The human mitochondrial proteins ISCA1, ISCA2, and IBA57 are essential for the generation of mitochondrial [4Fe-4S] proteins in a late step of Fe/S protein biogenesis. This process is important for mitochondrial physiology, as documented by drastic enlargement of the organelles and the loss of cristae membranes in the absence of these proteins.
Abstract
Members of the bacterial and mitochondrial iron–sulfur cluster (ISC) assembly machinery include the so-called A-type ISC proteins, which support the assembly of a subset of Fe/S apoproteins. The human genome encodes two A-type proteins, termed ISCA1 and ISCA2, which are related to Saccharomyces cerevisiae Isa1 and Isa2, respectively. An additional protein, Iba57, physically interacts with Isa1 and Isa2 in yeast. To test the cellular role of human ISCA1, ISCA2, and IBA57, HeLa cells were depleted for any of these proteins by RNA interference technology. Depleted cells contained massively swollen and enlarged mitochondria that were virtually devoid of cristae membranes, demonstrating the importance of these proteins for mitochondrial biogenesis. The activities of mitochondrial [4Fe-4S] proteins, including aconitase, respiratory complex I, and lipoic acid synthase, were diminished following depletion of the three proteins. In contrast, the mitochondrial [2Fe-2S] enzyme ferrochelatase and cellular heme content were unaffected. We further provide evidence against a localization and direct Fe/S protein maturation function of ISCA1 and ISCA2 in the cytosol. Taken together, our data suggest that ISCA1, ISCA2, and IBA57 are specifically involved in the maturation of mitochondrial [4Fe-4S] proteins functioning late in the ISC assembly pathway.
INTRODUCTION
Owing to the ancient character (Huber et al., 2003) of Fe/S clusters, homologues to the bacterial proteins dedicated to the formation of these cofactors are found in mitochondria of eukaryotes. In humans, Fe/S protein biogenesis (reviewed in Rouault and Tong, 2008, and Sheftel et al., 2010a) is initiated in the mitochondria with the removal of sulfur from cysteine by the pyridoxal phosphate–dependent cysteine desulfurase complex Nfs1-Isd11. The persulfide intermediate must be further reduced to sulfide, which is probably accomplished through a ferredoxin electron transport system comprising ferredoxin reductase and ferredoxin 2 (Sheftel et al., 2010b). Sulfide is then combined to form a [2Fe-2S] cluster with ferrous iron (Fe2+) on the scaffold protein IscU (Tong and Rouault, 2006). The molecular progression of nascent cluster transfer from IscU to target apoproteins has not been fully elucidated. However, the initial steps in this transfer require an Hsp70-based chaperone system (Knight et al., 1998; Uhrigshardt et al., 2010) and the monothiol Grx5 (Rodriguez-Manzaneque et al., 2002; Ye et al., 2010). Components involved in the later dissemination of de novo assembled clusters to specific apoproteins are only beginning to be characterized.
Among other components of Fe/S protein assembly, the isc and suf operons of bacteria encode the so-called A-type iron–sulfur cluster (ISC) proteins IscA and SufA, respectively, which are involved in the maturation of bacterial [4Fe-4S] proteins (Johnson et al., 2005; Tan et al., 2009). An additional member of this family, ErpA, is specifically involved in isoprenoid biosynthesis (Loiseau et al., 2007). The molecular function of these proteins in Fe/S biogenesis has only been partially resolved, and some differing views have been proposed. Several reports suggest the A-type ISC proteins act as alternative scaffolds for de novo Fe/S cluster synthesis (Krebs et al., 2001; Ollagnier-de-Choudens et al., 2001; Gupta et al., 2009), whereas others show that these proteins can bind iron with high affinity and may deliver the metal to the IscU scaffold (Ding and Clark, 2004; Ding et al., 2005; Wang et al., 2010; Lu et al., 2010).
Eukaryotes contain two A-type ISC proteins with conserved features in their primary structure, yet studies in yeast show that they perform nonredundant functions essential for respiratory growth (Jensen and Culotta, 2000; Kaut et al., 2000; Pelzer et al., 2000; Mühlenhoff et al., 2011). Only yeast Isa1, but not Isa2, can be replaced by the bacterial A-type ISC proteins. Yeast Isa1 and Isa2 are required for efficient maturation of the Fe/S proteins aconitase and succinate dehydrogenase (SDH). In addition, both of these proteins have been suggested to function in maintaining the activity of the radical SAM enzyme biotin synthase, even though they are dispensable for the incorporation of Fe/S clusters into the enzyme (Mühlenhoff et al., 2007). The human genome contains the Isa1 and Isa2 relatives ISCA1 and ISCA2, respectively. ISCA1 was recently shown to interact with IOP1 (also known as NARFL, the homologue to yeast Nar1) in a yeast two-hybrid screen (Song et al., 2009). Consequently, the same study implicated ISCA1 not only in mitochondrial, but also in cytosolic Fe/S protein production by showing a decrease in cytosolic aconitase (cAco; also known as iron-regulatory protein 1 [IRP1]) upon RNA interference (RNAi)–mediated ISCA1 depletion. In contrast, Isa1 and Isa2 from Trypanosoma brucei were recently shown to be required for the assembly of Fe/S clusters in mitochondrial but not cytosolic Fe/S proteins (Long et al., 2011). Yeast Isa1 and Isa2 function in concert with Iba57, which physically interacts with them (Gelling et al., 2008). Yeast cells depleted or devoid of Iba57 show virtually the same growth phenotypes and functional defects in Fe/S proteins as Isa1- or Isa2-deficient cells (Gelling et al., 2008). Furthermore, Escherichia coli cells devoid of an Iba57-related protein display defects in a subset of Fe/S proteins (Waller et al., 2010).
In an in silico gene expression analysis screen for human and mouse genes affecting heme biosynthesis, Nilsson et al. (2009) identified IBA57 and ISCA1, raising the question of whether these proteins function in heme biosynthesis or Fe/S cluster biogenesis. Targeted knockdown of these genes in zebrafish resulted in anemia. Of interest, previous studies demonstrated that deficiencies in two proteins of Fe/S protein biogenesis, ABCB7 (Allikmets et al., 1999) and GLRX5 (Wingert et al., 2005; Camaschella et al., 2007), can result in erythropoietic defects, indicating a connection between Fe/S protein synthesis and hemoglobin production. Because both these processes are relevant for several human diseases (Sheftel et al., 2010a; Ye and Rouault, 2010b), we attempted to better understand the physiological function of human ISCA1, ISCA2, and IBA57. We examined whether these proteins are directly involved in mitochondrial and cytosolic Fe/S protein biogenesis and/or in heme synthesis. Localization studies in HeLa cells identified these proteins in mitochondria. ISCA1 and ISCA2 were found to be required for the maturation of all tested mitochondrial [4Fe-4S] proteins, including the radical SAM enzyme lipoic acid synthase, whereas the [2Fe-2S] protein ferrochelatase and heme biosynthesis were unaffected. Cells deficient in IBA57 exhibited a similar phenotype to ISCA1- or ISCA2-depleted cells. Thus our study identifies these three proteins as members of the mitochondrial ISC assembly machinery. They perform a specific task late in the biosynthetic pathway of mitochondrial [4Fe-4S] proteins.
