Abstract
Histone chaperones can be broadly defined as histone-binding proteins that influence chromatin dynamics in an ATP-independent manner. Their existence reflects the importance of chromatin homeostasis and the unique and unusual biochemistry of the histone proteins. Histone supply and demand at chromatin is regulated by a network of structurally and functionally diverse histone chaperones. At the core of this network is a mechanistic variability that is only beginning to be appreciated. In this review, we highlight the challenges in determining histone chaperone mechanism and discuss possible mechanisms in the context of nucleosome thermodynamics. We discuss how histone chaperones prevent promiscuous histone interactions, and consider if this activity represents the full extent of histone chaperone function in governing chromatin dynamics.
Keywords: Histone, Histone Variant, Histone Chaperone, Nucleosome, Chromatin, Thermodynamics
1.0 Introduction
1.1 Features of the histone protein family
The histone protein family provides a set of components essential for the packaging and homeostasis of the eukaryotic genome. Histone proteins modulate virtually all DNA metabolic events by forming non-random interactions with the DNA. These interactions ultimately influence local and global genome accessibility, regulating the activity of nuclear enzymes such as DNA and RNA polymerases. Core histone protein family members include major-type (also described as canonical or replicative) histones H2A, H2B, H3 and H4, as well as variants thereof such as H2A.Z and centromeric H3 (cenH3). Two copies of the four major-type histone form a histone octamer, which wraps 146 base pairs of DNA to form the nucleosome (Figure 1A) [1]. Variant histones replace major-type histones in combinations to form variant-containing nucleosomes for specific biological contexts.
Fig. 1.
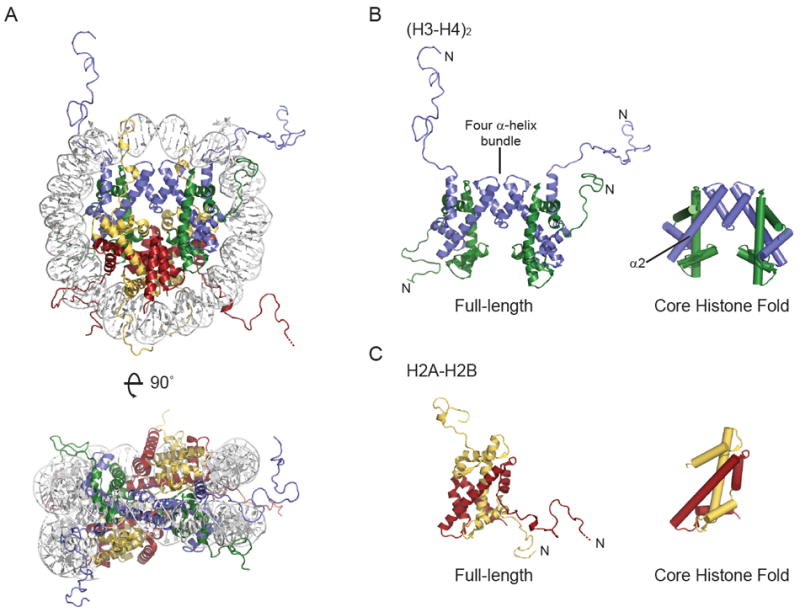
Structure of the major-type nucleosome (A), (H3-H4)2 tetramer (B) and H2A-H2B dimer (C). Cartoons were made using PDB 1KX5. DNA is white, H3 is blue, H4 is green, H2A is yellow and H2B is red. Missing residues at the N-terminus of H2B are shown by a dotted line. In B and C, an N indicates the proteins N-terminus, and the representation on the right shows only the core histone fold. The four α-helix bundle that mediates H3-H4 tetramer formation and the second α-helix of H3 are indicated.
