In quiescent cells, spatial regulation of specific proteins or RNA may have crucial functions for the entry into or exit from the stationary phase. The nuclear histone deacetylase Hos2 is observed to form a reversible cytoplasmic granule, and the formation of Hos2 granules depends on the small heat-shock protein Hsp42.
Abstract
One of many physiological adjustments in quiescent cells is spatial regulation of specific proteins and RNA important for the entry to or exit from the stationary phase. By examining the localization of epigenetic-related proteins in Saccharomyces cerevisiae, we observed the formation of a reversible cytosolic “stationary-phase granule” (SPG) by Hos2, a nuclear histone deacetylase. In the stationary phase, hos2 mutants display reduced viability. Additionally, they exhibit a significant delay when recovering from stationary phase. Hos2 SPGs also contained Hst2, a Sir2 homologue, and several stress-related proteins, including Set3, Yca1, Hsp26, Hsp42, and some known components of stress granules. However, Hos2 SPG formation does not depend on the formation of stress granules or processing bodies. The absence or presence of glucose is sufficient to trigger assembly or disassembly of Hos2 SPGs. Among the identified components of Hos2 SPGs, Hsp42 is the first and last member observed in the Hos2 SPG assembly and disassembly processes. Hsp42 is also vital for the relocalization of the other components to Hos2 SPGs, suggesting that Hsp42 plays a central role in spatial regulation of proteins in quiescent cells.
INTRODUCTION
Many microorganisms live under constant nutrient-limited conditions in the natural environment. When confronting such harsh circumstances, cells often become quiescent (Werner-Washburne et al., 1993). Although the quiescent state allows cells to survive in the short term, these cells will gradually lose their viability if the adverse conditions continue for a prolonged period of time, a process called chronological aging, which is distinct from the replicative aging observed in dividing cells in a nutrient-rich environment (Bitterman et al., 2003; Kaeberlein, 2010).
In the absence of nutrients, stationary-phase cells are maintained in a metabolically active, highly regulated, prosurvival state. In spite of their importance, the molecular mechanisms underlying physiological changes in quiescent cells are not fully understood. Yeast cells provide a good model for studying quiescence (MacLean et al., 2001; Gray et al., 2004; Kaeberlein, 2010). Log-phase yeast cells enter the diauxic shift when glucose is exhausted, and their metabolic activity is adjusted from fermentation to respiration. After nonfermentable carbon sources are exhausted, cells will progress through one to two divisions in a couple of days and eventually enter the stationary phase (Gray et al., 2004). Apart from ceasing cell growth and division, these quiescent yeast cells differ from proliferating cells in many aspects: they have more condensed chromosomes, drastically reduced transcription and translation rates, repressed mRNA degradation, thickened and less-permeable cell walls, and higher resistance to a variety of stresses, such as heat shock and starvation (Werner-Washburne et al., 1993). In higher organisms, chronological aging occurs in postmitotic cells, such as muscle and brain cells. Studies of nondividing quiescent yeast cells may also provide insights into chronological aging in higher organisms (MacLean et al., 2001; Martinez et al., 2004; Chen et al., 2005; Fabrizio et al., 2010).
Changing the subcellular distribution of proteins to achieve efficient protein interactions or intracellular activities is one of many physiological adjustments that occur when cells encounter environmental changes. Spatial regulation of proteins can be revealed by the formation of dynamic and functional assemblies under specific conditions. Stress granules and processing bodies (P-bodies) are two well-characterized cytoplasmic RNA granules in eukaryotic cells; the assembly of the former is induced by various stresses, including heat shock, starvation, ultraviolet irradiation, and oxidative stress, while the latter can also be observed in nonstressed cells (Anderson and Kedersha, 2009; Buchan and Parker, 2009; Thomas et al., 2011). Although both stress granules and P-bodies contain nontranslated mRNA and share some protein components, these two structures essentially have different constitutions and are likely to have different functions (Anderson and Kedersha, 2009; Buchan and Parker, 2009; Thomas et al., 2011). On the other hand, studies have shown that these assemblies interact with one another and that the formation of stress granules may depend on P-bodies under certain conditions (Anderson and Kedersha, 2008; Buchan et al., 2008).
The dynamic localization of cellular proteins and the formation of a subcellular architecture have also been observed in quiescent yeast cells (Narayanaswamy et al., 2009). During stationary phase, polarized actin cables and patches change their organizations into reversible granule-like structures called actin bodies (Sagot et al., 2006), and the nuclear proteasomes move to the nuclear periphery and subsequently form cytoplasmic proteasome storage granules (Laporte et al., 2008). Both structures readily disassemble when nutrients become available, suggesting that the formation of these structures is regulated.
In addition to forming granules, cells in nutrient-deprived environments undergo extensive changes in gene expression (Choder, 1991; Gasch et al., 2000; Gray et al., 2004; Ge et al., 2010). Epigenetic regulators are probably involved in this process. Recently, many proteins involved in epigenetic regulation have been found to affect replicative aging (Haigis and Guarente, 2006). In budding yeast, activation of the histone deacetylase Sir2 or the Sir2 homologues, Hst1 and Hst2, extends the life span (Guarente, 2000; Lamming et al., 2005), whereas activation of another histone deacetylase, Rpd3, has the opposite effect (Kim et al., 1999). However, research about the connection between epigenetics and chronological aging is still limited (Sandmeier et al., 2002; Camblong et al., 2007).
To better understand whether epigenetic-related proteins are also under spatial regulation in quiescent cells, we examined the localization of many epigenetic-related proteins at different stages of yeast cell growth. We observed that the formation of reversible cytoplasmic assemblies, which we named “stationary-phase granules” (SPGs), was a widespread phenomenon in quiescent cells. We show that one subset of these granules that contain the nuclear histone deacetylase Hos2 (Hos2 SPGs) plays an important role in stationary-phase cells. Haploid hos2 mutant cells lost their viability quickly in stationary phase, and the surviving cells took longer to exit from stationary phase. The Hos2 SPGs colocalized with starvation stress granules but not with actin bodies or proteasome storage granules. However, the formation of Hos2 SPGs was not affected by mutations interfering with the formation of stress granules or P-bodies. Deletion of HSP42, but not HSP26, completely abolished the formation of Hos2 SPGs and starvation stress granules, indicating the essential role of Hsp42 in spatial regulation of specific proteins in quiescent cells.
RESULTS
Widespread reorganization of proteins into cytoplasmic granules in stationary-phase yeast cells
A previous study has shown that many cytosolic proteins change their localization once yeast cells enter the stationary phase (Narayanaswamy et al., 2009). To better understand whether epigenetic-related proteins are also under spatial regulation in quiescent cells, we monitored the subcellular localizations of 20 nuclear and cytoplasmic green fluorescent protein (GFP)-fusion proteins over a 4-wk time period (Huh et al., 2003). These proteins comprised six histone deacetylases (Hda1, Hos2, Hst1, Hst2, Rpd3, and Sir2), two histone methyltransferases (Sds3 and Set2), four Sir2-interacting proteins (Net1, Rap1, Sir3, and Yku80), two nuclear pore complex–associated proteins (Nup49 and Mlp1), a histone protein (Htb1), an RNA polymerase II subunit (Rox3), a mitochondrial protein (Sod2), and three abundant cytosolic proteins (Sod1, Tdh2, and Yca1).
Most proteins we investigated changed their localization and formed cytoplasmic granules (SPGs), in quiescent cells. This result suggests that formation of cytoplasmic granules is a common behavior for proteins in quiescent yeast cells. These 20 proteins were divided into four groups according to their localization patterns during the progression of granule formation (summarized in Supplemental Table S1 and Figure S1). Different dynamics and morphologies between these granules suggest that they are probably distinct protein assemblies.