RESULTS
ISCA1, ISCA2, and IBA57 are localized to mitochondria
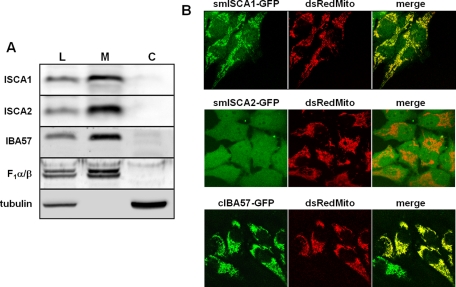
To determine the subcellular localization of ISCA1, ISCA2, and IBA57, specific antibodies were raised in rabbits. HeLa cells were fractionated by lysis with digitonin into a membrane pellet containing mitochondria and a cytosolic supernatant. Immunoblotting identified all three ISC proteins exclusively in the membrane pellet (Figure 1A, lane M). The alpha and beta subunits of mitochondrial F1 ATP synthase (F1α/β) and cytosolic tubulin served as markers for efficient fractionation. To further corroborate these localization data by an alternative experimental approach, we cotransfected HeLa cells with plasmids encoding C-terminally enhanced green fluorescent protein (EGFP)–tagged versions of ISCA1 (psmISCA1-GFP), ISCA2 (psmISCA2-GFP), or IBA57 (pcIBA57-GFP). As a control, a construct encoding mitochondria-targeted dsRed protein (pDsRed2-Mito) was cotransfected. In the cases of ISCA1 and IBA57, a vast majority of the green fluorescence was seen in mitochondria, colocalizing with the dsRed signal (Figure 1B). In contrast, smISCA2-GFP was present throughout the cells. This result is contradictory to the immunoblotting data and is likely due to a mislocalization of the protein due to either overexpression and/or GFP tagging. In support of this idea, when immunoblots from cells overexpressing psmISCA2-GFP were analyzed, a more slowly migrating band, probably representing the unprocessed precursor form of ISCA2-GFP, was present in cytosolic fractions (see discussion and Figure 3B, lane 11, later in the paper), indicating a hampered import into mitochondria upon overexpression. We conclude that all three ISC proteins are located in mitochondria.
FIGURE 1:
ISCA1, ISCA2, and IBA57 are localized to mitochondria. (A) HeLa cells were treated with digitonin and centrifuged at 15,000 × g to separate the cell lysate (L) into a membrane fraction containing mitochondria (M) and a cytosolic fraction (C). Localization of ISCA1, ISCA2, and IBA57 was analyzed by immunoblotting using antibodies raised against the respective proteins. Antibodies recognizing the α and β subunits of F1-ATP synthase (F1α/β) and tubulin served to estimate the efficiency of separating mitochondrial and cytosolic proteins, respectively. (B) HeLa cells were cotransfected twice with smISCA1-GFP, smISCA2-GFP, or cIBA57-GFP, together with mitochondria-targeted Discosoma sp. red protein (dsRedMito). Images of living cells were acquired by confocal microscopy.
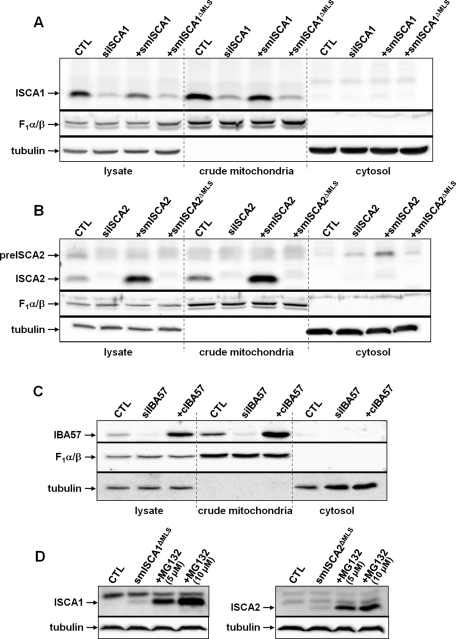
FIGURE 3:
Complementing proteins smISCA1, smISCA2 and cIBA57 functionally localize to mitochondria. HeLa cells were transfected three times as in Figure 2 with specific siRNAs as indicated. Cells were harvested 3 d after the third transfection and fractionated by digitonin treatment as in Figure 1. Cell extracts were analyzed by immunoblotting for the indicated proteins using F1α/β ATP synthase and tubulin as loading controls. Additional samples were cotransfected with the indicated siRNAs plus a vector encoding the corresponding ISC proteins for complementation testing (+smISCA1, +smISCA2, and +cIBA57). In the cases of ISCA1 (A) and ISCA2 (B) these vectors were resistant to RNAi due to silent mutations (sm). Cells were also cotransfected with smISCA1 or smISCA2 vectors lacking the coding information for the mitochondrial localization sequences (ΔMLS). In the case of IBA57 (C) no mutagenesis was necessary to create complementing IBA57 (cIBA57) since the used siRNA bound at the 3′ untranslated region. (D) HeLa cells were transfected with vectors smISCA1ΔMLS and smISCA2ΔMLS, respectively, and 48 h after transfection, a subset of cells was grown in the presence of 5 or 10 μM of the proteasome inhibitor MG132 for 16 h. Harvested cells were lysed, and cell extracts were subjected to immunostaining. preISCA2, putative precursor form of ISCA2.
RNAi-mediated depletion of ISCA and IBA57 proteins results in altered cell metabolism and abnormal mitochondrial ultrastructure
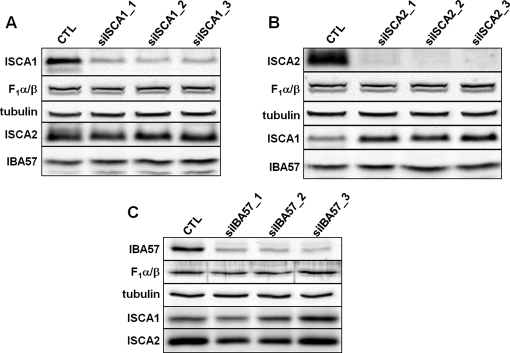
To analyze the physiological consequences of a deficiency in ISCA1, ISCA2, or IBA57, we used RNAi technology. Repeated electroporation-mediated transfection with small interfering RNA (siRNA) duplexes efficiently depleted both ISCA1, ISCA2, and IBA57 mRNAs (Supplemental Figure S1) and proteins (Figure 2) relative to control cells, whereas the levels of mitochondrial and cytosolic control proteins remained unchanged. Of interest, depletion of ISCA2—the protein most strongly affected by RNAi treatment in comparison to ISCA1 and IBA57—resulted in a concurrent increase in ISCA1 protein (Figure 2B). Similarly, depletion of ISCA1 resulted in an increase of ISCA2 levels, although one that was less pronounced as in the previous case (Figure 2A). Compared to ISCA1 and ISCA2, the depletion of IBA57 was relatively weak (Figure 2C). Only one of five tested siRNA sequences yielded a substantial depletion of IBA57. Nonetheless, a modest increase in ISCA1 levels was observed in cells depleted of IBA57 (Figure 2C).