Histone protein family members are defined by the presence of a core histone fold, composed of approximately 65 amino acids [2]. These amino acids adopt a three α-helix structure in the presence of a partner histone (Figure 1B, C), with the structural paralogues H3-H4 and H2A-H2B having quite distinct primary sequence. This contrasts to major-type histone orthologues, which are potentially the most conserved proteins identified to date. Conservation of H2A, H2B, H3 and H4 spans a vast evolutionary distance (including animals, plants and yeast), and extends beyond the core histone fold to peripheral α-helices and disordered N-terminal tails (also C-terminal in H2A). Higher eukaryotes have evolved variants of major-type H3 and H2A, while their hetero-dimerization partners H4 and H2B are invariant with few exceptions (reviewed in [3]). Yeast, for example, has a single major-type H3 (H3.3-like), while metazoans have one or two major-type H3 (H3.1 and H3.2 in mammals), as well as a replacement variant (H3.3) [4]. Present in all eukaryotes is a cenH3 variant that epigenetically defines the centromere [5]. These variants, as well as those of H2A, have high sequence similarity to their major-type counterpart, containing few amino acid substitutions or short insertions confined to specific histone surfaces. Conservation of histone protein family members alludes to heavy structural constraints imposed by nucleosome formation, and is evidence that histone sequence, structure and function are highly inter-dependent and major contributors to organism fitness.
The expression pattern of major-type histone genes is also conserved, although it is achieved through distinct mechanisms in different species [6]. Major-type histone genes, of which there are many isoforms in higher eukaryotes, are typically clustered, with expression peaking during S-phase of the cell cycle [7]. Stoichiometric and equal amounts of major-type histones are required to package the replicated DNA into nucleosomes. A conserved and highly regulated expression pattern testifies to the potential detriment of an unpackaged genome, even transiently. Variant histone genes, on the other hand, do not necessarily cluster with major-type histone genes and have varied expression patterns throughout the cell cycle [8]. Newly synthesized histones, particularly H3, H4 and their associated variants, can be discerned from old histones through a post-translational modification signature [9, 10].
Both major-type and variant histones are extensively post-translationally modified by diverse enzymatic activities including phosphorylation, acetylation, methylation and ubiquitination [11]. Histones can be a substrate as part of the nucleosome or when in a non-nucleosomal complex, depending on the enzyme and the location of the modified residue [12]. Post-translational modifications (ptms) occur predominantly in the lysine/arginine-rich histone tails, but also in the core histone fold [13]. A number of specific lysine acetylations directly influence histone biochemistry, particularly in the context of nucleosome assembly and stability [14, 15]. Many studies have been dedicated to identifying the enzymes involved in histone ptm and ptm-removal, as well as to deciphering the function of histone ptms alone and in combinations [16-18].
A third type of protein closely associated with chromatin metabolism is the linker histones. Despite their name, linker histones do not contain the core histone fold. They can however, associate stoichiometrically with nucleosomes and facilitate inter-nucleosome interactions and higher-order chromatin folding traditionally associated with gene silencing. Compared to major-type histones, little is known about the function and mechanism of linker histones (reviewed in [19]).
1.2 Prerequisites of nucleosome formation
The critical functions of major-type and variant histones in DNA metabolism rely on their ability to form a nucleosome. Numerous crystal structures have revealed that the histones in major-type and variant-containing nucleosomes adopt a similar, octameric arrangement (e.g. [1, 20]). Despite the lack of high-resolution structural evidence, deviant species of variant nucleosomes may be found in vivo [21]. The canonical nucleosome arrangement (Figure 1A) involves head-to-tail hetero-dimerization of the core histone folds of H2A and H2B, as well as H3 and H4. Two H3-H4 dimers further associate via a four α-helix bundle to form a (H3-H4)2 tetramer (Figure 1B). In the nucleosome, this (H3-H4)2 tetramer is flanked by a H2A-H2B dimer on either side. DNA is held around this histone octamer through multiple direct contacts [22]. These include at least one salt-bridge between a backbone phosphate of each DNA strand and each histone [22]. The histone disordered tails protrude between or in close proximity to the DNA gyres from the somewhat circular nucleosome core.
The structure of the nucleosome is in keeping with the many biochemical studies on recombinant histone proteins. Major-type histones, for example, cannot hetero-dimerize in any combination, with H2A-H2B and H3-H4 pairs being exclusive and obligate at low and high ionic strengths [23]. The H3-H4 dimer also exists in equilibrium with a (H3-H4)2 tetramer at physiological ionic strength in the absence of other protein or DNA factors [24, 25]. Contacts between the H2A-H2B dimer and (H3-H4)2 tetramer to form a stoichiometric octamer also occur in solution, albeit only at high ionic strength. High ionic strength, like the vicinity of polyanionic DNA, neutralizes some of the positive charges of the histones. Perhaps the integral observation in histone biochemistry is that salt gradients (high to low) of equimolar histone-DNA mixtures, induce nucleosome formation (Figure 2) [26]. Salt gradients circumvent the observed precipitation of histone-containing mixtures at physiological ionic strength [27]. This precipitation may be due to promiscuous ionic interactions between the highly basic histones and acidic DNA, histones and nucleosomes [28], or a result of histone conformational flexibility (S. D’Arcy, unpublished observation).
