Abstract
Columns containing immobilized low density lipoprotein (LDL) were prepared for the analysis of drug interactions with this agent by high-performance affinity chromatography (HPAC). R/S-Propranolol was used as a model drug for this study. The LDL columns gave reproducible binding to propranolol over 60 h of continuous use in the presence of pH 7.4, 0.067 M potassium phosphate buffer. Experiments conducted with this type of column through frontal analysis indicated that two types of interactions were occurring between R-propranolol and LDL, while only a single type of interaction was observed between S-propranolol and LDL. The first type of interaction, which was seen for both enantiomers, involved non-saturable binding; this interaction had an overall affinity (nKa) of 1.9 (± 0.1) × 105 M-1 for R-propranolol and 2.7 (± 0.2) × 105 M-1 for S-propranolol at 37 °C. The second type of interaction was observed only for R-propranolol and involved saturable binding that had an association equilibrium constant (Ka) of 5.2 (± 2.3) × 105 M-1 at 37 °C. Similar differences in binding behavior were found for the two enantiomers at 20 °C and 27 °C. This is the first known example of stereoselective binding of drugs by LDL or other lipoproteins. This work also illustrates the ability of HPAC to be used as a tool for characterizing mixed-mode interactions that involve LDL and related binding agents.
Keywords: Low density lipoprotein, Propranolol, Drug interactions, High-performance affinity chromatography, Stereoselective binding
Introduction
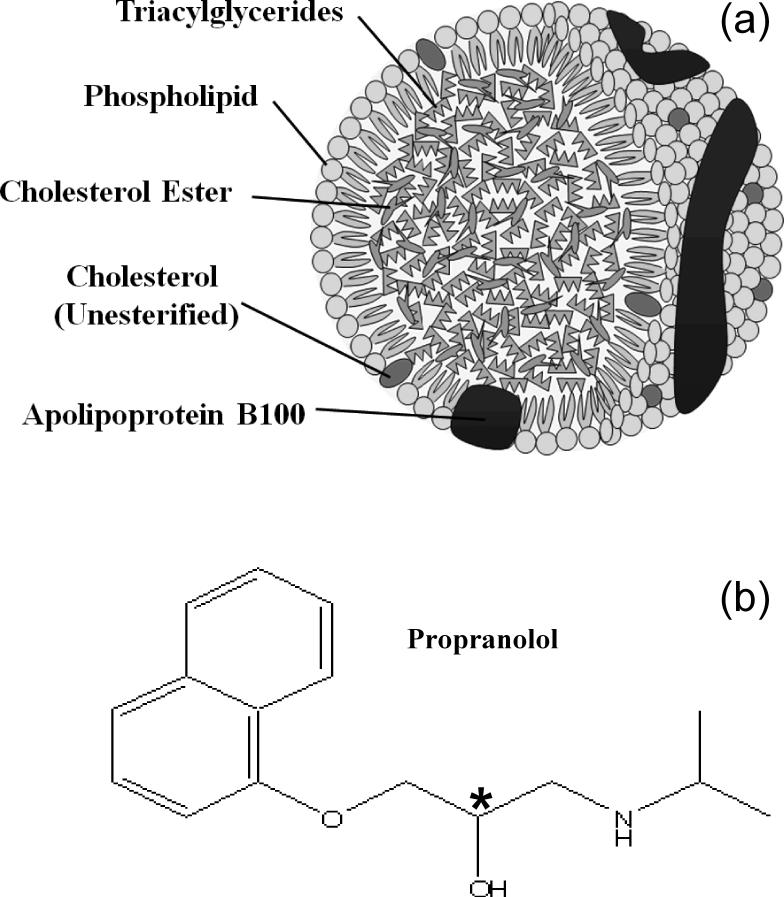
The interaction of drugs with serum proteins and other binding agents in blood is important in determining the apparent activity, distribution and pharmacokinetics of many pharmaceutical compounds in the body [1-7]. Low density lipoprotein (LDL) and related lipoproteins (e.g., high density lipoprotein, or HDL) are one group of binding agents that are known to interact with drugs and other solutes in serum [7-15]. As shown in Figure 1(a), LDL consists of a soluble macromolecular complex of proteins (i.e., apolipoproteins) and lipids that transports hydrophobic compounds and lipids such as cholesterols and triacylglycerols (e.g., triglycerides) [7-9]. LDL and related lipoproteins are also responsible for binding to several basic and neutral hydrophobic drugs [10]. In addition, LDL and HDL are often measured by clinical laboratories as indicators of the various fractions of cholesterol that are present in blood [8].
Figure 1.
Structures of (a) low density lipoprotein (LDL) and (b) propranolol. As shown in (a), the specific type of apolipoprotein that is present in LDL is apolipoprotein B100 (apoB100). Other lipoproteins have similar structures but may contain different apolipoproteins. In (b), the asterisk indicates the location of the chiral center in propranolol.
One drug that is known to bind to LDL and other lipoproteins is the chiral drug propranolol [16-19]. The structure of propranolol is provided in Figure 1(b). Prior methods used to examine the interactions of propranolol and other drugs with LDL and HDL have included equilibrium dialysis [16] and capillary electrophoresis carried out in a frontal analysis mode [17-19]. An alternative approach that will be examined in this report is high-performance affinity chromatography (HPAC). This method makes use of an HPLC column that contains an immobilized binding agent (e.g., LDL) to which a solution or sample of the drug of interest is applied. Based on the elution profile of the applied drug, it is possible to obtain information on the equilibrium constants and stoichiometry of the interactions that are occurring in the column. HPAC has been successfully used in numerous studies to examine the binding of drugs with serum proteins such as human serum albumin (HSA) and α1-acid glycoprotein (AGP) [3-6,20-26]. In a recent report it has been found that the same approach can be employed with columns containing the lipoprotein HDL [27]. Advantages of this approach include its speed, ease of automation, precision, and good correlation versus solution-phase results for HSA, AGP, and HDL [3,5,6,20,21,24,27].