FIGURE 2:
RNAi depletion of ISCA1, ISCA2, or IBA57 and the effects on the corresponding partner proteins. HeLa cells were transfected with (A) siISCA1, (B) siISCA2, or (C) siIBA57. Cells were harvested after 3 d of growth, and a fraction of the cells was retransfected. This procedure was performed twice. Cell lysates of all three transfection rounds (1–3) and of mock-transfected control cells (CTL) were examined by immunoblotting to compare respective protein levels of ISCA1, ISCA2, and IBA57. Immunostaining against F1α/β and tubulin served as loading controls.
The depletion of the ISCA and IBA57 proteins allowed us to analyze the physiological and functional consequences of their deficiencies. To control for the specificity of the observed effects, we first generated rescue constructs that express RNAi-resistant versions of the ISCA and IBA57 proteins. In the cases of ISCA1 and ISCA2, silent mutations were introduced into the regions targeted by the various siRNAs to generate the plasmids psmISCA1 and psmISCA2. For IBA57, only one siRNA sequence, whose target resided in an untranslated region of the mRNA, was used. Therefore no mutagenesis was needed to generate the rescue construct pcIBA57. Figure 3 shows the relative expression levels from these constructs in cells (lysate) after three transfections as detected by immunostaining with specific antibodies. The amount of plasmid-expressed smISCA1 was slightly lower than that of the endogenous protein, whereas smISCA2 and cIBA57 levels exceeded native levels of ISCA2 and IBA57, respectively. All plasmid-expressed proteins were exclusively localized to mitochondria, and no mature-sized products were detectable in the postmitochondrial supernatants (Figure 3). As already mentioned, we observed a weak band possibly corresponding to an unprocessed form of ISCA2 (Figure 3B, lane 11). To allow the investigation of the role of potentially cytosol-localized ISCA1 and ISCA2 by a functional complementation approach, we further transfected RNAi-treated HeLa cells with plasmids psmISCA1ΔMLS and psmISCA2ΔMLS, which led to overexpression of these proteins without their mitochondrial localization sequences. Successful production of the respective mRNAs was confirmed by quantitative real-time PCR (Supplemental Figure S1, A and B). In contrast to previously reported results (Song et al., 2009), deletion of the mitochondrial presequence of ISCA1 did not yield any detectable protein in either the postmitochondrial supernatant or the mitochondria, even when the cells were immediately trichloroacetic acid precipitated after harvesting (Figure 3A). This was also the case for truncated ISCA2. We speculate that these proteins are nonfunctional in the cytosol and thus may be efficiently degraded. This expectation was confirmed by inhibiting the proteasome with MG132 during growth of HeLa cells expressing the cytosolic versions of the ISCA proteins. Under these conditions, significant amounts of both ISCA proteins were detectable in the cell lysates (Figure 3D). The proteasomal degradation of the ISCA proteins in the cytosol argues against their function in this compartment.
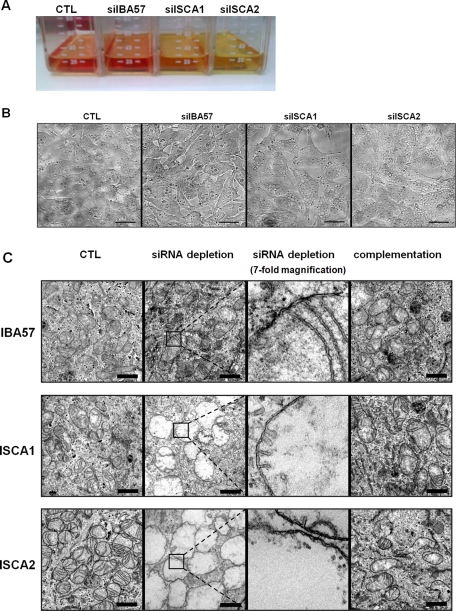
Following ISCA1 or ISCA2 depletion, we observed an acidification of the cell culture media, likely indicating a defect in respiratory function (Figure 4A; Sheftel et al., 2009). This was accompanied by a gross alteration in intracellular morphology manifested as large, vacuolar structures, giving the cells a spongiform appearance (Figure 4B). Because these structures seemed to resemble lipid droplets, we attempted to stain them with Sudan dyes (Lison and Dagnelie, 1935; Jones, 2008). No positive staining resulted in these experiments (unpublished data). We therefore examined the cells using transmission electron microscopy (EM). The ultrastructural analysis revealed that the large inclusions represented severely enlarged mitochondria (some measuring >5 μm in diameter; Figure 4C). No medium acidification and no gross microscopic alterations were observed for the IBA57-depleted cells; however, the EM analysis nonetheless revealed mitochondria that were generally deficient in cristae membranes (Figure 4C). This comparatively modest phenotype is consistent with the weaker depletion of IBA57 and with the less severe metabolic defects in the IBA57-deficient cells (see later discussion). Of importance, the ultrastructural alterations could be fully reversed by expression of a plasmid-encoded, RNAi-resistant ISCA or IBA57 protein, demonstrating the specificity of these effects. Together these data suggest that the ISCA and IBA57 proteins are required for the maintenance of normal mitochondrial morphology, especially for the formation of cristae membranes.
FIGURE 4:
A decrease in ISCA1, ISCA2, or IBA57 causes severe physiological and morphological changes. HeLa cells were transfected as depicted in Figure 3. (A) Culture media of cells thrice transfected with siISCA1, siISCA2, or siIBA57 or mock-treated control cells (CTL) are shown. The color change of the pH indicator (phenol red) from red toward yellow indicates a drop in pH of the media. (B) After the third transfection HeLa cells were seeded on coverslips and grown for 3 d. Cell morphology was then examined by light microscopy. Scale bars, 30 μm. (C) Three days after the final transfection, cells were fixed in culture dishes and evaluated by transmission electron microscopy. To better visualize the presence of the mitochondrial double membranes, the boxed areas in the respective siRNA-depleted samples are shown at sevenfold higher magnification in the third panel. Scale bars, 1 μm.
