfected with the empty vector or a vector containing the mutant 5-MCDG. In addition, the activity of DNA methyltransferase(s) in the nuclear extracts from kidney cells was not inhibited (hemimethylated substrate) by increasing concentrations of recombinant 5-MCDG (results not shown).
Figure 2.
Overexpression of 5-MCDG in human embryonal kidney cells allows the induction of the reporter gene β-galactosidase by ponasterone A from a methylated promoter-enhancer. (Top) The empty vector was stably transfected in the cell line containing the silent methylated reporter gene. (Middle) 5-MCDG was stably transfected into the cell line containing the silent reporter gene. (Bottom) Inactive 5-MCDG mutant was transfected as above. When the reporter gene was demethylated, it could be induced by ponasterone A (10 μM for 48 h).
Figure 4.
Western blot of nuclear extract with anti-5-MCDG (G⋅T mismatch glycosylase) antibodies. (Upper) Lanes: 1, nuclear extract from parent cells (293 M+); 2, nuclear extract from parent cells transfected with the inactive mutant of 5-MCDG; 3, nuclear extract from cells transfected with the empty vector; 4, nuclear extract from cells transfected with 5-MCDG cDNA. (Lower) Total protein loading with Ponceau staining before immunoreaction.
Figure 5.
Activity of 5-MCDG from nuclear extracts. Average values (triplicates) obtained by cutting out the specific band (27-mer) from the gel and measuring the radioactivity. (Inset) Separation of the reaction product (27-mer) on a sequencing gel (20% polyacrylamide). Lanes: 1, parent cell line; 2, cell line transfected with the empty vector; 3, cell line transfected with the inactive mutant of 5-MCDG; 4, cell line transfected with the 5-MCDG; 5, blank.
Overexpression of 5-MCDG in the Absence of the Inducer Ponasterone A Demethylates the Promoter Region of the Reporter Gene.
We first tested whether overexpression of 5-MCDG had any effect on overall genomic methylation. The values obtained by the modified procedure of the SssI methyltransferase accepting assay (30), expressed as pmol of labeled cytosine incorporated per μg of DNA, were 1.1 ± 0.02 for the cells not expressing 5-MCDG and 0.97 ± 0.01 for cells overexpressing 5-MCDG. These results clearly indicate that overexpression of 5-MCDG did not lead to genome-wide demethylation. However, because we detected restoration of lacZ induction in cells overexpressing 5-MCDG, the state of methylation of the transgene was tested from various clones that did or did not overexpress 5-MCDG and had not been exposed to ponasterone A. For this preliminary experiment, the isoschizomers MspI and HpaII were used. The results (Fig. 6) clearly show that, in the cell lines not overexpressing 5-MCDG (Vc2 and Vc3), the promoter of the transgene remains methylated (lanes 3 and 6, 500-bp band). In contrast, when 5-MCDG is overexpressed (HGc2, HGc5, and HGc7), HpaII sites were demethylated yielding DNA fragments of 180–300 bp (lanes 9, 12, and 15). Thus, overexpression of 5-MCDG, as tested in three clones, resulted in demethylation of the promoter region of the methylated reporter gene in the absence of the inducer ponasterone A.
Figure 6.
Southern blot of genomic DNA from clones of stably transfected kidney cells digested as follows. Lanes: D, BglII and AflII; M, BglII, AflII, and MspI; H, BglII, AflII, and HpaII. Vc2 and Vc3 are clones stably transfected with the empty vector. HGc2, HGc5, and HGc7 are clones stably transfected with 5-MCDG. (These last three clones stain positive for β-galactosidase activity after ponasterone A treatment). When the promoter-enhancer is demethylated, digestion with HpaII gives DNA fragments of 180–300 bp.
A more complete determination of the methylation status at all of the CpGs within the enhancer-promoter region was performed. The enhancer-promoter region was amplified from bisulfite-modified genomic DNA from various cell lines by using specific primers, and the individual PCR products were cloned and sequenced. The results (Fig. 7) show clearly that the promoter-enhancer from cells containing the empty vector or the inactive mutant of 5-MCDG remained essentially fully methylated. In contrast, the promoter-enhancer region from a cell line expressing high 5-MCDG activity became extensively demethylated. Demethylation occurred mainly in the promoter and the enhancer adjacent to the region immediately upstream. Further upstream, the enhancer remained methylated.