Fig. 2.
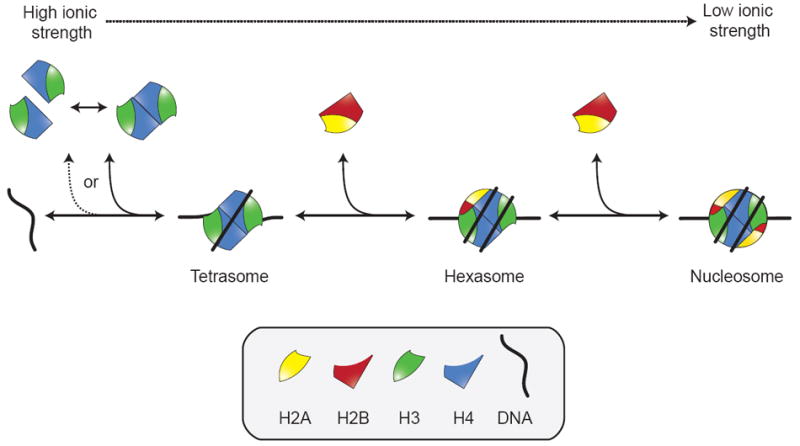
Model of sequential nucleosome assembly. The (H3-H4)2 tetramer (or two H3-H4 dimers) bind to DNA to form the tetrasome. This is followed by the addition of two H2A-H2B dimers. The intermediate, a tetrasome plus a single H2A-H2B dimer, is described as a hexasome. This process is reversible in salt gradient experiments as indicated. A similar sequence of events has been delineated for Nap1 nucleosome assembly in vitro.
Analysis of salt-dependent in vitro nucleosome reconstitution ultimately leads to a model of sequential nucleosome assembly (Figure 2). The sequential model postulates that the (H3-H4)2 tetramer (or two H3-H4 dimers) is first deposited onto DNA, followed by the addition of two H2A-H2B dimers. The ability of (H3-H4)2 tetramer to bind DNA at higher ionic strength than H2A-H2B dimer supports this model [29, 30]. While this model has been confirmed in vitro and is likely reversible [31, 32], evidence for its relevance in vivo is somewhat indirect [33, 34]. The distinct exchange rates of H2A-H2B (fast) and H3-H4 (slow) in chromatin are consistent with the model, although these may be a result of specific cellular variables and not generally applicable [35, 36]. It can also be argued that the path of the DNA makes it difficult to envision deposition of an intact histone octamer, the only other stable histone complex detected in vitro [37]. Cross-linked octamers, however, do assembly into nucleosomes with the same efficiency as uncross-linked histones in vitro [28, 38]. In the context of the sequential model, exchange of the central (H3-H4)2 tetramer, for example for incorporation of replication-independent H3 variants, would require the complete disassembly of the nucleosome.
In the cell, multiple steps have to precede the productive formation of nucleosomes (Figure 3). In particular, the histones must be synthesized and folded in the cytoplasm, before being imported into the nucleus and recruited to sites of deposition at the DNA. This flow of histones has to be highly facilitated and regulated to meet the supply and demand of DNA-templated processes. In detail, these prerequisites of nucleosome formation include:
Dimerization of newly synthesized H2A and H2B, as well as H3 and H4, in the cytoplasm. Folding of H2A-H2B and H3-H4 hetero-dimers likely involves traditional protein-folding chaperones such as Hsp70 and Hsp90 [39].
Nuclear import of H2A-H2B and H3-H4 (either as a dimer or a tetramer). While nuclear translocation makes use of standard import pathways, other proteins are likely to be involved and essential (see Pemberton et al., this issue).
Recruitment of H2A-H2B and H3-H4 to specific sites where nucleosomes can and are desired to form.
Prevention of non-specific interactions with other cellular proteins, DNA or RNA, as observed in vitro at physiological ionic strength [27, 40]. This is thought to be particularly important if histones are in excess compared to DNA and thus require storage.