ISCA and IBA57 protein depletion decreases mitochondrial Fe/S protein levels and enzyme activities
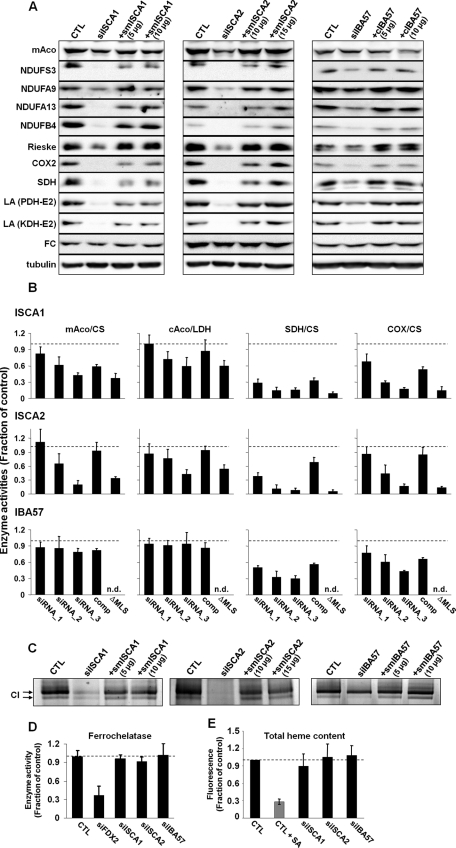
Defects in cristae membranes are characteristic for respiration-compromised mitochondria. Hence it seemed likely that the depletion of ISCA and IBA57 proteins elicited functional defects in cellular respiration. In human cells, the apoprotein levels of many Fe/S proteins are diminished when cells are unable to generate Fe/S clusters. These changes are detectable by immunoblotting. Therefore we examined the amounts of several Fe/S proteins after depletion of the ISCA or IBA57 proteins (Figure 5A). With the notable exception of ferrochelatase (FC; see later discussion) all tested mitochondrial Fe/S proteins (mitochondrial aconitase [mAco], SDH, several proteins of complex I, and Rieske Fe/S protein) were decreased substantially under deficiency of ISCA1 or ISCA2. The levels of control proteins lacking an Fe/S cluster, such as the α and β subunits of F1-ATP synthase, were not altered significantly (Figures 2 and 3). The substantial decrease of cytochrome c oxidase-2 (COX-2) of COX lacking Fe/S clusters cannot easily be explained, but a COX defect is also seen in Isa-depleted yeast cells (Mühlenhoff et al., 2011). This effect may well be a pleiotropic consequence of the specific loss of respiratory complexes I and II. The protein mAco, which was not as strongly diminished after ISCA depletion as other Fe/S proteins, was not affected by transfection with siRNA against IBA57. This may be attributed to the relatively inefficient depletion of IBA57 (Figure 2C). It is worthy to note that depletion of ISCA2 was stronger than that of ISCA1, which is congruent with the lower levels of mAco observed in ISCA2 siRNA-treated cells. The rescue constructs completely or at least partially restored the amounts of all proteins seen to decrease under depletion conditions, validating the specificity of the RNAi treatment and showing a functional complementation by the respective plasmid-encoded ISC proteins.
FIGURE 5:
Depletion of ISCA1, ISCA2, or IBA57 affects maturation of mitochondrial [4Fe-4S] proteins and cytosolic aconitase. HeLa cells were treated as described in Figure 3 with siRNAs and the indicated amounts of complementing plasmids. (A) Cell lysates of the third transfection (grown for 9 d) were examined by immunostaining with the indicated antibodies. Staining against tubulin served as a loading control. COX-2, cytochrome oxidase 2; CTL, mock-treated cells; FC, ferrochelatase; KDH, α-ketoglutarate dehydrogenase; LA, lipoic acid; mAco, mitochondrial aconitase; NDUFA9, subunit A9 of complex I; NDUFA13, subunit A13 of complex I; NDUFB4, subunit B4 of complex I; NDUFS3, subunit S3 of complex I; PDH, pyruvate dehydrogenase; Rieske, Fe/S protein of complex III; SDH, succinate dehydrogenase. (B) Harvested cells were fractionated by digitonin treatment and centrifugation (Figure 1A). Fractions were analyzed for the indicated enzyme activities by spectrophotometry. Citrate synthase (CS) and lactate dehydrogenase (LDH) activities were used to normalize mitochondrial and cytosolic enzyme quantifications, respectively. For each transfection set, values of treated cells were normalized to those of mock-treated control cells. The average (SD) of three sets of normalized values is presented. Suffix 1 to 3, number of transfections; comp, complementation construct after third transfection; ΔMLS, respective complementation construct lacking the mitochondrial localization sequence, after third transfection; n.d., not determined. (C) Cell pellets from B (third transfection) were mechanically lysed and subjected to blue native-PAGE. Complex I activity was determined using an in-gel activity assay. (D) After the third transfection, ferrochelatase enzyme activity was estimated by following the incorporation of 55Fe into deuteroporphyrin IX. The resultant 55Fe-heme formation was normalized to that of mock-treated control cells (CTL), and three data sets were averaged. The analysis includes cells RNAi depleted for human ferredoxin 2 (siFDX2; Sheftel et al., 2010b). (E) To measure the total heme content after three rounds of transfection, cell pellets were dissolved in 2 M oxalic acid and boiled for 30 min, which releases iron from heme, thus generating fluorescent protoporphyrin IX (Morrison, 1965; Ward et al., 1984). As a control, HeLa cells were treated with 0.1 mM of the heme biosynthesis inhibitor succinyl acetone (SA) and harvested after 3 d. Fluorescence emission was determined and normalized to that of control cells. Three independent data sets were averaged. Error bars represent SDs.
In addition to the measurement of Fe/S protein levels, the activity of the mitochondrial enzyme lipoic acid synthase can directly be followed by analyzing the levels of protein-bound lipoic acid by immunoblotting with an antibody that specifically recognizes this cofactor (Onder et al., 2006; Gelling et al., 2008). Human cells possess four lipoic acid–dependent enzymes, which are all localized in mitochondria: the E2 components of pyruvate dehydrogenase complex (DLAT; ∼70 kDa), 2-oxo-glutarate dehydrogenase complex (DLST; ∼42 kDa), and branched-chain α-ketoacid dehydrogenase complex (DBT; ∼46 kDa), as well as the H protein of the glycine cleavage system (GCSH; ∼14 kDa). In HeLa cells the anti–lipoic acid antibodies recognized two distinct bands that corresponded in electrophoretic mobility to PDH-E2 and KDH-E2 (Figure 5A). Depletion of ISCA1, ISCA2, or IBA57 resulted in strongly decreased detection of these bands, indicating a lower content in lipoic acid. Again, expression of the respective ISC proteins from the rescue plasmids restored the amount of lipoic acid–modified enzymes to normal. Together these results show that ISCA- and IBA57-depleted cells contain lower levels of mitochondrial Fe/S proteins, suggesting that these cells show a defect in the biogenesis of the Fe/S cofactor.
To further investigate the effects of ISCA or IBA57 depletion, we measured the enzyme activities of several other Fe/S proteins and non–Fe/S control proteins. Both ISCA1 and ISCA2 depletion significantly diminished the activities of mAco and SDH (Figure 5B). The activity of mitochondrial citrate synthase did not change appreciably and was used to normalize the data for mAco and SDH. It is somewhat surprising that there was also a marked decrease in COX activity, even though this enzyme does not contain any Fe/S cluster. All these activities were restored upon cotransfection with complementing psmISCA plasmids (comp), but no effect was obtained with the cytosol-targeted versions (ΔMLS). Consistent with the immunoblotting data, IBA57 deficiency did not affect mAco activity but significantly decreased SDH activity. We further estimated the enzymatic function of complex I (NADH-CoQ dehydrogenase) using in-gel activity measurements. Complex I activity was severely affected by ISCA1 or ISCA2 deficiency (Figure 5C). IBA57 depletion also decreased complex I activity but to a lesser degree. Taken together, these data clearly document a specific defect in mitochondrial Fe/S proteins after depletion of the ISCA and IBA57 proteins.