Hos2 proteins, but not all Set3/Hos2 complex components, relocalize into SPGs upon entry into quiescence
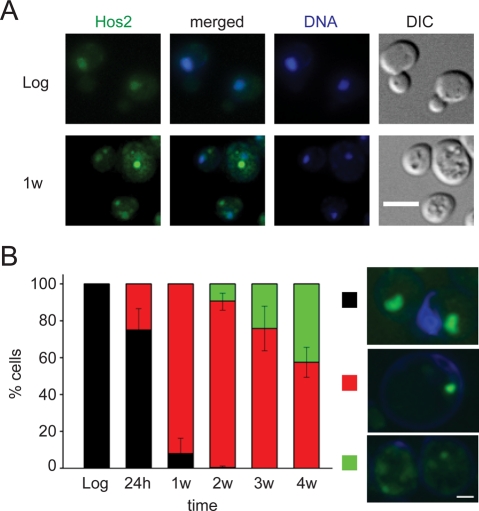
Previous studies have shown that the expression level of HOS2 is upregulated in cells during diauxic shift, early stationary phase, and certain stresses (DeRisi et al., 1997; Gasch et al., 2000). We decided to further investigate the function of the Hos2 SPGs in stationary phase (a group I SPG in Supplemental Table S1). The histone deacetylase Hos2 is an essential component of the Set3/Hos2 complex, which is involved in meiotic gene repression and is also required for the activation of many genes (Pijnappel et al., 2001; Wang et al., 2002). In log-phase cells, Hos2 is diffused in the nucleus (Figure 1A, top panels). As cells entered stationary phase, usually one or a few bright granules were visible in the cytoplasm, with only small amounts of Hos2 still remaining in the nucleus (Figure 1A, bottom panels). The proportion of cells displaying normal nuclear diffusion of Hos2 dramatically decreased after 7 d of growth, and the percentage of cells with diffuse cytoplasmic patches gradually increased at later time points (Figure 1B).
FIGURE 1:
Reorganization of Hos2 during stationary phase. (A) The deacetylase Hos2 is diffused in the nucleus in log-phase cells (top panels) but appears as a single bright Hos2 SPG in the cytosol upon entry into stationary phase (bottom panels). Cells were fixed and stained with 4′,6-diamidino-2-phenylindole (blue) to locate the nuclei. Scale bar: 5 μm. (B) Changes of Hos2 localization patterns in different growth stages. Cells were grown in YPD medium at 28°C and collected in log phase, diauxic shift (24 h), and stationary phase (1–4 wk). For each time point, the localization pattern of Hos2-GFP is classified as nuclear diffusion (black), single or a few bright Hos2 SPGs (red), or cytoplasmic patches (green). At least 125 cells were counted for each time point, and the mean values were calculated using data from three independent experiments. Typical images for each localization pattern are shown on the right. Hos2 proteins are shown in green. Error bars represent SD. Scale bar: 2 μm.
Two experiments were conducted to ensure that our observations represented the behavior of wild-type Hos2 protein. First, we tested whether the Hos2-GFP protein could rescue the hypersensitivity of hos2 mutants to secretory stress when grown on a tunicamycin-containing plate (Cohen et al., 2008). We observed that hos2 cells carrying the HOS2-GFP construct were as resistant to tunicamycin as wild-type cells, indicating that the Hos2-GFP protein is functional. Next we fused the Hos2 protein with a 13×Myc tag and examined the protein localization using immunostaining. A similar localization pattern was observed, indicating that our previous data were not due to a GFP-specific artifact (Figure S2).
To determine whether other components of the Set3/Hos2 complex were relocalized to the same SPGs, stationary-phase cells coexpressing Hos2-mCherry with Cpr1-, Hos4-, Set3-, Sif2-, or Snt1-GFP fusion proteins were examined following growth in yeast–peptone–dextrose (YPD) medium for 1 wk (Figure S3). Both Set3-GFP and Sif2-GFP reorganized into SPGs upon entry into quiescence, and a proportion of these SPGs colocalized with Hos2 SPGs (61% of Set3-GFP dots and 74% of Sif2-GFP dots, n = 81–87). In contrast, Hos4-GFP, Snt1-GFP, and Cpr1-GFP formed dots occasionally but did not colocalize with Hos2 SPGs. We also examined the formation of Hos2 SPGs in set3∆ mutant cells in which the Set3/Hos2 complex is defective. Deletion of SET3 did not affect Hos2 SPG formation, suggesting that the assembly of Hos2 SPGs is distinct from the Set3/Hos2 complex.
A hos2 mutation affects cell viability and exit from stationary phase in quiescent cells
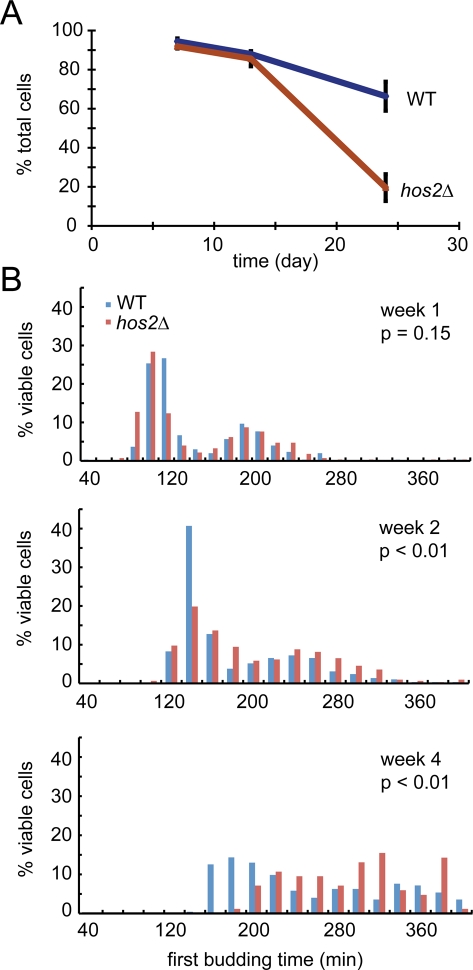
The Hos2 protein has been shown to function as a meiosis-specific repressor and an essential component of the secretory stress response (Pijnappel et al., 2001; Cohen et al., 2008). However, its role in cells during stationary phase is unknown. To address this question, we used in vivo time-lapse imaging to test whether a hos2 mutation would affect the survival or recovery of quiescent cells. Haploid wild-type and hos2 mutant cells were cultured in YPD medium for 4 wk. During this period, cells were collected at different time points and their recovery from stationary phase to log phase was examined (see Materials and Methods for details). At the early time points (7 and 13 d), both wild-type and hos2 mutant cells maintained a high degree of viability. However, the cell viability in hos2 mutant cells was dramatically reduced after 24 d in YPD medium (Figure 2A). The loss of viability was not simply due to acidification of the medium, as a constant pH was maintained throughout the 4-wk period (Burtner et al., 2009).
FIGURE 2:
Hos2 is important for cell viability and growth recovery in stationary-phase cells. Stationary-phase cell cultures were diluted in fresh YPD, and cell growth was monitored using time-lapse microscopy. (A) hos2 mutant cells exhibited lower viability after 24 d in stationary phase. Cells were monitored for 6 h after refeeding with nutrients, and only rebudding cells were counted as viable. At least 100 cells were counted for each experiment. Data represent the mean ± SEM of three biological replicates. (B) hos2 mutant cells resume mitosis more slowly after 13 d in stationary phase. The first rebudding time of each viable cell was recorded, and data from three independent biological repeats were pooled to generate the histograms. At least 200 cells were counted in each sample, except for the hos2 deletion strain in week 4 (n = 84). p values were calculated using the Mann-Whitney test.
We also analyzed the recovery process by determining the time required for cells to reenter the mitotic cell cycle after nutrient addition. In contrast to the viability assay, a significant difference between wild-type and hos2 mutant cells was detected in the 13-d samples (Mann-Whitney test, p < 0.01). The quiescent hos2 mutant cells took longer to reenter mitosis, and the delay increased as the cells aged (Figure 2B). We always observed two distribution patterns in the time of recovery, implying that the cell population was composed of two types of quiescent cells in stationary phase.
Hst2 and Yca1 colocalize with Hos2 SPGs during stationary phase
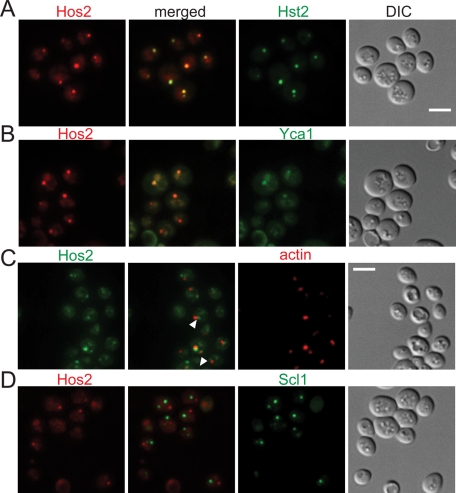
In our protein localization analyses, both Hst2-GFP and Yca1-GFP exhibited patterns similar to those of Hos2-GFP, suggesting they share similar characteristics during granule formation. This observation prompted us to investigate the possible association of Hos2 SPGs with either Hst2 or Yca1 protein. In 1-wk-old, nutrient-deprived cells containing Hos2 SPGs (1-wk cells), most Hos2 SPGs colocalized with Hst2-GFP (95 ± 5%, n = 194) and Yca1-GFP (91 ± 7%, n = 158; Figure 3, A and B). On the other hand, when we examined the Sir2 SPG, a cytoplasmic granule from another group (group II in Table S1), no significant colocalization with Hos2 SPGs or Yca1 SPGs was observed (Figure S4). These results suggest these proteins do not assemble by chance, and components of specific granules might be invariant.
