Pex11p plays a conserved role in peroxisome division. Although Pex11p is phosphorylated, the exact role of this modification was either unknown or confusing. Phosphorylation of Pichia pastoris Pex11p at serine 173 occurs at the peroxisome and is necessary for its interaction with Fis1p, a key protein of the peroxisome division complex.
Abstract
Peroxisome division is regulated by the conserved peroxin Pex11p. In Saccharomyces cerevisiae (Sc), induction of the phosphoprotein ScPex11p coincides with peroxisome biogenesis. We show that the ScPex11p homologue in Pichia pastoris (PpPex11p) is phosphorylated at serine 173. PpPex11p expression and phosphorylation are induced in oleate and coordinated with peroxisome biogenesis. PpPex11p transits to peroxisomes via the endoplasmic reticulum (ER). PpPex11p is unstable and ER restricted gin pex3Δ and pex19Δ cells, which are impaired in peroxisomal membrane protein biogenesis. In oleate medium, the P. pastoris mutants pex11A (constitutively unphosphorylated; S173A) and pex11D (constitutively phosphorylated; S173D) exhibit juxtaposed elongated peroxisomes (JEPs) and hyperdivided forms, respectively, although protein levels remain unchanged. In contrast with ScPex11p, the ER-to-peroxisome translocation in P. pastoris is phosphorylation independent, and the phosphorylation occurs at the peroxisome. We show that PpPex11p interacts with the peroxisome fission machinery via PpFis1p and is regulated by phosphorylation because PpPex11p and PpPex11Dp interact more strongly with PpFis1p than PpPex11Ap. Neither PpPex11p nor PpFis1p is necessary for peroxisome division in methanol medium. We propose a model for the role of PpPex11p in the regulation of peroxisome division through a phosphorylation-dependent interaction with the fission machinery, providing novel insights into peroxisome morphogenesis.
INTRODUCTION
Peroxisomes are predominantly responsible for two important functions in eukaryotic cells: 1) a variety of metabolic reactions involved in lipid metabolism (e.g., oxidation of very long chain and long, branched-chained fatty acids) and 2) quenching of peroxides and reactive oxygen species (Wanders and Tager, 1998; Wanders et al., 2001; Wanders and Waterham, 2006). Peroxisome biogenesis and enzymes are critical for the cell, as exemplified by the lethal defects manifested in humans and mice impaired in peroxisome biogenesis (Steinberg et al., 2006). The identification of various proteins—peroxins (encoded by PEX genes)—was achieved initially by genetic analysis of such peroxisome-deficient mutants in mammals and yeasts (Purdue and Lazarow, 2001; Weller et al., 2003).
Peroxisome division and metabolism respond to changes in the environment (Subramani, 1998). The fact that they are regulated by environmental cues makes them highly capable of content, number, and size alterations as required by the cell. Modulation of the peroxisome population and size is accomplished by coordinated biogenesis, division, and segregation to daughter cells (Fagarasanu et al., 2007). Peroxisomes and mitochondria share a common division machinery involving dynamin-related proteins (DRPs), which are involved in peroxisome fission (Hu, 2010). However, this division machinery must be activated differentially in an organelle-dependent manner on peroxisomes and mitochondria. Exactly how this is achieved was unknown and is the subject of this study.
Pex11p is involved in peroxisome division and proliferation. Mutant pex11Δ cells exhibit fewer but larger peroxisomes in comparison with the wild-type cells, whereas overexpression of Pex11p results in excessive peroxisome division, yielding numerous, small peroxisomes (Erdmann and Blobel, 1995; Marshall et al., 1995). On growth of yeast cells in peroxisome-inducing medium such as oleate, PEX11 gene expression increases 1000-fold as compared to steady-state levels in glucose medium (Karpichev and Small, 1998). In mammals, various isoforms of Pex11 mediate peroxisome division, which occurs in four steps: 1) insertion of Pex11p into the membrane, causing 2) elongation of the peroxisomal membrane, followed by 3) segregation of Pex11p into patches, leading to 4) recruitment of the division machinery for subsequent fission (Schrader et al., 1998; Subramani, 1998). DRPs play a crucial role in peroxisome fission because DRP mutants display peroxisomes with a beads-on-a-string phenotype, suggesting an indirect role of Pex11p upstream of division in which Pex11p either interacts with DRPs or modulates membrane curvature through lipid binding, eliciting peroxisome division (Barnett et al., 2000; Hoepfner et al., 2001; Opalinski et al., 2011b). Pex11p in Saccharomyces cerevisiae is phosphorylated at Ser-165/167 (Knoblach and Rachubinski, 2010). We initiated the present study to understand the mechanism by which Pex11p modulates division in Pichia pastoris and to answer whether its modification by phosphorylation has any role in this event.
In this study we show that P. pastoris (Pp) Pex11p is phosphorylated at Ser-173. Like its homologue ScPex11p, its expression and phosphorylation are strongly induced upon oleate (or methanol) induction. PpPex11p translocates from endoplasmic reticulum (ER) to peroxisomes in a Pex19p-dependent manner. Unlike its S. cerevisiae (Sc) homologue, the wild-type and mutant forms of PpPex11p are localized to peroxisomes following oleate induction, independent of their phosphorylation status. Peroxisome division is delayed in pex11A cells expressing only the unphosphorylated PpPex11p mutant, generating juxtaposed elongated peroxisomes (JEPs). Phosphorylation-dependent differential binding of PpPex11p with the fission machinery protein Fis1p yields novel insights regarding the role of Pex11p phosphorylation in peroxisome division.
RESULTS
As in S. cerevisiae and other organisms, Pex11p is involved in peroxisome division in P. pastoris. This is exemplified by the fact that P. pastoris cells lacking Pex11p have fewer and bigger peroxisomes when grown in oleate medium, indicative of a block in peroxisome division, and overexpression of Pex11p causes the appearance of numerous and smaller peroxisomes (Supplemental Figure S1). Pex11p is required for growth in oleate, where pex11∆ cells show an arrest in growth. However the absence of Pex11p has no significant effect on cell growth of P. pastoris or peroxisome division for cells grown in methanol medium (unpublished data).
