Abstract
Objective
Thyroid proteomics is a new direction in thyroid cancer research aiming at etiological understanding and biomarker identification for improved diagnosis.
Methods
Two-dimensional electrophoresis was applied to cytosolic protein extracts from frozen thyroid samples (ten follicular adenomas, nine follicular carcinomas, ten papillary carcinomas, and ten reference thyroids). Spots with differential expression were revealed by image and multivariate statistical analyses, and identified by mass spectrometry.
Results
A set of 25 protein spots significant for discriminating between the sample groups was identified. Proteins identified for nine of these spots were studied further including 14-3-3 protein beta/alpha, epsilon, and zeta/delta, peroxiredoxin 6, selenium-binding protein 1, protein disulfide-isomerase precursor, annexin A5 (ANXA5), tubulin alpha-1B chain, and α1-antitrypsin precursor. This subset of protein spots carried the same predictive power in differentiating between follicular carcinoma and adenoma or between follicular and papillary carcinoma, as compared with the larger set of 25 spots. Protein expression in the sample groups was demonstrated by western blot analyses. For ANXA5 and the 14-3-3 proteins, expression in tumor cell cytoplasm was demonstrated by immunohistochemistry both in the sample groups and an independent series of papillary thyroid carcinomas.
Conclusion
The proteins identified confirm previous findings in thyroid proteomics, and suggest additional proteins as dysregulated in thyroid tumors.
Introduction
Thyroid cancer constitutes the most prevalent endocrine malignancy and comprises a spectrum of indolent to highly aggressive tumor types derived from the thyroid follicular or calcitonin-producing cells (1, 2). Follicular thyroid carcinoma (FTC), papillary thyroid carcinoma (PTC), and follicular thyroid adenoma (FTA) originate from the follicular cell, the thyroid gland's most abundant structural unit (1). Improved diagnosis and prognostication of FTA, FTC, and PTC on preoperative fine needle aspiration biopsy (FNAB) are central issues in thyroid cancer research aiming at optimal treatment schemes for each individual patient. The FNAB sampling technique has been greatly facilitated by the use of ultrasonography, but conclusive distinction between FTA and FTC is not achieved in about 10–20% of cases (2). Therefore, the identification of molecular markers remains a key issue in thyroid cytology.
During the past few decades, significant progress has been achieved in defining the molecular etiology of thyroid cancer. Molecular genetic and cytogenetic studies have defined common activating events, such as PPARγ rearrangements in FTC, and rearrangements of RET or NTRK1 as well as BRAF mutations in PTC (3, 4). Gene expression profiling has revealed expression signatures associated with specific genetic abnormalities as well as with tumor phenotypes and clinical course (5, 6, 7, 8). However, it has so far not been possible to define a certain set of genes that can be simply assessed in daily diagnostic routine to unequivocally classify thyroid tumors (2).
More recently, proteomics (i.e. the study of the proteome) has been gaining ground in thyroid cancer research. Wilkins et al. (9) were the first to define the proteome as ‘the complete protein complement expressed by a genome’. However, since the proteome is highly dynamic, a more proper definition should also mention that this protein complement reflects a given cell at a specific time point and it includes all different isoforms and modifications (10). Only a minority of the variation in protein levels is reflected in differences at the mRNA level, emphasizing the importance of posttranscriptional regulation of protein abundances (11, 12).
Applying gel-based techniques, e.g. two-dimensional electrophoresis (2-DE) and mass spectrometric applications, protein profiles can be generated for comparative analyses of cancer subgroups. At this time, relatively few proteomic studies on thyroid tumors have been reported (13, 14, 15, 16, 17, 18). Using proteomics we have previously identified a high S100A6 expression in subcellular protein fractions of PTC as compared with FTC, FTA, and normal thyroid (19). In the present study we aimed to combine 2-DE profiles on the same study groups, with multivariate data analysis for prediction model building and identification of potential tumor markers.
Material and methods
Thyroid tissue samples
Fresh frozen and paraffin-embedded samples of tumors and reference thyroid tissues were obtained from 39 patients operated on for a thyroid tumor at the Karolinska University Hospital in Stockholm, Sweden. All samples were collected through the Karolinska Endocrine Tissue Biobank with informed consent from patients, and approval granted by the Local Ethics Committee. Clinical and histopathological details have been previously reported (19) and are summarized in Supplementary Table 1, see section on supplementary data given at the end of this article. In short, tumors were histopathologically classified as FTA (n=10), FTC (n=9), or PTC (n=10). All PTCs were of classic type, and no case of follicular or any other variant of PTC was included. Moreover, both PTCs and FTCs in this study were well-differentiated tumors. Reference thyroid tissue samples were nontumor tissue from the contralateral lobe of ten unrelated patients, who had undergone thyroidectomy for unilateral tumors. Frozen samples were used for 2-DE and western blotting, while paraffin-embedded samples from 30 out of these 39 cases were used for immunohistochemistry. An experienced histopathologist (A Höög) verified that all samples were representative for either reference tissue (100% nontumor thyroid cells) or the corresponding tumor (>90% tumor cells).
Moreover, formalin-fixed and paraffin-embedded tissue samples from 70 patients with PTC were collected at the Kyiv City Teaching Endocrinological Centre, Ukraine. These patients were 18 years of age or younger at the time of the nuclear accident at the Chornobyl nuclear power station, lived in nearby areas which were contaminated, and underwent thyroidectomy between 2004 and 2008. The Local Ethics Committee approved the collection and use of these samples.
All tumor cases studied were classified according to the latest criteria of the World Health Organization (1), and have been evaluated for possible presence of chronic lymphocytic thyroiditis (CLT).
Antibodies
Monoclonal anti-Lamin A/C, clone 636 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and monoclonal anti-prohibitin, clone II-14-10 (NeoMarkers, Fremont, CA, USA) were used as nuclear and cytoplasmic markers of accurate protein fractionations as described (20). The following antibodies were applied for verification of proteins identified in this study: monoclonal anti-14-3-3 β/ϵ/ζ (3C8): sc-59420 (Santa Cruz Biotechnology) against the beta, epsilon, and zeta isoforms of protein 14-3-3; monoclonal anti-PRX VI (1A11): sc-59671 (Santa Cruz Biotechnology) against peroxiredoxin 6 (PRX6); monoclonal anti-annexin V, clone AN5 (Sigma–Aldrich, Inc.) against annexin A5 (ANXA5); monoclonal anti-K-ALPHA-1, clone 4D1 (Sigma–Aldrich, Inc.) against tubulin alpha-1B chain (TUBA1B); monoclonal anti-A1AT, clone 1C2 (Sigma–Aldrich, Inc.) against alpha-1 antitrypsin precursor; affinity-isolated anti-SELENBP1 (Sigma–Aldrich, Inc.) against selenium-binding protein 1 (SELENBP1); and affinity-isolated anti-P4HB (ab1; Sigma–Aldrich, Inc.) against protein disulfide-isomerase precursor (PDIp). Monoclonal anti-β-actin, clone CA-15 (Sigma–Aldrich, Inc.) was used as control of equal protein loading in immunoblot analyses.