In this study, HPAC will be employed as a tool to study the interactions of propranolol with LDL. This work will adapt methods that have been previously used to examine drug interactions with immobilized HDL [27] and HSA or AGP [3-6,20-26]. After LDL has been immobilized to HPLC grade silica, the stability of the resulting columns will be examined through zonal elution studies. The same types of columns will then be used in frontal analysis experiments to examine the binding mechanisms for LDL with R- and S-propranolol at various temperatures. The results will be compared to data obtained in previous work using soluble LDL. This information should provide important clues regarding the nature of the interactions between R- and S-propranolol with LDL in the circulation. The results of this report should also make it possible to determine the potential advantages and limitations of using immobilized LDL columns with HPAC for drug binding studies, including those that involve chiral pharmaceutical agents.
Experimental
Chemicals
The human LDL (catalog number L7914, lot no. 036K1143), bovine serum albumin (BSA) and separate enantiomers of R- and S-propranolol were purchased from Sigma (St. Louis, MO, USA). The Nucleosil Si-500 silica (7 μm particle diameter, 500 Å pore size) was from Macherey Nagel (Düren, Germany). Reagents for the bicinchoninic acid (BCA) protein assay were from Pierce (Rockford, IL, USA). The total cholesterol assay kit was purchased from Wako (Richmond, VA, USA). All other chemicals were of the highest grades available. All solutions were prepared using water from a Nanopure purification system (Barnstead, Dubuque, IA, USA) that was filtered using Osmonics 0.22 μm nylon filters from Fisher Scientific (Pittsburgh, PA, USA).
Instrumentation
The chromatographic system utilized in zonal elution studies consisted of a PU-980 pump (Jasco, Tokyo, Japan), a LabPro injection valve (Rohnert Park, FL, USA), and a UV/Vis SpectroMonitor 3200 variable wavelength absorbance detector from LDC Thermoseparations (Riviera Beach, FL, USA). The chromatographic system utilized in the frontal analysis studies consisted of two 510 HPLC pumps (Waters, Milford, MA, USA), an F60-AL injection valve (Vici, Houston, TX, USA), a CH-500 column heater (Eppendorf, Hauppauge, NY, USA) and a Waters 2487 UV/Vis variable wavelength absorbance detector. Chromatographic data were collected using Waters Empower software and were processed using programs based on Labview 5.1 or 7.0 (National Instruments, Austin, TX, USA). Support materials were placed into columns using a slurry packer from Alltech (Deerfield, IL, USA).
Preparation of immobilized LDL
The Schiff base method was used to prepare the LDL silica, as based on an approach similar to the one developed in Ref. [27] for the immobilization of HDL. First, diol-bonded silica was prepared from Nucleosil Si-500 silica, as described previously [25]. A 0.3 g portion of the diol-bonded silica was placed into 6 mL of a 90:10 (v/v) mixture of acetic acid and water, to which was added 0.3 g periodic acid. This mixture was sonicated under vacuum for 20 min and shaken for over 2 h in the dark at room temperature. The resulting aldehyde-activated silica was washed four times with water and four times with pH 6.0, 0.10 M potassium phosphate buffer. The aldehyde silica was placed in 1 mL of pH 6.0, 0.10 M potassium phosphate buffer and sonicated for 5 min under vacuum. A 20 mg portion of sodium cyanoborohydride was added, followed by the addition of 5 mg LDL. This mixture was shaken in the dark at 4 °C for 7 days. The resulting LDL support was washed four times with pH 7.0, 0.10 M potassium phosphate buffer. A 5.2 mg portion of sodium borohydride was dissolved into 2 mL of pH 7.0, 0.10 M potassium phosphate buffer and slowly added to the LDL support to reduce any remaining aldehyde groups that were still present on the silica. This mixture was shaken for 90 min at room temperature. The final LDL support was washed six times with pH 7.0, 0.10 M potassium phosphate buffer and stored in the same pH 7.0 buffer at 4 °C until use. Diol silica was utilized as a control support in these studies.
The protein content of the LDL support was evaluated in triplicate by using a BCA protein assay [28] and BSA solutions prepared in pH 7.4, 0.067 M potassium phosphate buffer as the standards, with diol silica samples being used as the blanks. All samples and standard solutions were filtered through a 0.2 μm nylon filter prior to obtaining absorbance readings in this assay. The cholesterol content of the LDL support was evaluated in triplicate using a Wako Cholesterol E assay kit and diol silica as the blank [29]. The samples and standards were allowed to react in this assay according to the manufacturer's instructions and then filtered through a 0.2 μm nylon filter prior to absorbance measurements. Based on work with other agents such as HSA, the immobilization scheme that was used in this report is known to have good batch-to-batch reproducibility, producing a typical variation of only 5-10% in protein content for silica supports similar to those utilized in this study [4,24,25].
Chromatographic studies
The LDL support and control support were downward slurry packed at 3000 psi into 100 mm × 2.1 mm i.d. stainless steel columns using pH 7.4, 0.067 M potassium phosphate buffer as the packing solution. Separate LDL columns and supports were used for the zonal elution and frontal analysis studies, but were prepared from the same batch of LDL and silica and by using the same immobilization method. The columns were stored in pH 7.4, 0.067 M phosphate buffer at 4°C. Before each experiment, these columns were equilibrated at the specified temperature for a given study. All mobile phases were filtered through Osmonics 0.22 μm nylon filters and degassed under vacuum prior to use. The wavelength utilized to monitor the elution of R- or S-propranolol was 225 nm. The elution of sodium nitrate, which was employed as a non-retained solute, was monitored at 205 nm.