P. pastoris Pex11p is phosphorylated at Ser-173
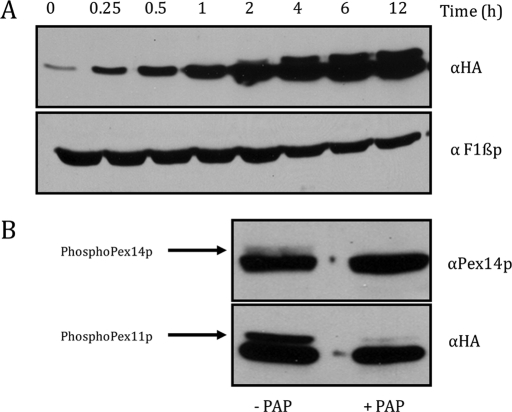
To characterize the nature of the Pex11p protein in P. pastoris, we undertook the analysis of its expression during peroxisome induction conditions on oleate medium. A plasmid expressing hemagglutinin (HA)-tagged PEX11 (Pex11p-2HA) from the PEX11 promoter was transformed into pex11Δ cells. The tagged version of Pex11p (Pex11p-2HA) complements the cellular growth and peroxisome morphology defects in pex11Δ cells (unpublished data). Protein abundance of Pex11p-2HA following incubation of the cells in oleate medium was determined by loading equal amounts of total protein extracts and detection by Western blotting analysis using anti-HA mouse antibody. In glucose medium (0 h), Pex11p-2HA expression was barely discernible, but the protein was induced in oleate medium. Incubation for 2 h or more in oleate showed the presence of slower-migrating upper band, which intensified concomitant with the lower band upon further incubation (Figure 1A). Protein abundance and phosphorylation of Pex11p-2HA following incubation in methanol medium are similar to those in oleate medium (unpublished data).
FIGURE 1:
Phosphorylation of Pex11p-2HA in oleate medium. (A) Time course of Pex11p-2HA with induction in oleate medium after growth in YPD. The 0.2 OD equivalent of total extract for the strain expressing Pex11p-2HA under its own promoter in pex11Δ cells taken at 0, 0.25, 0.5, 1, 2, 4, 6, and 12 h, respectively, was loaded and detected by Western blotting using anti-HA mouse antibody. Equal amounts of proteins were analyzed by Western blotting of the same aliquots with anti-F1βp rabbit antibody. (B) PAP treatment for cells expressing Pex11p-2HA under its own promoter in pex11Δ cells. Samples were incubated in PAP for 4 h after oleate induction (6 h). The 0.2 OD equivalent of extract was loaded and detected by Western blotting using anti-HA mouse antibody. Pex14p was used as a positive control for the assay and was detected by anti-Pex14p rabbit antibody. Cells not treated with PAP were used as negative control.
To determine whether the upper band corresponded to the phosphorylated form of Pex11p, potato acid phosphatase (PAP) treatment of the protein samples from oleate-grown cells was performed. Both the PAP-treated and the untreated samples were incubated for 4 h and analyzed by Western blotting using anti-HA mouse antibody. Pex14p, a protein known to be phosphorylated in oleate medium, was used as a positive control for the assay (Johnson et al., 2001). The untreated samples showed both (phosphorylated and dephosphorylated) bands for Pex11p-2HA and Pex14p. Both Pex11p-2HA and Pex14p lost the slower-migrating band in phosphatase-treated samples, confirming that the modification was indeed phosphorylation (Figure 1B).
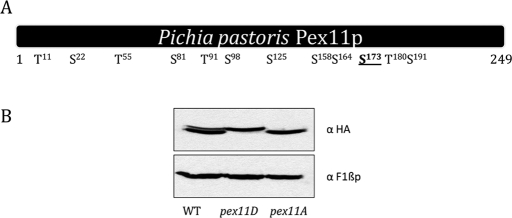
The putative phosphorylation sites in Pex11p were identified using the NetPhos 2.0 phosphorylation prediction server (Blom et al., 1999). Serine residues 22, 81, 98, 125, 158, 164, 173, and 191 and threonine residues 11, 55, 91, and 180 showed high probability for possible phosphorylation (Figure 2A). Individual point mutations were generated by site-directed mutagenesis (converting respective serine or threonine triplet codons to alanine), expressed in pex11Δ cells, and tested for the disappearance of the phosphorylation band. Of all the putative residues scanned, only Ser-173 when mutated lost the phosphorylation band, suggesting possible phosphorylation of Pex11p-2HA. Hence, the Pex11p-Ser173Ala mutant was referred to as the constitutively unphosphorylated (phospho) mutant. Similarly, a strain expressing a constitutively phosphorylated (phosphomimic) mutant of Pex11p-2HA was generated by mutating sequences encoding Ser-173 to Asp. The phosphomutant and phosphomimic mutants were designated as pex11A and pex11D, respectively. A strain expressing a wild-type copy of Pex11p with a C-terminal-2HA tag integrated into pex11Δ cells was designated as the wild type (SJS154). Both the mutants are derived from the wild-type plasmid containing Pex11p-2HA. Western blotting of wild-type cells expressing Pex11p-2HA, after overnight growth in yeast extract/peptone/dextrose (YPD) and transfer to oleate medium for 6 h, showed both (native and phosphorylated) bands, whereas the pex11D (SJS155) and pex11A (SJS156) strains showed only the upper (phosphorylated) or the lower (unphosphorylated) band, respectively (Figure 2B). Equal loading was observed for the loading control (F1βp) in all the strains.
FIGURE 2:
In vivo phosphorylation of Pex11p at Ser-173. (A) Schematic of putative phosphorylation sites of Pex11p (amino acids 1–249) as predicted by the NetPhos 2.0 server (www.cbs.dtu.dk/services/NetPhos). Respective positions of serine/threonine residues showing high probability for phosphorylation are shown. The Ser-173 site found to be responsible for the phosphorylation is underlined. (B) Cells expressing wild-type (WT) Pex11p-2HA, as well as Pex11Dp-2HA or Pex11Ap-2HA, respectively, were induced for 6 h in oleate medium. The 0.2 OD equivalent of total extract for all the strains was loaded and detected using anti-HA mouse antibody. Equal amounts of proteins were analyzed by Western blotting of the same aliquots with anti-F1βp rabbit antibody.
These data led us to conclude that Pex11p-2HA is phosphorylated at Ser-173 in P. pastoris.