Protein prefractionation
Subcellular protein fractions enriched for cytosolic and nuclear proteins were extracted from all 39 tissue samples according to our previously published protocol (20), quantified by Bradford assay (21), and verified by western blot analyses using anti-Lamin A/C and anti-prohibitin antibodies as previously reported for the majority of samples (19, 20).
Two-dimensional electrophoresis
Cytosolic protein extracts from all 39 thyroid tissue samples were separated by 2-DE, followed by silver staining using previously established experimental procedures (22). Briefly, for the first dimension (isoelectric focusing (IEF)) immobilized pH gradient (IPG) strips with a pH 4–7 range were used (17 cm, Bio-Rad). Seventy-five micrograms of protein were diluted in 300 μl rehydration buffer and this solution was applied on the strips. Active rehydration of the IPG strips and IEF was performed in a Protean IEF cell (Bio-Rad) according to the following program: step 1/7, 6 h at 50 V; step 2/7, 6 h at 60 V; step 3/7, 1 h at 60–500 V (linear); step 4/7, 1 h at 500 V; step 5/7, 2 h at 500–2000 V (linear); step 6/7, 1 h at 2000–8000 V (linear); and step 7/7, 5 h and 30 min at 8000 V (total of 53 000 V-h on average). Prior to the second dimension, all strips were incubated in equilibration buffer (50 mM Tris–HCl (pH 8.8), 69 mM SDS, 6 M urea, and 30% glycerol) containing 1% dithiothreitol (to achieve disulfide bonds reduction), and subsequently in equilibration buffer supplemented with 2.5% iodoacetamide (to achieve alkylation of SH-groups, thus preventing protein re- or misfolding). The protein separation was carried out in an ISO-DALT SDS–PAGE unit (GE Healthcare, Uppsala, Sweden) using 10–13% linear gradient acrylamide gels (1.5×200×230 mm, piperazine diacrylamide (PDA) as cross-linker) for an average total of 43 000 V-h.
Following the 2-DE all gels were silver stained according to a mass spectrometry compatible, modified protocol by Rabilloud et al. (23). All gels presenting high spot resolution, as well as absence of other signs of problematic protein separation (so called ‘streaking’), were scanned on a GS-710 flatbed scanner (Bio-Rad, Hercules, CA, USA) at a resolution of 105.8×105.8 μm. Spot detection, matching, and intensity measurements were carried out using PDQuest version 8.0 image analysis software (Bio-Rad).
2-DE data analysis and spot selection
Principal component analysis (24) and other diagnostic plots were used to assess the distribution and quality of data, the presence of outliers as well as the need of normalization or standardization. Based on the diagnostic plots we decided to include all samples and the data was normalized based on total intensity in valid spots (each spot's intensity was divided by the sum of intensities of all spots on the gel).
Differentially expressed individual proteins were identified by univariate statistical analysis. Fold-changes were calculated for the different subclasses (FTA, FTC, PTC, and reference thyroid), and compared using the nonparametric Wilcoxon's test. P values were adjusted using the Benjamini and Hochberg false discovery rate (FDR), taking multiple testing into account (25). The FDR cut-off value was set to 5%.
Spots present in at least 50% of the samples in one or more of the tumor subclasses (FTA, FTC, and PTC) were included in the multivariate analysis. Partial least squares discriminant analysis (PLS-DA) (26, 27) was utilized to build predictive models and to select gel spots that contribute to the distinction between the different sample groups (FTA–FTC and FTC–PTC). To generate the best predictive PLS model, the number of PLS components (latent variables) and spots in the model was optimized and the spots best distinguishing between the classes were identified. For this purpose, spots were ranked by the PLS-dependent variable importance on projection (VIP) score in this study and the most important spots were selected for prediction (28). The number of spots was decreased by 5% in each step, excluding the lowest-ranked spots, and the prediction success measures (geometric mean of sensitivity and specificity) were evaluated for the number of PLS components. The PLS modeling was performed within a bootstrap cross-validation to ascertain a stable variable selection and model optimization (29). The data was randomly divided into sets for training (80% of the samples) and testing (20% of the samples). The different PLS parameter settings were tested on the training set and the resulting success measures when applying the model to the test set were calculated. This was repeated 500 times and the mean success measures were collected and plotted. The optimal PLS parameter settings were decided as the minimal number of PLS components and spots still giving a good predictive power. The final set of spots was selected based on stability over bootstrap validation rounds (spots selected in at least 80% of bootstrap rounds were chosen for further evaluation and identification).
Protein digestion, peptide extraction, and mass spectrometry
Spots were excised manually and prepared for identification by mass spectrometry using previously described experimental procedures (22). Especially for faint spots, two or more gel plugs corresponding to the same protein spot were pooled from different gels. The excised gel spots were treated for in-gel digestion as follows: after removing the silver stain by Farmer's reagent (50 mM sodium thiosulphate/15 mM potassium ferricyanide), and extensive washing with water, gel plugs were treated with 50 mM ammonium bicarbonate (ambic) and then dried by neat acetonitrile (ACN). Porcine trypsin (modified, sequence grade from Promega) was added and incubation continued at 37 °C overnight. Digestion was terminated by using 10% trifluoroacetic acid and peptide retrieval was facilitated by mechanical vortex and sonication. The samples were desalted and concentrated using a micro-C18 ZipTip (Millipore, Billerica, MA, USA) and eluted directly onto the target plate using alpha-cyano-4-hydroxycinnamic acid in 75% ACN as matrix. Mass spectra for peptide mass fingerprinting were acquired in positive reflectron mode on an Ultraflex III TOF/TOF (Bruker Daltonics, Bremen, Germany). The instrument was optimized for the range 600–4500 m/z, following the manufacturer's instructions. The spectra were internally calibrated in quadratic mode using four autolytic peptides from trypsin (MH+842.51, 1045.56, 2211.10, and 3337.76 respectively) resulting in an error of <±0.02 Da. Searches for identities were done via the engine ProFound (The Rockefeller University and National Centre for Research Resources), applying the latest version of the NCBInr protein sequence database (NCBInr 2010/02/01) and according to the following conditions: taxonomy Homo sapiens; mass range 0–300 kDa; pI range 0–14; digestion by trypsin; missed cuts 1; C2H3ON-Cys as complete and methionine oxidation as partial modification; charge state was MH+; and mass tolerance 0.02 Da.
Western blot analyses
Protein fractions were resolved by SDS–PAGE in 16% Tricine gels, blotted onto 0.2 μm nitrocellulose membranes (Invitrogen), and blocked in 3% nonfat milk. Membranes were subsequently incubated overnight at 4 °C with the respective primary antibody at the following dilutions: anti-Lamin A/C at 1/2000; anti-prohibitin at 1/2000; anti-14-3-3 β/ϵ/ζ at 1/500; anti-PRX VI at 1/1000; anti-annexin V at 1/500; anti-K-ALPHA-1 at 1/2000; anti-A1AT at 1/2000; anti-SELENBP1 at 1/2000; anti-P4HB at 1/2000; and anti-β-actin at 1/16 000. HRP-conjugated goat anti-mouse or goat anti-rabbit were used as secondary antibodies at 1/12500 dilution. Membranes were exposed to Amersham Hyperfilm ECL film (GE Healthcare Limited, Buckinghamshire, UK).