Figure 7.
State of methylation of the promoter-enhancer region (600 bp) of the reporter gene in cell lines containing the empty vector (Top), the vector and 5-MCDG (Middle), and the vector and an inactive mutant of 5-MCDG (Bottom). For one cell line (HGC2 from Fig. 6), the bisulfite genomic sequencing of individual clones is shown. Solid circles are methylated CpGs, and vertical bars without circle are nonmethylated CpGs. TSS, transcription start site.
Overexpression of 5-MCDG Does Not Amplify the Reporter Gene.
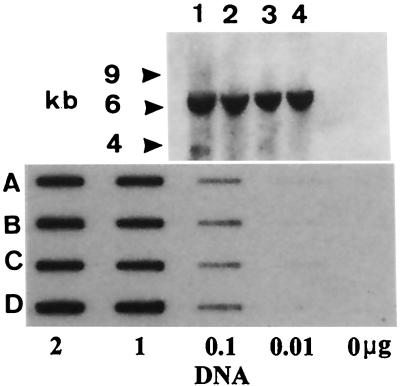
Because our experiments were conducted with stably transfected cell lines, it was possible that the selective pressure from antibiotics amplified the reporter gene (30). Thus, these results might be mimicked by a duplicated but nonmethylated copy of the reporter gene. To address this possibility, genomic DNA from the various cell lines was digested with EcoRI and probed for the lacZ structural gene. EcoRI cuts only once in the reporter gene construct with the second cut occurring in the flanking genomic sequence. The results (Fig. 8 Upper) show that, regardless of the cell lines tested, only a single fragment of about 6 kb was detected by the probe, indicating a single point of insertion.
Figure 8.
(Upper) Copy number determination of the integrated lacZ gene. Lanes: 1, parent cell line 293 M+; 2, mutant 5-MCDG; 3, cell line with empty vector; 4, cell line with 5-MCDG. Total genomic DNA was digested with EcoRI. EcoRI cuts only once in the integrated construct with a second cut generating the 6-kb fragment in surrounding genomic DNA. In Southern blots, the coding region of lacZ was used as a labeled probe. (Lower) Under the antibiotic selection of transformants, there is no amplification of the reporter gene. Slot blots of total genomic DNA (cleaved with EcoRI) where the coding region of lacZ was used as labeled probe. Rows: A, cell line with 5-MCDG; B, cell line with the empty vector; C, cell line with the mutant 5-MCDG; D, parent cell line 293 M+. 293 M+ is the parent cell line and is transfected with the mutant of 5-MCDG, the empty vector, or 5-MCDG.
An end-point titration of genomic DNA by slot blot hybridization (Fig. 8 Lower) clearly showed that there was no change in the gene dosage of the reporter gene, despite prolonged cell culture in the presence of antibiotics.
In the Cells Overexpressing 5-MCDG, There Is an Association Between RXRα and 5-MCDG.
Because we did not observe genome-wide demethylation in cells overexpressing 5-MCDG, we explored other possible mechanism for the specific targeting of DNA demethylation to the promoter-enhancer. Chambon and colleagues (33) have shown that there is a physical interaction between G⋅T mismatch DNA glycosylase and the retinoid receptors. Because the enhancer of the reporter gene in our system had tandem repeats of retinoid–ecdysone receptor binding sites, we tested whether such a physical interaction between the hormone receptor and the glycosylase occurred. Nuclear extracts from the cell lines containing the empty vector or vectors overexpressing 5-MCDG or the mutant of 5-MCDG were tested for such an interaction.
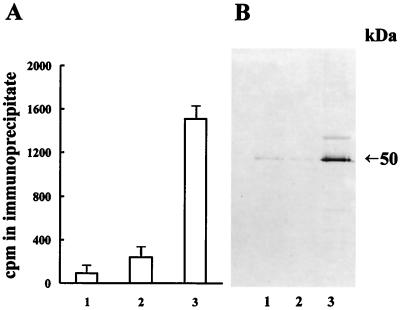
The results (Fig. 9A) show that an antibody directed against 5-MCDG precipitated significant retinoic acid binding activity in the nuclear extracts from cells overproducing 5-MCDG. The identity of the retinoic acid binding activity in the immunoprecipitates was further analyzed on Western blots. Fig. 9B shows clearly that the immunoprecipitate from cells overproducing 5-MCDG reacted with anti-RXRα antibodies. Identical results were obtained in the presence or absence of the ligand retinoic acid, indicating that the association of 5-MCDG with retinoid receptor is ligand independent. The reciprocal experiment could not be carried out because, under our experimental conditions, the retinoid receptor antibody did not precipitate the labeled retinoid receptor present in nuclear extracts. Thus these results strongly suggest that there is a physical interaction between the retinoid receptor and 5-MCDG. Additional tests with radioactively labeled ecdysone and ecdysone receptor antibodies failed to detect an association of the ecdysone binding protein with 5-MCDG (data not shown).