Wild-type and mutant Pex11p are induced during peroxisome induction on oleate
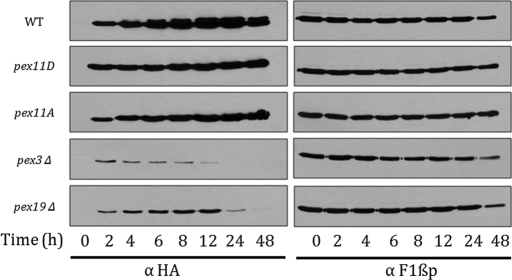
To compare the relative protein abundance of Pex11p-2HA in wild-type and mutant strains, the time course of induction in oleate medium (from 0 to 48 h) was performed. Equal amounts of total protein (as reflected by F1βp) were loaded and detected using Western blotting. As observed before, expression of Pex11p-2HA protein was repressed in glucose (YPD) medium for wild type, as well as for the mutants. On induction of peroxisome biogenesis, the wild type and the phosphorylation mutants of Pex11p-2HA were robustly expressed. After 2–4 h of induction, the slower-migrating phosphorylated species was observed for wild-type Pex11p-2HA, suggesting a possible role of phosphorylation in regulation of peroxisome biogenesis. Even though the relative protein abundance of Pex11p-2HA in wild type and phosphorylation mutants was similar and increased upon oleate induction, the pex11A strain lacked the slower-migrating band, whereas in pex11D, only the slower-migrating band was observed. Pex11p-2HA was found to be unphosphorylated and unstable in cells lacking peroxisomes—pex3Δ as well as pex19Δ (Figure 3). This is not surprising, given the roles of Pex3p and Pex19p in peroxisomal membrane protein biogenesis and Pex11p-2HA trafficking to peroxisomes (Agrawal et al., 2011). In addition, the dephosphorylated state of Pex11p-2HA in peroxisome-deficient mutants suggested that Pex11p-2HA might be phosphorylated on peroxisomes.
FIGURE 3:
Time course of Pex11p-2HA expression in wild-type and mutant strains. Cells expressing wild-type Pex11p-2HA under its own promoter in pex11Δ, pex3Δ, and pex19Δ cells or expressing Pex11Dp/Pex11Ap from the PEX11 promoter in pex11Δ cells were grown in YPD and transferred to oleate. Aliquots were taken at 0, 2, 4, 6, 8, 12, 24, and 48 h, respectively, and 0.2 OD equivalent of the extracts was loaded and detected using anti-HA mouse antibody. Equal amounts of proteins were analyzed by Western blotting of the same aliquots with anti-F1βp rabbit antibody.
Pex11p traffics from the ER to peroxisome in a Pex19p-dependent manner
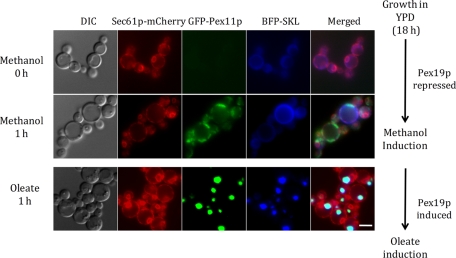
Pulse chase of green fluorescent protein (GFP)–Pex11p from a peroxisome-deficient to a peroxisome-rich state was monitored to determine whether Pex11p could be accumulated at the ER and chased to peroxisomes in a Pex19p-dependent manner. We generated a P. pastoris strain expressing GFP-Pex11p from the methanol-inducible alcohol oxidase (AOX1) promoter and Pex19p from the oleate-inducible thiolase (POT1) promoter in a pex19Δ strain background. The strain also harbored an ER (Sec61p-mCherry) and peroxisomal markers (blue fluorescent protein [BFP]–SKL). The AOX1 promoter is repressed in oleate and glucose media, whereas the POT1 promoter is repressed in methanol and glucose media, making them ideal candidates for the inducible-promoter assay (Kos et al., 1995; Sears et al., 1998). Cells were grown to log phase in YPD and transferred to methanol medium. Here GFP-Pex11p accumulated with time (1 h) at the ER, as suggested by partial colocalization with Sec61p-mCherry (Figure 4, top). These cells, in which Pex19p expression was repressed, represent the peroxisome-deficient state. The cells were then shifted to oleate medium, in which the expression of GFP-Pex11p was turned off and that of Pex19p was induced from the oleate-inducible thiolase promoter. Under these conditions, the ER-localized GFP-Pex11p disappeared and reappeared at structures (at 1 h) that are peroxisomes as judged by colocalization of GFP-Pex11p with the peroxisome marker BFP-SKL (Figure 4, bottom). This pulse-chase study demonstrates that Pex11p, with the help of Pex19p, translocates from the ER to peroxisomes upon induction of biogenesis, as already described (Agrawal et al., 2011). As an additional control, we also repeated this experiment by including a 1-h incubation of the cells in YPD (to shut off the AOX1 promoter, as described in Sears et al., 1998) between the growth in methanol and the transfer to oleate medium (unpublished data). Similar results were obtained, in that the ER-localized GFP-Pex11p was chased to peroxisomes in oleate.
FIGURE 4:
Pulse chase of Pex11p in cells shifted from a peroxisome-deficient to a peroxisome-containing state. Cells expressing GFP-Pex11p from the AOX1 promoter, Pex19 from the POT1 promoter in a pex19Δ background, the peroxisome marker (BFP-SKL from the AOX1 promoter), and an ER marker (Sec61p-mCherry from its own promoter) were used. After growth in YPD, cells were grown in methanol medium for 1 h. In this medium, Pex19p was repressed (as it is expressed from POT1 promoter, which is induced in oleate medium), creating a peroxisome-deficient state. GFP-Pex11p expressed from the AOX1 promoter was induced on methanol. The cells were washed and transferred to oleate medium, in which Pex19p was expressed, creating a peroxisome-induction state, but GFP-Pex11p was repressed. Cells were grown in this medium for 1 h. Microscopy was performed for the cells in methanol and oleate media. ER and peroxisome localization was confirmed by colocalization with Sec61p-mCherry and BFP-SKL proteins, respectively. The top two lanes represent peroxisome-deficient, and the bottom two lanes represent peroxisome-induced, states of the cells. DIC, differential interference contrast. Scale bar, 5 μm.
Pex11p trafficking from ER to peroxisome is independent of its phosphorylation state
In S. cerevisiae, it was suggested that the trafficking of Pex11p from the ER to peroxisome was dependent on its phosphorylation status. Despite the claims that phosphorylation was required for ScPex11p transit from the ER to peroxisome, Pex11Ap was confined to peroxisomes (and did not colocalize with ER) in YPD and oleate media. On the other hand, Pex11Dp was found at the ER–peroxisome interface. Wild-type ScPex11p was found in both ER and the peroxisome compartments in oleate, but the distribution changed in YPD, where it colocalized only with the Pot1p-positive peroxisomes (Knoblach and Rachubinski, 2010).