Immunohistochemistry
Four micrometer paraffin sections were deparaffinized, rehydrated, and heated in a microwave oven in citrate buffer (pH 6) for 20 min. Hydrogen peroxide 0.3% in water (30 min incubation in room temperature (RT)) and avidin (1 h, RT) were used for blocking the endogenous hyperoxidase activity and biotin respectively. All sections were then blocked in 1% BSA (20 min, RT) followed by primary antibody incubation (overnight incubation in a moist chamber at 4 °C). The monoclonal antibodies (MAB) anti-14-3-3 β/ϵ/ζ and anti-annexin V were each used at a dilution of 1/250. Experimental conditions were optimized after testing various antibody dilutions as well as incubation solely with either secondary antibody or avidin–biotin complex (ABC). The ABC method (Vectastain Elite kit; Vector Laboratories, Burlingame, CA, USA) was applied for 30 min to visualize antibody–antigen binding, followed by 6 min incubation with diaminobenzedine tetrahydrochloride and counterstaining with hematoxylin. Incubation in the absence of a primary antibody was used as negative control. Immunohistochemical staining was evaluated by four of the authors (A Höög, A Dinets, C Larsson, and A Sofiadis). Images were captured using a ProgRes C12 Plus camera and the ProgRes Capture Pro 2.5 Software program (Jenoptik, Jena, Germany). The staining was scored concerning both the proportion of stained cells (negative, <25, 26–50, and >50%) and the signal intensity (negative, weak, moderate, and strong).
Results
2-DE profiling and spot selection
Cytosolic protein extracts of tissue samples from FTC, FTA, PTC, and reference thyroid were separated by 2-DE. Using PDQuest an average of 800 protein spots were detected and matched between the 39 sample gels included in this study and the virtual master gel, with a matching rate between individual gel and master gel of 95–99%. (In PDQuest, master gel is a virtual gel which depicts each spot's position and pixel intensity gathering information from all separate gels within one experiment.) Univariate analyses revealed at least twofold increased or decreased intensity for 99 spots between the FTA and FTC groups (P<0.05, Wilcoxon's rank test), and for 27 spots between the FTC and PTC groups (P<0.05, Wilcoxon's rank test).
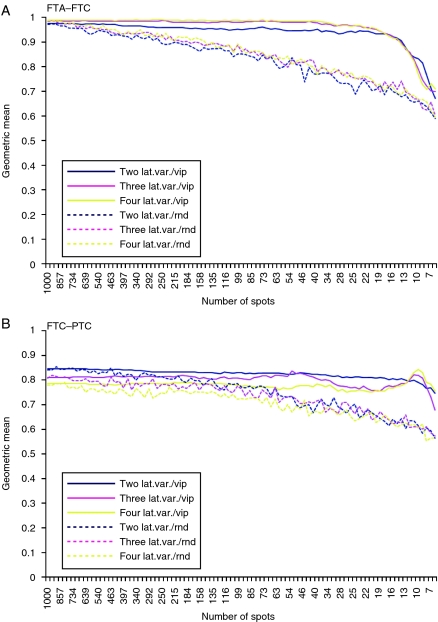
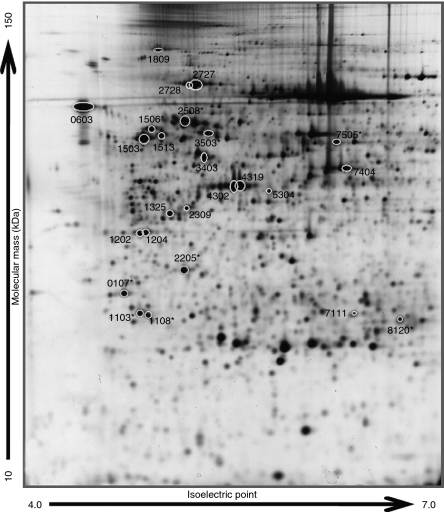
In multivariate analysis by PLS-DA, two predictive models were built for comparison between FTA and FTC and between FTC and PTC. Both models displayed a better predictive power using VIP selected spots compared with randomly selected ones (Fig. 1). The FTA–FTC model presented slightly better overall predictive performance than the FTC–PTC model. The selection of spots for further validation was based on the least number of spots still maintaining a good predictive power as well as stability over bootstrap validation rounds; spots selected in at least 80% of the 500 bootstrap rounds were considered. This resulted in 25 spots for the FTA–FTC model and 19 spots for the FTC–PTC model. After visual inspection, one spot from the FTC–PTC model (spot number 0401, data not shown) was excluded from the investigation, as this was an elongated, vaguely defined spot very close to the precipitation line at the acidic edge of the 2-DE gel, rendering any further analysis unreliable. The 18 resulting spots in the FTC–PTC model all overlapped to the FTA–FTC model (Table 1). An example of a 2-DE gel with the 25 spots of interest is shown in Fig. 2.
Figure 1.
Graphical presentation of the performance of the PLS-DA models built for the comparison between FTA and FTC (A) and between FTC and PTC (B). The continuous lines correspond to spots selected after vip-score ranking, whereas dotted lines correspond to random spot selection. (lat.var., latent variable; vip, variable importance on projection; and rnd, random).
Table 1.
Identities of all spots displaying stability and good performance in distinguishing between thyroid tumors according to the study's partial least squares discriminant analysis (PLS-DA) models.