In contrast to all other Fe/S enzymes analyzed here previously, we detected no change in FC protein levels under limiting ISCA or IBA57 concentrations (Figure 5A). We therefore directly measured the FC catalytic activity by incubating cell pellets with deuteroporphyrin and 55Fe in the presence of ascorbate followed by organic extraction of heme (Lange et al., 1999; Song et al., 2009). FC activity was not compromised when the ISCA or IBA57 proteins were depleted (Figure 5D). As a control, depletion of Fdx2, which was previously shown to elicit a decrease in FC protein amount (Sheftel et al., 2010b), reduced FC activity to ∼30% of mock-transfected cells. Consistent with these in vitro data, total heme levels of HeLa cells did not change in ISCA- and IBA57-depleted cells (Figure 5E). On treatment with succinyl-acetone, an inhibitor of heme biosynthesis, a fourfold lower heme content was found. These data unambiguously show that the ISCA and IBA57 proteins are not required for FC function in heme synthesis, suggesting normal maturation of its [2Fe-2S] cluster. In contrast, the ISC component Fdx2, which functions in early stages of Fe/S protein biogenesis and thus is required for maturation of all cellular Fe/S proteins, is also essential for maturation of this [2Fe-2S] protein. Our data are therefore compatible with the idea that the ISCAs and IBA57 proteins act later in the ISC assembly pathway and may specifically function in [4Fe-4S] protein maturation.
A role of the ISCA and IBA57 proteins for Fe/S protein maturation in the cytosol?
Previous studies identified (overproduced) ISCA1 in the cytosol and consequently assigned a role to the protein in the maturation of cytosolic IRP1 and xanthine oxidase Fe/S proteins (Song et al., 2009). Because we did not detect ISCA and IBA57 proteins in this compartment under physiological conditions (see earlier discussion), we examined the effect of depletion of these proteins on the maturation of cytosolic Fe/S proteins. The protein levels of dihydropyrimidine dehydrogenase (DPYD; Schnackerz et al., 2004) and glutamate phosphoribosylpyrophosphate amidotransferase (GPAT; Martelli et al., 2007; Sheftel et al., 2010b) are indicative of the amounts of Fe/S clusters associated with these proteins. DPYD and GPAT levels were not significantly altered by any of the RNAi treatments for ISCA and IBA57 protein depletion, in contrast to the severe effect upon depletion of NBP35 or ISCU (Figure 6A and Supplemental Figure 2). Similarly, no significant decrease of cytosolic IRP1 protein levels was observed. A clear (twofold to threefold) decrease in cytosolic aconitase activity (cAco; normalized to lactate dehydrogenase) was detected when cells were treated with siRNA against ISCA1 or ISCA2 but not IBA57 (Figure 5B). To examine whether this diminished activity could be rescued by overexpressing ISCA1 or ISCA2 in the cytosol, we cotransfected the cells with the corresponding ΔMLS rescue plasmids lacking the coding information for their mitochondrial presequence. The cytosolic versions of these ISC assembly proteins were unable to restore cAco activity, whereas the full-length ISCA proteins almost fully reverted the cAco activity back to control levels.
FIGURE 6:
Depletion of ISCA1, ISCA2, or IBA57 affects IRP1 but not other cytosolic Fe/S proteins. HeLa cells were transfected as in Figure 5A. (A) Cell lysates of the third transfection were examined by immunoblotting using the indicated antibodies. Tubulin served as a loading control. (B) Cell lysates from A were analyzed for IRP1-binding activity to the iron-responsive element of ferritin mRNA by electrophoretic mobility shift assay. Because IRP1 and IRP2 possess the same running behavior, anti-IRP2 antibodies were used to supershift the corresponding protein. Treatment with 2% β-mercaptoethanol (β-ME) was used to reveal the maximal IRP1-binding capacity.
When cAco loses its Fe/S cluster, the protein assumes a conformation that confers a high affinity for mRNA stem-loop structures known as iron-responsive elements (IREs; Rouault, 2006; Wallander et al., 2006). Referred to as “IRP1 activity,” the amount of IRP1-bound IRE in this state can be estimated by conducting electrophoretic mobility shift assays (EMSAs) using radiolabeled IRE-containing mRNA as a probe. As depicted in Figure 6B, ISCA1, ISCA2, or IBA57 depletion resulted in an increase in IRP1-IRE binding activity that was complemented by overexpression of the respective RNAi-resistant ISC genes from the rescue constructs. Treatment of the lysates with 2% β-mercaptoethanol allowed visualization of the total amount of IRP1 present. Consistent with the immunoblots, no changes in total IRP1 protein levels were observed. Thus the EMSA measurements fully corroborated the observations made for the enzyme activities of IRP1. Taken together, the analyses of three different cytosolic Fe/S proteins demonstrate differential effects of the ISCA and IBA57 protein deficiencies. On the one hand, the normal levels of DPYD and GPAT suggest no general function of these mitochondrial ISC proteins in cytosolic Fe/S protein maturation. On the other hand, there is a clear decrease in mature cAco upon their depletion.
Effect of ISCA and IBA57 protein depletion on key proteins of iron homeostasis
The activation of the IRE-binding activity of IRP1 upon ISCA and IBA57 protein depletion suggested consequences on proteins of cellular iron metabolism. Transferrin receptor (TfR), the gateway for iron import into most cells, harbors IRE stem loops in its mRNA's 3X′ untranslated region (UTR), endowing its mRNA greater stability upon IRP binding. On the contrary, an IRE present in the 5′UTR of ferritin (Ft) mRNA blocks translation of the protein under elevated IRP activity, allowing reciprocal regulation of Ft, which fits to Ft's function as an iron-storage protein. As expected, ISCA protein–deficient cells, with their greater IRP1 activity, have increased TfR and decreased Ft levels (Figure 6A). Likely due to the limited depletion efficiency, IBA57-deficient cells showed only weak effects on TfR and Ft levels. We therefore conclude that ISCA and IBA57 depletion results in detectable effects on cAco/IRP1 maturation, which, in turn, leads to a dysregulation of two key proteins of cellular iron metabolism. Finally, we analyzed the levels of IRP2 upon ISCA or IBA57 depletion. The amount of IRP2 slightly decreased, yet only under ISCA2 deficiency (Figure 6A). This decrease is likely a consequence of the increased cellular iron uptake due to higher transferrin receptor levels. Of note, this result was opposite to what is seen for depletion of ISC assembly proteins, which severely affects cytosolic Fe/S protein biogenesis (such as FDX2; see, e.g., Sheftel et al., 2010b).