FIGURE 3:
Hos2 SPGs colocalize with Hst2 and Yca1, but not with actin bodies and proteasome storage granules. (A and B) Hos2-mCherry colocalizes with Hst2-GFP (95 ± 5%, n = 194) and Yca1-GFP (91 ± 7%, n = 158) in stationary-phase cells. (C) Actin bodies are often partially colocalized with or adjacent to Hos2 SPGs (indicated by white arrowheads; 46 ± 17%, n = 81). Stationary-phase cells expressing Hos2-GFP were fixed and stained with Texas Red-X phalloidin to detect actin bodies. (D) Hos2 SPGs do not colocalize with proteasome storage granules (Scl1-GFP). The numbers in parentheses represent the mean ± SD of the proportion of total Hos2 dots that colocalize with (or partially colocalize with or are adjacent to, as seen in (C)) the other proteins in three biological replicates. Scale bars: 5 μm.
Hos2 SPG in quiescent cells is a reversible structure
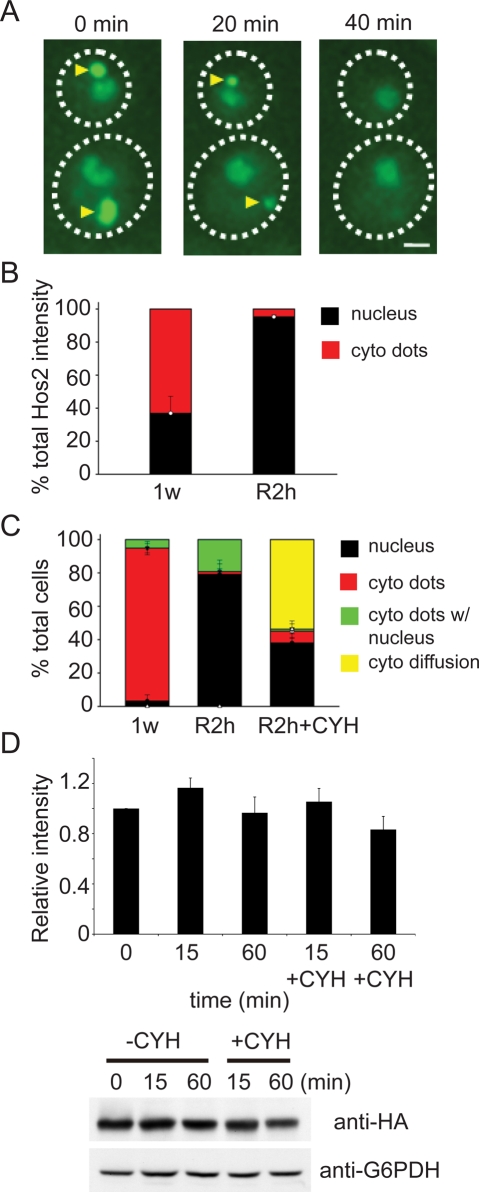
We next asked whether the SPGs in quiescent yeast cells are simply aggregates or specific, regulated structures. If the SPGs are formed by impaired or denatured proteins, it is unlikely that these proteins will be relocalized back to the nucleus when cells resume mitosis. On the other hand, a reversible granule suggests specific functions in quiescence. To clarify this issue, we monitored the localization of Hos2-GFP during the recovery process of 1-wk cells. After refeeding with nutrients, most Hos2 SPGs quickly dissolved, and the characteristic pattern of log-phase cells could be observed within 40 min (Figure 4A). Interestingly, no cells with Hos2 SPGs showed signs of rebudding, suggesting that disassembly of Hos2 SPGs may be crucial for cells to reenter mitosis.
FIGURE 4:
Hos2 granules are reversible upon nutrient replenishment. (A) Time-lapse images of two recovering cells. Hos2-GFP–expressing cells were grown to stationary phase and then cultivated on a Lab-Tek chambered coverglass with fresh medium. Hos2 SPGs (yellow arrowheads) quickly disassembled once nutrients were replenished. Scale bar: 2 μm. (B) Changes in the distribution of Hos2-GFP before (1w) and after (R2h) recovery from stationary phase. The fluorescence intensity of Hos2-GFP in Hos2 SPGs and nuclei was quantified (see Materials and Methods and Figure S5 for details), and the ratios between these two intensities are shown. Error bars represent SD. (C) Frequency of cells that have different Hos2-GFP localization patterns before and after recovery from stationary phase. Stationary-phase cells were preincubated with (R2h + CYH) or without (R2h) cycloheximide for 1 h, washed, resuspended in fresh YPD medium with (R2h + CYH) or without (R2h) cycloheximide for 2 h, and then at least 100 cells were analyzed in each experiment. Data represent the mean ± SD of three biological replicates. (D) Hos2 is not degraded during disassembly of Hos2 SPGs. One-week stationary-phase cells carrying 3×HA-tagged Hos2 were resuspended in fresh medium and collected at different time points with or without cycloheximide (35 μg/ml). Protein lysates were hybridized with antibodies against HA or G6PDH. Relative ratios of Hos2/G6PDH intensity are shown. The signal intensity of the Western blot was quantified; all the ratios were normalized to the ratio of the 0 min sample. One example of the Western blot is shown. Data represent the mean ± SEM of three biological replicates.
In 1-wk cells, more than 60% of cellular Hos2 protein was found in cytoplasmic SPGs. By contrast, Hos2 protein was nearly exclusively restricted to the nucleus after 2 h of replenishment with nutrients (Figures 4B and S5). In addition, Hos2 SPGs could be detected in over 95% of 1-wk cells. After 2-h refeeding, the percentage of cells containing Hos2 SPGs was drastically reduced to ∼20% (Figure 4C). In those cells that still contained Hos2 SPGs, the size and intensity of the granules were significantly decreased. We also measured the total amount of Hos2 in recovering cells. No obvious change was detected during the process of recovery, suggesting the Hos2 protein in SPGs was not degraded, but rather was redistributed from SPGs to the nucleus (Figure 4D).
When 2-wk cells were examined, we found that Hos2 SPGs took longer to disassemble in the recovering cells, which was consistent with our observation that older cells had a longer delay in reentering mitosis (Figure 2B). In the recovery experiments, we also monitored the other Hos2 SPG components, Hst2 and Yca1. Both proteins showed patterns similar to those of Hos2 (unpublished data).
Protein synthesis is not required for Hos2 SPG disassembly
To determine whether the recovered proteins were preexisting or newly synthesized, we transferred 1-wk cells into fresh medium in the presence of cycloheximide, an inhibitor of protein synthesis. After 2 h, Hos2 redistributed into the nucleus in nearly 40% of cells, indicating that nuclear Hos2 protein in the recovering cells was not newly synthesized (Figure 4, C and D). Furthermore, even in the presence of cycloheximide, only a small proportion of cells still contained Hos2 SPGs, suggesting that disassembly of these granules was translation-independent. However, unlike the typical nuclear diffusion observed in cells without cycloheximide treatment, Hos2 molecules in many drug-treated cells (56%) remained in the cytosol without a significant change in total protein quantity. It is therefore likely that interference with protein translation leads to inefficient translocation of proteins back into the nucleus.