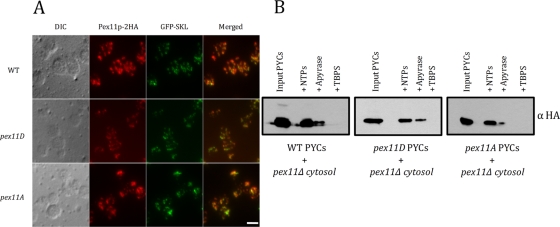
To study the localization of Pex11p in P. pastoris, immunofluorescence analysis of oleate-induced cells (strains expressing wild-type and mutant Pex11p-2HA) was performed. Both wild-type and mutant forms of Pex11p-2HA colocalized with structures containing GFP-SKL, suggesting proper peroxisome localization (Figure 5A). The important difference between ScPex11p and PpPex11p is that the latter is repressed in YPD and expressed (with phosphorylation, after ∼2 h) in oleate, whereas ScPex11p is not only expressed but also phosphorylated in YPD.
FIGURE 5:
Pex11p trafficking in wild-type and mutant cells from the ER to peroxisomes. (A) Immunofluorescence microscopy for paraformaldehyde-fixed wild-type, pex11D, or pex11A cells was performed. GPF-SKL was expressed from the GAP promoter. Cells were grown in YPD and transferred to oleate medium (6 h) for induction. Anti-HA rat primary antibody, as well as Alexa Fluor 568–conjugated, goat anti-rat secondary antibody, were used for detection. Scale bar, 5 μm. (B) ER budding assay. After washing with TBPS, permeabilized wild-type and mutant cells were incubated with cytosol from pex11Δ cells and an ATP-regenerating system at 20°C for 90 min. After centrifugation, the pellet was resuspended in SDS-sample buffer and analyzed by Western blotting with anti-HA mouse antibody. Sample treated with apyrase, as well as samples without the ATP-regenerating system, served as negative control.
A second method for confirming proper exit of Pex11p-2HA was used in which the budding of preperoxisomal vesicles from the ER was compared in wild-type and mutant strains. Wild-type and mutant permeabilized yeast cells (PYCs) were respectively incubated with pex11Δ cytosol for 1.5 h at 20°C. The budding of ER-derived vesicles was not affected in wild-type, pex11D, or pex11A cells. In wild-type cells, Pex11p-2HA was phosphorylated, whereas in pex11D or pex11A cells, only the phosphorylated and unphosphorylated forms were observed, respectively. In the controls, the budding of vesicles containing Pex11p-2HA was ATP dependent (apyrase) and was abolished when the cytosol was substituted by tert-butyl-bicyclophospho-orthionate (TBPS) buffer (Figure 5B).
These data collectively show that the phosphorylation state of PpPex11p plays no role in its trafficking from the ER to peroxisomes.
Phosphorylation mutants of Pex11p cause hyperdivided and juxtaposed elongated peroxisomes
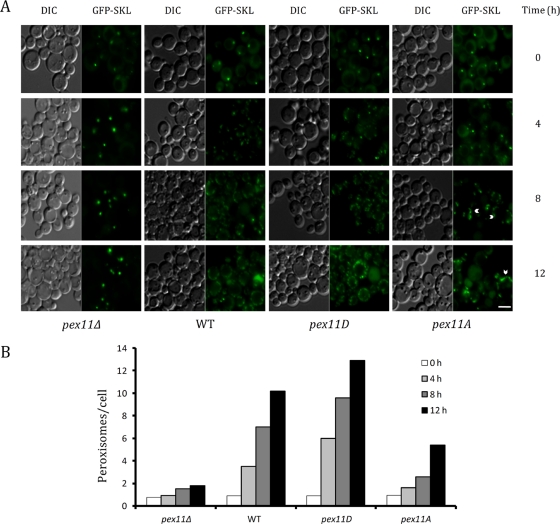
Because the Pex11p phosphorylation mutants showed no difference in trafficking to peroxisomes, we examined the roles of constitutively phosphorylated and constitutively unphosphorylated forms on peroxisome formation. We conducted morphological analysis of peroxisome biogenesis by growing wild-type and mutant cells (harboring GFP-SKL) in glucose, followed by peroxisome induction in oleate medium. Morphology was monitored by fluorescence microscopy over a period of 12 h. Peroxisomes were seen as 1 or 2 puncta/cell in YPD (0 h). Within 4 h after transfer, peroxisomes were dividing. Variations in peroxisome morphologies in the wild-type and mutant cells were observed and became more divergent with further incubation. Morphological distinctions were apparent by 8–12 h. In wild-type cells, peroxisomes proliferated and divided with passing time. In the pex11Δ cells, peroxisomes were enlarged and clustered around 8–12 h. Peroxisomes in pex11D cells divided at a faster rate than in wild-type cells, resulting in a higher number of peroxisomes per cell. In pex11A cells, peroxisomes were juxtaposed and elongated (Figure 6A). Morphometric analysis of the average number of peroxisomes per cell confirmed the observed differences, showing that peroxisome number decreased in pex11A and increased in pex11D at 8 and 12 h as compared with wild-type cells. A decrease in the number of peroxisomes in pex11Δ as compared with wild-type cells was representative of clustering. Per cell, on average, pex11D cells had ∼9 peroxisomes, compared with 1 or 2 peroxisomes in pex11A cells at 8 h. The pex11Δ cells had 2 peroxisomes, compared with 7 peroxisomes in wild-type cells (Figure 6B). Although Pex11p-2HA was phosphorylated in both oleate and methanol media, the requirement of Pex11 was visible only in oleate medium, in which pex11A cells showed JEPs, whereas in methanol there was no difference in peroxisome morphology compared relative to the wild-type cells (unpublished data).
FIGURE 6:
Morphometric analyses of peroxisomes in wild-type and mutant strains. (A) Fluorescence microscopy was performed for pex11Δ, wild-type (WT), pex11D, or pex11A cells expressing GFP-SKL from the GAP promoter. After growth in YPD, cells were transferred to oleate medium and incubated for 12 h. Samples were taken at 0, 4, 8, and 12 h and analyzed by fluorescence microscopy. Arrowheads represent JEPs found in pex11A cells. Scale bar, 4 μm. (B) Graphical representation of the average number of peroxisomes per cell for pex11Δ, wild-type, pex11D, and pex11A cells. Peroxisome counts were performed on 50 randomly recorded cells per time point per strain. Tubulated peroxisomes are counted as single peroxisomes.