DISCUSSION
Our study presents the identification and functional characterization of important members of the mitochondrial ISC assembly machinery termed ISCA1, ISCA2, and IBA57. We show that these proteins are localized and functional inside mitochondria and represent important components for mitochondrial biogenesis. Their depletion in cell culture caused striking alterations in mitochondrial morphology, including a vast enlargement of the organelles and a loss of cristae membranes. Such a phenotype has been previously observed in cases of mitochondrial DNA (mtDNA) mutations (Nass, 1984; Chang et al., 2004; Gdynia et al., 2010). This eye-catching effect is explained by a molecular function of these proteins in the maturation of several mitochondrial Fe/S proteins carrying a [4Fe-4S] cluster, including complexes I and II of the respiratory chain. The functional defect of these complexes and of aconitase usually contributes to the loss of cristae membranes and of mtDNA (Stevens, 1981; Chen et al., 2005). We previously documented similar yet distinct morphological changes of cells depleted of the core ISC assembly machinery components Nfs1, ferredoxin 2, or the complex I–specific Fe/S maturation factor Ind1 (Biederbick et al., 2006; Sheftel et al., 2009, 2010b). The biogenesis of the [2Fe-2S] protein ferrochelatase, on the other hand, occurred independent of the ISCA and IBA57-depleted proteins, and the heme content was unchanged, excluding a role of these proteins in heme synthesis. Hence we propose that the ISCA and IBA57 proteins play a dedicated role in the biogenesis of [4Fe-4S] clusters and/or their insertion into apoproteins and not in heme synthesis. Recent studies provide compelling evidence for an essential requirement of bacterial and yeast relatives of human ISCA1, ISCA2, and IBA57 for efficient maturation of [4Fe-4S] proteins (Tan et al., 2009; Mühlenhoff et al., 2011). We therefore conclude that the dedicated function of the Isa and Iba57 proteins is conserved from bacteria to human. Nevertheless, there are characteristic differences between the various A-type ISC proteins in the different organisms, showing that the specificity of these proteins for their Fe/S protein targets may vary. For instance, Isa2/ISCA2, which can clearly be distinguished in its primary sequence from Isa1/ISCA1 and is unique for eukaryotes, cannot be replaced by bacterial A-type ISC proteins, in contrast to Isa1/ISCA1 (Mühlenhoff et al., 2011). Bacterial ErpA is highly specific for the maturation of [4Fe-4S] proteins in isoprene biosynthesis, whereas IscA and SufA have a broader target spectrum (Loiseau et al., 2007).
Little has been known about human ISCA1 and ISCA2 or their mutual partner protein IBA57 (Gelling et al., 2008). Inspired by the results of a high-throughput screen, a recent study found a small proportion of cellular ISCA1 in a cytosolic fraction. From these results a role of ISCA1 in cytosolic Fe/S protein maturation was proposed (Song et al., 2009), and yet a lysis of mitochondria during cell fractionation was not excluded. Similar to the results in our report, Song et al. (2009) observed a decrease in both mitochondrial and cytosolic aconitase activity in cells depleted of ISCA1. However, we were unable to rescue cytosolic aconitase activity in ISCA-deficient cells by overexpressing the respective ISCA proteins in the cytosol. Moreover, despite a strong depletion of ISCA1 (or ISCA2) in HeLa cells, our experiments failed to elicit appreciable decreases in the extramitochondrial [4Fe-4S] proteins GPAT and DPYD. Both proteins are highly sensitive tools for measuring Fe/S protein biogenesis, and their protein levels are strongly compromised under all instances of core mitochondrial or cytosolic Fe/S protein biogenesis deficiency (Stehling et al., 2004, 2008; Martelli et al., 2007; Sheftel et al., 2010b). Furthermore, Song et al. (2009) demonstrate that the acute recovery rate of cAco activity, following peroxide treatment, is unaffected by ISCA1 depletion, suggesting that the cytosolic repair of reactive oxygen species (ROS)–damaged cAco does not require a cytosolic ISCA1 isoform. Finally and most convincing, we were able to detect overproduced ISCA1 or ISCA2 in the cytosol of HeLa cells, but only when proteasomal activity was compromised by the inhibitor MG132. These experiments show that the ISCA proteins became synthesized in the cytosol and yet were efficiently degraded in this compartment. We therefore reason that these proteins are not located outside mitochondria, and we propose that the effect on IRP1 activity upon ISCA protein depletion might be an indirect phenotype.
The conclusion that ISCA and IBA57 proteins are not directly involved in the maturation of extramitochondrial Fe/S proteins is supported by recent findings in yeast and trypanosomes (Long et al., 2011; Mühlenhoff et al., 2011). In yeast, growth conditions were observed that lead to defects in cytosolic Fe/S proteins (Kaut et al., 2000; Pelzer et al., 2000). How can these effects be explained? Isa/ISCA depletion elicits a potent inhibition of several complexes of the mitochondrial electron transport chain. Such a condition is expected to increase the levels of ROS in the cells (Herrero et al., 2008), which are known to particularly destabilize labile Fe/S clusters, for instance, that of aconitase or yeast Leu1 (Brazzolotto et al., 1999; Pantopoulos, 2004; Mühlenhoff et al., 2011). Consistent with this notion, depletion of human Ind1—an assembly factor specifically required for the Fe/S protein maturation of complex I—also elicited a modest, likely indirect decrease in cytosolic aconitase activity (Sheftel et al., 2009).
Depletion of ISCA and IBA57 proteins in HeLa cells affects the citric acid cycle in four different enzymes: the two Fe/S proteins aconitase and succinate dehydrogenase and the lipoic acid–dependent enzymes pyruvate dehydrogenase and α-ketoglutarate dehydrogenase. The functional defect of these enzymes was proposed to decrease the efficiency of the citric acid cycle and in turn to diminish the level of succinyl-CoA, a precursor of porphyrins and heme (Ye and Rouault, 2010a). It therefore was surprising that HeLa cells depleted in these three ISC proteins contain a normal amount of heme. What might be the explanation for this observation? As mentioned, heme synthesis catalyzed by the [2Fe-2S] protein ferrochelatase appears to be unaffected in these deficiencies. Moreover, it is evident that the net production of succinyl-CoA in the ISC protein–depleted situation must occur independent of a functional citric acid cycle and instead is maintained by other biosynthetic reactions. Several such pathways are known (Lehninger et al., 2008). First, succinyl-CoA can be synthesized from the branched-chain amino acids isoleucine and valine. However, this requires the activity of branched-chain ketoacid dehydrogenase, another mitochondrial lipoic acid–dependent enzyme, which therefore should also be inefficient. A second possibility is the conversion of propionyl-CoA derived from either methionine or odd-numbered fatty acids to succinyl-CoA via methyl-malonyl-CoA. This sequence of reactions is independent of the citric acid cycle and hence may produce enough succinyl-CoA to satisfy the requirement for normal heme biosynthesis.
We observed a puzzling decrease in COX activity and protein (COX-2) under ISCA- and IBA57-depletion conditions in both human and yeast cells (Mühlenhoff et al., 2011) despite the fact that COX does not contain Fe/S clusters. As discussed earlier, this effect is not caused by a compromised heme biosynthesis (see also Long et al., 2011). Similarly, it is unlikely to be the result of an impaired heme A production, since this reaction depends on the [2Fe-2S] ferredoxin Yah1, which is unaffected in its maturation in Isa-Iba57–deficient yeast cells and possibly in respective human cells (Sheftel et al., 2010b; Mühlenhoff et al., 2011). A reasonable explanation for the cytochrome c oxidase deficiency may therefore be the overall failure of ISCA- and IBA57-deficient cells to assemble respiratory complexes I and II and their supercomplexes, consequently eliciting a pleiotropic effect on other members of the electron transfer chain. This interpretation may also explain the decrease of the [2Fe-2S]-carrying Rieske Fe/S protein of complex III. In Isa-depleted yeast cells the [2Fe-2S] cluster can still be inserted at wild-type efficiency in the matrix, excluding a direct function of the Isa proteins in [2Fe-2S] cluster insertion into this protein (Mühlenhoff et al., 2011). This may be similar in human cells, and yet complex III assembly fails due to the defects in complexes I and II. In striking contrast to ISCA-IBA57 deficiency, RNAi depletion of Ind1 caused a potent decrease in complex I activity and protein without affecting COX function (Sheftel et al., 2009). Although this reemphasizes the specificity of Ind1 for complex I assembly, we cannot exclude the possibility that other, yet-to-be-identified [4Fe-4S] proteins are involved in a more direct manner in COX maturation.