Glucose is a critical factor determining assembly and disassembly of Hos2 SPGs
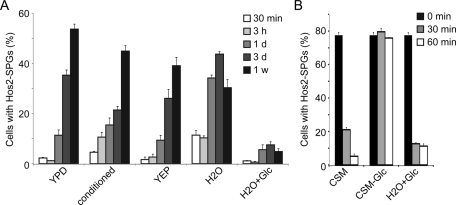
A previous study showed that metabolic status controls the entry into and exit from the quiescent state (Laporte et al., 2011). To understand which nutrients or chemicals trigger assembly of Hos2 SPGs, log-phase cells were resuspended in conditioned medium (from 2-wk cultures), yeast–peptone medium (YEP, rich medium but lacking any carbon source), H2O, or H2O plus 2% glucose. Cells were able to form Hos2 SPGs in the conditioned medium, YEP, or H2O, but not in H2O plus 2% glucose (Figure 5A). These results suggest that depletion of glucose is sufficient to induce the Hos2 SPG formation. Nonetheless, the kinetics of Hos2 SPG formation in different conditions is not exactly the same, suggesting that some other factors also may be involved. We also tested the effect of glucose on disassembly of Hos2 SPGs. Stationary-phase cells were resuspended in H2O plus 2% glucose, complete synthetic medium (CSM), and CSM-glucose, and the Hos2 SPGs were monitored. Hos2 SPGs quickly disassembled within 1 h of suspension in H2O plus 2% glucose or CSM, but did not disassemble in CSM-glucose (Figure 5B). Altogether, our results indicate that glucose is a critical factor for assembly or disassembly of Hos2 SPGs.
FIGURE 5:
Glucose is a critical factor for assembly and disassembly of Hos2 SPGs. (A) Depletion of glucose is sufficient to induce the Hos2 SPG formation. Log-phase cells were resuspended in conditioned medium (from 2-wk cultures), YEP, H2O or H2O + 2% glucose, and the formation of Hos2 SPGs was examined after 1 wk. (B) Glucose triggers disassembly of Hos2 SPGs. Stationary-phase cells were resuspended in H2O + 2% glucose, CSM, or CSM-glucose, and the Hos2 SPGs were monitored at different time points. At least 100 cells were counted at each time point. Data represent the mean ± SEM of three biological replicates.
Hos2 SPGs do not colocalize with actin bodies or proteasome storage granules
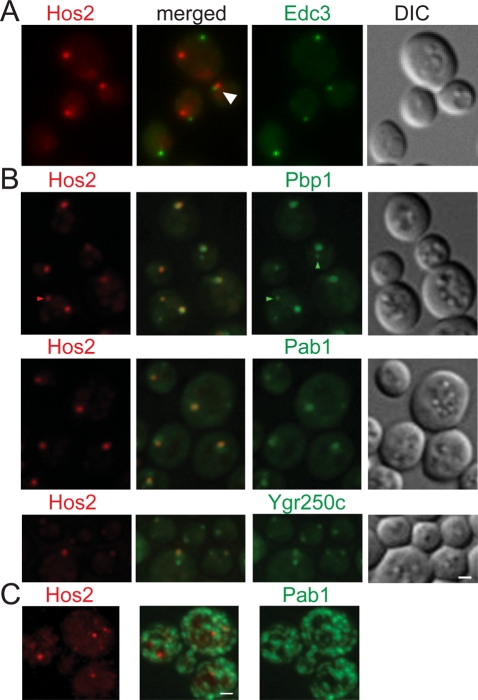
The actin cytoskeleton is central for intracellular protein localization. F-actin and several actin-binding proteins have been found to assemble into cytoplasmic structures called actin bodies in quiescent cells (Sagot et al., 2006). Actin localization during stationary phase was examined in cells expressing Hos2-GFP. We found that Hos2 SPGs did not colocalize with actin bodies but that many Hos2 SPGs (46 ± 17%, n = 81) partially overlapped with or were located adjacent to actin bodies (Figure 3C). Similar results were obtained for Hst2 and Yca1, but not for Scl1 (10 ± 6%, n = 342), a component of proteasome storage granules, or Edc3 (9 ± 3%, n = 370), a component of P-bodies.
Proteasomes in yeast are also known to reorganize during the stationary phase into cytoplasmic structures called proteasome storage granules (Laporte et al., 2008). We found that Hos2 did not colocalize with Scl1, indicating that Hos2 SPGs and proteasome storage granules are distinct structures (Figure 3D).
Hos2 proteins colocalize with stress granules during stationary phase but not under heat-shock conditions
It has been reported that the formation of P-bodies and stress granules is triggered by entry into stationary phase in yeast cells (Teixeira et al., 2005; Brengues and Parker, 2007). We therefore tested whether there was any interaction between Hos2 SPGs and these two structures. Our results showed that Edc3, a central component of P-bodies, did not colocalize with the Hos2 SPG in 1-wk cells. However, we also observed that a proportion of Hos2 SPGs were adjacent to the Edc3 signal (38 ± 7%, n = 329), implying an interaction between these two types of granules (Figures 6A and S6). By contrast, Hos2 SPGs colocalized with the components of stress granules (Figure 6B), Pab1 (86 ± 6%, n = 204), Pbp1 (99 ± 1%, n = 202), and Ygr250c (96 ± 2%, n = 219). The formation of stress granules is greatly reduced in pbp1 mutant cells (Buchan et al., 2008). However, we found that deletion of PBP1 did not affect the formation of Hos2 SPGs in quiescent cells (Figure 7A).
FIGURE 6:
Hos2 SPGs do not colocalize with P-bodies but colocalize with stress granules during stationary phase. (A) Hos2 SPGs do not colocalize with P-bodies (Edc3-GFP), but a proportion of Hos2 SPGs (38 ± 7%, n = 329) are interacting with Edc3 dots (white arrowhead). (B) Hos2-mCherry colocalizes with the components of stress granules, Pab1-GFP (86 ± 6%, n = 204), Pbp1-GFP (99 ± 1%, n = 202), and Ygr250c-GFP (96 ± 2%, n = 219). Additional small dots of Hos2, Pbp1, and Pab1 are occasionally found in some cells (red and green arrowheads). Ygr250c normally forms two dots, one of which colocalizes with Hos2 SPGs. The numbers in parentheses represent the mean ± SD of the proportion of total Hos2 dots that colocalize with the other proteins in three biological replicates. (C) Heat shock–induced stress granules are morphologically distinct from those formed during stationary phase and do not colocalize with Hos2 foci. Scale bars: 2 μm.
FIGURE 7:
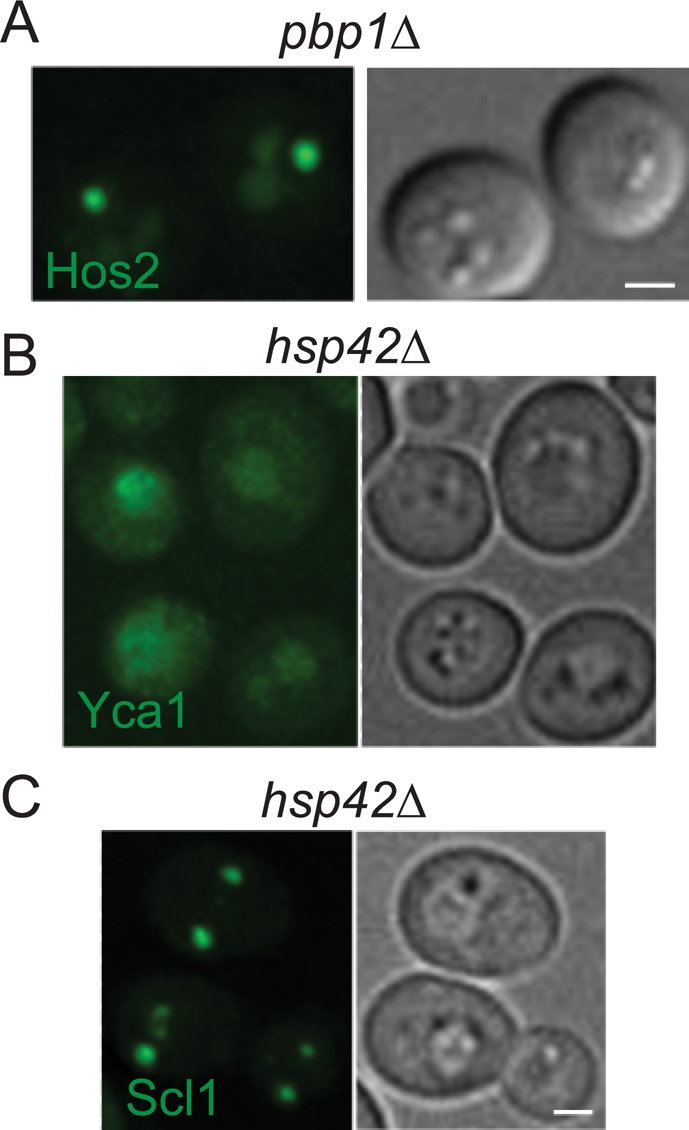
SPG formation in different mutants. (A) Formation of Hos2 SPGs is not affected in pbp1∆ mutant cells. (B) Yca1 is unable to form granules in stationary-phase hsp42∆ cells. (C) Formation of proteasome storage granules (Scl1-GFP) is not affected in hsp42∆ mutant cells. Scale bars: 2 μm.