Peroxisomes in pex11D cells mimicked the overexpression phenotype of Pex11p, in which the peroxisomes hyperproliferate. In pex11A cells, peroxisomes were tubulated and elongated, giving rise to juxtaposed-elongated peroxisomes (JEPs). JEPs suggest a possible function of Pex11p proteins in driving membrane tubulation. Since the arrest was before the peroxisome fission step, a possible role of Pex11p phosphorylation in recruitment of the peroxisome division machinery was hypothesized.
Pex11p interaction with the fission machinery protein Fis1p is differentially regulated by its phosphorylation state
The morphological analysis of pex11A cells showed the presence of JEPs. To test whether these structures were formed due to incomplete peroxisome fission, we looked for possible interactions of Pex11p with fusion machinery proteins. Recently, Fis1p, a tail-anchored protein, was shown to be shared between peroxisomes and mitochondria for the recruitment of other division proteins such as Mdv1p, Caf4p, and Dnm1p (Pan and Hu, 2011). As in other organisms, Fis1p is localized to peroxisomes and mitochondria in P. pastoris, and fis1∆ cells showed elongated peroxisomes as compared with wild-type cells, which showed normal, oleate-induced peroxisomes (unpublished data).
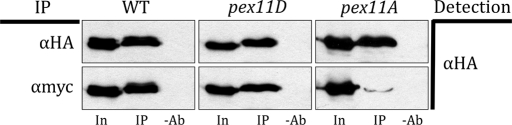
We studied the interaction of wild-type and mutant HA-tagged Pex11p with myc-tagged Fis1p. In the strains expressing wild-type Pex11p-2HA or Pex11Dp-2HA and Fis1p-myc, immunoprecipitation of Pex11p/Pex11Dp-2HA coimmunoprecipitated Fis1p-myc. In contrast, immunoprecipitation of Pex11Ap-2HA failed to efficiently coimmunoprecipitate Fis1p-myc, suggesting only weak interaction with Fis1p-myc by unphosphorylated Pex11p. Immunoprecipitation in the absence of the antibody did not yield any signal (Figure 7). The blots were somewhat overexposed for equal amounts of time to see whether any residual Pex11p-2HA (especially in the pex11A cells) might have been immunoprecipitated. At lower exposure times, however, a relatively smaller amount of Pex11p-2HA was coimmunoprecipitated with Fis1p-myc.
FIGURE 7:
Phosphorylation-dependent interaction of Pex11p-2HA with Fis1p. Coimmunoprecipitation of proteins in cells expressing Pex11p-2HA, Pex11Dp, or Pex11Ap from the PEX11 promoter along with Fis1p-myc from its own promoter in a pex11Δ background. Either anti-HA mouse or anti-myc mouse antibody was used to pull down Pex11p-2HA and Fis1p-myc, respectively. For detection, anti-HA rat primary and goat anti-rat secondary antibodies were used. A pull-down without any antibody was used as a negative control. The 0.2 OD equivalent of input and 5 OD equivalents of immunoprecipitated samples were loaded for Western blotting. In, input; IP, immunoprecipitation with respective antibody; −Ab, immunoprecipitation without antibody.
Taken together, the coimmunoprecipitation analyses, as well as the fluorescence microscopy data, provide strong evidence that Pex11p interacts physiologically with Fis1p of the peroxisome division machinery in a phosphorylation-dependent manner.
DISCUSSION
This article addresses the expression, subcellular localization, and functional role of phosphorylation of P. pastoris Pex11p. First, the expression of Pex11p is repressed in growth medium containing glucose and induced heavily in oleate (Figure 3). Second, in a peroxisome-deficient state, Pex11p is localized to a subdomain of the ER and is targeted to peroxisomes upon induction of peroxisome biogenesis (Figure 4). Third, phosphorylation of Pex11p is induced after 2 h of oleate induction at Ser-173 (Figures 1 and 2). Although the overall sequence identity between ScPex11p and PpPex11p is low (∼33%), the location of the phosphorylation (Ser-165 and/or Ser-167 in S. cerevisiae and Ser-173 in P. pastoris) sites is conserved in both yeasts. Although Pex11p is induced and phosphorylated on both methanol and oleate, we observed no effect of the deletion of PpPEX11 or PpFIS1 (unpublished data) when cells were grown in methanol. Fourth, phosphorylation of PpPex11p occurs on peroxisomes and is not responsible for translocation of PpPex11p from the ER to peroxisomes (Figure 5). This observation is in contrast with the situation in ScPex11p, in which phosphorylation occurred at the ER and was reported to play a major role in peroxisome targeting (Knoblach and Rachubinski, 2010). Fifth, morphometric analyses of antagonistic phosphorylation mutants of Pex11p show phenotypes with hyperproliferated and juxtaposed-elongated peroxisomes (Figure 6). Finally, a physiologically relevant interaction of Pex11p with Fis1p confirms the results obtained by morphometric analyses and establishes a central role for Pex11p phosphorylation in the recruitment of Fis1p, a key component of the peroxisome division machinery (Figure 7).
We offer several possibilities for why Pex11p and Fis1p are not required for peroxisome division of cells grown in methanol. The first is that some other proteins (such as Pex25p and/or Pex27p belonging to the Pex11p protein family) substitute for Pex11p to stimulate peroxisome division. Second, another DRP, such as Vps1p, may bypass the Pex11p and Fis1p requirement during division. Finally, the interaction of Pex11p and Fis1p may be insufficient in this medium to induce peroxisome division. Experiments are underway to test these hypotheses.
In contrast to the previous studies, our results provide greater clarity regarding the subcellular site of PpPex11p phosphorylation and the role of this modification in the recruitment and activation of peroxisome division, which may provide insights into how Fis1p is modulated selectively on other subcellular compartments as well. The relevance of such insights stems from the fact that the conserved Fis1p interacts with dynamin-like proteins, whose mutation in humans causes lethality due to defects in peroxisomal and mitochondrial fission (Waterham et al., 2007).