The zonal elution experiments that were used to evaluate the stability of the LDL columns were carried out using a mobile phase that consisted of a pH 7.4, 0.067 M potassium phosphate buffer. This mobile phase was continuously applied to a 100 mm × 2.1 mm i.d. column containing the LDL support over the course of 60 h at 37 °C. A 20 μL portion of 50 μM R-propranolol was injected onto an LDL column and a control column at 1.0 mL/min under these conditions while the retention of the injected drug was monitored. The column void time, which was 23.2 (± 0.5) s at 1.0 mL/min, was determined by injecting a 20 μL sample of 50 μM sodium nitrate onto the LDL column and control column. The retention time for each peak was found by using its central moment, as determined by utilizing Peakfit 4.12 software (Systat Software, San Jose, CA).
The frontal analysis studies were carried out in triplicate by using 100 mm × 2.1 mm i.d. LDL columns or control columns. These studies were conducted at 20 °C, 27 °C or 37 °C and at 1.0 mL/min in the presence of a mobile phase that consisted of pH 7.4, 0.067 M potassium phosphate buffer. A total of nine solutions containing 0.2-25 μM R- or S-propranolol in the mobile phase were applied to the LDL column and control column. Similar propranolol concentrations have been used in previous frontal analysis studies with HDL columns [27]. The lower end of this range corresponded to approximately the lowest concentration at which the breakthrough times for R- and S-propranolol could reliably be measured on the chromatographic system. The upper end of this concentration range overlapped with the propranolol concentrations that have been used in CE/frontal analysis studies [17] while still providing a signal that was within the linear response of the detector for the HPAC system.
The retained drug in the frontal analysis experiments was eluted by passing only pH 7.4, 0.067 M potassium phosphate buffer through the column prior to the next frontal analysis experiment. The amount of drug that was required to saturate the LDL column or control column was determined by integration of the resulting breakthrough curve [3] by using Labview 5.1. Corrections were made for the void time and non-specific binding of propranolol to the support by subtracting the breakthrough time of the control column from that of LDL column at each concentration of applied drug, as described in previous work with propranolol on similar HDL columns [27]. For instance, based on this correction, the non-specific binding of R- or S-propranolol to the support made up approximately 12% of the total binding noted on the LDL column at an applied analyte concentration of 25 μM. This extent of non-specific binding was easily corrected by the approach used in this study [27]. The precision of the frontal analysis measurements varied between ±0.1% and ±14% at all the conditions that were examined, with typical precisions for most experiments being ±4-5% or less and with the precision at even the lowest tested concentrations being ±1-7%.
Results and Discussion
General properties of the LDL support
The composition of the LDL support was examined by using both a BCA protein assay and a cholesterol assay. Based on the protein assay, the support was found to contain 6.9 (± 0.4) mg protein per gram silica. This amount corresponded to 27.7 (± 1.6) mg or 12 (± 1) nmol of LDL per gram silica, as based on an average molar mass of 2.3 × 106 g/mol for LDL and a typical apolipoprotein content of 25% for LDL particles [17]. The cholesterol content of the same support was 7.2 (± 1.1) mg cholesterol per gram silica. This cholesterol level corresponded to 16 (± 2.4) mg of LDL per gram silica, as based on a typical cholesterol content of 45% for LDL particles [7,8,10,14,17]. A similar difference in the estimated lipoprotein levels when using the cholesterol assay versus the protein assay has been noted in a previous study employing immobilized HDL [27]. The nKa values reported later in this report were determined using the amount of immobilized LDL based on the protein assay, with nKa values based on the cholesterol assay being 40% lower.
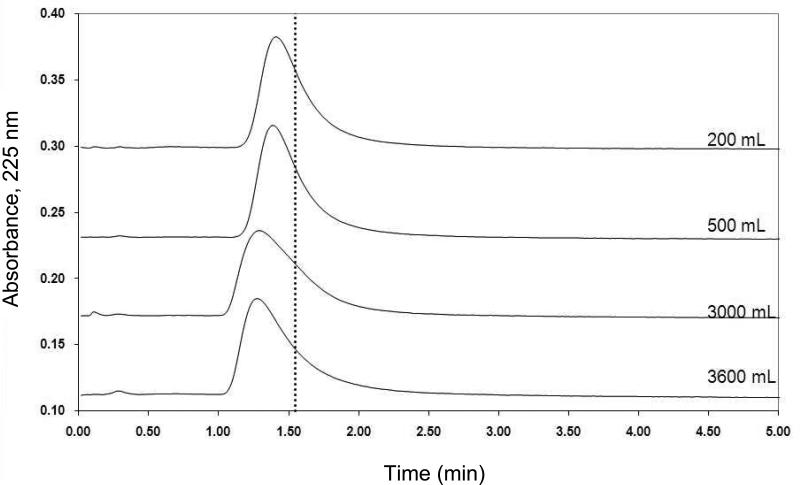
The stability of this lipoprotein support in a chromatographic system was examined by using zonal elution experiments in which periodic injections of R-propranolol were made onto an LDL column under controlled temperature and flow rate conditions. Figure 2 shows the observed retention for 20 μL injections of 50 μM R-propranolol as a function of time for the LDL column when this column was used at 1 mL/min for over 60 h at 37 °C. The first injection of 50 μM R-propranolol occurred after 10 mL of mobile phase (i.e., a time of 10 min at 1.0 mL/min) had been passed through the freshly prepared LDL column. Over the next 60 h of continuous use, the LDL column gave reproducible retention for R-propranolol with a variation of only ± 2%. The average retention time under these conditions was 90.5 (± 1.8) s, which corresponded to a retention factor of 2.90 (± 0.06) (Note: comparable retention has been noted in prior work with propranolol on HPAC columns containing immobilized HDL) [27]. The amount of mobile phase that was passed through the LDL column during this period of time was 3.6 L, which corresponded to 1.6 × 104 column volumes. This result indicated that the LDL column was sufficiently stable for drug binding studies under such conditions. A similar conclusion was reached in prior work examining the stability of immobilized HDL in HPAC columns [27].
Figure 2.
