Yos9p is involved in ER-associated degradation (ERAD) of misfolded glycoproteins. This study shows that Yos9p is required for ER retention of ERAD substrates by targeting them to the Hrd1p E3 ligase. This ER retention is independent of the glycan degradation signal on substrates and is separable from the later degradation step.
Abstract
The endoplasmic reticulum (ER) has an elaborate quality control system, which retains misfolded proteins and targets them to ER-associated protein degradation (ERAD). To analyze sorting between ER retention and ER exit to the secretory pathway, we constructed fusion proteins containing both folded carboxypeptidase Y (CPY) and misfolded mutant CPY (CPY*) units. Although the luminal Hsp70 chaperone BiP interacts with the fusion proteins containing CPY* with similar efficiency, a lectin-like ERAD factor Yos9p binds to them with different efficiency. Correlation between efficiency of Yos9p interactions and ERAD of these fusion proteins indicates that Yos9p but not BiP functions in the retention of misfolded proteins for ERAD. Yos9p targets a CPY*-containing ERAD substrate to Hrd1p E3 ligase, thereby causing ER retention of the misfolded protein. This ER retention is independent of the glycan degradation signal on the misfolded protein and operates even when proteasomal degradation is inhibited. These results collectively indicate that Yos9p and Hrd1p mediate ER retention of misfolded proteins in the early stage of ERAD, which constitutes a process separable from the later degradation step.
INTRODUCTION
Quality control mechanisms of the endoplasmic reticulum (ER) monitor the folding states of proteins and retain immature proteins in the ER. Terminally misfolded proteins are targeted to degradation pathways known as ER-associated protein degradation (ERAD; Vember and Brodsky, 2008). Misfolded proteins are recognized and targeted to the ubiquitin ligase complexes for ubiquitination, which is followed by degradation by proteasome in the cytosol. In yeast Saccharomyces cerevisiae, the Hrd1p complex consisting of Hrd1p, Hrd3p, Usa1p, and Der1p ubiquitinates ERAD substrates with misfolded luminal domains or transmembrane lesions (Vashist and Ng, 2004; Carvalho et al., 2006). Hrd1p forms an oligomer that is likely involved in the movement of ERAD substrate proteins across the ER membrane (Carvalho et al., 2010).
N-Linked glycans have been shown to function as degradation signals in ERAD of some misfolded glycoproteins. Yos9p is a lectin-like ERAD factor, which is required for ERAD of misfolded glycoproteins (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). Yos9p forms complexes with an ER luminal Hsp70, BiP, as well as with Hrd3p (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006) and has been proposed to function in recognition of glycan ERAD signals formed by the mannosidase Mnl1p/Htm1p (Jakob et al., 2001; Nakatsukasa et al., 2001; Quan et al., 2008; Clerc et al., 2009; Sakoh-Nakatogawa et al., 2009; Gauss et al., 2011).
Although less efficiently than correctly folded proteins, some misfolded proteins can leave the ER and subsequently come back to the ER for ERAD or move further along the secretory pathway (Caldwell et al., 2001; Vashist et al., 2001; Hirayama et al., 2010). The ER exit of misfolded proteins depends on their intrinsic ER exit signals because removal of exit signals results in their ER retention (Kawaguchi et al., 2010). Addition of a strong ER exit signal or inactivation of the ERAD pathway leads to increased Golgi delivery of some ERAD substrates (Kincaid and Cooper, 2007). Sorting of misfolded proteins between ERAD and ER exit pathways is thus affected by the balance between the operational activities of the two pathways. Nevertheless, removal of intrinsic ER exit signals does not simply stimulate ERAD of misfolded proteins (Kawaguchi et al., 2010), leaving the mechanism of ER retention of misfolded proteins for ERAD unclear.
In the present study, we analyzed the sorting of misfolded proteins between the ERAD and secretory pathways by constructing new model proteins that can be sorted to both ERAD and vacuolar transport pathways. We found that sorting efficiency of the model ERAD substrate proteins to the ER retention for ERAD is correlated well with their efficiency to interact with Yos9p. Inhibition of proteasomal degradation in ERAD by treating cells with a proteasome inhibitor did not affect the ER retention efficiency of misfolded proteins, suggesting that ER retention is not coupled with the later degradation step in ERAD. We also found that Hrd1p has an activity to retain misfolded proteins in the ER. Yos9p is required for targeting of misfolded proteins to Hrd1p for ER retention independent of the glycan ERAD signals. We thus propose new roles of Yos9p and Hrd1p in ER retention of misfolded proteins for ERAD.
RESULTS
Carboxypeptidase Y/mutant CPY fusion proteins containing both folded and misfolded units show distinct fates
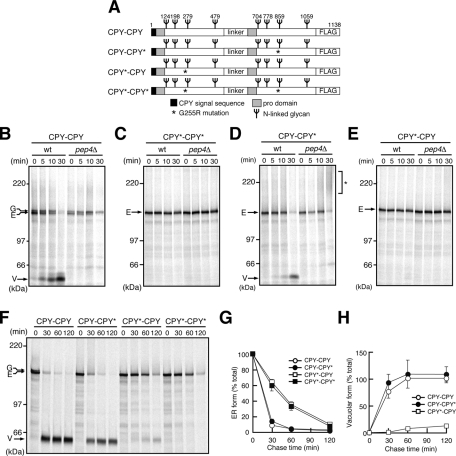
To assess the effects of alteration in the balance between the signals for ERAD and ER exit (or vacuolar targeting), we constructed a series of fusion proteins: mutants of carboxypeptidase Y (CPY; CPY-CPY, CPY–mutant CPY [CPY*], CPY*-CPY, and CPY*-CPY*) consisting of properly folded vacuolar CPY and/or its misfolded mutant CPY* with the G255R mutation (Finger et al., 1993; Figure 1A). The two CPY or CPY* domains are linked in tandem via a 68-residue linker segment that consists of the streptavidin-binding peptide (SBP; Keefe et al., 2001) and triple-hemagglutinin (3HA) epitope tag. Whereas the N-terminal unit contains the signal sequence of CPY, the C-terminal one lacks the signal sequence but contains the 3×FLAG epitope tag at the C-terminus. These fusion proteins possess eight N-linked glycosylation sites and two sites for PEP4-dependent processing in the vacuole past the prosequences.
FIGURE 1:
CPY-CPY and CPY-CPY* are efficiently transported to the vacuole, whereas CPY*-CPY and CPY*-CPY* are retained in the ER. (A) Schematic representations of the fusion proteins used in this study. (B–E) Wild-type (wt) and pep4Δ cells expressing CPY-CPY (B), CPY*-CPY* (C), CPY-CPY* (D), or CPY*-CPY (E) were pulse labeled with 35S-amino acids for 10 min and chased for the indicated times at 30°C. Fusion proteins were recovered from cell extracts by immunoprecipitation using anti-CPY antibodies and analyzed by SDS–PAGE and radioimaging. Arrows indicate the positions of the ER form (E), Golgi form (G), and vacuolar form (V) of the fusion proteins. The asterisk in D shows the hyperglycosylated form of CPY-CPY*. (F) Wild-type cells expressing CPY-CPY, CPY-CPY*, CPY*-CPY, or CPY*-CPY* were analyzed by pulse-chase experiments as in B. (G, H) Relative amounts of the ER (G) and the vacuolar (V) forms of CPY*-CPY in F were quantified by radioimaging. Error bars represent SDs from three independent experiments. The amount of the ER form at 0 min was set to 100%.