| PLS-DA model | Univariate analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot number | Protein names | Accession no. | FTA–FTC | FTC–PTC | Expected mass (kDa) | Expected pI | Empirical mass (kDa) | Empirical pI | Matched peptides | Min. seq. coverage (%) | Profound expectation valuea | FTA vs FTC over-(↑)/under (↓)-expression (adjusted P value) | FTC vs PTC over-(↑)/under (↓)-expression (adjusted P value) |
| 0107 | 14-3-3 ϵ | NP006752.1 | + | + | 29.3 | 4.6 | 30 | 4.6 | 15/53 | 61 | 3.1×10−7 | ↑ (0.008) | ↓ (0.036) |
| 0603 | Calreticulin precursor | NP004334.1 | + | + | 48.2 | 4.3 | 69 | 4.4 | 13/65 | 49 | 6.2×10−7 | ↑ (0.134) | ↓ (0.415) |
| 1103 | 14-3-3 ζ/δ | NP003397.1 | + | + | 27.9 | 4.7 | 27 | 4.7 | 18/50 | 49 | 4.7×10−12 | ↑ (0.008) | ↓ (0.013) |
| 1108 | 14-3-3 β/α | NP003395.1 | + | − | 28.2 | 4.8 | 27 | 4.7 | 13/64 | 43 | 1.4×10−9 | ↑ (0.016) | ↓ (0.102) |
| 1202 | hsp gp96 prec. | AAK74072.1 | + | + | 90.4 | 4.7 | 40 | 4.7 | 14/66 | 16 | 9.5×10−4 | ↑ (0.016) | ↓ (0.098) |
| 1204 | hsp gp96 prec. | + | + | 90.4 | 4.7 | 40 | 4.7 | 8/52 | 10 | 4.3×10−1 | ↑ (0.016) | ↑ (0.629) | |
| 1325 | hsp gp96 prec. | + | − | 90.4 | 4.7 | 43 | 4.8 | 11/37 | 13 | 1.6×10−4 | ↑ (0.030) | ↓ (0.442) | |
| 1503 | hsp gp96 prec. | + | + | 90.4 | 4.7 | 57 | 4.7 | 16/40 | 19 | 2.1×10−10 | ↑ (0.026) | ↑ (0.994) | |
| 1506 | PDIp or thyroid hormone-binding protein precursor | NP000909.2 | + | + | 57.5 | 4.8 | 59 | 4.7 | 21/76 | 44 | 6.5×10−6 | ↑ (0.033) | ↓ (0.318) |
| 1513 | Endoplasmin precursor | NP003290.1 | + | + | 92.7 | 4.8 | 57 | 4.8 | 16/44 | 17 | 7.7×10−7 | ↑ (0.029) | ↓ (0.530) |
| 1809 | + | + | 92.7 | 4.8 | 93 | 4.8 | 24/56 | 28 | 2.8×10−10 | ↑ (0.016) | ↓ (0.340) | ||
| 2205 | ANXA5 | NP001145.1 | + | + | 36.0 | 5.0 | 36 | 5.0 | 15/60 | 42 | 2.8×10−13 | ↑ (0.016) | ↓ (0.029) |
| 2309 | Endoplasmin precursor | NP003290.1 | + | − | 92.7 | 4.8 | 44 | 5.0 | 12/34 | 14 | 2.1×10−4 | ↑ (0.013) | ↓ (0.076) |
| 2508 | A1AT | NP000286.3 | + | − | 46.9 | 5.4 | 62 | 5.0 | 20/59 | 42 | 2.6×10−7 | ↑ (0.623) | ↑ (0.279) |
| 2727 | 78 kDa glucose-regulated protein precursor | NP005338.1 | + | − | 72.4 | 5.1 | 78 | 5.1 | 32/68 | 52 | 3.0×10−14 | ↑ (0.182) | ↓ (0.181) |
| 2728 | + | + | 72.4 | 5.1 | 78 | 5.1 | 30/58 | 45 | 4.5×10−16 | ↑ (0.661) | ↑ (0.872) | ||
| 3403 | Protein disulfide-isomerase A6 precursor | NP005733.1 | + | + | 48.5 | 5.0 | 51 | 5.1 | 12/48 | 36 | 6.9×10−13 | ↑ (0.011) | ↓ (0.056) |
| 3503 | TUBA1B | NP006073.2 | + | + | 50.8 | 4.9 | 57 | 5.2 | 11/25 | 39 | 1.1×10−12 | ↑ (0.016) | ↓ (0.054) |
| 4302 | ACTB protein | AAH12854.1 | + | + | 40.5 | 5.6 | 47 | 5.4 | 14/84 | 49 | 2.2×10−1 | ↑ (0.995) | ↓ (0.903) |
| 4319 | + | + | 40.5 | 5.6 | 47 | 5.4 | 10/47 | 30 | 2.7×10−2 | ↑ (0.047) | ↓ (0.877) | ||
| 5304 | Creatine kinase B-type | NP001814.2 | + | − | 42.9 | 5.3 | 47 | 5.6 | 11/25 | 36 | 2.0×10−9 | ↓ (0.615) | ↑ (0.013) |
| 7111 | hsp β1 | NP001531.1 | + | + | 22.8 | 6.0 | 27 | 6.2 | 6/25 | 30 | 4.9×10−4 | ↓ (0.077) | ↑ (0.230) |
| 7404 | ALB protein | AAH41789.1 | + | − | 48.6 | 6.0 | 50 | 6.2 | 12/37 | 35 | 2.0×10−13 | ↑ (0.270) | ↑ (0.629) |
| 7505 | SELENBP1 | NP003935.2 | + | + | 52.9 | 5.9 | 55 | 6.1 | 15/44 | 31 | 1.1×10−10 | ↓ (0.016) | ↑ (0.036) |
| 8120 | PRX6 | NP004896.1 | + | + | 25.1 | 6.0 | 26 | 6.8 | 10/25 | 40 | 2.0×10−9 | ↓ (0.008) | ↑ (0.094) |
14-3-3 ϵ, 14-3-3 epsilon; 14-3-3 ζ/δ, 14-3-3 zeta/delta; 14-3-3 β/α, 14-3-3 beta/alpha; hsp gp96 prec., heat shock protein gp96 precursor; PDIp, protein disulfide-isomerase precursor; ANXA5, annexin A5; A1AT, alpha-1 antitrypsin precursor; TUBA1B, tubulin alpha-1B chain; hspβ1, Heat shock protein beta-1; SELENBP1, selenium-binding protein 1; PRX6, peroxiredoxin 6.
The simple interpretation of an expectation value is the number of matches that would be expected to have a particular score, if the matches were completely random. Therefore, the smaller the expectation value, the more likely that a particular match is a true match, rather than a random one.
Figure 2.
Image of a silver-stained 2-DE gel. The 25 spots selected from the FTA–FTC model are indicated by circles (total overlap with the 18 spots from the FTC–PTC model). The numbers shown correspond to the standard spot number automatically assigned to each spot by the image analysis software (PDQuest). Spot numbers followed by an asterisk refer to the nine spots selected for further validation.
MALDI-TOF-MS: protein identification
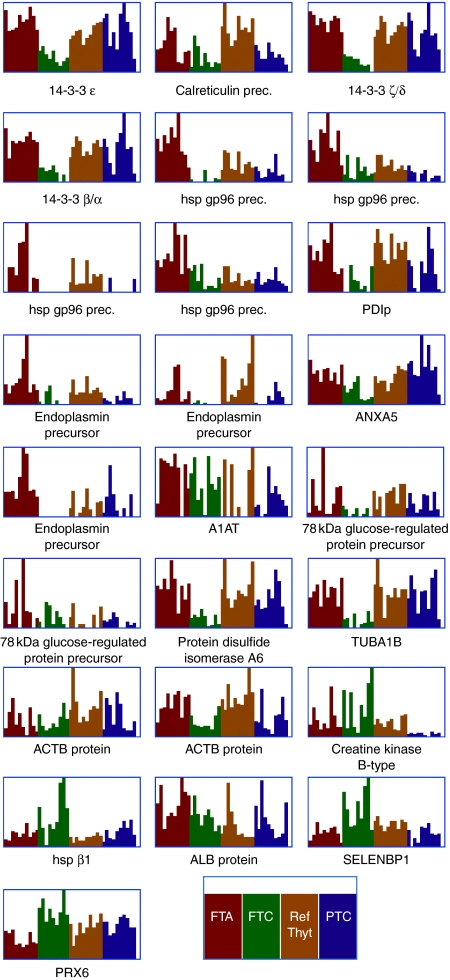
Gel plugs for the selected 25 protein spots were excised, analyzed by MALDI-TOF-MS, and the resulting spectra were matched against the NCBInr database. The resulting protein identifications are summarized in Table 1, and Fig. 3 compares corresponding individual spot intensities between samples in the four study groups. Nine of these proteins were selected for further analyses – namely 14-3-3 isoforms: β/α, ϵ, and ζ/δ, ANXA5, TUBA1B, PRX6, α1-antitrypsin precursor (A1AT), SELENBP1, and PDIp. The remaining protein identifications constituted of heat shock proteins, calreticulin, protein disulfide isomerase A6, ACTB protein, endoplasmin precursor, creatine kinase B-type, 78 kDa glucose-regulated protein precursor, and albumin.