In summary, our study characterizes ISCA1, ISCA2, and IBA57 proteins as crucial components of the mitochondrial ISC assembly machinery. Our present work and a recent investigation in yeast (Mühlenhoff et al., 2011) show that these proteins are specifically required for the generation of [4Fe-4S] clusters and their dedicated insertion into mitochondrial apoproteins. Potential diseases arising from mutations in these genes are expected to elicit defects in mitochondrial respiration and in lipoic acid–dependent proteins (Cameron et al., 2011; Navarro-Sastre et al., 2011).
MATERIALS AND METHODS
Materials
siRNAs were purchased from Applied Biosystems (Foster City, CA; sequences are in Supplemental Table S1). For all assays performed, two separate scrambled siRNAs resulted in identical effects as mock transfections, which served as controls in all experiments. Polyclonal antibodies were raised in rabbits using purified proteins produced in E. coli. The remaining antibodies used were against F1α/β, Rieske, COX-2 (H. Schägger and I. Wittig), TfR (Zymed, San Francisco, CA), Ft (ICN Biomedical, Irvine, CA), tubulin (clone DM1α; Sigma-Aldrich, St. Louis, MO), SDH, NDUFS3, NDUFA9, NDUFB4, and NDUFA13 (MitoSciences, Eugene, OR), lipoic acid (Calbiochem, La Jolla, CA), DPYD (Santa Cruz Biotechnology, Santa Cruz, CA), IRP2 and IRP1 (R. Eisenstein), GPAT (H. Puccio), mAco (L. Szweda), and ferrochelatase (T. and H. A. Dailey). Cell culture reagents were purchased from PAA Laboratories (Pasching, Austria). All other reagents were purchased from Carl Roth (Karlsruhe, Germany) unless specified otherwise. HeLa cells were grown in DMEM containing 4.5 g/l glucose and supplemented with 8% fetal calf serum, 1 mM glutamine, and 1% penicillin–streptomycin.
Plasmids
All oligonucleotides used, including siRNA sequences, are provided in Supplemental Table S1. Human cDNA clones for ISCA1, ISCA2 (imaGenes, Source BioScience LifeScience, Nottingham, United Kingdom), and IBA57 (Invitrogen, Carlsbad, CA) were purchased. The IBA57 clone was missing the first three codons, which were added by PCR amplification during subcloning. For recombinant expression, PCR amplification was used to clone the genes without their predicted mitochondrial targeting sequences into pETDuet-1 (Novagen, EMD4Biosciences, Gibbstown, NJ), which incorporates a hexahistidine tag. C43 (DE3) E. coli already harboring a plasmid encoding the GroES-GroEL chaperone system (plasmid kindly supplied by M. Hayer-Hartl, Martinsried, Germany) were used for recombinant expression. The proteins were purified using a nickel-nitrilotriacetic acid column and, if necessary, contaminating chaperone proteins were removed by size-exclusion chromatography. For rescue experiments using ISCA1 and IBA57, both full-length cDNAs (plasmids pISCA1-GFP and pIBA57-GFP) and clones with the stop codon removed (pISCA1 and pIBA57) were cloned into pEGFP-N3 (Clontech, Mountain View, CA). Because the siRNA sequence used for IBA57 targeted the 3′ untranslated region, no mutagenesis was required to generate a rescue construct (pcIBA57). For ISCA1 (psmISCA1), mutagenesis was performed in those coding regions targeted by the siRNA to introduce silent changes in the gene sequence according to described methods (Zheng et al., 2004). To generate the ISCA2 rescue mutant (psmISCA2; Supplemental Table S1), the gene was ordered from GenScript (Piscataway, NJ) and subcloned into pEGFP-N2. For all rescue constructs, versions without a stop codon before the EGFP tag were also generated (psmISCA1-GPF, psmISCA2-GFP, pcIBA57-GFP). pDsRed2-Mito was from Clontech.
Miscellaneous procedures
Previously described methods were used for fractionating HeLa cells using digitonin, carrying out electroporation transfection, assaying the activities of mitochondrial and cytosolic aconitase, complex I, SDH, COX, citrate synthase, and LDH, and determining IRP-binding activity (Stehling et al., 2009). Immunoblots to detect ISCA1 and ISCA2 were generated from tricine-SDS-PAGE gels (Schägger, 2006). All bar graphs represent the average of at least three independent sets of transfections.
Supplementary Material
Acknowledgments
We thank R. Rösser for expert technical support, S. Molik for antibody generation, and an anonymous reviewer for suggesting the use of MG132. This work was generously supported by funds from the Deutsche Forschungsgemeinschaft (SFB 593 and TR1, Gottfried-Wilhelm Leibniz Program, and GRK 1216), the von Behring-Röntgen Stiftung, the Max-Planck Gesellschaft, the Fonds der Chemischen Industrie (R.L.), the Alexander-von-Humboldt Stiftung, the Fonds de Recherche en Santé du Québec, and the Canadian Institutes of Health Research (A.D.S.).
Abbreviations used:
- cAco
cytosolic aconitase
- COX
cytochrome c oxidase
- DPYD
dihydropyrimidine dehydrogenase
- EM
electron microscopy
- FC
ferrochelatase
- GPAT
glutamate phosphoribosylpyrophosphate amidotransferase
- IRE
iron-responsive element
- IRP
iron regulatory protein
- ISC
iron-sulfur cluster
- LDH
lactate dehydrogenase
- mAco
mitochondrial aconitase
- RNAi
RNA interference
- SAM
S-adenosyl methionine
- SDH
succinate dehydrogenase
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0772) on February 9, 2012.