Each CPY/CPY* fusion protein was expressed in the CPY-deficient yeast prc1∆ mutant under the regulation of the PRC1 promoter. Trypsin digestion showed that the CPY and CPY* units in those fusion proteins were folded and misfolded, respectively (Supplemental Figure S1). We followed the fates of the newly synthesized CPY/CPY* fusion proteins by pulse-chase analyses in vivo. CPY-CPY was detected as a 150-kDa ER form with a cleaved signal sequence and with ER glycosylation, which was then converted to a 154-kDa species and subsequently to a 60-kDa species during 30 min of chase in wild-type cells (Figure 1B). The 154-kDa species is the Golgi form of CPY-CPY, as inhibition of the ER-to-Golgi transport by the sec12-4 mutation blocked its generation (Supplemental Figure S2). The 60-kDa species, which was not generated in pep4Δ cells lacking vacuolar Pep4 protease (Figure 1B, pep4Δ), arose from the vacuolar-processed fragments of CPY-CPY. The time course of the maturation process of CPY-CPY was similar to that of CPY (Nishikawa et al., 1994), indicating that CPY-CPY is transported normally to the vacuole as efficiently as CPY.
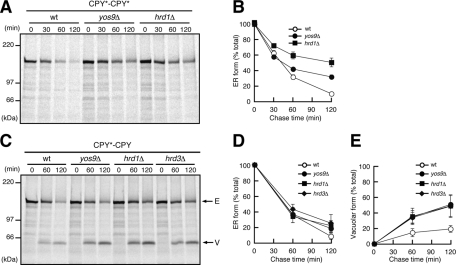
In contrast to CPY-CPY, CPY*-CPY* remained as the ER form even after 30 min of chase (Figure 1C), and its decrease was characterized by a half-life of ∼40 min in longer chase experiments (Figure 1, F–H). The ER form of CPY*-CPY* was stabilized significantly in the ERAD-defective yos9Δ and hrd1Δ mutants (Figure 2, A and B) as observed for CPY*-3HA (Supplemental Figure S3), indicating that CPY*-CPY* is sorted to the ERAD pathway in the ER. The fates of CPY-CPY and CPY*-CPY* are thus the same as those of CPY and CPY*, respectively, suggesting that tandem connection of the same CPY or CPY* units itself does not affect the fates of the resultant fusion proteins.
FIGURE 2:
CPY*-CPY* and CPY*-CPY are degraded by ERAD. (A) Wild-type, yos9Δ. or hrd1Δ cells expressing CPY*-CPY* were analyzed by pulse-chase experiments as in Figure 1. (B) Relative amounts of CPY*-CPY* were quantified by radioimaging. Error bars represent SDs from three independent experiments. The amount of CPY*-CPY* at 0 min was set to 100%. (C) Wild-type, yos9Δ, hrd1Δ, or hrd3Δ cells expressing CPY*-CPY were pulse chased and analyzed. Arrows indicate the position of the ER (E) or the vacuolar (V) forms of CPY*-CPY. (D, E) Relative amounts of the ER (D) and the vacuolar (E) forms of CPY*-CPY in C were quantified as in Figure 1.
Although CPY-CPY* and CPY*-CPY contain both folded and misfolded units, their fates were quite different. We observed PEP4-dependent generation of the vacuolar form of CPY-CPY* within 30 min of chase (Figure 1D). We also observed formation of smear species of CPY-CPY* in the pep4Δ mutant (Figure 1D, asterisk), which probably arose from hyperglycosylation in the CPY* unit in the Golgi (Spear and Ng, 2003), as the smear species were converted into a single 135-kDa form after removal of N-linked glycans by endoglycosidase H treatment (Supplemental Figure S4). These results collectively show that CPY-CPY* was efficiently transported to the vacuole via the Golgi. The vacuole form of CPY-CPY* was mainly produced from the N-terminal CPY unit, since removal of a N-glycosylation site from the N-terminal CPY unit but not from the C-terminal CPY* unit reduced the molecular weight of its vacuole form (Supplemental Figure S5). The misfolded CPY* unit was probably not resistant to vacuolar proteases in wild-type cells. In contrast, CPY*-CPY remained as the ER form after 30 min of chase (Figure 1E). The vacuole form of CPY*-CPY, which was mainly produced from the C-terminal CPY unit (Supplemental Figure S5), appeared only after 60 min of chase and then increased to ∼20% of total labeled CPY*-CPY after the 120-min chase (Figure 1, F and H). The ER form of CPY*-CPY was moderately stabilized by deletion of the YOS9, HRD1, or HRD3 gene, which also made generation of the vacuole form more prominent (Figure 2, C–E). Therefore CPY*-CPY preferably stays in the ER for degradation instead of being transported to the vacuole.
CPY-CPY* and CPY*-CPY are differently recognized by Yos9p
The foregoing results suggest that CPY-CPY* and CPY*-CPY are differently recognized by the ER quality control system for sorting between the ER retention for ERAD and the ER exit for vacuolar targeting via the Golgi. Therefore we sought to find which ERAD component is responsible for the differential recognition between CPY-CPY* and CPY*-CPY in the ER. BiP is a molecular chaperone Hsp70 in the ER and binds preferentially to misfolded ERAD substrates to maintain their solubility for efficient ERAD (Nishikawa et al., 2001). When we tested interactions of BiP with CPY/CPY* fusion proteins by coimmunoprecipitation, BiP bound to folded CPY-CPY only poorly as compared with the fusion proteins containing CPY* units (Figure 3A). However, binding efficiency of BiP is nearly the same for CPY-CPY*, CPY*-CPY, and CPY*-CPY*, so that the observed difference in the fates between pulse-labeled CPY*-CPY and CPY-CPY* cannot be ascribed to their interactions with BiP.
FIGURE 3:
BiP binds equally to CPY-CPY* and CPY*-CPY, whereas Yos9p binds to CPY*-CPY more efficiently than CPY-CPY*. Cell extracts were prepared from wild-type cells transformed with an empty vector or a vector expressing CPY-CPY, CPY-CPY*, CPY*-CPY, or CPY*-CPY* as indicated and subjected to immunoprecipitation with anti-FLAG agarose. (A) Cell lysates (Input) and immunoprecipitated materials (IP: FLAG) were analyzed by SDS–PAGE and immunoblotting with anti-CPY (IB: αCPY) and anti-BiP (IB: αBiP) antibodies. Relative amounts of BiP coprecipitated with the fusion proteins were quantified. Error bars represent SDs from three independent experiments. The amount of BiP coprecipitated with CPY*-CPY* was set to 100%. (B) Cell lysates (Input) and immunoprecipitated materials (IP: FLAG) were analyzed by SDS–PAGE and immunoblotting with anti-CPY (IB: αCPY) and anti-Yos9p (IB: αYos9p) antibodies. Relative amounts of Yos9p coprecipitated with the fusion proteins were quantified. The amount of Yos9p coprecipitated with CPY*-CPY* was set to 100%.