Many other stresses, including heat shock, are known to induce the formation of stress granules (Buchan and Parker, 2009). To clarify whether Hos2 also relocalizes into stress granules upon heat shock, we incubated log-phase cells at 37°C or 46°C. No alteration in the localization of Hos2 was observed at 37°C for up to 2 h. On the other hand, robust heat shock at 46°C induced the formation of cytoplasmic punctate foci of Hos2 within 15 min (Figure 6C). The pattern of these punctate foci was different from that observed during quiescence, as they consisted of a larger number of tiny foci scattered in the cytoplasm, rather than one to two large, bright dots. Although heat shock–induced stress granules also display a punctate pattern (Grousl et al., 2009), there was no apparent colocalization between these two types of foci (Figure 6C). These results suggest that Hos2 SPGs (and starvation stress granules) have distinct structures from previously characterized heat shock–induced stress granules.
Heat-shock proteins Hsp26 and Hsp42 colocalize with Hos2 SPGs in quiescent cells
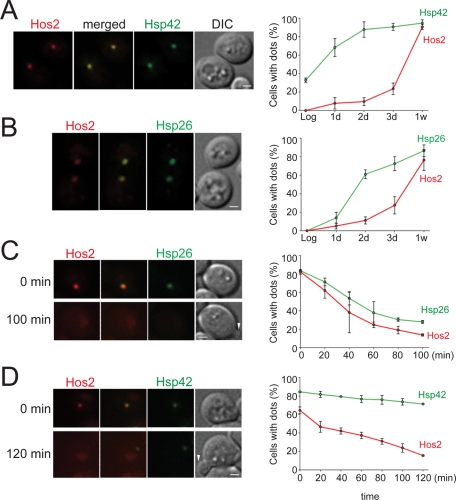
Molecular chaperones are essential for the assembly and maintenance of many multiprotein complexes (Ellis and Minton, 2006; Dezwaan and Freeman, 2008; Gong et al., 2009). Previous studies have shown that the expression levels of two small heat-shock proteins, Hsp26 and Hsp42, are highly induced in stationary phase (DeRisi et al., 1997; Gasch et al., 2000). Thus we decided to examine whether these two chaperones play a role in the formation of Hos2 SPGs. In log-phase cells, both Hsp26 and Hsp42 proteins diffused in the cytosol. On entry into stationary phase, these proteins aggregated into one or a few bright dots. When protein localization was examined in cells expressing both Hos2-mCherry and Hsp26-GFP or Hsp42-GFP, we observed that Hos2 SPGs were highly colocalized with Hsp26 (90 ± 8%, n = 500) and Hsp42 (99 ± 1%, n = 1053; Figure 8, A and B). This result suggests that both Hsp26 and Hsp42 participate in the formation of Hos2 SPGs.
FIGURE 8:
Hsp26 and Hsp42 colocalize with Hos2 SPGs. Cells expressing Hos2-mCherry and Hsp42-GFP or Hsp26-GFP were grown to different stages, and the kinetics of SPG assembly and disassembly were analyzed. (A and B) Both Hsp42-GFP (99 ± 1%, n = 1053) and Hsp26-GFP (90 ± 8%, n = 500) colocalize with Hos2-mCherry in stationary-phase cells. Hsp42 and Hsp26 form SPGs earlier than Hos2-mCherry (n = 78–305). Data represent the mean ± SD of three biological replicates. Scale bars: 1 μm. (C and D) Hos2 protein is released earlier from SPGs than Hsp26 and Hsp42 proteins upon nutrient replenishment. Far left, time-lapse images of cells recovering from stationary phase. Both Hos2-mCherry and Hsp26-GFP dots disappear within 100 min (n = 72), but most Hsp42-GFP dots remain unchanged (n = 136). Far right, quantitative analyses of time-lapse images. Data represent the mean ± SD of two biological replicates. White arrowheads indicate emerging buds. Scale bars: 2 μm.
We subsequently compared the time course of Hos2 SPG formation in cells collected during a 1-wk interval. The proportion of cells exhibiting Hsp26 or Hsp42 dots increased rapidly within the first 3 d of culturing (Figure 8, A and B). In particular, the percentage of cells with Hsp42 dots was nearly identical at 2 d and at 1 wk, indicating that almost all Hsp42 observed in 1-wk cells had already formed within the first 2 d. In contrast to the Hsp26 and Hsp42 signals, which escalated rapidly, Hos2 SPGs formed slowly during the first 2 d of growth; however, formation of Hos2 SPGs significantly increased afterward (Figure 8, A and B). These data reveal that Hsp26 and Hsp42 assemble into SPGs before Hos2 during stationary phase.
Next we examined the behavior of Hsp26 and Hsp42 proteins during cell recovery. After 1-wk cells were provided with nutrients, Hsp26 dots gradually disappeared within 100 min, with a rate similar to that of the disappearance of Hos2 dots (Figure 8C). In contrast, most of the Hsp42 signal was sustained for the duration of the 120-min recovery time (during which more than 50% of cells rebudded; Figure 8D). Although cells did not rebud until Hos2 had fully relocated, the onset of rebudding did not require the disassembly of the Hsp42 granules. Overall, Hsp26 and Hsp42 granules assembled earlier and disassembled later than Hos2 SPGs, raising the possibility that these two chaperones might serve as scaffolding proteins in the assembly of Hos2 SPGs.
Formation of Hos2 SPGs and stress granules during stationary phase is Hsp42-dependent
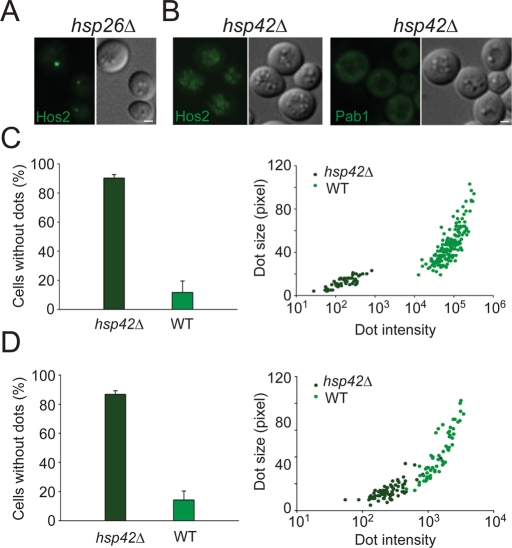
The localization pattern of Hsp26 and Hsp42 prompted us to investigate the effect of these two chaperones on the formation of Hos2 SPGs. As shown in Figure 9A, hsp26∆ cells exhibited no defects in Hos2 SPG formation. In contrast, Hos2 relocalized from the nucleus to the cytoplasm in hsp42 mutant cells, but was unable to assemble into typical SPGs during stationary phase (Figure 9B). The proportion of quiescent cells possessing detectable cytoplasmic Hos2 SPGs was reduced from 88 ± 8% in wild-type cells to 10 ± 2% in hsp42∆ cells (Figure 9C). Even though some dot-like structures could still be observed in a minority of hsp42∆ cells, their average size and fluorescence intensity were reduced when compared with wild-type cells (Figure 9C). In hsp42∆ cells, Yca1 also failed to reorganize into SPGs during stationary phase (Figure 7B). Only 16 ± 7% of quiescent mutant cells had detectable Yca1 SPGs, in contrast to 65 ± 7% of wild-type cells. Again, the dot size and dot intensity of Yca1-GFP were reduced.
FIGURE 9:
Formation of Hos2 SPGs depends on Hsp42. (A) Hos2 SPG formation is not affected in stationary-phase hsp26∆ cells. (B) Hos2 and Pab1 (a component of stress granules) are unable to form granules in stationary-phase hsp42∆ cells. Scale bars: 2 μm. (C) hsp42∆ cells display a severe defect in the formation of Hos2 SPGs. Left, most hsp42∆ cells do not form Hos2 SPGs (hsp42∆, n = 253, two experiments; WT, n = 235, two experiments). Right, dot-like structures of Hos2 that formed in hsp42∆ cells are drastically reduced in size and intensity (arbitrary units; hsp42∆, n = 50, two experiments; WT, n = 164, two experiments). Each data point represents a measurement from a single cell. (D) hsp42∆ cells also display a severe defect in the formation of stationary-phase stress granules (Pab1-GFP). Left, most hsp42∆ cells do not form stress granules (hsp42∆, n = 649, two experiments; WT, n = 401, two experiments). Right, dot-like structures of Pab1 that formed in hsp42∆ cells are drastically reduced in size and intensity (hsp42∆, n = 76, two experiments; WT, n = 60, two experiments). Scale bars: 2 μm.