Chromatograms showing the change in R-propranolol retention as a function of mobile phase volume for pH 7.4, 0.067 M phosphate buffer as it was passed through a 100 mm × 2.1 mm i.d. LDL column at 1 mL/min and 37 °C. The dashed vertical line is provided for reference and shows the central moment of the peak for R-propanolol at the beginning of this experiment (Note: there was no significant change in the position of this central moment during the course of this study).
Frontal analysis studies
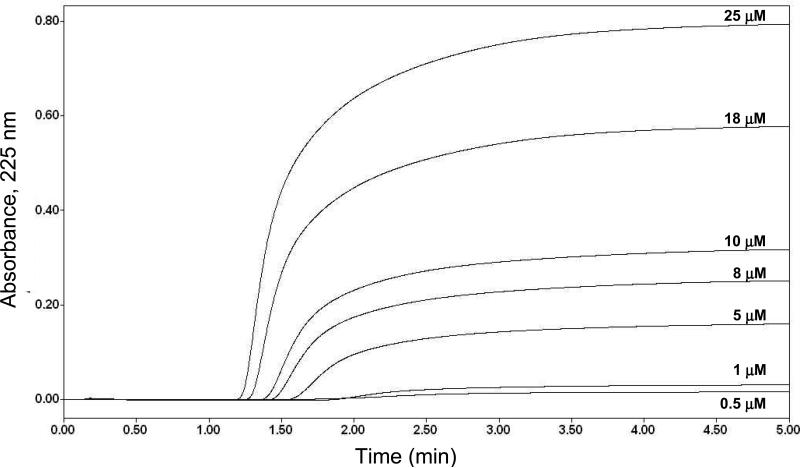
Frontal analysis studies were next conducted to evaluate the binding of R- or S-propranolol to the immobilized LDL support. Figure 3 shows typical frontal analysis breakthrough curves that were obtained on a new LDL column. These results were collected within the period of time during which the LDL support was noted to be stable in the previous section (i.e., when less than 3600 mL of mobile phase had passed through the column). The moles of applied analyte (mLapp) that were required to reach the mean position of each breakthrough curve were then used along with the known concentration of the applied drug ([D]) to generate binding isotherms and to fit the data to various binding models to determine the types of interactions that were occurring between LDL and R- or S-propranolol.
Figure 3.
Typical frontal analysis results obtained for the application of R-propranolol to a 100 mm × 2.1 mm i.d. LDL column at concentrations of 0.5, 1, 5, 8, 10, 18, or 25 μM; these results were obtained at 1.0 mL/min and 27°C in the presence of pH 7.4, 0.067 M phosphate buffer.
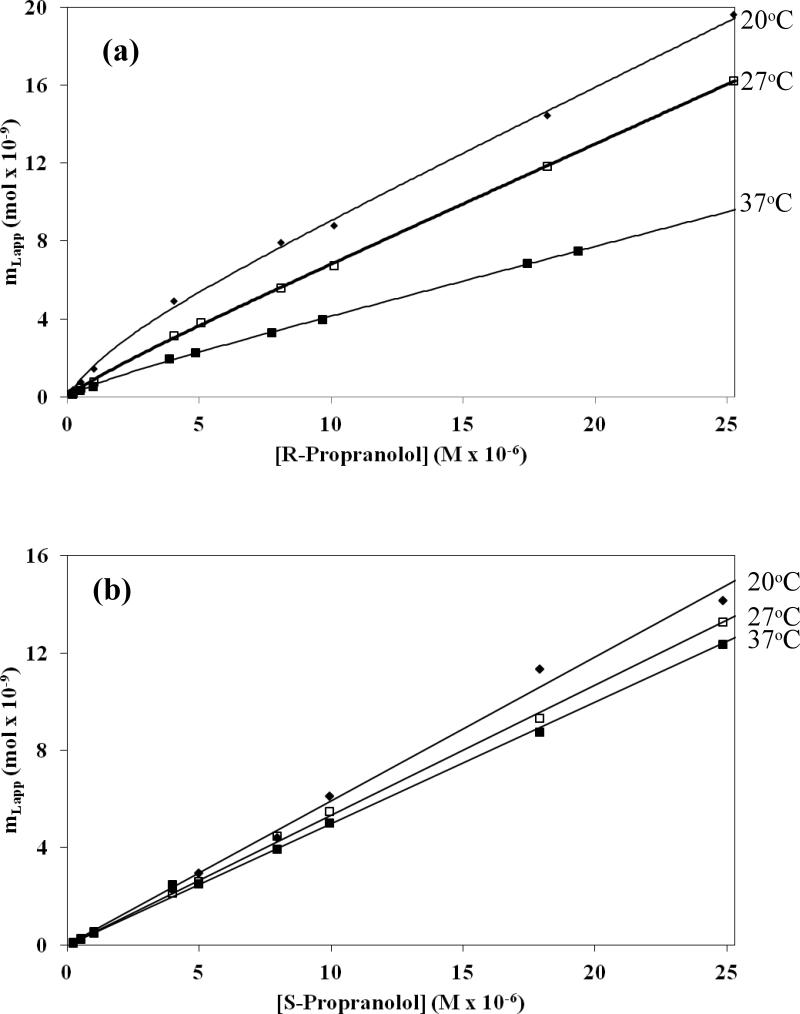
Plots of mLapp versus [D] for R- and S-propranolol were prepared using the frontal analysis data. Examples of such graphs are given in Figure 4. These plots were fit to equations representing four different binding models, as summarized in Table 1 and Eqs. 1-4. For instance, a model involving saturable binding at a single type of site (Eq. 2) was examined because this may have been present if a drug were undergoing site specific binding to apolipoproteins on the surface of LDL [16-18,27]. A saturable two-site binding model (Eq. 4) was used to depict site-specific binding of a drug to multiple locations, as might occur if apolipoproteins contained two distinct binding regions for the applied drug. Another possible interaction that was considered was one in which non-saturable binding (Eq. 1) was present between the drug and interior hydrophobic core of LDL [16-18,27]. A mixed-mode model was also considered in which a combination of saturable sites and a group of non-saturable interactions were present (Eq. 3), as noted earlier for various drugs with HDL [17]. The corresponding association equilibrium constants, binding capacities, or overall affinities that were obtained for each model are summarized in the Electronic Supplementary Material in Tables S1 and S2. The quality of each fit was examined and compared by using the correlation coefficients for the fits, the overall residual values, and the distribution of the data about the best-fit line for each model. The parameters for the models that provided the best-fits to the data are given in Table 2.