Positioning of H2A-H2B and H3-H4 into a nucleosome configuration.
Eviction and exchange of nucleosomal histones. This pertains to the incorporation or removal of a variant histone from a pre-existing nucleosome.
Fig. 3.
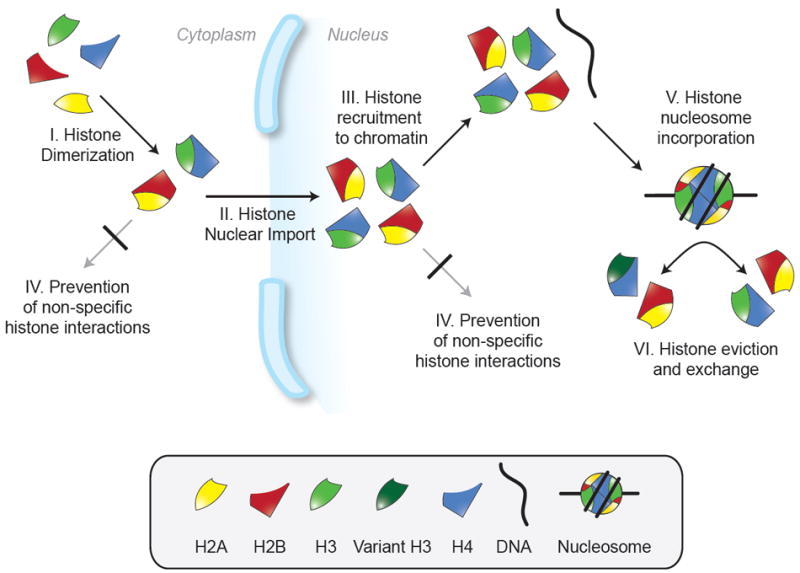
Prerequisites of nucleosome formation and contexts of histone chaperone activity. Histone chaperones may facilitate nucleosome formation by being involved in some or all of the processes I-VI indicated. Histones may be transferred between chaperones to complete all these processes in a regulated manner.
An important functional class of proteins involved in many of these processes, especially IV-VI, are the histone chaperones.
2.0 Histone Chaperones
2.1 A framework of histone chaperone function
The term ‘chaperone’ was first applied to a protein involved in nucleosome formation in the late seventies [27]. Studies by Laskey and colleagues identified the acidic protein NPM in Xenopus laevis egg extracts able to bind histones and facilitate nucleosome formation in an ATP-independent manner [27, 41]. It is remarkable that these two properties remain the only shared and thus defining characteristics of histone chaperones today. It should be reinforced that histone chaperones possess both histone-binding and ATP-independent nucleosome assembly, but are not stable constituents of the final nucleosome product. This distinguishes them from proteins with histone ptm-recognition domains, such as bromodomains, as well as from ATP-dependent nucleosome remodelers. Histone association is routinely assayed both in vitro and in vivo using standard techniques, while nucleosome assembly is frequently only assessed by in vitro experiments on linear or circular DNA. A protein involved in any one of the aforementioned prerequisites II-VI is likely to have in vitro nucleosome assembly activity (Figure 3). Experiments involving yeast genetics or cell-free extracts are more stringent tests of so-called direct nucleosome assembly in vivo.
Efforts by many laboratories employing diverse experimental strategies have identified conserved histone chaperones involved in some or all of I-VI in yeast and metazoans (see [42] for a review of the functions of specific histone chaperones). Cytosolic H2A-H2B and H3-H4, for example, are bound by Nap1 and NASP, Asf1 respectively, and might remain bound during translocation to the nucleus [39, 43-46]. Nap1 and Asf1 also function in the nucleus, where the histones are channeled into distinct pathways associated with DNA metabolic events. These events are typically classified as DNA replication-dependent or independent. CAF1 is thought to be a replication-dependent chaperone, while HIRA activity is replication-independent [45, 47, 48]. Many chaperones are functionally linked to transcriptional regulation, such as FACT, which facilitates eviction of H2A-H2B from the nucleosome [49-52]. NPM represents a storage chaperone insofar as it binds the large pool of histones that will be used for packaging newly replicated DNA in the X. laevis egg [41]. Homologous NPM chaperones also act as sinks for histones removed during sperm development and fertilization [53, 54]. Linker histone chaperone activity has been described as an addition feature of Nap1, NASP and NPM1, potentially regulating higher order chromatin structure [55-58]. Histone chaperones thus function in an array of cellular events, broadly categorized by the aforementioned I-VI (Figure 3).