Figure 3.
Graphical representation of individual spot intensities for the 25 spots identified by PLS-DA across all separate gels included in the study. Below each graph is given either the full or the abbreviated name of the corresponding protein identified by MALDI-TOF-MS (see Table 1). FTA, follicular thyroid adenoma; FTC, follicular thyroid carcinoma; Ref thyr, reference thyroid; and PTC, papillary thyroid carcinoma.
Model of nine selected protein spots: predictive power
To test the predictive power of the nine selected protein spots in the FTA–FTC PLS-DA model, a fivefold cross-validation was applied to all samples (FTA–FTC). Moreover, we applied a four- and threefold cross-validation receiving exactly the same result (data not shown). A PLS-DA model was built on the 25 protein spots identified, the nine selected spots as well as nine randomly picked spots from the list of 25. The resulting average prediction success over five cross-validation rounds showed that the nine selected spots performed as well as the full set of 25 spots (geometric mean=1, positive predictive value=1). The nine randomly picked spots out of the 25 yielded slightly worse prediction success (geometric mean=0.94, positive predictive value=0.93). The same control PLS-DA model was built in the case of the FTC–PTC comparison giving similar results; 0.90 for the full set of 19 spots, 0.94 for the seven spots overlapping to the FTA–FTC model and 0.88 for seven spots randomly picked out of the set of the above-mentioned 19 spots.
Model of nine selected protein spots: validation of protein expression
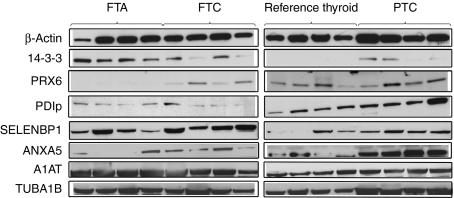
The expression of proteins corresponding to the nine selected spots was qualitatively verified by western blot analyses of four samples per group (Fig. 4). For ANXA5, TUBA1B, PRX6, A1AT, SELENBP1, and PDIp unique antibodies were applied, while the 14-3-3 isoforms β/ϵ/ζ were detected with a single MAB directed against all three isoforms.
Figure 4.
Western blots illustrating expression of the nine selected proteins in samples from each study group (reference thyroid, FTA, FTC, and PTC). β-Actin was used as control of protein loading and quality in all analyses.
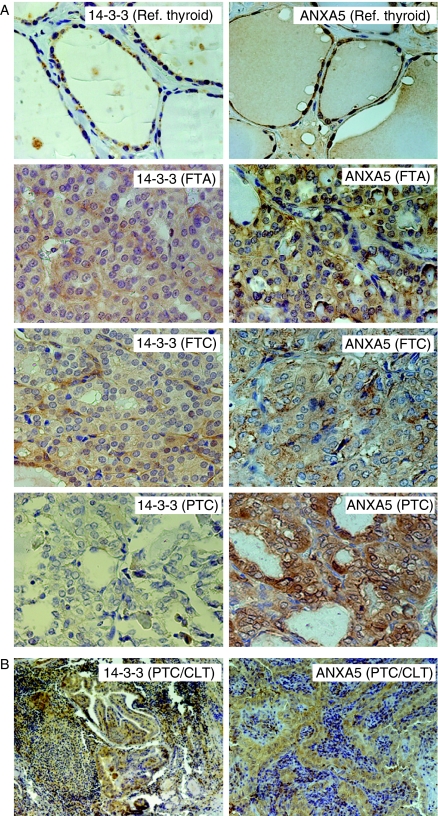
Finally, the expression of two proteins, namely 14-3-3 β/ϵ/ζ and ANXA5, was further studied by immunohistochemistry (Fig. 5). 14-3-3 was expressed in the cytoplasm of reference thyroid and tumor cells. In the latter, positive staining was observed in 33% of FTA samples, 67% of FTC, and 80% of PTC. For ANXA5 positive staining, mainly located in the cells' cytoplasm, was observed in all FTA and PTC samples as well as in 89% of FTC. Given the observation of subsets of lymphocytes positive for 14-3-3 in PTC, we further analyzed 70 PTC samples with presence/absence of CLT. This showed weak to strong 14-3-3 expression in varying proportions of the cells in 44/68 PTC cases, with positive and negative CLT areas. ANXA5 showed weak to strong expression in 50–100% of the tumor cells in 66/68 PTC cases, all with negative lymphocytes.
Figure 5.
Immunohistochemical analysis of protein expression for 14-3-3 (isoforms: β/α, ϵ and ζ/δ) and ANXA5. (A) Photomicrographs in 40× magnification of paraffin sections where predominantly cytosolic staining is visualized for 14-3-3 and ANXA5 in the different sample groups. (B) Photomicrographs in 10× magnification for 14-3-3 and 16× magnification for ANXA5 analysis on paraffin sections of PTC with CLT visualizing non-stained lymphocytes together with positively stained tumor cells.
Discussion
We here report the identification of a set of protein spots with differential expression in multivariate comparisons of FTC–FTA and FTC–PTC. Subcellular prefractionated protein samples from a relatively high number of samples (nine or ten per group) representing the four thyroid tissue groups of reference thyroid, FTA, FTC, and PTC were included in the study. In addition, we applied multivariate statistical analysis for building a stable predictive model. The usefulness of prefractionation in thyroid proteomics e.g. for increased resolution, has been previously reported for gel-based and other types of profiling (14, 20, 30). In addition, the high levels of thyroglobulin in thyroid tissues have been recognized as a possible problem in thyroid proteomic studies (30). In our study the typical signs of thyroglobulin described by Krause et al. (30) were not observed in the 2-DE gels (Fig. 2). A likely explanation for this difference is that thyroglobulin-containing vesicles were not fractionated together with cytoplasmic proteins using the applied protocol for sample fractionation. Hence, thyroglobulin is not expected to have seriously affected the 2-DE analysis in this study.