REFERENCES
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum Mol Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- Biederbick A, Stehling O, Rösser R, Niggemeyer B, Nakai Y, Elsässer HP, Lill R. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol. 2006;26:5675–5687. doi: 10.1128/MCB.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazzolotto X, Gaillard J, Pantopoulos K, Hentze MW, Moulis JM. Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J Biol Chem. 1999;274:21625–21630. doi: 10.1074/jbc.274.31.21625. [DOI] [PubMed] [Google Scholar]
- Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TM, Chi CS, Tsai CR, Lee HF, Li MC. Paralytic ileus in MELAS with phenotypic features of MNGIE. Pediatr Neurol. 2004;31:374–377. doi: 10.1016/j.pediatrneurol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- Ding B, Smith ES, Ding H. Mobilization of the iron centre in IscA for the iron-sulphur cluster assembly in IscU. Biochem J. 2005;389:797–802. doi: 10.1042/BJ20050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Clark RJ. Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem J. 2004;379:433–440. doi: 10.1042/BJ20031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdynia G, et al. Danger signaling protein HMGB1 induces a distinct form of cell death accompanied by formation of giant mitochondria. Cancer Res. 2010;70:8558–8568. doi: 10.1158/0008-5472.CAN-10-0204. [DOI] [PubMed] [Google Scholar]
- Gelling C, Dawes IW, Richhardt N, Lill R, Mühlenhoff U. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol. 2008;28:1851–1861. doi: 10.1128/MCB.01963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de CS. Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J Am Chem Soc. 2009;131:6149–6153. doi: 10.1021/ja807551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Ros J, Belli G, Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Huber C, Eisenreich W, Hecht S, Wachtershauser G. A possible primordial peptide cycle. Science. 2003;301:938–940. doi: 10.1126/science.1086501. [DOI] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC. Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol Cell Biol. 2000;20:3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Jones ML. Lipids. In: Bancroft JD, Gamble M, editors. Theory and Practice of Histological Techniques. Philadelphia: Churchill Livingstone; 2008. pp. 187–215. [Google Scholar]
- Kaut A, Lange H, Diekert K, Kispal G, Lill R. Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J Biol Chem. 2000;275:15955–15961. doi: 10.1074/jbc.M909502199. [DOI] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- Lange H, Kispal G, Lill R. Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J Biol Chem. 1999;274:18989–18996. doi: 10.1074/jbc.274.27.18989. [DOI] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry, San Francisco: WH. Freeman; 2008. [Google Scholar]
- Lison L, Dagnelie J. Methods nouvelles de coloration de la myéline. Bull Histol Appl Physiol Pathol Technique Microsc. 1935;12:85–91. [Google Scholar]
- Loiseau L, Gerez C, Bekker M, Ollagnier-de CS, Py B, Sanakis Y, Teixeira de MJ, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Changmai P, Tsaousis AD, Skalicky T, Verner Z, Wen YZ, Roger AJ, Lukes J. Stage-specific requirement for Isa1 and Isa2 proteins in the mitochondrion of Trypanosoma brucei and heterologous rescue by human and Blastocystis orthologues. Mol Microbiol. 2011;81:1403–1418. doi: 10.1111/j.1365-2958.2011.07769.x. [DOI] [PubMed] [Google Scholar]
- Lu J, Bitoun JP, Tan G, Wang W, Min W, Ding H. Iron-binding activity of human iron-sulfur cluster assembly protein hIscA1. Biochem J. 2010;428:125–131. doi: 10.1042/BJ20100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A, Wattenhofer-Donze M, Schmucker S, Bouvet S, Reutenauer L, Puccio H. Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum Mol Genet. 2007;16:2651–2658. doi: 10.1093/hmg/ddm163. [DOI] [PubMed] [Google Scholar]
- Morrison GR. Fluorometric microdetermination of heme protein. Anal Chem. 1965;37:1124–1126. doi: 10.1021/ac60228a014. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerl MJ, Flauger B, Pirner HM, Balser S, Richhardt N, Lill R, Stolz J. The ISC proteins Isa1 and Isa2 are required for the function but not for the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:495–504. doi: 10.1128/EC.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Richter N, Pines O, Pierik AJ, Lill R. Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J Biol Chem. 2011;286:41205–41216. doi: 10.1074/jbc.M111.296152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass MM. Analysis of methylglyoxal bis(guanylhydrazone)-induced alterations of hamster tumor mitochondria by correlated studies of selective rhodamine binding, ultrastructural damage, DNA replication, and reversibility. Cancer Res. 1984;44:2677–2688. [PubMed] [Google Scholar]
- Navarro-Sastre A, et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am J Hum Genet. 2011;89:656–667. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 2009;10:119–130. doi: 10.1016/j.cmet.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. Iron-sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J Biol Chem. 2001;276:22604–22607. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- Onder O, Yoon H, Naumann B, Hippler M, Dancis A, Daldal F. Modifications of the lipoamide-containing mitochondrial subproteome in a yeast mutant defective in cysteine desulfurase. Mol Cell Proteomics. 2006;5:1426–1436. doi: 10.1074/mcp.M600099-MCP200. [DOI] [PubMed] [Google Scholar]
- Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann NY Acad Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- Pelzer W, Mühlenhoff U, Diekert K, Siegmund K, Kispal G, Lill R. Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron-sulfur proteins. FEBS Lett. 2000;476:134–139. doi: 10.1016/s0014-5793(00)01711-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Schnackerz KD, Dobritzsch D, Lindqvist Y, Cook PF. Dihydropyrimidine dehydrogenase: a flavoprotein with four iron-sulfur clusters. Biochim Biophys Acta. 2004;1701:61–74. doi: 10.1016/j.bbapap.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010a;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Mühlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA. 2010b;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel AD, Stehling O, Pierik AJ, Netz DJ, Kerscher S, Elsässer HP, Wittig I, Balk J, Brandt U, Lill R. Human ind1, an iron-sulfur cluster assembly factor for respiratory complex I. Mol Cell Biol. 2009;29:6059–6073. doi: 10.1128/MCB.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Tu Z, Lee FS. Human ISCA1 interacts with IOP1/NARFL and functions in both cytosolic and mitochondrial iron-sulfur protein biogenesis. J Biol Chem. 2009;284:35297–35307. doi: 10.1074/jbc.M109.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Elsässer HP, Brückel B, Mühlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- Stehling O, Netz DJ, Niggemeyer B, Rösser R, Eisenstein RS, Puccio H, Pierik AJ, Lill R. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol Cell Biol. 2008;28:5517–5528. doi: 10.1128/MCB.00545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Sheftel AD, Lill R. Controlled expression of iron-sulfur cluster assembly components for respiratory chain complexes in mammalian cells. Methods Enzymol. 2009;456:209–231. doi: 10.1016/S0076-6879(08)04412-1. [DOI] [PubMed] [Google Scholar]
- Stevens B. Mitochondrial structure. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 471–488. [Google Scholar]
- Tan G, Lu J, Bitoun JP, Huang H, Ding H. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem J. 2009;420:463–472. doi: 10.1042/BJ20090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010;19:3816–3834. doi: 10.1093/hmg/ddq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller JC, et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc Natl Acad Sci USA. 2010;107:10412–10417. doi: 10.1073/pnas.0911586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang H, Tan G, Si F, Liu M, Landry AP, Lu J, Ding H. In vivo evidence for the iron-binding activity of an iron-sulfur cluster assembly protein IscA in Escherichia coli. Biochem J. 2010;432:429–436. doi: 10.1042/BJ20101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH, Jordan I, Kushner JP, Kaplan J. Heme regulation of HeLa cell transferrin receptor number. J Biol Chem. 1984;259:13235–13240. [PubMed] [Google Scholar]
- Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Rouault TA. Erythropoiesis and iron sulfur cluster biogenesis. Adv Hematol. 2010a;2010 doi: 10.1155/2010/329394. pii: 329394 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Rouault TA. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry. 2010b;49:4945–4956. doi: 10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.