The sheer number of histone chaperones and histone-binding proteins identified to date supports the long-held view of there being little or no free histones at any given time or locus in the cell. Implicit in the idea that most histones are nucleosomal or chaperone-bound, is the transfer of histones from one chaperone to another (a ‘hand-off’). This would allow a specific chaperone to perform a subset of I-VI (Figure 3). The recent description of distinct, ordered, complexes associated with newly synthesized histones suggests that this notion is applicable to H3-H4 [39]. Biochemical analysis of H3-H4 purified from either the cytoplasm or nucleus, failed to detect free or unbound histones [39, 44, 45, 59, 60]. Rather, H3-H4 was found in different histone chaperone-containing complexes. Details of the temporal hierarchy of these complexes, as well as a similar study for H2A-H2B, are eagerly awaited. The localization of bona fide chaperones in both the nucleus and the cytoplasm (e.g. [46]), as well as the often stoichiometric amounts of histone chaperones and histones [45, 59], also attest to a small or absent population of free histones in the cell.
2.2 Histone chaperone structural diversity
Given the common task, a surprising number of unrelated structural motifs are found throughout known histone chaperones. While many full-length or multimeric histone chaperone structures are outstanding, several have been solved (e.g. [61-65]). Structures of Nap1, Asf1 and NPM are illustrative examples. Nap1 adopts a homo-dimeric earmuff fold that is characterized by a non-coiled coil motif and two composite α/β domains [61]. Asf1 and NPM, however, have β-sandwich architectures with immunoglobulin-like and jelly roll topologies respectively [62, 63]. The Nap1 fold has only been found in functionally similar homologues such as Vps75 and SET [66, 67], while Asf1 and NPM folds have been observed in proteins with varied functions. Further, in both solution and in the crystal, Nap1 is an obligate homo-dimer, while Asf1 is a monomer and NPM is a homo-pentamer or decamer [62, 63]. These oligomeric states are thought to persist in vivo. Hetero-oligomeric chaperones also exist with examples including FACT, composed of SSRP1 and SPT16; and CAF1, composed of p150, p60 and RbAp48, in humans. The latter two subunits adopt yet another structural motif, a β-propeller fold [68, 69]. This diversity in basic structure and oligomeric state suggests that each histone chaperone evolved independently to serve a specialized function in the cell. Different folds may facilitate particular facets of histone chaperone activity (such as I-IV, Figure 3), with each facet being achieved in multiple ways.
Although histone chaperone structure is varied, one commonality somewhat widespread is the presence of regions rich in glutamic and aspartic acids. These acidic stretches are often found near the chaperones C-terminus and are presumably disordered, at least in the absence of a binding partner. A shortlist of chaperones containing such acidic regions includes Nap1, Asf1, Rtt106, Chz1, NPM, DAXX (not at the C-terminus) and the SPT16 subunit of FACT [70]. Unlike the folded domains of histone chaperones, these acidic stretches aren’t necessarily conserved. Fungal Asf1, for example, contains these stretches, while human Asf1 does not [62]. Similarly, the length and composition of these stretches varies between the many Nap1-like proteins in humans, as well as between yeast and Drosophila melanogaster Nap1. The latter is intriguing as the acidic stretches in D. melanogaster Nap1 contain sites of polyglutamylation [71]. This ptm may compensate for a less acidic composition.
2.3 Perspective
An important principle emerges from these introductory remarks; histone chaperones are mechanistically diverse. Mechanistic diversity reflects the many types of histone proteins (major-type, variant and linker), the many contexts of ‘chaperoning’ activity, and the structural heterogeneity of histone chaperone proteins. These physical and contextual variances ultimately make it extremely difficult to define histone chaperones from a mechanistic standpoint. In fact, the most apt and encompassing mechanistic description remains unchanged since the founding article by Laskey and colleagues over 30 years ago [27]. Laskey recognized that the complete instruction manual for nucleosome assembly in vitro is contained in the histones and DNA. He thus proposed that histone chaperones function by preventing promiscuous histone-DNA interactions. The challenge is to determine precisely how histone chaperones prevent these interactions and if this is the true extent of histone chaperone activity in vivo. The histone chaperone family contains mechanistically-defined, as well as contextually-defined, sub-groups. The remainder of this article will identify key obstacles in determining histone chaperone mechanism and will propose potential mechanisms based on the thermodynamic properties of nucleosome assembly and disassembly, and histone chaperone-histone interactions.