Our 2-DE results were analyzed by two different statistical methods. While the univariate analysis shows which spots are differently expressed at a certain significance level (5% FDR), the multivariate analysis is performed to identify the minimum set of spots giving good predictive performance. For the FTA–FTC comparison, 99 spots were identified by the univariate analysis, whereas 25 were selected by PLS-DA. The same figures for the FTC–PTC comparison were 27 and 19 respectively. This might be related to a varying degree of common shared characteristics (i.e. protein content) between FTA–FTC and FTC–PTC respectively. In total, 25 spots were identified which corresponded to 24 different proteins. Some of these have been previously described as frequently detected in proteomic studies, such as hsp β1, 78 kDa glucose-regulated protein (hsp 70 kDa 5), and calreticulin (31). A subset of nine selected protein spots was shown to yield high predictive power for the FTC–FTA and FTC–PTC models by PLS-DA, and expression of the corresponding proteins was further validated by western blot analyses. Among these, ANXA5, A1AT, ACTB, SELENBP1, PRX6, and 14-3-3 isoform sigma (σ) have been detected in proteomic profilings of thyroid tissues or by screening of thyroid tumors (13, 30, 32, 33).
The proteins identified in this study constitute candidates that could potentially become thyroid cancer markers in daily clinical praxis. It is worth mentioning that, even if some of these proteins seemed to be individually insignificant after the univariate statistical analysis, they were chosen within our PLS-DA multivariate model because of their contribution to the model's stability and good predictive performance as a set of proteins. Some of these proteins have also been implicated in human cancer including thyroid.
Protein 14-3-3 isoforms β/α, ϵ, and ζ/δ were identified in this study as significant in the FTC–FTA comparison and 14-3-3 isoforms ϵ and ζ/δ were also significant in the FTC–PTC comparison. Furthermore, 14-3-3 expression in tumor cell cytoplasm was demonstrated by immunohistochemistry. It has recently been reported that 14-3-3 proteins are involved in fate-determining cell functions like survival or apoptosis signaling, tumor suppression, and cell growth, mostly by integrating various intracellular cues (34). In thyroid cancer, two studies of the related 14-3-3 isoform σ have shown a nearly exclusive expression of this protein in PTC (either of conventional type or follicular-variant PTC), indicating that it plays an important role in the development of this particular type of tumor (35, 36). Moreover, Lal et al. (37) have shown that the expression of 14-3-3(σ) in thyroid cancer cells is ruled by the CpG island hypermethylation. The present study provides evidence that three other members of the 14-3-3 protein family are expressed in the cytoplasm of thyroid tumor cells. In 2-DE comparisons, 14-3-3 was found at lower levels in FTC (Fig. 3), suggesting that 14-3-3 is a candidate for inclusion in a panel of markers for differentiation between FTA–FTC or FTC–PTC. However, further studies are warranted to study the expression and biological role of different 14-3-3 isoforms including β/α, ϵ, and ζ/δ as well as the σ isoform in thyroid tumors.
ANXA5 is a 36 kDa protein which has been mostly used in combination with radionuclide labeling for the in vivo detection of apoptosis (38). Evidence for its possible relation to different cancer forms has been shown in recent proteomic studies (39, 40, 41). In this study ANXA5 was detected in both the FTC–FTA and FTC–PTC comparisons. The expression was verified by western blot analysis and also shown to be located in cytoplasm without staining of lymphocytes, which is often present especially in PTC.
SELENBP1, a protein which was first identified in humans in 1997 by Chang et al. (42), has been reported to participate in cell functions and processes like aging, lipid metabolism, protein transportation within the Golgi apparatus, cell growth, and toxification/detoxification processes (43). Decreased levels of SELENBP1 expression have been reported in lung adenocarcinoma (44, 45), colorectal adenocarcinoma (46), ovarian cancer (47), and gastric cancers (48). Recently, Silvers et al. (49) provided evidence showing that SELENBP1 mRNA and protein levels decrease as nondysplastic Barrett's esophagus progresses to Barrett's esophagus with high-grade dysplasia and esophageal adenocarcinoma. We observed lower levels of protein spots corresponding to SELENBP1 in PTC in comparison with reference thyroid tissue (Fig. 3), a finding that is in concordance with Brown et al. (13).
PRXs constitute a set of enzymes that serves as part of a cell's antioxidant system responsible for maintaining an appropriate level of reactive oxygen species (ROS). Zhang et al. (50) recently brought up the issue of the use of PRXs as targets in cancer radiotherapy, as they appear to be important not only for the cell's detoxification from ROS, but also for its proliferation and survival. PRX6, which is included in our panel of suggested markers for thyroid tumors, has been reported to be overexpressed in a variety of tumors such as malignant mesothelioma (51), esophageal carcinoma (52), oligodendroglioma, (53) and breast cancer (41, 54). Thyroid-related studies on PRXs have so far not concerned PRX6 (55, 56). In this study we observed low intensities for protein spots corresponding to PRX6 in FTA in relation to FTC, PTC, and reference thyroid tissue, and although based on a small number of cases, western blot analysis (Fig. 4) also suggested that PRX6 could be underexpressed in FTA. These observations would suggest a possible role for PRX6 as a complementary marker mainly for the distinction of FTA.
PDIp is the constitutively expressed β subunit of prolyl 4-hydroxylases (P4Hs), which not only keeps the hydroxylase's α subunits in solution, but is also responsible for maintaining them in a nonaggregated, catalytically active form (57). Among other functions, P4Hs are important for cellular responses to hypoxia (decreased oxygen supply) through the interaction with hypoxia-induced factors (HIFs) (58, 59). Recently, Hellman et al. (22) reported the downregulation of PDIp in cervical carcinoma in comparison with vaginal cancer. The lower intensities of spots corresponding to PDIp observed in FTC and PTC as compared with thyroid tissue or FTA (Fig. 3) would be in agreement with a role in the deregulated HIF mechanism, which renders cancerous cells able to survive and proliferate in hypoxic conditions.
To summarize, this study has identified a set of proteins with differential expression in follicular and papillary thyroid tumors. A quantitative multimarker assay, such as multiplexed ELISA or multiple reaction monitoring (MRM) assay, could be developed and applied to test the ability of our PLS model to separate tumor groups. The strength with MRM-based assays is that the same peptides that lead to the identification of proteins from a gel spot could potentially be selected for quantitation using targeted proteomics, hence eliminating difficulties with cross-reactivity of antibodies. Upon verification in an independent tumor material, the clinical utility of proteins like 14-3-3, PRX6, ANXA5, SELENBP1, or PDIp should be evaluated in prospective studies, preferably on cytology since the ultimate goal is to apply this knowledge in FNAB specimens.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/EJE-11-0856.
Acknowledgements
The authors would like to thank Lisa Ånfalk for her excellent technical assistance in tissue sample handling and Johan Lengqvist, Hanna Eriksson, Maria Pernemalm, Sara Ståhl, and Elena Ossipova for all practical and intellectual contributions on mass spectrometry.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This study was financially supported by the Swedish Cancer Society, the Swedish Research Council, the Cancer Society in Stockholm, the Gustav V Jubilee Foundation, the Stockholm County Council, Karolinska Institutet, and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine.
References
- Tumours of the thyroid and parathyroid. In WHO Classification of Tumours: Pathology and Genetics of Tumours of Endocrine Organs, pp 49–133. Eds RA DeLellis, RV Lloyd, PU Heitz & C Eng. Lyon: IARC Press, 2004.