The above data, together with our prior results showing colocalization of stress granules and Hos2 SPGs in stationary-phase cells, led us to question whether Hsp42 is also critical for the assembly of starvation stress granules. We observed that Pab1 failed to assemble into granules in most hsp42∆ quiescent cells (Figure 9, B and D). In the few cells that contained visible stress granules, both the dot size and dot intensity were largely decreased (Figure 9D), similar to those seen in Hos2 SPGs.
To examine whether Hsp42 is required for the formation of other types of SPGs, we checked for the formation of proteasome storage granules using Scl1-GFP–expressing cells. As shown in Figure 7C, deletion of HSP42 did not cause any defect in the formation of proteasome storage granules in quiescence. Our results indicate that the small heat-shock protein Hsp42 plays a central and specific role in Hos2 SPG assembly.
Finally, we asked whether deletion of HOS2 would affect the formation of SPGs or P-bodies. In hos2 mutant cells, we did not observe any obvious defect in the formation of Hsp42, Hsp26, Pbp1, Pab1, and Dcp2 dots (Figure S7). Nonetheless, it remains possible that Hos2 has some subtle effects on these structures that cannot be detected by our current assays.
DISCUSSION
Spatial regulation of proteins in quiescent cells
Our study shows a widespread reorganization of nuclear and cytoplasmic proteins into various cytoplasmic SPGs in quiescent yeast cells, indicating a unique pattern of spatial and temporal regulation. Although it is well known that mitotic cells have evolved precise spatial regulation of many proteins, the spatial aspects of quiescence are underappreciated. Several previous studies have reported similar phenomena in yeast and bacteria (Wilkinson et al., 1998; Patel et al., 2004; Sagot et al., 2006; Laporte et al., 2008; Narayanaswamy et al., 2009; Tudisca et al., 2010). Cytoplasmic structures, such as stress granules or P-bodies, are also observed in a wide range of eukaryotic cells (Buchan and Parker, 2009). It is possible that some of these SPGs performing specific functions in quiescent cells are conserved structures in different organisms.
It is surprising to observe that many nuclear proteins accumulate in cytoplasmic SPGs. Quiescent yeast cells have a reduced overall transcriptional activity and condensed chromosomes. The nucleus probably undergoes a certain level of “shutdown” to conserve energy in quiescence (Choder, 1991; Gasch et al., 2000; Gray et al., 2004; Ge et al., 2010). It is possible that quiescent cells sequester some transcription factors and other nuclear proteins with diverse activities in cytoplasmic compartments to provide a means for quick response to environmental changes. Alternatively, the export and subsequent cytoplasmic sequestration of unnecessary nuclear proteins may provide a means to maintain nuclear integrity when the nucleus encounters hostile conditions.
In quiescent yeast cells, the compartmentalization process of nuclear proteins is highly ordered. Newly formed SPGs are generally juxtaposed with the nucleus, while they move farther away from the nucleus in later stages. This finding implies that during stationary phase, nuclear proteins pass through the nuclear envelope and immediately form assemblies instead of diffusing freely in the cytoplasm. Likewise, SPGs move to the vicinity of the nucleus prior to disassembly, which may accelerate redistribution of proteins.
Assembly and disassembly of Hos2 SPGs
The cytoplasm is a very crowded environment, consisting of many soluble and insoluble proteins, RNA species, and other macromolecules (Minton, 2001). Environmental stresses may result in an even more congested state, causing proteins to aggregate spontaneously (Ellis and Minton, 2006). We argue that the formation of Hos2 SPGs does not simply result from nonspecific protein aggregation. First, Hos2 SPGs are reversible structures that disassemble immediately when nutrients are replenished. Protein aggregation usually cannot be reversed in such a manner (Narayanaswamy et al., 2009; Thomas et al., 2011). Second, the assembly and disassembly of Hos2 SPGs are regulated by the presence of glucose and also exhibit specific kinetics and patterns, both of which are not common characteristics of nonspecific protein aggregation (An et al., 2008; Erickson and Lykke-Andersen, 2011). Third, we observed that Hos2 SPGs colocalize only with starvation stress granules, but not with other previously identified granule-like structures. It is likely that Hos2 SPGs share some structures (and functions) with stress granules, even though their components do not overlap exactly.
Molecular chaperones are known to be involved in many protein-folding processes (Sakahira et al., 2002; Gong et al., 2009). However, their assisting role in assembling folded proteins into functional oligomeric structures is less well studied (Ellis, 2006). In this paper, we demonstrated that deletion of the molecular chaperone Hsp42 dramatically reduced the formation of Hos2 SPGs (and starvation stress granules). How does Hsp42 function in the formation of these granules? Small heat-shock proteins, such as Hsp42 and Hsp26, assemble into remarkably versatile dynamic oligomers with diverse subunit numbers and architectures (Haslbeck et al., 2005; Sun and MacRae, 2005). One can imagine that the chaperone activity of Hsp42 may merely function to facilitate the assembly of Hos2 SPGs. However, the specific kinetics and colocalization pattern of Hsp42 and Hos2 and the strong dependence of Hos2 SPG formation on Hsp42 suggest a more direct role for Hsp42.
We speculate that Hsp42 may function as a scaffolding molecule that promotes protein–protein interactions (Eyles and Gierasch, 2010; Stengel et al., 2010). Under this hypothesis, the stationary phase induces the remodeling of Hsp42 oligomeric architectures; small Hsp42 oligomers reorganize to form larger oligomers and further agglomerate into Hos2 SPGs. Binding to clients (i.e., early recruited proteins, such as Hsp26) can facilitate the formation of this complex. We propose that the chaperone activities of Hsp42 and Hsp26 ensure the fidelity of the assembly process by preventing improper interactions and providing protection for the constitutive proteins. Interestingly, Hsp42 has been shown to interact with some potential stress granule or P-body components, such as Lsm12, Mkt1, and Sgt2 in previous large-scale studies (Gavin et al., 2006; Collins et al., 2007; Costanzo et al., 2010).
Many heat-shock proteins have been proposed as actors in the formation of stress granules (Cuesta et al., 2000; Gilks et al., 2004; Suzuki et al., 2009). It is likely that a sophisticated chaperone network coordinates the formation of different granules under various stresses. Moreover, since Hsp26 and Hsp42 are exclusively present in Hos2 SPGs, it will be interesting to see whether other granule-like structures utilize different sets of chaperones in their assemblies.
Functions of Hos2 SPGs
The negative correlation between the presence of Hos2 SPGs and cell rebudding suggests a specific role for Hos2 SPGs in the regulation of quiescence. We observed that deletion of HOS2 reduced the viability and delayed the recovery of quiescent cells. Evidence from other studies also indicates that other components of Hos2 SPGs are critical for stationary-phase cells. A large-scale screen found that deletion of SET3 or SIF2 shortens the chronological life span of mutant cells (Powers et al., 2006). The viability of aged stationary-phase cultures is compromised when Yca1 is mutated (Herker et al., 2004). Mutations in Hsp26 or Hsp42 cause accelerated aging phenotypes in early stationary-phase yeast cells (Haslbeck et al., 2004). During entry to, maintenance of, and exit from quiescence, cells may require wholesale reprogramming of regulatory networks and remodeling of intracellular structures (Gray et al., 2004). It is possible that the Hos2 deacetylase activity is involved in one or multiple steps during quiescence regulation.
How is the formation of Hos2 SPGs correlated to the structural and functional adaptations of quiescent cells? Hos2 SPGs may serve as temporary depositories for the storage of proteins under harsh conditions, as has been suggested for actin bodies, assemblies of metabolic enzymes, and proteasome storage granules (Sagot et al., 2006; Laporte et al., 2008; Narayanaswamy et al., 2009). When the stresses are relieved, the stored proteins can be released and readily perform their functions, accelerating the exit from quiescence and return to proliferation.