A lectin-like protein Yos9p is known to recognize the glycan signals of misfolded ERAD substrates (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005). When analyzed by coimmunoprecipitation, affinities of the CPY/CPY* fusion proteins for Yos9p were in the order of CPY*-CPY* > CPY*-CPY > CPY-CPY* > CPY-CPY (Figure 3B), which correlated well with the order of sorting efficiency to the ER retention or prevention from the vacuolar transport (Figure 1, B–H). CPY*-CPY*, which interacted with Yos9p most efficiently, was preferentially degraded by ERAD (Figure 2A). On the other hand, CPY*-CPY, which was recognized by Yos9p less efficiently than CPY*-CPY*, was targeted to both the ERAD and vacuolar transport pathways (Figure 2C). CPY-CPY*, as well as CPY-CPY, efficiently exited the ER and reached the vacuole (Figure 1, F and H). Although Yos9p could cooperate with BiP to facilitate substrate targeting to the Hrd1p complex (Carvalho et al., 2006; Denic et al., 2006), the present results suggest that Yos9p, not BiP, plays a primary role in substrate sorting to the ER retention or the ERAD pathway.
We next analyzed the effects of direct inhibition of the ER exit on ERAD and on Yos9p binding of the CPY/CPY* fusion proteins. The cargo-sorting receptor Erv29p is responsible for the ER exit of CPY, whose depletion causes impaired vacuolar trafficking of CPY (Belden and Barlowe, 2001). When we suppressed the ER exit of CPY-CPY* by deletion of the ERV29 gene, the ER form of CPY-CPY* indeed underwent Hrd1p-dependent degradation (Supplemental Figure S6, A and B). In parallel, depletion of Erv29p slightly increased the interactions of Yos9p with CPY-CPY* but not with CPY*-CPY (Supplemental Figure S6C).
Involvement of Yos9p and Hrd1p in active retention of misfolded proteins in the ER
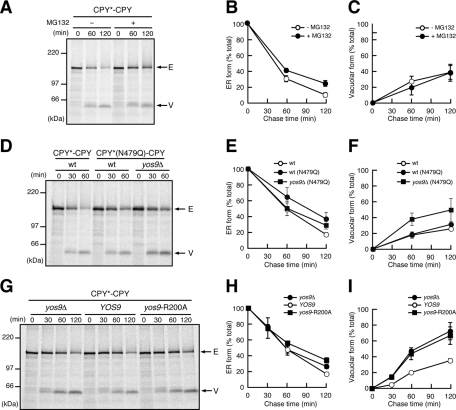
As was shown, depletion of the ERAD factor Yos9p, Hrd1p, or Hrd3p resulted in enhanced vacuolar transport of CPY*-CPY. We thus asked whether vacuolar transport of CPY*-CPY is accelerated by inactivation of ERAD in general. Although carbobenzoxyl-leucinyl-leucinyl-leucinal (MG132) treatment, which inhibits proteasomal degradation of ERAD substrates including CPY*, stabilized CPY*-CPY in the ER, generation of the vacuolar form of CPY*-CPY was not affected (Figure 4, A–C). Therefore inhibition of proteasomal degradation, a later step of the ERAD pathway, does not affect sorting of CPY*-CPY between the ER retention and ER exit for the vacuole.
FIGURE 4:
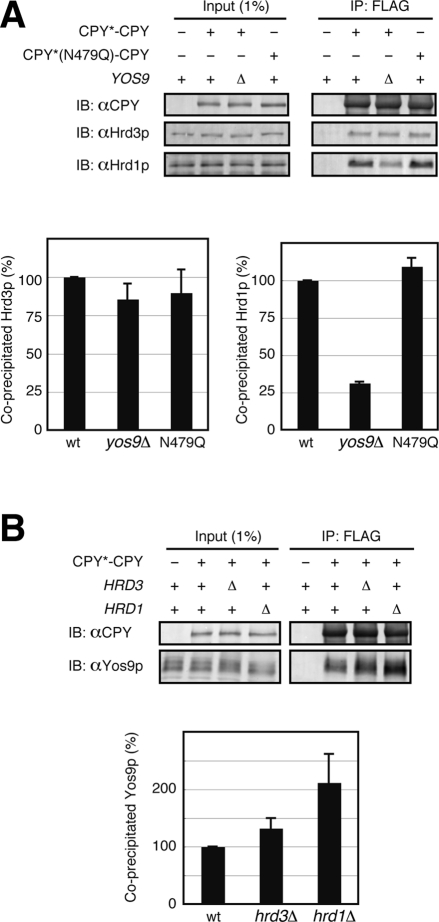
Yos9p is required for ER retention of CPY*-CPY. (A) Wild-type cells expressing CPY*-CPY were analyzed by pulse-chase experiments in the presence (+) or the absence (−) of 100 μM MG132 as in Figure 1. Arrows indicate the position of the ER (E) or the vacuolar (V) forms of CPY*-CPY. (B, C) Relative amounts of the ER (B) and the vacuolar (C) forms of CPY*-CPY in A are shown as in Figure 1. (D) Wild-type cells expressing CPY*-CPY or CPY*(N479Q)-CPY and yos9Δ cells expressing CPY*(N479Q)–CPY were analyzed by pulse-chase experiments. (E, F) Relative amounts of the ER (E) and the vacuolar (F) forms of the fusion proteins in D. (G) yos9Δ cells expressing CPY*-CPY were transformed with an empty vector or a vector expressing Yos9p or Yos9p-R200A and analyzed by pulse-chase experiments. (H, I) Relative amounts of the ER (H) and the vacuolar (I) forms of CPY*-CPY in G.
N-glycans function as degradation signals in ERAD of misfolded glycoproteins, including CPY*. We thus asked whether deletion of those glycan degradation signals in misfolded proteins affects their ER retention. Among the four N-linked glycans in CPY*, only the N-glycan attached to Asn-479 functions as a degradation signal, and CPY* variants lacking this glycan signal were degraded poorly as compared with CPY* with all four N-linked glycans (Kostova and Wolf, 2005; Spear and Ng, 2005). Removal of the glycan degradation signal in the N-terminal CPY* unit of CPY*-CPY by the N479Q mutation stabilized CPY*-CPY in the ER, yet it did not enhance formation of its vacuolar form (Figure 4, D–F). However, the YOS9 deletion accelerated formation of the vacuolar form of CPY*(N479Q)-CPY, as in the case of CPY*-CPY (Figure 2, C–E). Coimmunoprecipitation analyses showed that Yos9p binds to CPY*(N479Q)-CPY with efficiency similar to that of CPY*-CPY (Supplemental Figure S7A). Therefore ER retention of CPY*-CPY does not depend on glycan degradation signals but requires recognition by Yos9p.