- Damante G, Scaloni A, Tell G. Thyroid tumors: novel insights from proteomic studies. Expert Review of Proteomics. 2009;6:363–376. doi: 10.1586/epr.09.51. [DOI] [PubMed] [Google Scholar]
- Lui WO, Zeng L, Rehrmann V, Deshpande S, Tretiakova M, Kaplan EL, Leibiger I, Leibiger B, Enberg U, Hoog A, Larsson C, Kroll TG. CREB3L2-PPARgamma fusion mutation identifies a thyroid signaling pathway regulated by intramembrane proteolysis. Cancer Research. 2008;68:7156–7164. doi: 10.1158/0008-5472.CAN-08-1085. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Nikiforov YE. Molecular genetics of thyroid cancer: implications for diagnosis, treatment and prognosis. Expert Review of Molecular Diagnostics. 2008;8:83–95. doi: 10.1586/14737159.8.1.83. [DOI] [PubMed] [Google Scholar]
- Barden CB, Shister KW, Zhu B, Guiter G, Greenblatt DY, Zeiger MA, Fahey TJ., III Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clinical Cancer Research. 2003;9:1792–1800. [PubMed] [Google Scholar]
- Finley DJ, Zhu B, Barden CB, Fahey TJ., III Discrimination of benign and malignant thyroid nodules by molecular profiling. Annals of Surgery. 2004;240:425–436. doi: 10.1097/01.sla.0000137128.64978.bc. discussion 436–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzanti C, Zeiger MA, Costouros NG, Umbricht C, Westra WH, Smith D, Somervell H, Bevilacqua G, Alexander HR, Libutti SK. Using gene expression profiling to differentiate benign versus malignant thyroid tumors. Cancer Research. 2004;64:2898–2903. doi: 10.1158/0008-5472.CAN-03-3811. [DOI] [PubMed] [Google Scholar]
- Lui WO, Foukakis T, Liden J, Thoppe SR, Dwight T, Hoog A, Zedenius J, Wallin G, Reimers M, Larsson C. Expression profiling reveals a distinct transcription signature in follicular thyroid carcinomas with a PAX8–PPAR(gamma) fusion oncogene. Oncogene. 2005;24:1467–1476. doi: 10.1038/sj.onc.1208135. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Pasquali C, Appel RD, Ou K, Golaz O, Sanchez JC, Yan JX, Gooley AA, Hughes G, Humphery-Smith I, Williams KL, Hochstrasser DF. From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology. 1996;14:61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Mann M. Proteomics. Annual Review of Genomics and Human Genetics. 2004;5:267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Molecular & Cellular Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Brown LM, Helmke SM, Hunsucker SW, Netea-Maier RT, Chiang SA, Heinz DE, Shroyer KR, Duncan MW, Haugen BR. Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Molecular Carcinogenesis. 2006;45:613–626. doi: 10.1002/mc.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Karger S, Schierhorn A, Poncin S, Many MC, Fuhrer D. Proteomic profiling of cold thyroid nodules. Endocrinology. 2007;148:1754–1763. doi: 10.1210/en.2006-0752. [DOI] [PubMed] [Google Scholar]
- Moretz WH, III, Gourin CG, Terris DJ, Xia ZS, Liu Z, Weinberger PM, Chin E, Adam BL. Detection of papillary thyroid carcinoma with serum protein profile analysis. Archives of Otolaryngology – Head & Neck Surgery. 2008;134:198–202. doi: 10.1001/archoto.2007.34. [DOI] [PubMed] [Google Scholar]
- Netea-Maier RT, Hunsucker SW, Hoevenaars BM, Helmke SM, Slootweg PJ, Hermus AR, Haugen BR, Duncan MW. Discovery and validation of protein abundance differences between follicular thyroid neoplasms. Cancer Research. 2008;68:1572–1580. doi: 10.1158/0008-5472.CAN-07-5020. [DOI] [PubMed] [Google Scholar]
- Wang JX, Yu JK, Wang L, Liu QL, Zhang J, Zheng S. Application of serum protein fingerprint in diagnosis of papillary thyroid carcinoma. Proteomics. 2006;6:5344–5349. doi: 10.1002/pmic.200500833. [DOI] [PubMed] [Google Scholar]
- Krause K, Jessnitzer B, Fuhrer D. Proteomics in thyroid tumor research. Journal of Clinical Endocrinology and Metabolism. 2009;94:2717–2724. doi: 10.1210/jc.2009-0308. [DOI] [PubMed] [Google Scholar]
- Sofiadis A, Dinets A, Orre LM, Branca RM, Juhlin CC, Foukakis T, Wallin G, Hoog A, Hulchiy M, Zedenius J, Larsson C, Lehtio J. Proteomic study of thyroid tumors reveals frequent up-regulation of the Ca2+-binding protein S100A6 in papillary thyroid carcinoma. Thyroid. 2010;20:1067–1076. doi: 10.1089/thy.2009.0400. [DOI] [PubMed] [Google Scholar]
- Forsberg L, Larsson C, Sofiadis A, Lewensohn R, Hoog A, Lehtio J. Pre-fractionation of archival frozen tumours for proteomics applications. Journal of Biotechnology. 2006;126:582–586. doi: 10.1016/j.jbiotec.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Hellman K, Alaiya AA, Becker S, Lomnytska M, Schedvins K, Steinberg W, Hellstrom AC, Andersson S, Hellman U, Auer G. Differential tissue-specific protein markers of vaginal carcinoma. British Journal of Cancer. 2009;100:1303–1314. doi: 10.1038/sj.bjc.6604975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T, Vuillard L, Gilly C, Lawrence JJ. Silver-staining of proteins in polyacrylamide gels: a general overview. Cellular and Molecular Biology. 1994;40:57–75. [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P. Principal component analysis. Chemometrics and Intelligent Laboratory Systems. 1987;2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, Statistical Methodology. 1995;57:289–300. [Google Scholar]
- Geladi P, Kowalski B. Partial least-squares regression – a tutorial. Analytica Chimica Acta. 1986;185:1–17. doi: 10.1016/0003-2670(86)80028-9. [DOI] [Google Scholar]
- Wold S, Sjöstrom M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems. 2001;58:109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- Wold S, Johansson E & Gocchi M. PLS – partial least-squares projections to latent structures. In 3D QSAR in Drug Design; Theory, Methods and Applications, pp 523–550. Ed HK Dordrecht. The Netherlands: Kluwer Academic Publishers, 1993.