To cope with starvation, nonselective autophagy is activated in yeast cells (Cebollero and Reggiori, 2009; Nakatogawa et al., 2009). A protective mechanism that can prevent cells from degrading proteins vital for stress response or quiescence exit is important. Indirect evidence from a previous study shows that cytoplasmic protein granules are not designated to undergo autophagy (Narayanaswamy et al., 2009), suggesting that these granules formed in quiescence are shelters from autophagic degradation. In agreement with this study, the protein level of Hos2 in Hos2 SPGs is fairly stable throughout the first few weeks of the stationary phase (Figure 10; unpublished data). By contrast, a majority of ribosomal subunits that do not form cytoplasmic assemblies during stationary phase are degraded by autophagy upon nutrient deprivation (Kraft et al., 2008). Markedly, Hos2-GFP and Yca1-GFP signals are principally enriched in vacuoles in hsp42∆ cells when they fail to form SPGs (Figures 9B and 7B).
FIGURE 10:
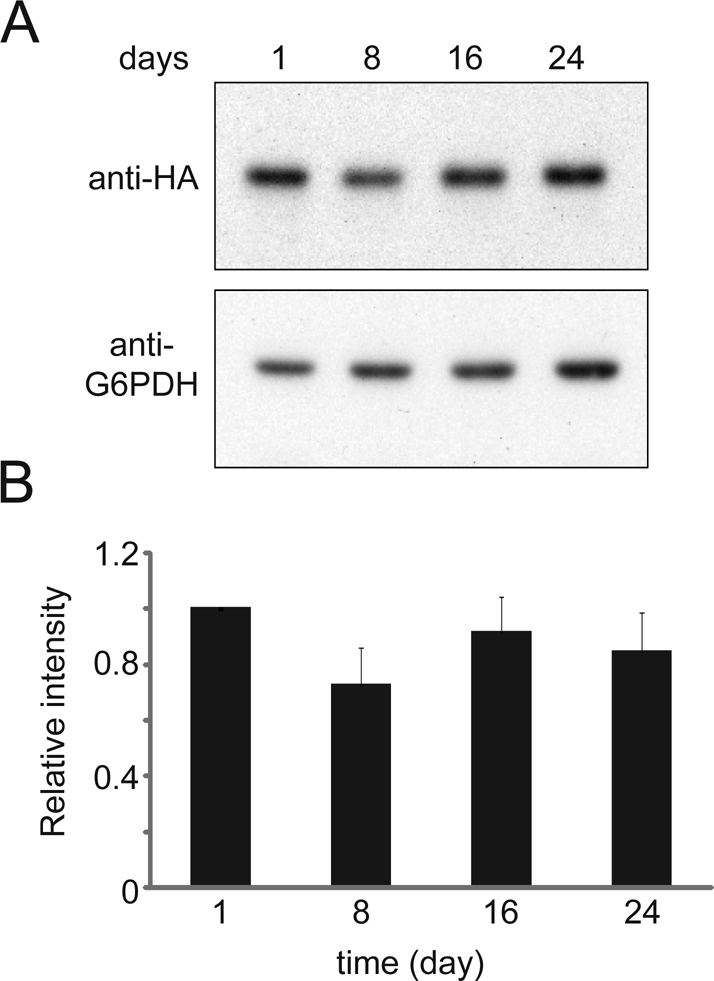
Hos2 protein is stable throughout the first few weeks of stationary phase. (A) Cells carrying 3×HA-tagged Hos2 were grown in YPD medium at 28°C and collected after 1, 8, 16, and 24 d. Protein lysates were hybridized with antibodies against HA or G6PDH. (B) Relative ratios of Hos2/G6PDH intensity. The signal intensity of the Western blot was quantified, and all the ratios were normalized to the ratio of the 1-d sample.
Formation of Hos2 SPGs may also remove proteins from their original locations. The Tup1-Ssn6 corepressor, a Hos2-interacting complex, has been shown to repress the expression of a diverse set of genes, including 10% of the genes that are induced by at least twofold after the diauxic shift (DeRisi et al., 1997; Davie et al., 2003). It is possible that the Hos2 in Hos2 SPGs sequesters the Tup1-Ssn6 complex from the nucleus and thus derepresses the stationary-phase genes.
Alternatively, relocalization of proteins into Hos2 SPGs may be linked to the regulation of stress responses. The formation of Hos2 SPGs vastly increases the local concentration of proteins and provides a compartment in which they are in proximity to substrates, which may coordinate and facilitate biochemical reactions when metabolites become limited and the cell interior becomes more crowded. Yca1 has been suggested as playing a role in protein quality control. Deletion of YCA1 results in an increased level of protein aggregation and autophagic bodies (Lee et al., 2010). Stress granules are also involved in monitoring mRNA quality and translation. It is possible that the Hos2 SPG functions as a quality control center that can determine which proteins or RNAs need to be degraded, repaired, or stored in quiescent cells. Although different quality control pathways already exist in log-phase cells, it is more critical to coordinate these pathways when cells face harsh conditions.
Posttranslational modification of colocalized components in Hos2 SPGs provides an economical, rapid, and reversible means to adjust protein function during stress. A previous study showed that a cytoplasmic deacetylase, HDAC6, functions in coordinating other stress granule components during granule formation. Impairing the activities of HDAC6 abolishes stress granule formation (Kwon et al., 2007). Although deletion of HOS2 shows no obvious effects on the formation of Hsp42 SPGs or stationary-phase stress granules, these results do not rule out the possibility that Hos2 and Hst2 have some regulatory roles in the structure or dynamics of these granules. This will be a critical area for future studies.
The widespread reorganization of nuclear and cytoplasmic proteins into reversible assemblies indicates that it is a universal mechanism for coping with starvation. This phenomenon further strengthens the idea that quiescent cells are exquisitely organized and dynamic, rather than homogeneous and static. Protein deacetylases have been shown to play an important role in regulating various biological pathways (Mellert and McMahon, 2009; Imai and Guarente, 2010; Guan and Xiong, 2011). Our results provide evidence that localization of protein deacetylases, such as Hos2 and Hst2, is highly regulated and may contribute to specific regulation in quiescent cells. By further dissecting the functions of Hos2 SPGs and protein deacetylase activities in quiescent cells, we may be able to uncover innovative pathways that cells use to fight chronological aging.
MATERIALS AND METHODS
Yeast strains and growth conditions
All Saccharomyces cerevisiae haploid strains used in this study are listed in Table S2. C-terminally GFP- or mCherry-tagged strains were obtained either from a chromosomal GFP-tagged library (Huh et al., 2003) or constructed. The GFP or mCherry tags were inserted in-frame at the C-terminus of the coding region of a gene, as described previously (Howson et al., 2005), and constructs were verified by PCR. All fusion proteins were expressed under their endogenous promoters.
Cells were cultured at 28°C with aeration in liquid YPD or CSM media for 2.5–3 h (log phase), 24 h (diauxic shift), or 1–4 wk (stationary phase) before being examined by microscopy. Distilled water was added regularly to compensate for loss from evaporation. For recovery, stationary-phase cells were collected by centrifugation, washed twice with 1× PBS buffer, and resuspended in YPD, 2× CSM, or 2× CSM supplemented with 5% YPD. Cycloheximide (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 100 μg/ml to inhibit protein synthesis when needed.
Microscopy
Yeast cells grown to different phases were harvested by centrifugation at 12,000 × g and resuspended in 1× PBS buffer or CSM medium (for log-phase cells only). The incubation of cells in 1× PBS buffer did not influence the localization patterns of GFP fusion proteins for as long as 7 h. For calcofluor staining of cell walls, an aliquot of a stock solution (20 mg/ml) of calcofluor white (Sigma-Aldrich) was added to the cell suspensions to a final concentration of 200 μg/ml; this was followed by incubation for 5 min at room temperature. Cells were then washed twice with 1× PBS buffer or CSM medium. For staining of DNA, live or fixed cells were incubated with Hoechst 33258 (0.5 μg/ml; Invitrogen, Carlsbad, CA) in distilled water for 10 min at room temperature and then washed twice. For actin body staining, cells were first fixed directly in the growth medium by incubation with 0.025% glutaraldehyde for 3 min, washed three times with 1× PBS buffer, resuspended in 1× PBS buffer, further fixed by incubation with 4% paraformaldehyde for 30 min, washed three times with 1× PBS buffer, resuspended in 1× PBS buffer, and incubated for 60 min with 0.5 U of Texas Red-X phalloidin (Invitrogen). Subsequently, cells were washed five times with 1× PBS buffer.