Yos9p has a mannose 6-phosphate receptor homology (MRH) domain, which was suggested to mediate recognition of the glycan signals for ERAD (Hosokawa et al., 2010). Purified recombinant Yos9p has an ability to bind to the glycan degradation signals, yet this binding is abolished by the R200A mutation in the MRH domain of Yos9p (Quan et al., 2008). The isolated MRH domain of OS-9, a mammalian counterpart of Yos9p, was found to directly interact with the ERAD signal glycans (Satoh et al., 2010). To examine the roles of the Yos9p MRH domain in the ER retention of misfolded proteins or prevention from the vacuolar traffic, we expressed the R200A mutant of Yos9p (Yos9p-R200A) in the yos9Δ strain. Coimmunoprecipitation analyses showed that Yos9p-R200A efficiently interacted with CPY*-CPY (Supplemental Figure S7B), which is consistent with previous reports (Bhamidipati et al., 2005; Denic et al., 2006). However, expression of Yos9p-R200A did not suppress the enhanced formation of the CPY*-CPY vacuolar form in the yos9Δ mutant (Figure 4, G–I), indicating that the MRH domain is essential for the ER retention of CPY*-CPY. It is probable that ER retention of CPY*-CPY requires the function of Yos9p other than its binding ability to ERAD substrates, for example, interactions with the Hrd1p complex.
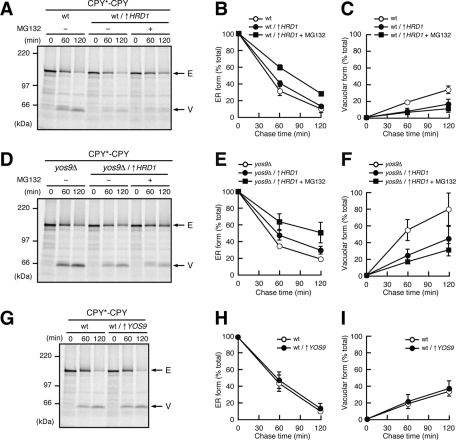
To test the possibility that Hrd1p plays roles in ER retention of misfolded proteins, we overexpressed Hrd1p from the GAL1 promoter and found that the Hrd1p overexpression significantly suppressed the vacuolar delivery of CPY*-CPY (Figure 5, A–C). Because suppression of the vacuolar transport of CPY*-CPY was observed even when proteasomal degradation was inhibited by MG132 (Figure 5, A–C), the enhanced ER retention was not due to the enhanced later degradation step in ERAD by Hrd1p overexpression. Overexpression of Hrd1p also suppressed the vacuolar delivery of CPY*-CPY in the yos9Δ mutant both in the presence and absence of MG132, indicating that Hrd1p overexpression bypassed the Yos9p requirement (Figure 5, D–F). These results collectively suggest that Hrd1p itself has an activity to retain misfolded proteins in the ER even in the absence of Yos9p. In contrast, overexpression of Yos9p did not suppress the vacuolar transport of CPY*-CPY (Figure 5, G–I), suggesting that the amount of Yos9p does not limit the capacity for ER retention of misfolded proteins.
FIGURE 5:
Hrd1p has an activity to retain misfolded proteins in the ER. (A) Wild-type cells expressing CPY*-CPY transformed with an empty vector or a vector expressing Hrd1p from the GAL1 promoter (↑HRD1) were grown in media containing 2% galactose and analyzed by pulse-chase experiments in the presence (+) or the absence (−) of 100 μM MG132 as in Figure 1. Arrows indicate the position of the ER (E) or the vacuolar (V) forms of CPY*-CPY. (B, C) Relative amounts of the ER (B) and the vacuolar (C) forms of CPY*-CPY in A were quantified as in Figure 1. (D) yos9Δ cells expressing CPY*-CPY transformed with an empty vector or a vector expressing Hrd1p from the GAL1 promoter (↑HRD1) were grown in media containing 2% galactose and analyzed by pulse-chase experiments in the presence (+) or the absence (−) of 100 μM MG132. (E, F) Relative amounts of the ER (E) and the vacuolar (F) forms of the fusion proteins in D)were quantified as in Figure 1. (G) Wild-type cells expressing CPY*-CPY transformed with an empty vector or a vector expressing Yos9p from a multicopy plasmid (↑YOS9) were analyzed by pulse-chase experiments. (H, I) Relative amounts of the ER (H) and the vacuolar (I) forms of CPY*-CPY in G were quantified as in Figure 1.
Coimmunoprecipitation experiments revealed the physical interaction between CPY*-CPY and Hrd1p, which was inhibited by ∼70% by deletion of the YOS9 gene (Figure 6A), and deletion of HRD1 led to accumulation of CPY*-CPY at the level of Yos9p (Figure 6B). Therefore Yos9p functions in targeting of CPY*-CPY to Hrd1p for ER retention. A CPY*-CPY variant containing the N479Q mutation interacted with Hrd1p as efficiently as CPY*-CPY, indicating that targeting of CPY*-CPY to Hrd1p by Yos9p is independent of the glycan degradation signal (Figure 6A). Nevertheless, the Yos9p-R200A was defective in the targeting of CPY*-CPY to Hrd1p (Supplemental Figure S7C). Coimmunoprecipitation experiments also showed the interaction between Hrd3p and Yos9p. Consistent with the previous reports that Hrd3p interacts with ERAD substrates upstream of Yos9p (Gauss et al., 2006; Denic et al., 2006; Stanley et al., 2011), the interaction between CPY*-CPY and Hrd3p was hardly affected by deletion of the YOS9 gene (Figure 6A). It should be noted that deletion of YOS9 affected the amount of Hrd1p but not Hrd3p coprecipitated with CPY*-CPY, although the yos9 mutation does not affect the interaction between Hrd1p and Hrd3p (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006). This probably reflects the possibility that most of Hrd3p was dissociated from Hrd1p in our solubilization condition (unpublished data). In contrast, depletion of HRD3 did not decrease the amount of Yos9p coprecipitated with CPY*-CPY (Figure 6B). Overexpression of Hrd3p from the GAL1 promoter suppressed the vacuolar transport of CPY*-CPY (Figure 7, A–C), which was accompanied by the increased interaction of CPY*-CPY with Hrd1p (Figure 7D). We also observed increased interaction between Yos9p and CPY*-CPY by overexpression of Hrd3p, suggesting that Hrd3p acts upstream of Yos9p to target misfolded proteins to Hrd1p.
FIGURE 6:
Yos9p is required for targeting of CPY*-CPY to Hrd1p. (A) Cell lysates were prepared from wild-type or yos9Δ cells transformed with a vector alone, a vector expressing CPY*-CPY, or a vector expressing CPY*(N479Q)-CPY and subjected to immunoprecipitation with anti-FLAG agarose. Cell lysates (Input) and immunoprecipitated materials (IP: FLAG) were analyzed by SDS–PAGE and immunoblotting with anti-CPY (IB: αCPY), anti-Hrd3p (IB: αHrd3p), and anti-Hrd1p (IB: αHrd1p) antibodies. Relative amounts of Hrd3p or Hrd1p coprecipitated with the fusion proteins were quantified. Error bars represent SDs from three independent experiments. The amount of Hrd3p or Hrd1p coprecipitated with CPY*-CPY in wild-type lysate was set to 100%. (B) Cell lysates were prepared from wild-type cells transformed with a vector alone or wild-type, hrd1Δ, or hrd3Δ cells transformed with a vector expressing CPY*-CPY and subjected to immunoprecipitation as in Figure 3B.