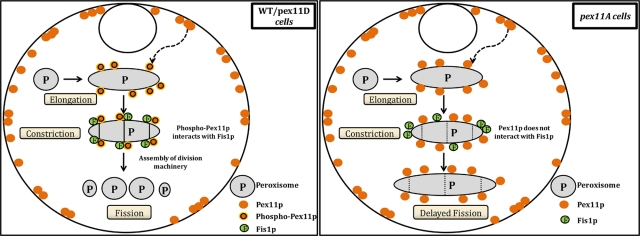
The results obtained from the present study allow us to postulate a working model for the control of peroxisome division in P. pastoris cells (Figure 8). For wild-type as well as pex11D cells, upon induction of peroxisome biogenesis, Pex11p is heavily expressed and shuttled to peroxisomes via the ER in a Pex19p-dependent manner. Here it gets phosphorylated, which in turn activates the interaction of Pex11p with Fis1p directly or indirectly. Fis1p then recruits other division machinery proteins. This completes the assembly of the peroxisome fission complex that encircles and constricts the peroxisome membrane, causing division. Similarly, in pex11A cells, upon induction of peroxisome biogenesis, Pex11Ap is heavily expressed and shuttled to peroxisome via the ER in a Pex19p-dependent manner. However, since the phosphorylation site is mutated, Pex11p cannot interact with Fis1p and consequently fails to further recruit the fission complex, giving rise instead to an intermediate state of JEP (Figure 8).
FIGURE 8:
Model describing Pex11p-phosphorylation–mediated peroxisomal division in the yeast Pichia pastoris. In wild-type and pex11D cells, Pex11p translocates from the ER to peroxisomes in a Pex19p-dependent manner. Pex11p gets phosphorylated on peroxisomes, and this activates interaction with Fis1p, causing the recruitment of the peroxisome division machinery and peroxisome fission. In pex11A cells, Pex11p translocates from the ER to peroxisomes in a Pex19p-dependent manner. Because the phosphorylation site is mutated, Pex11p is not phosphorylated and hence cannot activate interaction with Fis1p or recruitment of the peroxisome division machinery, creating JEPs. Pex19p-mediated translocation of Pex11p is represented by a dashed, curved arrow. Dotted vertical lines represent impaired division.
Mitochondria and peroxisomes share their fission machinery. In S. cerevisiae, DRPs such as Vps1p and Dnm1p regulate peroxisome abundance (Kuravi et al., 2006). The recruitment of Dnm1p requires anchoring of Fis1p to the membrane (Mozdy et al., 2000). Fis1p is a tail-anchored protein with dual localization to membranes with low ergosterol content such as peroxisomes and mitochondria (Kemper et al., 2008). Loss of Fis1p inhibits both peroxisomal and mitochondrial division and forms elongated organelles (Motley et al., 2008). In addition, ectopic expression of Pex11p from yeast, plant, or human causes JEP formation, suggesting the evolutionary conserved function of Pex11p proteins in tubulation. In the same study, overexpression of human Fis1p (but not Drp1p) was shown to be sufficient to fragment observed JEPs into normal-shaped peroxisomes, demonstrating the Pex11p-induced JEPs to be the intermediates in membrane proliferation, with Fis1p being the limiting factor for peroxisome constriction and division (Koch et al., 2010). Similar structures (JEPs) were observed with Pex11Ap in the present study, when it was unable to interact efficiently with Fis1p. This observation suggests that the interaction of Pex11p with Fis1p, leading to recruitment of fission complex, precedes the peroxisome division step.
Of interest, P. pastoris cells completely lacking Pex11p exhibit fewer and larger peroxisomes, but no JEP, which suggests that Pex11Ap still has a role, perhaps in peroxisome tubulation, but not in peroxisome constriction and division. This possibility is suggested by the work on Pex11p in Penicillium chrysogenum demonstrating the presence of an N-terminal amphipathic helix, which causes membrane remodeling, leading to tubulation of vesicles. This property is conserved from yeast to humans (Opalinski et al., 2011a).
The N-terminal region of Mdv1p (and its paralogue Caf4p) interacts with Fis1p, whereas its WD40 repeats interact with Dnm1p, making it an adaptor forming a functional Fis1p-Mdv1p/Caf4p-Dnm1p complex (Naylor et al., 2006). In S. cerevisiae, both the paralogues are present, whereas in P. pastoris, only one homologue (accession number XP_002490961) of Mdv1p/Caf4p is found. Similarly, a homologue of ScDnm1p is also present in P. pastoris (accession number XP_002492979). The presence of corresponding homologues of Fis1p, Mdv1p/Caf4p, and Dnm1p in P. pastoris and the fact that phosphorylated Pex11p interacts with Fis1p suggests that they all are members of the same pathway for peroxisome division. To conclude, Pex11p interaction with Fis1p was previously shown in plants and mammals, but the mode of regulation was unknown (Lingard et al., 2008). Here we successfully extend this study to yeast showing phosphorylation dependence for the regulation of division.
In S. cerevisiae, the involvement of cyclin-dependent kinase Pho85p was shown to increase Pex11p phosphorylation. Furthermore, it was hypothesized that since Pho85p binds multiple cyclins as well as increases Pex11p phosphorylation, it might be able to explain a connection between cell cycle progression and peroxisome division (Knoblach and Rachubinski, 2010). It is surprising that in P. pastoris, overexpression of Pho85p or Rim15p (substrate of Pho85p kinase), as well as the deletion of PHO85, had no effect on the levels of phosphorylated Pex11p (unpublished data), suggesting that a Pho85p-independent (or redundant) signaling pathway is responsible for Pex11p phosphorylation. It would be interesting for future studies to identify the kinase phosphorelay acting upstream of Pex11p.
In S. cerevisiae, Pex11p was shown to homodimerize via the Cys-3 residue, and mutation of this residue to alanine inhibited dimerization, causing numerous and small peroxisomes. The study suggested the monomeric form to be the active species (Marshall et al., 1996). However, P. pastoris Pex11p lacks cysteine residues in its protein sequence. Therefore, an alternate mechanism for activation of Pex11p to induce division is necessary. Here we suggest a phosphorylation-mediated activation of Pex11p for division.