For live-cell imaging, yeast cells were spotted onto an agarose pad and immediately imaged at 28°C. For time-lapse studies, log- or stationary-phase cells were harvested by centrifugation, washed twice with 1× PBS buffer, resuspended in 200 μl 1× PBS buffer, and then added to an Attofluor cell chamber (Invitrogen) or a 4-well Lab-Tek chambered coverglass (Thermo Fisher Scientific, Rochester, NY). The cells were immobilized on concanavalin A–coated coverslips by incubating them for 5–10 min. Unattached cells were gently washed off with 1× PBS buffer. The immobilized cells were cultivated in the chamber with prewarmed medium and observed immediately. To distinguish between original stationary-phase cells and newly emerged buds, stationary-phase cells were first labeled using NHS-Rhodamine (Sigma-Aldrich) before being resuspended in fresh media.
Cells were observed using a DeltaVision platform (Applied Precision, Issaquah, WA) with an Olympus IX70 wide-field microscope equipped with a 100×, 1.4 numerical aperture Plan-Apochromat objective and a Coolsnap HQ camera or a Cascade II 512 electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ). To optimize the accuracy of protein colocalization, each Z-stack was acquired with mCherry signals immediately followed by GFP signals. The gain was 510, and binning was 1 × 1. All cells were imaged at 28°C. The same exposure time was used for both GFP and mCherry fusion proteins. Cells were optically sectioned into multiple slices with a spacing of 160–200 nm (the focus step size was 300–400 nm for long-term, time-lapse experiments), and captured images were deconvolved using the DeltaVision softWoRx 3.7.1 restoration system.
Image analysis
Unless otherwise specified, images are maximal projections of Z-stacks. However, for quantification of signal intensities, sum projections of Z-stacks were used. Intensity was measured using ImageJ (http://rsbweb.nih.gov/ij), and the background intensity was subtracted. When counting the number of protein foci, we examined at least 100 cells from three independent experiments by inspecting Z-sections of each cell and further confirmed using three-dimensional projections. To analyze and quantify the intensities and spatial distributions of GFP and mCherry signals, we used ImageJ and the three-dimensional graph function of softWoRx 3.7.1. For quantifying colocalization, we used JACoP (Bolte and Cordelieres, 2006) as a plug-in for ImageJ. Cell Counter (http://rsbweb.nih.gov/ij/plugins/cell-counter.html) was also used as a plug-in for ImageJ to count and categorize assemblies and cells.
Measurement of the rebudding ability and the first rebudding time of stationary-phase cells
Stationary-phase cell cultures were diluted 50-fold in YPD, and 2 μl of diluted culture was spotted onto YPD agarose (2%) pads. The cells on the YPD agarose pads were then placed into a sealed humidity chamber to avoid shrinkage of the pads and were subsequently observed by microscopy. The humidity chamber was made from a plastic Petri dish in which the center part of the lid was removed and replaced with a glass coverslip (24 × 60 mm). Time-lapse movies were taken, and the subsequent image analysis was done using ImageJ. In this experiment, we observed that most cells that failed to rebud in the first 6 h after refeeding of nutrients would not rebud at a later time point. Thus, for measuring cell viability, only cells that rebudded within 6 h following nutrient replenishment were counted as viable. We also found that counting rebudding cells by microscopy is a more reliable method for cell viability, as some yeast strains formed cell aggregates in stationary phase, which would skew measurements when the colony-forming assay was performed. At least 100 cells from three independent experiments were counted. The time point at which each viable cell started to rebud was recorded. Data from three independent biological repeats were pooled and used for the histograms. At least 200 cells were counted in each sample, except for the hos2 deletion strain in week 4 (n = 84).
Immunoblot analysis
Yeast cells were cultured in YPD medium at 28°C for 4 wk. Cells were collected at different time points and lysed for extraction of proteins under alkaline conditions (Kushnirov, 2000). About 6.2 × 107 yeast cells were harvested by centrifugation, washed, treated with NaOH for 5 min at room temperature, and boiled in 1 ml of SDS loading buffer for 3 min. Protein lysate was centrifuged at 10,000 × g for 5 min, and 5 μl of the supernatant was loaded per lane. SDS–PAGE was performed according to a standard protocol (Laemmli, 1970). Immunoblotting with anti-hemagglutinin (anti-HA; Covance, Princeton, NJ) and anti–glucose-6-phosphate dehydrogenase (anti-G6PDH; Sigma) was performed according to the recommended protocols from the manufacturers.
Supplementary Material
Acknowledgments
We thank Chen-Kung Chou, Sze-cheng Lo, and members of the Leu lab for helpful discussions and Andrew Murray, Scott C. Schuyler, and Shao-Win Wang for comments on the manuscript. We thank H. Kuhn and A. Pena for manuscript editing. We thank the Imaging Core of Institute of Molecular Biology for microscopy support. This work was supported by Academia Sinica of Taiwan (grant no. AS23-17) and the National Science Council of Taiwan (grant no. NSC99-2321-B-001-031).
Abbreviations used:
- anti-G6PDH
anti-glucose-6-phosphate dehydrogenase
- anti-HA
anti-hemagglutinin
- CSM
complete synthetic medium
- GFP
green fluorescent protein
- P-bodies
processing bodies
- SPGs
stationary-phase granules
- YEP
yeast–peptone
- YPD
yeast–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0752) on February 15, 2012.
REFERENCES
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1793:1413–1421. doi: 10.1016/j.bbamcr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ding Q, Keller JN. The stationary phase model of aging in yeast for the study of oxidative stress and age-related neurodegeneration. Biogerontology. 2005;6:1–13. doi: 10.1007/s10522-004-7379-6. [DOI] [PubMed] [Google Scholar]
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Mallory MJ, Strich R, Yao TP. Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot Cell. 2008;7:1191–1199. doi: 10.1128/EC.00059-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Davie JK, Edmondson DG, Coco CB, Dent SY. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem. 2003;278:50158–50162. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem. 2006;387:485–497. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- Erickson SL, Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J Cell Sci. 2011;124:293–297. doi: 10.1242/jcs.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles SJ, Gierasch LM. Nature's molecular sponges: small heat shock proteins grow into their chaperone roles. Proc Natl Acad Sci USA. 2010;107:2727–2728. doi: 10.1073/pnas.0915160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ge H, Wei M, Fabrizio P, Hu J, Cheng C, Longo VD, Li LM. Comparative analyses of time-course gene expression profiles of the long-lived sch9Δ mutant. Nucleic Acids Res. 2010;38:143–158. doi: 10.1093/nar/gkp849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T, et al. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–649. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson R, Huh WK, Ghaemmaghami S, Falvo JV, Bower K, Belle A, Dephoure N, Wykoff DD, Weissman JS, O'Shea EK. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp Funct Genomics. 2005;6:2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RE, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci USA. 2010;107:13348–13353. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean M, Harris N, Piper PW. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast. 2001;18:499–509. doi: 10.1002/yea.701. [DOI] [PubMed] [Google Scholar]
- Martinez MJ, et al. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol Biol Cell. 2004;15:5295–5305. doi: 10.1091/mbc.E03-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert HS, McMahon SB. Biochemical pathways that regulate acetyltransferase and deacetylase activity in mammalian cells. Trends Biochem Sci. 2009;34:571–578. doi: 10.1016/j.tibs.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HV, Vyas KA, Li X, Savtchenko R, Roseman S. Subcellular distribution of enzyme I of the Escherichia coli phosphoenolpyruvate:glycose phosphotransferase system depends on growth conditions. Proc Natl Acad Sci USA. 2004;101:17486–17491. doi: 10.1073/pnas.0407865101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I, Pinson B, Salin B, Daignan-Fornier B. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol Biol Cell. 2006;17:4645–4655. doi: 10.1091/mbc.E06-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci USA. 2002;99(suppl 4):16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 2002;21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JL. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci USA. 2010;107:2007–2012. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Minami M, Suzuki M, Abe K, Zenno S, Tsujimoto M, Matsumoto K, Minami Y. The Hsp90 inhibitor geldanamycin abrogates colocalization of eIF4E and eIF4E-transporter into stress granules and association of eIF4E with eIF4G. J Biol Chem. 2009;284:35597–35604. doi: 10.1074/jbc.M109.036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudisca V, Recouvreux V, Moreno S, Boy-Marcotte E, Jacquet M, Portela P. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur J Cell Biol. 2010;89:339–348. doi: 10.1016/j.ejcb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR, Wallace M, Morphew M, Perry P, Allshire R, Javerzat JP, McIntosh JR, Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.