FIGURE 7:
Effect of overexpression of Hrd3p on ER retention of CPY*-CPY. (A) Wild-type cells expressing CPY*-CPY transformed with an empty vector or a vector expressing Hrd3p from the GAL1 promoter (↑HRD3) were grown in media containing 2% galactose and analyzed by pulse-chase experiments as in Figure 1. Arrows indicate the position of the ER (E) or the vacuolar (V) forms of CPY*-CPY. (B, C) Relative amounts of the ER (B) and the vacuolar (C) forms of CPY*-CPY in A were quantified as in Figure 1. (D) Cell lysates were prepared from wild-type cells or cells overexpressing Hrd3p from the GAL1 promoter (↑HRD3), which were transformed with an empty vector or a vector expressing CPY*-CPY, and subjected to immunoprecipitation with anti-FLAG agarose. Cell lysates (Input) and immunoprecipitated materials (IP: FLAG) were analyzed by SDS–PAGE and immunoblotting with anti-CPY (IB: αCPY), anti-Hrd1p (IB: αHrd1p), and anti-Yos9p (IB: αYos9p) antibodies. Relative amounts of Hrd1p and Yos9p coprecipitated with the fusion proteins were quantified. Error bars represent SDs from three independent experiments. The amount of Hrd1p or Yos9p coprecipitated with CPY*-CPY in wild-type lysate was set to 100%.
Overexpression of protein disulfide isomerase–inhibited ERAD and vacuolar delivery of CPY*-CPY
We also analyzed involvement of other ER-resident chaperones in ER retention of CPY*-CPY. Lhs1p is another member of the Hsp70 family in the yeast ER with ∼30% similarity to BiP, which functions as a nucleotide exchange factor for BiP (Steel et al., 2004). We previously showed that depletion of Lhs1p does not affect ERAD of CPY* (Nishikawa et al., 2001). Again, depletion of Lhs1p did not affect stability or vacuolar transport of CPY*-CPY (Supplemental Figure S8, A–C).
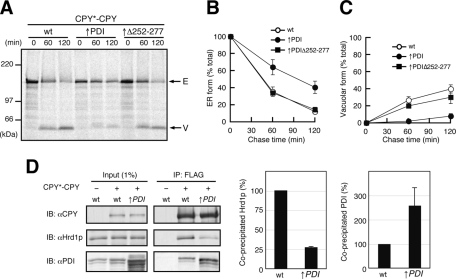
Protein disulfide isomerase (PDI) is an oxidoreductase in the ER, which functions in formation and rearrangement of disulfide bonds of its substrate proteins (Gilbert 1997). PDI also has a chaperone-like function and is involved in ERAD of misfolded proteins, including CPY* (Gillece et al., 1999). To test the possible involvement of PDI in ER retention of CPY*-CPY, we overexpressed PDI from a multicopy plasmid (approximate sevenfold overproduction as judged from immunoblotting) and found that the vacuolar delivery of CPY*-CPY was significantly suppressed (Figure 8, A and C). In contrast, overexpression of PDIΔ252-277, a PDI mutant defective in the peptide-binding activity (Gillece et al., 1999), did not inhibit vacuolar transport of CPY*-CPY. We also found that overexpression of PDI but not PDIΔ252-277 significantly stabilized the ER form of CPY*-CPY (Figure 8, A and B), which was accompanied by the reduced interaction of Hrd1p with CPY*-CPY (Figure 8D). Interaction between PDI and CPY*-CPY was increased approximately threefold, suggesting that overexpressed PDI trapped CPY*-CPY through its chaperone-like activity, thereby sequestering it from both ERAD and vacuolar transport pathways.
FIGURE 8:
Overexpression of PDI inhibited both ERAD and vacuolar transport of CPY*-CPY. (A) Wild-type cells expressing CPY*-CPY transformed with an empty vector or a multicopy vector expressing PDI (↑PDI) or PDIΔ252-277 (↑Δ252-277) were analyzed by pulse-chase experiments as in Figure 1. Arrows indicate the position of the ER (E) or the vacuolar (V) forms of CPY*-CPY. (B, C) Relative amounts of the ER (B) and the vacuolar (C) forms of CPY*-CPY in A were quantified as in Figure 1. (D) Cell lysates were prepared from wild-type cells or cells overexpressing PDI from a multicopy plasmid (↑PDI), which were transformed with an empty vector or a vector expressing CPY*-CPY, and subjected to immunoprecipitation with anti-FLAG agarose. Cell lysates (Input) and immunoprecipitated materials (IP: FLAG) were analyzed by SDS–PAGE and immunoblotting with anti-CPY (IB: αCPY), anti-Hrd1p (IB: αHrd1p), and anti-PDI (IB: αPDI) antibodies. Relative amounts of Hrd1p and PDI coprecipitated with the fusion proteins were quantified. Error bars represent SDs from three independent experiments. The amount of Hrd1p and PDI coprecipitated with CPY*-CPY* in wild-type lysate was set to 100%. Relative amount of PDI in cell lysates was also quantified.
DISCUSSION
Accumulated evidence suggests that the ERAD and ER exits of misfolded proteins compete with each other. However, the mechanism of ER retention of ERAD substrates remained unclear because ER retention and later steps in the ERAD process are usually coupled with each other. In the present study, we assessed ER retention of misfolded CPY/CPY* fusion proteins, especially CPY*-CPY, by monitoring their vacuolar traffic, which competes with ER retention, independent of the later steps in ERAD. We thus found that the two ERAD factors Yos9p and Hrd1p play essential roles in ER retention of misfolded proteins. The ER retention depends on Yos9p and Hrd1p and correlates well with the interactions of Yos9p, but not BiP, with substrate proteins. The interaction between CPY*-CPY and Hrd1p, as well as ER retention of CPY*-CPY, is impaired in yeast cells lacking Yos9p (Figures 4, G–I, and 6A). However in the absence of Yos9p, overexpression of Hrd1p can still recover the ER retention of CPY*-CPY (Figure 5F), suggesting that Hrd1p itself has an activity to interact with misfolded proteins and retain them in the ER. Therefore, efficient ER retention of misfolded proteins requires the recognition of misfolded proteins and their delivery to Hrd1p by Yos9p (Figure 9), although Yos9p is not essential but increases efficiency of this process mediated by Hrd1p. Because inhibition of degradation in ERAD in general does not increase vacuolar traffic of CPY*-CPY, degradation and ER retention of misfolded proteins are separable processes, and ER retention becomes irreversible past a certain step in ERAD. ER retention is likely committed by Yos9p recognition and delivery of misfolded proteins to Hrd1p.
FIGURE 9:
A model for ER retention of CPY*-CPY by ERAD factors. (A) Yos9p likely functions in the committed step of ER retention of CPY*-CPY in addition to its role in the glycan-dependent degradation step. (B) CPY*-CPY is missorted to ER exit in the absence of Yos9p.