MATERIALS AND METHODS
Strains, plasmids, and media
Various P. pastoris strains and plasmids used in this study are listed in Table 1. The strains were cultured at 30°C for all experiments. Various media were used as described previously (Yan et al., 2008): YPD (1% yeast extract, 2% peptone, 2% glucose), oleate medium (0.67% yeast nitrogen base without amino acids, 0.02 g/l each of l-histidine and l-arginine, 0.1% yeast extract, 0.2% oleic acid, and 0.02% Tween-40) and SD (0.67% yeast nitrogen base without amino acids, 2% glucose, and 0.02 g/l each of l-histidine or l-arginine).
TABLE 1:
Pichia pastoris strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| PPY12 | arg4, his4 | Gould et al. (1992) |
| SJS141 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (S-173 to D) (HIS4), FIS1::FIS1-MYC (ARG4) | This study |
| SJS152 | pex11Δ:: ZeoR, PGAP::GFP-SKL (ARG4), his4 | This study |
| SJS154 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (HIS4), PGAP::GFP-SKL (ARG4) | This study |
| SJS155 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (S-173 to D) (HIS4), PGAP::GFP-SKL (ARG4) | This study |
| SJS156 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (S-173 to A) (HIS4), PGAP::GFP-SKL (ARG4) | This study |
| SJS159 | pex3Δ:: ZeoR, PEX11::PEX11-2HA (HIS4), arg4 | This study |
| SJS160 | pex19Δ:: ZeoR, PEX11::PEX11-2HA (HIS4), arg4 | This study |
| SJS161 | pex11Δ:: ZeoR, PGAP-Pex11-2HA(HIS4), Pgap-GFP-SKL (ARG4) | This study |
| SJS171 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (S-173 to A) (HIS4), FIS1::FIS1-MYC (ARG4) | This study |
| SJS175 | pex11Δ:: ZeoR, PEX11::PEX11-2HA (HIS4), FIS1::FIS1-MYC (ARG4) | This study |
| SJS210 | pex19Δ:: ZeoR, SEC61::mCHERRY-SEC61 (HygR), PAOX1::BFP-SKL (KanR), PAOX1::GFP-PEX11 (ARG4), PPOT1::BPEX19 (HIS4) | This study |
| SKF13 | PPY12: pex19Δ:: ZeoR, arg4, his4 | Snyder et al. (1999) |
| SMY261a | PPY12: PGAP::GFP-SKL (ARG4), his4 | M. Yan |
| SMY278 | PPY12: pex11Δ:: ZeoR, arg4, his4 | M. Yan |
| SMY3 | PPY12: pex3Δ:: ZeoR, arg4, his4 | M. Yan |
Cloning of P. pastoris PEX11
The sequence of the open reading frame (ORF) corresponding to RPPA08527 from the ERGO database (courtesy of Integrated Genomics, Mount Prospect, IL) showed homology with ScPEX11 and was designated as PpPEX11 based on its function and the localization of the encoded protein (see Results). The same sequence is also deposited in the National Center for Biotechnology Information P. pastoris database with accession number XP_002491415. This ORF was amplified using PCR from genomic DNA of P. pastoris strain PPY12, and its sequence was confirmed. The pex11A and pex11D mutants were made using site-directed mutagenesis to convert the phosphorylation site S173 to A or D, respectively.
Phosphatase treatment
Briefly, cells were lysed with acid-washed glass beads and centrifuged at 8000 × g for 1 min at 4°C. After boiling the cells for 5 min, 100 μl of cell-free lysate was incubated with 400 μl of potato acid phosphatase (Kemper et al., 2008) buffer (50 mM 2-(N-morpholino)ethanesulfonic acid [pH 6.0], 1 mM dithiothreitol, containing protease inhibitor cocktail [ Karpichev and Small, 1998] [Sigma-Aldrich, St. Louis, MO] and 1 mM phenylmethylsulfonyl fluoride) with 10 U of PAP (Sigma-Aldrich) for 4 h. The PAP was not added to untreated samples. Pex14p was used as a positive control. The reaction was stopped by adding100 μl of 6× SDS sample buffer and boiling at 100°C for 7 min.
Fluorescence microscopy
For colocalization studies, cells were grown on YPD to an optical density (OD) of 1.2–2.0 and switched to oleate medium during exponential phase. Images were captured using a Plan Apochromat 100×, 1.40 numerical aperture, oil immersion objective on a motorized fluorescence microscope (Axioskop 2 MOT plus; Carl Zeiss, Jena, Germany) coupled to a monochrome digital camera (AxioCam MRm; Carl Zeiss) and processed using AxioVision software (version 4.5; Carl Zeiss). Peroxisomal and ER markers, GFP-SKL (pKSN133) and Sec61p-mCherry (pKSN256), respectively, as well as Pex3p-GFP (pJCF533), were provided courtesy of Kanae Noda and Jean-Claude Farré of our lab.
For the examination of peroxisome morphology following oleate induction, cells were first grown in YPD, switched to oleate medium as mentioned before, and imaged at 0, 4, 8, and 12 h. The average of the total number of peroxisomes/cell for 50 cells was calculated and plotted.
For pulse-chase experiments, cells were first grown on YPD (OD 2.0), induced in methanol medium for 1 h (for expression of GFP-Pex11p), and then switched to oleate medium for induction of Pex19p for 1 h. Cells were imaged every hour (in both methanol and oleate medium).
For immunofluorescence analyses samples were prepared as described previously (Yan et al., 2008), with some modifications. Briefly, paraformaldehyde-fixed cells were spheroplasted with Zymolyase 20T (Seikagaku, Tokyo, Japan) and postfixed with acetone at −20°C after adhering them to poly-l-lysine–coated glass slides. Samples were incubated with blocking buffer (4.3 mM Na2HPO4, 1.4 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, 1% skim milk, 0.1% bovine serum albumin, 0.1% n-octyl glucoside) for 0.5 h and incubated overnight with primary anti-HA rat (1:3000) antibody at 4°C. Samples were washed with blocking buffer and incubated with Alexa Fluor 568–conjugated anti–rat immunoglobulin G goat (1:200) antibody (Molecular Probes, Invitrogen, Carlsbad, CA). After 1 h of incubation in the dark, samples were washed with blocking buffer. A drop of mounting medium (95% glycerol, 0.1% p-phenylenediamine) was added, and a coverslip was then placed over the sample for imaging.