Figure 4.
Frontal analysis results for the binding of (a) R- and (b) S-propranolol to an LDL column at 20 °C, 27 °C, and 37 °C. These results were obtained in the presence of pH 7.4, 0.067 M phosphate buffer. Other conditions are given in the text. The best-fit parameters are summarized in Table 2. The precision of binding capacities that were acquired to form these plots were within the range of ±0.1 to ±14%, with typical values for most experiments being in the range of ±4-5% or less.
Table 1.
Binding models used to fit frontal analysis data for R-propranolol and S-propranolol on LDL columns
| Binding model | Predicted responsea | |
|---|---|---|
| Non-saturable interaction | mLapp = mL1Ka[D] | (1) |
| Single group of saturable sites | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) | (2) |
| Two interactions, saturable site + non-saturable | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) + mL2Ka[D] | (3) |
| Two groups of saturable sites | mLapp = (mL1Ka1[D])/(1 + Ka1[D]) + (mL2Ka2[D])/(1 + Ka2[D]) | (4) |
Symbols: mLapp, moles of applied analyte required to reach the mean position of the breakthrough curve; mL1, total moles of active binding site 1; Ka1, association equilibrium constant for saturable binding of the analyte to the ligand at site 1; [D], concentration of the applied drug; mL2, total moles of active binding site 2; Ka2, association equilibrium constant for saturable binding of the analyte to the ligand at site 2; Ka, association equilibrium constant for the analyte in a non-saturable interaction.
Table 2.
Binding parameters obtained for R- and S-propranolol on an LDL columna
| Enantiomer & binding model | Temperature (°C) | mL1 (mol) | Ka1 (M-1) | nKa (M-1)b | Correlation Coefficient (r) |
|---|---|---|---|---|---|
| R-Propranolol, Two interactions: saturable + non-saturable | 20 | 9.0 (± 3.2) × 10-10 | 4.3 (± 3.4) × 105 | 3.2 (± 0.1) × 105 | 0.9998 |
| 27 | 3.0 (± 0.9) × 10-9 | 4.4 (± 2.7) × 105 | 3.5 (± 0.2) × 105 | 0.9992 | |
| 37 | 7.5 (± 1.5) × 10-10 | 5.2 (± 2.3) × 105 | 1.9 (± 0.1) × 105 | 0.9998 | |
| S-Propranolol, Single interaction, non-saturable | 20 | - | - | 3.2 (± 0.1) × 105 | 0.9973 |
| 27 | - | - | 2.8 (± 0.1) × 105 | 0.9995 | |
| 37 | - | - | 2.7 (± 0.2) × 105 | 0.9999 | |
The numbers in parentheses represent a range of ± 1 S.D. All of these results were measured in pH 7.4, 0.067 M potassium phosphate buffer.
In the value for nKa, (i.e., the overall affinity for a non-saturable interaction), the values of mL1 and mL2 in Eqs. 1 and 3 of Table 1 were converted into the moles of analyte or drug per binding agent (n) by dividing the best-fit result for mL1Ka or mL2Ka by the estimated moles of LDL. This latter value was obtained by using the protein content of the LDL support, a typical protein content for LDL of 25% (w/w), and an average molar mass for LDL of 2.3 × 106 g/mol [17]. The better precision noted for nKa versus Ka1 for R-propranolol probably reflects the greater contribution made by the non-saturable versus saturable interactions at many of the drug concentrations used in this study.
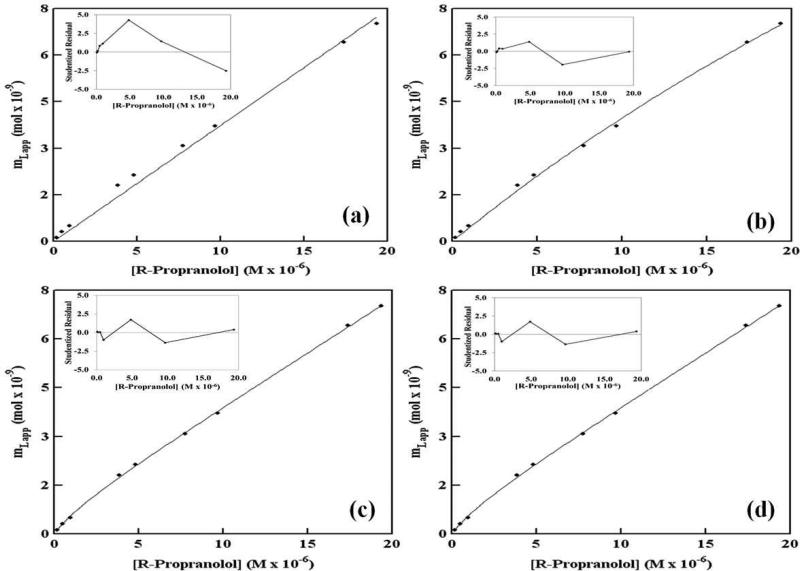
R-Propranolol was found to undergo multiple types of interactions with the immobilized LDL, as demonstrated through a comparison of the various binding models in Figure 5(a-d). R-Propranolol showed the best fit to a mixed-mode model that involved both saturable sites and non-saturable binding. It should be noted that the mixed-mode model for the saturable and nonsaturable interactions, as shown in Figure 5(d), gave the same correlation coefficient and residual plot as the two-site saturable model, as given in Figure 5(c) (e.g., values for r of 0.9998). However, the best-fit equilibrium constants that were obtained for these two models resulted in a much better precision for the saturable/non-saturable model (e.g., see the standard deviations listed in the Electronic Supplementary Material and Table S1 for Ka1, mL1 and nKa in this model versus those values listed for the best-fit parameters in a model based on two groups of saturable sites). The apparent similarities in these two fits can be explained by the fact that Eq. 4 in Table 1 for the two-site saturable model approaches Eq. 3 for a model based on saturable/non-saturable interactions as the term Ka2[D] in the denominator of Eq. 4 becomes much less than one. Under these conditions, the term for the second interaction in Eq. 4 is now mathematically equivalent to the non-saturable term in Eq. 3. This type of situation was noted to occur in the fit of the two-site saturable model to the R-propranolol/LDL data and explains the large uncertainly that resulted for this fit. This observation also explains why the residual plots in Figures 5(c-d) for these two cases were so similar, because they were actually describing the same overall model in which there was a relatively high affinity saturable site and lower affinity, essentially non-saturable binding.