Figure 9.
Antibodies against 5-MCDG precipitate a complex containing RXRα. (A) Immunoprecipitation of a labeled retinoic acid binding protein with anti-5-MCDG. Bars: 1, nuclear extract of cells transfected with the empty vector; 2, with the inactive mutant 5-MCDG; 3, with the active 5-MCDG. Data are the mean ± SD (n = 3). (B) Western blot of the immunoprecipitates of A with RXRα antibodies.
Discussion
In our system, the overexpression of 5-MCDG does not lead to a genome-wide demethylation. This is hardly surprising because a massive action of 5-MCDG on a methylated genome could possibly lead to widespread DNA instability by creating numerous abasic sites, which require repair. For this reason, an alternative mechanism is probably responsible for global DNA demethylation (3). It is particularly true for the early global demethylation of the paternal genome occurring in blastocytes (3). The absence of significant global demethylation despite glycosylase overexpression suggests the role of other factors in determining the occurrence of demethylation at specific sites. Our results show a clear bias for demethylation at the promoter region itself rather than the upstream enhancer region that is primarily required for hormone induction, a further indication of the nonrandom nature of demethylation. Furthermore, the demethylation observed in our constructs was determined from cells that had not been exposed to ponasterone A (corresponding to virtually undetectable LacZ expression). Thus, in our system, DNA demethylation is necessary but not sufficient to activate the reporter gene.
In the presence or absence of the ligand retinoic acid, there is a physical association of retinoid receptors and the G⋅T mismatch DNA glycosylase (33), which also has 5-methylcytosine DNA glycosylase activity (25). The binding of the hormone receptor to 5-MCDG might be the signal that targets the glycosylase to methylated DNA, although the presence of other factors cannot be excluded. Our results as shown in Fig. 9 demonstrate that such an association between 5-MCDG and RXRα is taking place.
As already stated above (see Fig. 7), demethylation of the reporter gene occurs mainly at the minimal heat shock promoter and at a few sites in the adjacent enhancer. A similar situation was observed during the course of demethylation of avian vitellogenin gene after estradiol treatment of immature chickens (13). For the avian vitellogenin gene, the demethylation target was close to the estrogen response element and downstream of the transcription initiation site. Proceeding from the binding site of the glycosylase, demethylation could be either processive or distributive. We have shown (24) that the purified chicken embryo enzyme is distributive and not processive. Thus it is unlikely that a passive DNA demethylation is caused by 5-MCDG out-competing the maintenance methyltransferase for binding to the hemimethylated substrate. In vitro experiments showed that the activity of purified DNA methyltransferase 1 from HeLa cells or from human embryonic kidney cells is not inhibited by increasing concentrations of recombinant 5-MCDG (J.-P.J., unpublished results).
We have also shown (24, 25) that the purified enzyme and the recombinant enzyme preferentially use hemimethylated DNA as substrate. This reaction requires a minimum of one round of DNA replication in the absence of maintenance methylation for the generation of hemimethylated sites. In the present case, it would be interesting to know how many other genes become demethylated and reactivated when 5-MCDG is overexpressed. Although this is not yet known, differential hybridization to DNA microarrays should provide further examples. Because steroid hormones and retinoic acid trigger the demethylation of specific genes (ref. 34 and references therein and ref. 35), it would also be of interest to know whether the demethylation of DNA occurs through the same mechanism. Demethylation by 5-MCDG may involve the interaction with other macromolecules. For example, in the highly purified demethylation complex, in addition to 5-MCDG, CpG-rich RNA (36) and the p68 RNA helicase (37) were also detected. This observation and the finding that p68 RNA helicase can be associated with the estrogen receptor (38) suggest some interesting possibilities. For example, is there a functional link between these macromolecules and the in vivo demethylation of genes regulated by steroid hormones?