In vitro ER budding assay
The assay was performed as described earlier (Agrawal et al., 2011). Briefly, permeabilized cells were washed twice with TBPS buffer (115 mM potassium acetate, 2.5 mM magnesium acetate, 0.25 M sorbitol, protease inhibitor cocktail [PIC], and 25 mM hydroxyethyl-piperazine ethanesulfonic acid, pH 7.2). The budding reaction contained 4.5 OD600/25 μl of the PYCs, 1 mg of the S1 fraction, and an ATP-regenerating system (1 mM ATP, 0.1 mM GTP, 20 mM creatine phosphate, 0.2 mg/ml creatine phosphate kinase [Sigma-Aldrich]) in a 100-μl total reaction volume. The reaction mixture was incubated at 20°C for 90 min and terminated by chilling the samples on ice. To deplete samples of ATP, apyrase (Sigma-Aldrich) was added instead of the ATP-regenerating system. After the reaction, PYCs were pelleted by centrifugation at 13,000 rpm for 1 min. The reaction supernatant (RS) of two reactions was pooled and spun at 200,000 × g for 1 h. The pellet (RS 200 kgP) was resuspended in SDS sample buffer, heated, and analyzed by Western blotting. Wild-type and mutant PYCs were incubated with cytosol from pex11Δ cells.
Immunoprecipitation
Samples were prepared as described previously (Yan et al., 2008), with some modifications. Cells were grown on YPD to an OD of 1.0–1.2 and switched to oleate medium during exponential phase. A 100 OD600 amount of the oleate-grown cells were resuspended in 3 ml of immunoprecipitation (Hoepfner et al., 2001) buffer (20 mM Tris-HCl, pH 7.5, 0.3 M NaCl, 1 mM EDTA, PIC [Sigma-Aldrich]) supplemented with 0.5% NP-40 and lysed by vortexing with acid-washed glass beads. The lysate was subjected to centrifugation at 20,000 × g for 10 min. A 1-ml amount of lysate was incubated with 20 μl of anti-HA mouse (Covance, Berkeley, CA) or anti-myc mouse (Abcam, Cambridge, MA) monoclonal antibody overnight at 4°C. Then, 200 μl of the GammaBind G Sepharose beads (GE Healthcare, Piscataway, NJ) prewashed in immunoprecipitation (IP) buffer was added to the lysate and incubated further for 2 h. The beads were then washed thrice with 2 ml of IP buffer and boiled in 200 μl of SDS loading buffer. Samples (free of beads) were analyzed by Western blotting. The 5 OD equivalents of the immunoprecipitated and 0.2 OD equivalents of input samples for each of the wild-type, pex11D, and pex11A strains were loaded and analyzed by Western blotting using anti-HA rat antibody.
Supplementary Material
Acknowledgments
We thank Integrated Genomics for access to the ERGO database and Jean-Claude Farré, Mingda Yan, and Kanae Noda for plasmids and strains. This work was supported by a MERIT award to S.S. (National Institutes of Health DK 41737).
Abbreviations used:
- BFP
blue fluorescent protein
- DRP
dynamin-related protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- IP
immunoprecipitation
- JEP
juxtaposed-elongated peroxisome
- OD
optical density
- ORF
open reading frame
- PAP
potato acid phosphatase
- PIC
protease inhibitor cocktail
- Pp
Pichia pastoris
- PYC
permeabilized yeast cells
- Sc
Saccharomyces cerevisiae
- TBPS
tert-butyl-bicyclophospho-orthionate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0782) on February 15, 2012.
REFERENCES
- Agrawal G, Joshi S, Subramani S. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2011;108:9113–9118. doi: 10.1073/pnas.1018749108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett P, Tabak HF, Hettema EH. Nuclear receptors arose from pre-existing protein modules during evolution. Trends Biochem Sci. 2000;25:227–228. doi: 10.1016/s0968-0004(00)01579-6. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol. 2001;155:979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. Molecular basis of peroxisome division and proliferation in plants. Int Rev Cell Mol Biol. 2010;279:79–99. doi: 10.1016/S1937-6448(10)79003-1. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Snyder WB, Cereghino JL, Veenhuis M, Subramani S, Cregg JM. Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast. 2001;18:621–641. doi: 10.1002/yea.711. [DOI] [PubMed] [Google Scholar]
- Karpichev IV, Small GM. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6560–6570. doi: 10.1128/mcb.18.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci. 2008;121:1990–1998. doi: 10.1242/jcs.024034. [DOI] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem. 2010;285:6670–6680. doi: 10.1074/jbc.M109.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, Brocard C. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123:3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- Kos W, Kal AJ, van Wilpe S, Tabak HF. Expression of genes encoding peroxisomal proteins in Saccharomyces cerevisiae is regulated by different circuits of transcriptional control. Biochim Biophys Acta. 1995;1264:79–86. doi: 10.1016/0167-4781(95)00127-3. [DOI] [PubMed] [Google Scholar]
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell. 2008;20:1567–1585. doi: 10.1105/tpc.107.057679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Dyer JM, Quick ME, Goodman JM. Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J Cell Biol. 1996;135:123–137. doi: 10.1083/jcb.135.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. Pmp27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- Opalinski L, Kiel JA, Williams C, Veenhuis M, van der Klei IJ. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2011a;30:5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Veenhuis M, van der Klei IJ. Peroxisomes: membrane events accompanying peroxisome proliferation. Int J Biochem Cell Biol. 2011b;43:847–851. doi: 10.1016/j.biocel.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Pan R, Hu J. The conserved fission complex on peroxisomes and mitochondria. Plant Signal Behav. 2011;6:870–872. doi: 10.4161/psb.6.6.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- Sears IB, O'Connor J, Rossanese OW, Glick BS. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–790. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Snyder WB, Faber KN, Wenzel TJ, Koller A, Luers GH, Rangell L, Keller GA, Subramani S. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol Biol Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Jansen GA, Skjeldal OH. Refsum disease, peroxisomes and phytanic acid oxidation: a review. J Neuropathol Exp Neurol. 2001;60:1021–1031. doi: 10.1093/jnen/60.11.1021. [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Tager JM. Lipid metabolism in peroxisomes in relation to human disease. Mol Aspects Med. 1998;19:69–154. doi: 10.1016/s0098-2997(98)00003-x. [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Waterham HR. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim Biophys Acta. 2006;1763:1707–1720. doi: 10.1016/j.bbamcr.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Weller S, Gould SJ, Valle D. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet. 2003;4:165–211. doi: 10.1146/annurev.genom.4.070802.110424. [DOI] [PubMed] [Google Scholar]
- Yan M, Rachubinski DA, Joshi S, Rachubinski RA, Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol Biol Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.