3.0 Major Challenges in Determining Histone Chaperone Mechanism
3.1 The interaction between histones and their chaperones
For many histone chaperones, a missing link in the elucidation of mechanism is the absence of structural detail regarding their interaction with histones. While numerous chaperone-only structures are available, they often lend little insight into the mode of histone-binding. Major-type co-structures solved to date include human and yeast Asf1 with H3-H4 [72-74], and the homologous p46/p55 subunits of human and D. melanogaster CAF1 respectively with a H4 peptide [68, 75]. Even though p46 paralogue p48 assembles cenH3 nucleosomes in vitro [21], the significance of the latter in terms of histone chaperone mechanism is vague. This is because p46/p55 is a member of several complexes involved in histone biochemistry [76]. It is interesting to note however that the insertion of the H4 histone fold α-helix into the binding groove of p46/p55 requires a conformational rearrangement in the H3-H4 heterodimer. In the Asf1-H3-H4 co-structure, Asf1 binds at the H3 interface required for (H3-H4)2 tetramer formation (Figure 4B). The C-terminal β-strand of H4 also extends away from the H3-H4 dimer and inserts into a hydrophobic pocket in Asf1. In the nucleosome or histone octamer, this strand folds back and aligns with the C-terminal β-strand of H2A (Figure 1). Both large and small conformational differences in the co-structures attest to a degree of plasticity in the arrangement of histone protein secondary structures, particularly when the histones are non-nucleosomal. Such plasticity, as well as the involvement of intrinsically unstructured regions, may account for the lack of co-structures available for the many other histone chaperones.
Fig. 4.
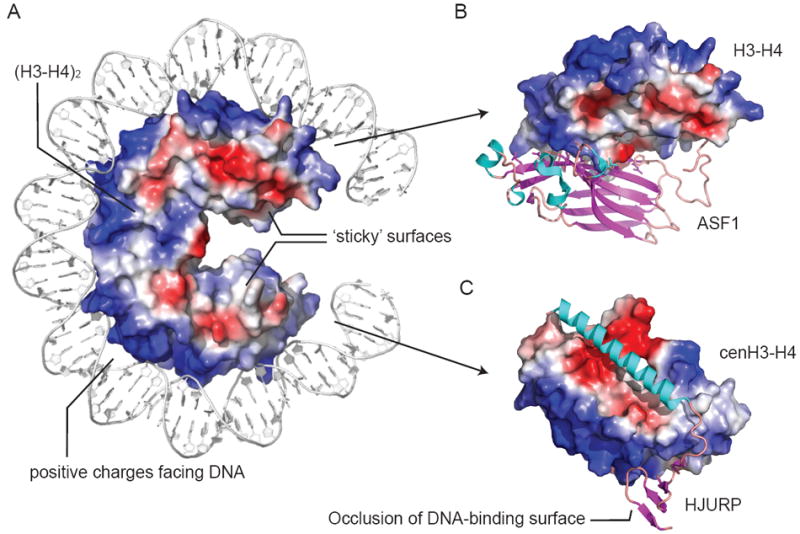
Nature of histone-DNA (A) and histone-histone chaperone (B, C) interactions. A shows a vacuum electrostatic surface charge of (H3-H4)2 with 72 base pairs of DNA from PDB 1KX5. Basic surfaces occur along the path of DNA. The opposite surface is involved in inter-histone interactions and is more hydrophobic. The four α-helix bundle of the (H3-H4)2 tetramer is buried in A. B shows Asf1 (cartoon) preventing (H3-H4)2 tetramer formation by interfering with four α-helix bundle formation (PDB 2HUE). C shows HJURP (cartoon) binding along α2 of cenH3, disrupting (cenH3-H4)2 tetramer formation. HJURP also blocks DNA binding through a C-terminal β-sheet domain (PDB 3R45). In B and C, the H3-H4 dimers are shown as surface in an identical orientation to A.