Acknowledgments
We thank Mrs. Y. C. Jost for typing the manuscript. Many thanks also go to Mr. P. Müller for the synthesis of the oligonucleotides. We are grateful to Prof. P. Chambon who generously provided the polyclonal antibody directed against RXRα. Finally, many thanks to Prof. von Wettstein who has always shown a keen interest in our work and who communicated this report. This work was sponsored by Novartis Research Foundation.
Abbreviations
- 5-MCDG
5-methylcytosine DNA glycosylase
- RXRα
retinoid X receptor α
References
- 1.Howlett S K, Reik W. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 2.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Nature (London) 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 3.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 4.Monk M, Boubelik M, Lehnert S. Development (Cambridge, UK) 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 5.Sanford J P, Clark H J, Chapman V M, Rossant J. Genes Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- 6.Shemer R, Kafri T, O'Connell A, Eisenberg S, Breslow J L, Razin A. Proc Natl Acad Sci USA. 1991;88:11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 8.Jaenisch R. Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 9.Rougier N, Bourc'his D, Gomes D M, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mermod J J, Bourgeois S, Defer N, Crépin M. Proc Natl Acad Sci USA. 1983;80:110–114. doi: 10.1073/pnas.80.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilks A, Cozens P, Mattaj J, Jost J P. Proc Natl Acad Sci USA. 1982;79:4252–4255. doi: 10.1073/pnas.79.14.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgan V, Elbrecht A, Goldman P, Lazier C B, Deeley R. J Biol Chem. 1982;257:14453–14460. [PubMed] [Google Scholar]
- 13.Saluz H P, Jiricny J, Jost J P. Proc Natl Acad Sci USA. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo K, Silke J, Georgier O, Marti P, Giorannini N, Rungger D. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh C L. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh C L. Curr Opin Genet Dev. 2000;10:224–228. doi: 10.1016/s0959-437x(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. Nature (London) 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 18.Cedar H, Verdine G L. Nature (London) 1999;397:568–569. doi: 10.1038/17492. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe A P, Jones P L, Wade P A. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S S. J Mol Biol. 2000;302:1–7. doi: 10.1006/jmbi.2000.4046. [DOI] [PubMed] [Google Scholar]
- 21.Gjerset R A, Martin D W. J Biol Chem. 1982;257:8581–8583. [PubMed] [Google Scholar]
- 22.Razin A, Szyf M, Kafri T, Roll M, Giloh H, Scarpa S, Carotti D, Cantoni G L. Proc Natl Acad Sci USA. 1986;83:2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vairapandi M, Duker H J. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jost J P, Siegmann M, Sun L, Leung R. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 25.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost J P. Proc Natl Acad Sci USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. . (First Published April 25, 2000; 10.1073/pnas.100107597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost J P. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan J J, Wiebauer K, Jiricny J. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 28.No D, Yao T P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jost J P, Jost Y C. J Biol Chem. 1994;269:10040–10043. [PubMed] [Google Scholar]
- 30.Schmitt F, Oakeley E J, Jost J P. J Biol Chem. 1997;272:1534–1540. doi: 10.1074/jbc.272.3.1534. [DOI] [PubMed] [Google Scholar]
- 31.Raizis A M, Schmitt F, Jost J P. Anal Biochem. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y L, Sun L, Jost J P. Nucleic Acids Res. 1996;24:2718–2722. doi: 10.1093/nar/24.14.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. J Biol Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 34.Jost J P, Schwarz S, Oakeley E J. Curr Top Steroid Res. 1998;1:103–110. [Google Scholar]
- 35.Burbelo P D, Horikoshi S, Yamada Y. J Biol Chem. 1990;265:4839–4843. [PubMed] [Google Scholar]
- 36.Jost J P, Frémont M, Siegmann M, Hofsteenge J. Nucleic Acids Res. 1997;25:4545–4550. doi: 10.1093/nar/25.22.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jost J P, Schwarz S, Hess D, Angliker H, Fuller-Pace F, Stahl H, Thiry S, Siegmann M. Nucleic Acids Res. 1999;27:3245–3252. doi: 10.1093/nar/27.16.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]