Figure 5.
Examination according to various binding models of frontal analysis data obtained for R-propranolol on an LDL column at 37 °C. The models used in this analysis were as follows: (a) non-saturable interactions, (b) a single group of saturable sites, (c) two separate groups of saturable sites, and (d) a group of non-saturable interactions plus a group of saturable sites. The insets show the residual plots for the fit of each model to the experimental data, as based on studentized residual values that are expressed in relative standard deviation units. These results were obtained in the presence of pH 7.4, 0.067 M phosphate buffer. The correlation coefficients (n = 9) were as follows: (a) 0.9959, (b) 0.9992, (c) 0.9998, and (d) 0.9998. The precision of these results was in the same range as described for Figure 4. The results for all of these fits are summarized in the Electronic Supplementary Material.
A similar comparison made for S-propranolol indicated that this drug gave the best fit to a model for a single type of interaction involving non-saturable binding. For the S-propranolol/LDL interactions, each model depicted in Table 1 gave essentially the same correlation coefficient and residual plot (e.g., values for r greater than 0.9985). However, the best-fit equilibrium constants that were obtained for models other than the non-saturable binding model had large variations in their values (e.g., see the standard deviations listed in the Electronic Supplementary Material and Table S2 for the estimated equilibrium constants and binding capacities of these models versus the values listed for nKa in a model based on only non-saturable binding). This can be explained by the fact that Eqs. 2-4 in Table 1 for one-site saturable binding, saturable/non-saturable binding and two-site saturable binding and can all approach Eq. 1 for a model based only on non-saturable interactions. This latter situation occurs when Ka1[D] or Ka2[D] become much smaller than one in the denominators of Eqs. 2-4 and as the term for site-specific binding in Eq. 3 becomes much smaller than the term for non-saturable interactions. It was found through this approach that the data for S-propranolol at all of the temperatures examined consistently gave the best-fit with Eq. 1 and a non-saturable model. The same conclusion was reached when using double-reciprocal plots of 1/mLapp versus 1/[D]. With this second group of plots a linear response was again obtained for S-propranolol, as would be predicted for a non-saturable model but not for one that involved any saturable interactions [27].
Equilibrium binding constants and temperature studies
Table 2 summarizes the best-fit results that were obtained for R- and S-propranolol with LDL according to the most representative models for these interactions. For R-propranolol, its saturable interaction with LDL had an association equilibrium constant (Ka1) of 5.2 (± 2.3) × 105 M-1 at 37 °C, which represented relatively high affinity binding. This binding was probably occurring between R-propranolol and apolipoprotein B100 (apoB100) on LDL, as suggested by a comparison of the measured binding capacity of LDL during this interaction versus the moles of apoB100 that were present in the LDL column. For instance, the total moles of these binding sites were consistently in the range of 0.7 to 3.0 nmol for R-propranolol between 20 °C and 37 °C (average, 1.6 nmol). This value was in the same range as the amount of LDL that was estimated to be present in the column (i.e., 1.9 (± 0.1) nmol), where each LDL particle typically contains one apoB100 molecule [7]. Similar saturable binding of propranolol with apolipoproteins has been suggested in prior work with immobilized HDL [27].
The second type of interaction seen for R-propranolol with LDL had an overall affinity (nKa) of 1.9 (± 0.1) × 105 M-1 at 37 °C, which represented non-saturable binding. This second type of binding was believed to occur between R-propranolol and phospholipids or the non-polar core of LDL, as suggested in previous work examining the binding of R/S-propranolol and other drugs with both LDL and HDL [16,17,27]. A comparable overall affinity of 2.7 (± 0.2) × 105 M-1 was obtained in this study for the non-saturable binding of S-propranolol with LDL at 37 °C.
The effect of temperature on the interactions between R- and S-propranolol and LDL was also examined. As shown in Table 2, it was found that a moderate change in temperature did not have a significant effect on the equilibrium constants, binding capacities, or binding models that were obtained for R- and S-propranolol with LDL between 20 °C and 37 °C. At each temperature that was employed in this current report, the binding models listed in Table 2 gave correlation coefficients for R- and S-propranolol that were greater than 0.998. The Ka1 values measured for the saturable binding of R-propranolol with LDL varied from only 4.3-5.2 × 105 M-1 over this temperature range, and the nKa values for non-saturable binding by this enantiomer were in the range of 1.9-3.5 × 105 M-1. The nKa values for non-saturable binding by S-propranolol with LDL were in a similar range of 2.7-3.2 × 105 M-1 under the given temperature conditions.