Although CPY*-CPY was preferentially sorted to the ERAD pathway, CPY-CPY* was transported to the vacuole as efficiently as CPY-CPY in spite of the presence of the misfolded CPY* unit. Yos9p recognizes CPY-CPY* less efficiently than CPY*-CPY, indicating that not only the presence of a misfolded domain, but also the position of the misfolded domain is important for recognition by Yos9p. In the present case, the N-terminally placed misfolded CPY* unit was recognized by Yos9p more efficiently than the C-terminally placed one. This may perhaps connected with the fact that secretory proteins enter the ER lumen from their N-termini. In this scenario, recognition of misfolded domains starts as early as the protein translocation process. Further study will be needed to test this possibility.
Although BiP is required for ERAD and can interact with proteins with misfolded domains (Nishikawa et al., 2001), BiP binding is not sufficient for sorting of proteins to degradation by ERAD. Only proteins efficiently interacting with Yos9p are efficiently retained in the ER and degraded by ERAD. Because BiP contributes to maintaining solubility of misfolded proteins in the ER (Nishikawa et al., 2001), misfolded proteins interacting with both BiP and Yos9p are likely sorted to the ERAD pathway. Depletion of Lhs1p, a nucleotide exchange factor for BiP, did not affect the fate of CPY*-CPY (Supplemental Figure S8), which also supports the interpretation that BiP does not function in the sorting process.
PDI is another ER-resident chaperone required for ERAD of misfolded proteins, including CPY*. However, overexpression of PDI but not PDIΔ252-277 inhibited both ERAD and vacuolar transport of CPY*-CPY, which was accompanied by significant increase in the interaction of CPY*-CPY with PDI (Figure 8). Preferential binding of chaperone PDI to CPY*-CPY could simply render it unavailable for further sorting to ERAD or ER exit pathway. Because PDI functions in oxidative protein folding, interaction between PDI and CPY*-CPY might take place in the early step of the ER quality control. CPY*-CPY trapped by PDI was probably retained in the ER by the action of the C-terminal ER retention signal of PDI.
Recent cross-link experiments suggested that Hrd3p functions upstream of Yos9p (Stanley et al., 2011). Consistent with this, we found that overexpression of Hrd3p suppressed vacuolar transport of CPY*-CPY by enhancing interaction of CPY*-CPY with Yos9p and Hrd1p (Figure 7). However, depletion of Hrd3p did not decrease the amount of Yos9p coprecipitated with CPY*-CPY (Figure 6B). Because depletion of Hrd3p causes decrease in the Hrd1p level as well (Plemper et al., 1999; Gardner et al., 2000; Gauss et al., 2006; unpublished data), the observed enhancement of the vacuolar transport of CPY*-CPY in the hrd3Δ mutant could have merely arisen from decreased Hrd1p (Figure 2, C and E). Perhaps the decreased interaction between CPY*-CPY and Yos9p caused by Hrd3p depletion may be compensated by the opposite effect due to decrease in the Hrd1p level.
It was unexpected that, although the N-linked glycan attached to Asn479 of CPY*-CPY (Figure 4E) functions as a degradation signal as in the case of CPY* (Kostova and Wolf, 2005; Spear and Ng, 2005), efficient ER retention of CPY*-CPY did not require this degradation signal (Figure 4F). Coimmunoprecipitation experiments showed that CPY*(N479Q)-CPY interacted with both Yos9p and Hrd1p as efficiently as CPY*-CPY (Figure 6A and Supplemental Figure S7A). How can we reconcile those divergent results on the role of the degradation signal in ER retention and ERAD? Perhaps, whereas Yos9p can target CPY*-CPY to Hrd1p for ER retention in a glycan ERAD signal–independent manner, Yos9p may be also involved in the later step at the level of the Hrd1p complex for decoding the glycan signal for degradation. Although the glycan degradation signal is dispensable for ER retention of CPY*-CPY, Yos9p-R200A containing a mutation in the MRH domain, which was suggested to mediate the glycan signal recognition, is defective in this process (Figure 4I). Yos9p-R200A efficiently interacted with CPY*-CPY (Supplemental Figure S7B), but the interaction between CPY*-CPY and Hrd1p was significantly impaired in the yos9-R200A mutant cells (Supplemental Figure S7C). The MRH domain of Yos9p may be required for targeting of misfolded proteins to Hrd1p by, for example, interacting with components of the Hrd1p complex.
OS-9 is a mammalian orthologue of Yos9p, and involvement of OS-9 variants OS-9.1 and OS-9.2 in ER retention of misfolded proteins was reported (Bernasconi et al., 2008). However, mechanisms of Yos9p and the OS-9 variants functioning in the ER retention of misfolded protein seem to be different. Whereas overexpression of OS-9 variants enhances ER retention of misfolded proteins (Bernasconi et al., 2008), overproduction of Yos9p itself does not increase retention efficiency of CPY*-CPY in the ER (Figure 5I). The MRH domain of the OS-9 variants but not of Yos9p is dispensable for the ER retention of misfolded proteins. It will be interesting to ask whether Hrd1p orthologues are involved in the OS-9 variants–mediated ER retention process in mammalian cells.
In summary, by using CPY* fusion proteins as new model ERAD substrates, we dissected the process mediated by Yos9p in ERAD and revealed a novel role of Yos9p in the early steps of ERAD, that is, ER retention of misfolded proteins by delivering them to Hrd1p. Revealing the precise role of Yos9p in the later step of ERAD, most likely in cooperation with the Hrd1p complex, is open to future studies.
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this study are SEY6210 (MATα ura3 leu2 trp1 his3 lys2 suc2; Robinson et al., 1988) and SNY1062 (MATα pep4::TRP1 ura3 leu2 trp1 his3 lys2 suc2; Nishikawa et al., 2008). Gene disruption using Candida glabrata HIS3 (CgHIS3), TRP1 (CgTRP1; Kitada et al., 1995), or kanMX4 (Wach et al., 1994) was performed as described by Sakumoto et al. (1999). The erv29::LEU2 allele was constructed using the LEU2 gene of pJJ283 (Jones and Prakash, 1990). The sec12-4 mutant gene was cloned by PCR using the genomic DNA of MBY10-7AD (MATa/MATα secl2-4/secl2-4 ura3/ura3 trpl/trpl his3/his3 his4/his4 leu2/leu2; Nishikawa et al., 1994) as a template. The amplified DNA was introduced into the polylinker site of pRS306 (Sikorski and Hieter, 1989). The resulting plasmid was used to introduce the sec12-4 allele as described before (Nishikawa et al., 2001). The constructed strains were as follows: HNY2 (MATα prc1::CgHIS3 ura3 leu2 trp1 his3 lys2 suc2), HNY9 (MATα prc1::CgHIS3 pep4::TRP1 ura3 leu2 trp1 his3 lys2 suc2), HNY20 (MATα prc1::CgHIS3 hrd1::CgTRP1 ura3 leu2 trp1 his3 lys2 suc2), HNY22 (MATα prc1::CgHIS3 yos9::CgTRP1 ura3 leu2 trp1 his3 lys2 suc2), HNY23 (MATα prc1::CgHIS3 erv29::LEU2 ura3 leu2 trp1 his3 lys2 suc2), HNY24 (MATα prc1::CgHIS3 sec12-4 ura3 leu2 trp1 his3 lys2 suc2), HNY101 (MATα prc1::CgHIS3 hrd3::kanMX4 ura3 leu2 trp1 his3 lys2 suc2), HNY116 (MATα prc1::CgHIS3 lhs1::kanMX4 ura3 leu2 trp1 his3 lys2 suc2), and INY25 (MATα prc1::CgHIS3 erv29::LEU2 hrd1::kanMX4 ura3 leu2 trp1 his3 lys2 suc2).