- Efron B. Estimating the error rate of a prediction rule – improvement on cross-validation. Journal of the American Statistical Association. 1983;78:316–331. doi: 10.2307/2288636. [DOI] [Google Scholar]
- Krause K, Schierhorn A, Sinz A, Wissmann JD, Beck-Sickinger AG, Paschke R, Fuhrer D. Toward the application of proteomics to human thyroid tissue. Thyroid. 2006;16:1131–1143. doi: 10.1089/thy.2006.16.1131. [DOI] [PubMed] [Google Scholar]
- Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- Srisomsap C, Subhasitanont P, Otto A, Mueller EC, Punyarit P, Wittmann-Liebold B, Svasti J. Detection of cathepsin B up-regulation in neoplastic thyroid tissues by proteomic analysis. Proteomics. 2002;2:706–712. doi: 10.1002/1615-9861(200206)2:6%3C706::AID-PROT706%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Giusti L, Iacconi P, Ciregia F, Giannaccini G, Donatini GL, Basolo F, Miccoli P, Pinchera A, Lucacchini A. Fine-needle aspiration of thyroid nodules: proteomic analysis to identify cancer biomarkers. Journal of Proteome Research. 2008;7:4079–4088. doi: 10.1021/pr8000404. [DOI] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends in Cell Biology. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Nakamura Y, Kakudo K, Kuma K, Miyauchi A. Caveolin-1 and 14-3-3 sigma expression in follicular variant of thyroid papillary carcinoma. Pathology, Research and Practice. 2005;201:545–549. doi: 10.1016/j.prp.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Ito Y, Miyoshi E, Uda E, Yoshida H, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Matsuura N, Kakudo K, Kuma K, Miyauchi A. 14-3-3 Sigma possibly plays a constitutive role in papillary carcinoma, but not in follicular tumor of the thyroid. Cancer Letters. 2003;200:161–166. doi: 10.1016/S0304-3835(03)00282-9. [DOI] [PubMed] [Google Scholar]
- Lal G, Padmanabha L, Provenzano M, Fitzgerald M, Weydert J, Domann FE. Regulation of 14-3-3sigma expression in human thyroid carcinoma is epigenetically regulated by aberrant cytosine methylation. Cancer Letters. 2008;267:165–174. doi: 10.1016/j.canlet.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg FG. In vivo detection of apoptosis. Journal of Nuclear Medicine. 2008;49(Suppl 2):81S–95S. doi: 10.2967/jnumed.107.045898. [DOI] [PubMed] [Google Scholar]
- Alfonso P, Canamero M, Fernandez-Carbonie F, Nunez A, Casal JI. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. Journal of Proteome Research. 2008;7:4247–4255. doi: 10.1021/pr800152u. [DOI] [PubMed] [Google Scholar]
- Srisomsap C, Sawangareetrakul P, Subhasitanont P, Chokchaichamnankit D, Chiablaem K, Bhudhisawasdi V, Wongkham S, Svasti J. Proteomic studies of cholangiocarcinoma and hepatocellular carcinoma cell secretomes. Journal of Biomedicine & Biotechnology. 2010;2010:437143. doi: 10.1155/2010/437143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongwatchara P, Promwikorn W, Srisomsap C, Chokchaichamnankit D, Boonyaphiphat P, Thongsuksai P. Differential protein expression in primary breast cancer and matched axillary node metastasis. Oncology Reports. 2012;26:185–191. doi: 10.3892/or.2011.1266. [DOI] [PubMed] [Google Scholar]
- Chang PW, Tsui SK, Liew C, Lee CC, Waye MM, Fung KP. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. Journal of Cellular Biochemistry. 1997;64:217–224. doi: 10.1002/(SICI)1097-4644(199702)64:2%3C217::AID-JCB5%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxidants & Redox Signaling. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang H, Miller CT, Thomas DG, Gharib TG, Misek DE, Giordano TJ, Orringer MB, Hanash SM, Beer DG. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. Journal of Pathology. 2004;202:321–329. doi: 10.1002/path.1524. [DOI] [PubMed] [Google Scholar]
- Li LS, Kim H, Rhee H, Kim SH, Shin DH, Chung KY, Park KS, Paik YK, Chang J. Proteomic analysis distinguishes basaloid carcinoma as a distinct subtype of nonsmall cell lung carcinoma. Proteomics. 2004;4:3394–3400. doi: 10.1002/pmic.200400901. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang HJ, You KT, Kim SH, Lee KY, Kim TI, Kim C, Song SY, Kim HJ, Lee C. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics. 2006;6:3466–3476. doi: 10.1002/pmic.200500629. [DOI] [PubMed] [Google Scholar]
- Huang KC, Park DC, Ng SK, Lee JY, Ni X, Ng WC, Bandera CA, Welch WR, Berkowitz RS, Mok SC, Ng SW. Selenium binding protein 1 in ovarian cancer. International Journal of Cancer. 2006;118:2433–2440. doi: 10.1002/ijc.21671. [DOI] [PubMed] [Google Scholar]
- He QY, Cheung YH, Leung SY, Yuen ST, Chu KM, Chiu JF. Diverse proteomic alterations in gastric adenocarcinoma. Proteomics. 2004;4:3276–3287. doi: 10.1002/pmic.200300916. [DOI] [PubMed] [Google Scholar]
- Silvers AL, Lin L, Bass AJ, Chen G, Wang Z, Thomas DG, Lin J, Giordano TJ, Orringer MB, Beer DG, Chang AC. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clinical Cancer Research. 2010;16:2009–2021. doi: 10.1158/1078-0432.CCR-09-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Letters. 2009;286:154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- Kinnula VL, Lehtonen S, Sormunen R, Kaarteenaho-Wiik R, Kang SW, Rhee SG, Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. Journal of Pathology. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakanishi T, Hiramatsu M, Mabuchi H, Miyamoto Y, Miyamoto A, Shimizu A, Tanigawa N. Proteomics-based approach identifying autoantibody against peroxiredoxin VI as a novel serum marker in esophageal squamous cell carcinoma. Clinical Cancer Research. 2006;12:6415–6420. doi: 10.1158/1078-0432.CCR-06-1315. [DOI] [PubMed] [Google Scholar]
- Park CK, Kim JH, Moon MJ, Jung JH, Lim SY, Park SH, Kim DG, Jung HW, Cho BK, Paek SH. Investigation of molecular factors associated with malignant transformation of oligodendroglioma by proteomic study of a single case of rapid tumor progression. Journal of Cancer Research and Clinical Oncology. 2008;134:255–262. doi: 10.1007/s00432-007-0282-1. [DOI] [PubMed] [Google Scholar]
- Karihtala P, Mantyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clinical Cancer Research. 2003;9:3418–3424. [PubMed] [Google Scholar]
- Gerard AC, Many MC, Daumerie C, Knoops B, Colin IM. Peroxiredoxin 5 expression in the human thyroid gland. Thyroid. 2005;15:205–209. doi: 10.1089/thy.2005.15.205. [DOI] [PubMed] [Google Scholar]
- Yanagawa T, Ishikawa T, Ishii T, Tabuchi K, Iwasa S, Bannai S, Omura K, Suzuki H, Yoshida H. Peroxiredoxin I expression in human thyroid tumors. Cancer Letters. 1999;145:127–132. doi: 10.1016/S0304-3835(99)00243-8. [DOI] [PubMed] [Google Scholar]
- Kivirikko KI, Myllyharju J. Prolyl 4-hydroxylases and their protein disulfide isomerase subunit. Matrix Biology. 1998;16:357–368. doi: 10.1016/S0945-053X(98)90009-9. [DOI] [PubMed] [Google Scholar]
- Poon E, Harris AL, Ashcroft M. Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert Reviews in Molecular Medicine. 2009;11:e26. doi: 10.1017/S1462399409001173. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Annals of Medicine. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.