The nKa values measured for R- and S-propranolol for their non-saturable binding with LDL were approximately five- to nine-fold higher than values of 3.7-4.1 × 104 M-1 that have been measured at pH 7.4 and between 4 °C and 37 °C for the same type of interaction of these enantiomers with immobilized HDL [27]. However, this result was expected based on the results of previous studies that have compared drug binding by LDL versus HDL [16,17] and the fact that LDL has a much larger portion of hydrophobic components (i.e., cholesterol and triacylglycerides) than HDL [7-9,17]. Although LDL and HDL also contain different types of apolipoproteins [7], the Ka1 values measured for the saturable binding of R-propranolol with LDL were similar to values of 1.4-1.9 × 105 M-1 that have been estimated for saturable binding by the same solute with HDL [27]. In addition, the range of Ka1 and nKa values estimated in this study agreed with the range of 1-4 × 105 M-1 in overall affinities that have been reported at pH 7.4 and 25 °C to 37 °C for R- and S-propranolol with soluble LDL when using only a non-saturable binding model [16,17].
The fact that R- and S-propranolol consistently followed different binding models in this report supports the observation that the binding of propranolol to LDL is stereoselective. This stereoselectivity was created by the fact that only the R-enantiomer had significant saturable binding to LDL under the conditions of this study (e.g., as probably took place at apoB100). In earlier studies with HDL, both R- and S-propranolol were found to undergo two types of interactions: a saturable interaction that probably involved apolipoproteins and a non-saturable, partition-like interaction. However, no chiral selectivity has been observed for R/S-propranolol with HDL [27]. Prior work examining the binding of propranolol with LDL has either used a racemic preparation of this drug [16] or has found no significant differences in the binding of R- and S-propranolol to LDL when using only a non-saturable binding model [17]. This latter result is not surprising given the similarity in the binding constants that were seen in this present report for the saturable and/or non-saturable interactions of R- and S-propranolol with LDL.
The fact that stereoselective binding was seen in this report for R- and S-propranolol with LDL but not in prior work with HDL that also used a mixed-mode model [27] can be explained by the fact that different types of apolipoproteins are present in HDL and LDL. For instance, HDL may contain up to 5-6 apolipoproteins per particle, with these apolipoproteins consisting of ApoA1, ApoA2, ApoE and ApoC [7]. LDL contains only one apolipoprotein molecule per particle (i.e., ApoB100) [7]. The ability of apoB100 to specifically bind to hormones and drug-like compounds has been previously noted for a number of steroids, including 17-β-estradiol, testosterone, and progesterone [30-32]. However, the results of this study are believed to be the first instance in which stereoselective interactions with apoB100, apolipoproteins, or lipoprotein particles have been observed.
Conclusions
In this report, LDL was immobilized in chromatographic columns and examined for use in drug binding studies, with R- and S-propranolol being used as model solutes for this work. It was found that an LDL column could be used over at least 60 h of continuous operation without any significant loss of retention for R-propranolol. The use of this type of column in frontal analysis studies indicated that R- and S-propranolol had two distinct types of interactions with LDL. Each of these drug enantiomers had non-saturable binding with LDL, which was believed to be due to interactions with the phospholipids or the non-polar core of LDL. A second type of interaction was observed only with R-propranolol and involved saturable binding. This saturable interaction had an association equilibrium constant of 4.3-5.2 × 105 M-1 at 20 °C to 37 °C. The overall affinities for the non-saturable interactions of R- and S-propranolol were similar and ranged from 1.9-3.5 × 105 M-1 at 20 °C to 37 °C. These values showed good agreement with binding constants that have been reported for propranolol when using a similar mixed-mode model for immobilized HDL [27] and with the overall affinities that have been measured for soluble LDL based on a non-saturable model [16,17].
The differences seen in the binding of R- and S-propranolol to LDL, particularly with regard to the presence or absence of measurable saturable interactions, indicated that this binding was stereoselective. It is well known that chiral selectivity can occur as drugs bind to serum proteins such as HSA and AGP [3,4,33,34]. However, this report is the first known example of chiral selectivity being seen for LDL or any other lipoprotein. The stoichiometry of the saturable interactions for R-propranolol with LDL suggested that this stereoselectivity was due to the binding of R-propranolol to apoB100. Thus, this work is also the first possible example of chiral interactions that involve the binding of a drug with an apolipoprotein. Future work with similar HPAC columns could be carried out using zonal elution and competition studies [3,4,24,25] to further examine the interactions of R- and S-propranolol on LDL, as well as the possible interaction of R-propranolol with other solutes known to bind to apoB100 (e.g., testosterone, 17-β-estradiol, and progesterone) [30-32].
This report illustrated the ability of HPAC to be used as a tool for characterizing mixed-mode interactions that involve LDL and related binding agents. As noted in prior work with HDL columns [27], this approach can provide analysis times of only a few minutes per run (e.g., see examples in Figures 2-3). The LDL columns developed in this report were also sufficiently stable to be used for hundreds of experiments. This feature produced a significant reduction versus current methods in the amount of ligand that was needed for a large number of experiments. For instance, the CE/frontal analysis studies in Ref. [17] were conducted using 150 nmol LDL per analysis. In contrast, one column in this current report contained 350 nmol LDL and was typically utilized for more than 160 experiments, or an average of less than 2.2 nmol LDL per analysis. Use of the same LDL columns for multiple studies made it possible to minimize run-to-run variations. In addition, it was possible to use these columns with standard HPLC detectors to provide good limits of detection during the binding studies [3,4,27]. The combined result of these advantages was the ability to quickly obtain precise data over a variety of drug concentrations, which helped lead to the identification of mixed-mode interactions and stereoselective binding between LDL and R- or S-propranolol. These same features should make similar HPAC columns and methods valuable in future studies examining the binding of other drugs and solutes with LDL or alternative lipoproteins.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health under grant R01 GM 044931 and was performed in facilities renovated with support under grant RR015468-01.