Plasmid construction
Construction of a series of fusion genes expressing CPY and/or CPY* fused in tandem via a linker sequence was performed as follows. A 0.1-kb DNA fragment encoding SBP (Keefe et al., 2001) was amplified from pRCCHJ SBP-1 (Kida et al., 2007) by PCR and inserted into the KpnI site of pSK-3HA (Sato and Wada, 1997) to generate pHN6. A 0.25-kb DNA fragment encoding the SBP fused to three copies of the HA epitope (SBP-3HA) was amplified by PCR. A DNA fragment corresponding to nucleotides −666–1596 of the PRC1 gene was amplified by PCR from pTSY1000 (Stevens et al., 1986). The resulting two PCR products were mixed and used as templates to amplify a 2.5-kb DNA fragment encoding CPY-SBP-3HA by PCR, which was subsequently cloned into pRS316 (Sikorski and Hieter, 1989) to generate pHN23. The plasmid pHN24, which contains a DNA fragment encoding CPY*-SBP-3HA, was constructed as described using pJJ244prc1-1 (Nishikawa et al., 2001) instead of pTSY1000. A DNA fragment corresponding to nucleotides 61–2090 of the PRC1 gene was amplified from pTSY1000 by PCR and cloned into pBluescriptII SK(+). A DNA fragment encoding the 3×FLAG tag was introduced between nucleotides 1596 and 1597 of the PRC1 gene, and the resulting plasmid containing encoding CPY-3×FLAG was named pHN27. A DNA fragment encoding CPY*-3×FLAG was constructed as described, using pJJ244prc1-1, and the resulting plasmid was named pHN28. A DNA fragment of pHN27 encoding CPY-3×FLAG was inserted into pHN23 or pHN24 to generate pHN31 or pHN33, respectively. A DNA fragment of pHN28 encoding CPY*-3×FLAG was inserted into pHN23 or pHN24 to generate pHN32 or pHN34, respectively. The constructed pHN31, pHN32, pHN33, or pHN34 was used to express CPY-CPY, CPY-CPY*, CPY*-CPY, or CPY*-CPY*, respectively, in yeast cells. The N479Q mutation was introduced into pHN23 or pHN24 by PCR mutagenesis, generating plasmid pHN23(N479Q) or pHN24(N479Q), respectively. The N1059Q mutation was introduced into pHN27 or pHN28 by PCR mutagenesis, generating plasmid pHN27(N1059Q) or pHN28(N1059Q), respectively. A DNA fragment of pHN27(N1059Q) encoding CPY(N1059Q)-3×FLAG was inserted into pHN24 to generate pHN39, which expresses CPY*-CPY(N1059Q) in yeast cells. A DNA fragment of pHN27 encoding CPY-3×FLAG was inserted into pHN24(N479Q) to generate pHN40, which expresses CPY*(N479Q)-CPY in yeast cells. A DNA fragment of pHN28(N1059Q) encoding CPY*(N1059Q)-3×FLAG was inserted into pHN23 to generate pHN101, which expresses CPY-CPY*(N1059Q) in yeast cells. A DNA fragment of pHN28 encoding CPY*-3×FLAG was inserted into pHN23(N479Q) to generate pHN102, which expresses CPY(N479Q)-CPY* in yeast cells. To construct a plasmid for expression of CPY*-3HA in yeast cells, a DNA fragment corresponding to residues 28–532 of CPY* open reading frame (ORF) was amplified by PCR and inserted into the polylinker sites of p416GPD (Mumberg et al., 1995), and then a DNA fragment for 3HA tag followed by a stop codon was inserted just after the 532nd codon of CPY* ORF. A DNA fragment corresponding to residues 465–532 of CPY* ORF, 3HA tag, and the CYC1terminator was amplified by PCR and inserted into the NcoI and XhoI sites of pHN24 to generate pZL4.
A 2.5-kb DNA fragment containing the YOS9 ORF with 500–base pair upstream and 400–base pair downstream regions was amplified by PCR and cloned into pRS315 and pYO325 (Qadota et al., 1992) to generate pRS315-YOS9 and pYO325-YOS9, respectively. The R200A mutation was introduced into the YOS9 ORF by PCR mutagenesis, and the resulting plasmid was named pRS315-YOS9R200A. A 2.2-kb DNA fragment containing the HRD1 ORF with 560–base pair downstream region or a 3.0-kb DNA fragment containing the HRD3 ORF with 500–base pair downstream region was amplified by PCR and cloned into p414GAL1 (Mumberg et al., 1994) to generate p414GAL1-HRD1 or p414GAL-HRD3, respectively. A 2.6-kb DNA fragment containing the PDI ORF with 500–base pair upstream and 500–base pair downstream regions was amplified by PCR and cloned into pYO324 (Qadota et al., 1992) to generate pYO324-PDI. The PDIΔ252-277 mutation was introduced by PCR mutagenesis, resulting in the plasmid pYO324-PDIΔ252-277.
Pulse-chase experiments
For metabolic labeling, yeast cells were grown in sulfate-free minimal medium containing 0.1 mM (NH4)2SO4 and 0.5% casamino acids to an early log phase. A total of 5 × 107 cells was harvested, washed once with distilled water, and suspended in 2.5 ml of sulfate-free minimal medium (Nishikawa and Nakano, 1991). After incubation at 30°C for 1 h, cells were labeled with 2 MBq of Expre35S35S Protein Labeling Mix (PerkinElmer, Waltham, MA) for 10 min. Chase was initiated by addition of an equal volume of minimal medium containing 0.5% casamino acids, 2 mM (NH4)2SO4, 0.5 mM cysteine, and 0.54 mM methionine. At each time point, 1 ml of cell suspension was taken and mixed with 1 ml of ice-cold NaN3 to terminate metabolic labeling. Preparation of cell extracts and immunoprecipitation with anti-CPY antiserum were performed as described previously (Nishikawa et al., 1990). A 5-μl amount of anti-CPY antiserum was added to the cell extracts, equivalent to 1×107 cells. When indicated, MG132 (Peptide Institute, Osaka, Japan) was added to the labeling medium at 100 μM for 60 min prior to the labeling. For endoglycosidase H treatment, immunoprecipitated materials were treated with 8.3 U/μl endoglycosidase H (New England BioLabs, Ipswich, MA) for 5 h at 37°C according to the manufacturer's instruction. The samples were analyzed by SDS–PAGE and radioimaging with a Typhoon 9200 image analyzer and ImageQuant software (GE Healthcare, Piscataway, NJ).