References
- 1.Otagiri M. Drug Metab Pharmacokinet. 2005;20:309–323. doi: 10.2133/dmpk.20.309. [DOI] [PubMed] [Google Scholar]
- 2.Bertucci C. Domenici E Curr Med Chem. 2002;9:1463. doi: 10.2174/0929867023369673. [DOI] [PubMed] [Google Scholar]
- 3.Patel S, Wainer IW, Lough JW. In: Handbook of Affinity Chromatography. 2nd ed Hage DS, editor. CRC Press; Boca Raton: 2006. pp. 663–683. [Google Scholar]
- 4.Hage DS, Jackson A, Sobansky M, Schiel JE, Yoo MY, Joseph KS. J Sep Sci. 2009;32:835–853. doi: 10.1002/jssc.200800640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xuan H, Hage DS. Anal Biochem. 2005;346:300–310. doi: 10.1016/j.ab.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 6.Mallik R, Xuan H, Guiochon G, Hage DS. Anal Biochem. 2008;376:154–156. doi: 10.1016/j.ab.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonas A. In: Biochemistry of Lipids, Lipoproteins, and Membranes. Vance DE, Vance JE, editors. Elsevier; Amsterdam: 2002. pp. 483–504. [Google Scholar]
- 8.Barklay M. In: Blood Lipids and Lipoproteins: Quantitation, Composition, and Metabolism. Nelson GJ, editor. John Wiley; New York: 1972. pp. 587–603. [Google Scholar]
- 9.Wasan KM, Cassidy SM. J Pharm Sci. 1998;87:411–424. doi: 10.1021/js970407a. [DOI] [PubMed] [Google Scholar]
- 10.Harmony JAK, Aleson AL, McCarthy BM. In: Biochemistry and Biology of Plasma Lipoproteins. Scanu AM, Spector AA, editors. Marcel Dekker; New York: 1986. pp. 403–430. [Google Scholar]
- 11.Mbewu AD, Durrington PN. Atherosclerosis. 1990;85:1–14. doi: 10.1016/0021-9150(90)90177-k. [DOI] [PubMed] [Google Scholar]
- 12.Durrington PN. In: Lipoproteins and Lipids. Durrington PN, editor. Wright; London: 1989. pp. 255–277. [Google Scholar]
- 13.Havel RJ, Kane JP. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. McGraw-Hill; New York: 1995. pp. 1129–1138. [Google Scholar]
- 14.Skipski VR. In: Blood Lipids and Lipoproteins Quantitation, Composition, and Metabolism. Nelson GJ, editor. John Wiley; New York: 1972. pp. 471–583. [Google Scholar]
- 15.Kwong TC. Clin Chim Acta. 1985;151:193–216. doi: 10.1016/0009-8981(85)90082-8. [DOI] [PubMed] [Google Scholar]
- 16.Glasson S. Mol Pharmacol. 1980;17:187–191. [PubMed] [Google Scholar]
- 17.Ohnishi T, Mohamed NAL, Shibukawa A, Kuroda Y, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. J Pharm Biomed Anal. 2002;27:607–614. doi: 10.1016/s0731-7085(01)00569-6. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed NAL, Kuroda Y, Shibukawa A, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. J Chromatogr A. 2000;875:447–453. doi: 10.1016/s0021-9673(99)01288-1. [DOI] [PubMed] [Google Scholar]
- 19.Mohamed NAL, Kuroda Y, Shibukawa A, Nakagawa T, El Gizawy S, Askal HF, El Kommos ME. J Pharm Biomed Anal. 1999;21:1037–1043. doi: 10.1016/s0731-7085(99)00197-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, Wainer IW. J Chromatogr B. 2008;870:22–26. doi: 10.1016/j.jchromb.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollósy F, Valkó K, Hersey A, Nunhuck S, Kéri G, Bevan C. J Med Chem. 2006;49:6958–6971. doi: 10.1021/jm050957i. [DOI] [PubMed] [Google Scholar]
- 22.Buchholz L, Cai CH, Andress L, Cleton A, Brodfuehrer J, Cohen L. Eur J Pharm Sci. 2002;15:209–215. doi: 10.1016/s0928-0987(01)00219-6. [DOI] [PubMed] [Google Scholar]
- 23.Loun B, Hage DS. Anal Chem. 1994;66:3814–3822. doi: 10.1021/ac00093a043. [DOI] [PubMed] [Google Scholar]
- 24.Hage DS, Chen J. In: Handbook of Affinity Chromatography. Hage DS, editor. CRC Press/Taylor & Francis; New York: 2006. pp. 595–628. [Google Scholar]
- 25.Tweed S, Loun B, Hage DS. Anal Chem. 1997;69:4790–4798. doi: 10.1021/ac970565m. [DOI] [PubMed] [Google Scholar]
- 26.Mallik R, Xuan H, Hage DS. J Chromatogr A. 2007;1149:294–304. doi: 10.1016/j.chroma.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Sobansky M, Hage DS. Anal Biochem. 2010;397:107–114. doi: 10.1016/j.ab.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Allain CC, Poon LS, Chan CSG, Richmond W, Fu PC. Clin Chem. 1974;20:470–475. [PubMed] [Google Scholar]
- 30.Brunelli R, Greco G, Barteri M, Krasnowska EK, Mei G, Natella F, Pala A, Rotella S, Ursini F, Zichella L, Parasassi T. FASEB J. 2003;17:2127–2129. doi: 10.1096/fj.02-1181fje. [DOI] [PubMed] [Google Scholar]
- 31.Wang AJ, Vainikka K, Witos J, D'Ulivo L, Cilpa G, Kovanen PT, Oorni K, Jauhiainen M, Riekkola ML. Anal Biochem. 2010;399:93–101. doi: 10.1016/j.ab.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 32.D'Ulivo L, Chen J, Meinander K, Oorni K, Kovanen PT, Riekkola ML. Anal Biochem. 2008;383:38–43. doi: 10.1016/j.ab.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Patel S, Wainer IW, Lough JW. In: Handbook of Affinity Chromatography. 2nd ed Hage DS, editor. CRC Press; Boca Raton: 2006. pp. 571–592. [Google Scholar]
- 34.Allenmark S. Chromatographic Enantioseparation: Methods and Applications. 2nd ed Ellis Horwood; New York: 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.