Vacuolar transport efficiency of the fusion proteins was calculated based on the numbers of methionine and cysteine residues in their ER and vacuole forms and ratio of 35S-labeled methionine and cysteine in Expre35S35S Protein Labeling Mix (Met:Cys = 77:23). The ER form of each fusion protein contains 13 methionine and 22 cysteine residues. The vacuolar-processed form of each CPY unit contains 5 methionine and 11cysteine residues. Because the vacuolar form of CPY-CPY was produced from both CPY units, it contains 10 methionine and 22 cysteine residues. In contrast, the vacuolar forms of CPY-CPY* and CPY*-CPY were produced mostly from the CPY unit but not from the CPY* unit (Supplemental Figure S5). We calculated their vacuolar transport efficiencies based on the assumption that they contain 5 methionine and 11 cysteine residues.
For protease digestion of labeled proteins, 35S-labeled cells were disrupted by agitation with glass beads in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH, pH 7.4, and 50 mM NaCl. Proteins were extracted by incubating the cell lysates in 50 mM HEPES-KOH, pH 7.4, 50 mM NaCl, and 2% Triton X-100 for 20 min on ice, which was followed by centrifugation at 22,000 × g for 10 min at 4°C. The cleared extracts were treated with 25 μg/ml trypsin (Sigma-Aldrich, St. Louis, MO) for 15 min on ice. The reaction was terminated by addition of soybean trypsin inhibitor (Sigma-Aldrich) to 250 μg/ml, and proteins were recovered by precipitation using trichloroacetic acid. Proteins in the pellet were solubilized in 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1% SDS and subjected to immunoprecipitation with anti-CPY antibodies or anti–FLAG M2 agarose beads (Sigma-Aldrich). Proteins that were not recovered with anti-FLAG agarose beads were subjected to the second round of immunoprecipitation with anti-CPY antibodies. Immunoprecipitated materials were analyzed as described.
Coimmunoprecipitation analysis
Yeast cells grown to a mid-log phase were harvested by centrifugation, washed with 10 mM NaN3, and suspended in 50 mM HEPES-KOH, pH 6.8, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM CaCl2, and protease inhibitor cocktail. Cells were disrupted by agitation with glass beads, followed by addition of Triton X-100 at 1%. Insoluble materials were removed by centrifugation at 22,000 × g for 10 min at 4°C. The resulting protein extracts were incubated with anti–FLAG M2 agarose for 2 h at 4°C. Proteins recovered with agarose beads were analyzed by SDS–PAGE and immunoblotting using anti-CPY, anti-Yos9p (a gift from D. Ng, National University of Singapore, Singapore), anti-Hrd3p (a gift from T. Sommer, Max-Delbrück-Center for Molecular Medicine, Berlin, Germany), anti-Hrd1p, or anti-BiP antibodies. Cy5-Labeled anti-rabbit immunoglobulin G antibody (GE Healthcare) was used as the secondary antibody. Proteins were visualized and quantified with a Storm 860 or a Typhoon 9200 image analyzer and ImageQuant software (GE Healthcare). To calculate coimmunoprecipitation efficiency, the amounts of coprecipitated proteins were normalized to the amounts of precipitated CPY/CPY* fusion proteins.
Supplementary Material
Acknowledgments
We thank D. Ng for anti-Yos9p antibody; T. Sommer for anti-Hrd3p antibody; M. Sakaguchi A. Nakano Y. Ohya, and Y. Wada for plasmids; L. Zhou for assistance in plasmid constructions; K. Nakatsukasa for critically reading the manuscript; and members of the Endo lab for discussion. This study was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
Abbreviations used:
- CPY
carboxypeptidase Y
- CPY*
a mutant of CPY
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- HA
hemagglutinin
- MG132
carbobenzoxyl-leucinyl-leucinyl-leucinal
- MRH
mannose 6-phosphate receptor homology
- SBP
streptavidin-binding peptide
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0722) on February 1, 2012.
REFERENCES
- Belden WJ, Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Pertel T, Luban J, Molinari M. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J Biol Chem. 2008;283:16446–16454. doi: 10.1074/jbc.M802272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R, Kanehara K, Carvalho P, Ng DT, Aebi M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell. 2011;24:782–793. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- Gilbert HF. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Römisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol. 1999;147:1443–1456. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama H, Seino J, Kitajima T, Jigami Y, Suzuki T. Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J Biol Chem. 2010;285:12390–404. doi: 10.1074/jbc.M109.082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kato K. The role of MRH domain-containing lectins in ERAD. Glycobiology. 2010;20:651–660. doi: 10.1093/glycob/cwq013. [DOI] [PubMed] [Google Scholar]
- Jakob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kawaguchi S, Hsu C-L, Ng DTW. Interplay of substrate retention and export signals in endoplasmic reticulum quality control. PLoS ONE. 2010;5:e15532. doi: 10.1371/journal.pone.0015532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr Purif. 2001;23:440–446. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Kincaid MM, Cooper AA. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol Biol Cell. 2007;18:455–463. doi: 10.1091/mbc.E06-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Yamaguchi E, Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995;165:203–206. doi: 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci. 2005;118:1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae, comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5797–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T. Mnl1p, an α-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J Biol Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Hirata A, Endo T. Nuclear inner membrane fusion facilitated by yeast Jem1p is required for spindle pole body fusion but not for the first mitotic nuclear division during yeast mating. Genes Cells. 2008;13:1185–1195. doi: 10.1111/j.1365-2443.2008.01236.x. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Hirata A, Nakano A. Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol Biol Cell. 1994;5:1129–1143. doi: 10.1091/mbc.5.10.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Nakano A. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim Biophys Acta. 1991;1093:135–143. doi: 10.1016/0167-4889(91)90114-d. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Umemoto N, Ohsumi Y, Nakano A, Anraku Y. Biogenesis of vacuolar membrane glycoproteins of yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:7440–7448. [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112:4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- Qadota H, Ishii I, Fujiyama A, Ohya Y, Anraku Y. RHO gene products, putative small GTP-binding proteins, are important for activation of the CAL1/CDC43 gene product, a protein geranylgeranyltransferase in Saccharomyces cerevisiae. Yeast. 1992;8:735–741. doi: 10.1002/yea.320080906. [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae, isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Nishikawa S, Endo T. Roles of protein-disulfide isomerase-mediated, disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J Biol Chem. 2009;284:11815–11825. doi: 10.1074/jbc.M900813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumoto N, et al. A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast. 1999;15:1669–1679. doi: 10.1002/(SICI)1097-0061(199911)15:15<1669::AID-YEA480>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Sato MH, Wada Y. Universal template plasmid for introduction of the triple-HA epitope sequence into cloned genes. Biotechniques. 1997;23:254–256. doi: 10.2144/97232bm17. [DOI] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 2010;40:905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear ED, Ng DTW. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear ED, Ng DTW. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley AM, Carvalho P, Rapoport TA. Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking. FEBS Lett. 2011;585:1281–1286. doi: 10.1016/j.febslet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- Stevens TH, Rothman JH, Payne GS, Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986;102:1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DTW. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vember SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.