The insight provided by the Asf1-H3-H4 structure is testimony to the information that may be gleaned from co-structures. Asf1 binding to an obligate H3-H4 dimer has implications for the sequential model of nucleosome assembly both in vivo and in vitro, mechanisms of nucleosome disassembly, histone ptm (as Asf1, like Vps75, activates histone acetyltransferase Rtt109 [77]) and a potential H3-H4 hand-off (see [42] for a more complete discussion). Overall, however, the Asf1-H3-H4 co-structure clearly indicates that binding to histone chaperones and nucleosome formation, are mutually exclusive. This idea is echoed in other co-structures available involving variant histones. These include Chz1 with H2A.Z-H2B [64], and HJURP/Scm3 with cenH3-H4 [78-80]. HJURP and Scm3 are cenH3-H4-specific chaperones in human and yeast respectively. Histone-binding by HJURP/Scm3 is incompatible with the histone-DNA interactions observed in the nucleosome (Figure 4C). Like Asf1, HJURP and Scm3 preclude (cenH3-H4)2 tetramer formation, albeit through a different mechanism. In HJURP-cenH3-H4, the interface required for (cenH3-H4)2 tetramer formation is sterically occluded, while in Scm3-cenH3-H4 it is also distorted. Another common feature of these co-structures is the presence of extensive hydrophobic interactions at the histone interface. This reinforces that histone chaperones are likely to be doing more than just mimicking salt.
The co-structures of HJURP and Scm3 with cenH3-H4 have also yielded insight into histone chaperone discrimination of H3 variants. Both HJURP and Scm3 contain a long α-helix that runs anti-parallel to the central α-helix of the cenH3 histone fold. Overall affinity is achieved through interlinking hydrophobic residues flanked by salt bridges, reminiscent of a coiled-coil interaction. Variant-specificity is conferred through small highly conserved alterations in the hydrophobic surface, and also through neighboring loop-loop contacts, spanning the region of cenH3 disparate in sequence to major-type H3. It is interesting to note that variations in the same hydrophobic surface occur in mammalian variants H3.1, H3.2 and H3.3. As this region does not make crucial contacts in the histone octamer, it may have evolved novel functions more easily. In metazoans, H3.1 is specifically chaperoned by CAF1, while H3.3 is specifically chaperoned by HIRA and DAXX [44, 45, 60]. It remains an exciting field of study to determine how structurally and mechanistically histone chaperones combine the discrimination of histone variants with their cognate nucleosome assembly activity (see Hamiche et al., this issue).
In the absence of other co-structures, a lot of emphasis has also been placed on the fact that Asf1 binds H3-H4 via the exposed edge of a β-sheet. Such β-sheets have been proposed to be primary histone-binding sites as numerous other histone chaperones contain them (e.g. Nap1 and NPM) [72]. Based on biochemical and biophysical approaches, however, this hypothesis has not held up in all cases. Mutagenesis of a Nap1-like protein, for example, confirms that the β-sheets are not likely involved in H2A-H2B recognition [66]. The histone chaperone family likely employs a variety of histone-recognition motifs and the lack of co-structures currently hinders the determination of mechanism. Co-structures are required to identify any commonalities among histone chaperones, as well as to explain the molecular basis (or lack thereof) of specificity for a particular histone (major-type, variant or linker). Co-structures may also reveal if the widespread acidic stretches can adopt ordered structures in the presence of histones.
3.2 Applicability of in vitro results in vivo
Another challenge in determining histone chaperone mechanism is the interpretation of factor-dependent in vitro nucleosome reconstitution assays. These assays determine if a particular factor promotes nucleosome formation in histone-DNA mixtures at less than 350 mM salt and pH 7.5 to 8.0. These ionic and pH conditions allow recapitulation of general chromatin properties observed in vivo. If precipitation is avoided and nucleosomes are formed, then the factor is said to have nucleosome assembly activity and consequently thought to deposit histones in vivo. Such assembly activity is often assumed to stem from direct deposition of histones onto DNA by the histone chaperone. A recent thermodynamic study of Nap1-mediated nucleosome assembly however, reinforces that such a direct mechanism is not required in vitro or in vivo (see also section 4.3) [14]. The results of this study and others (e.g. [32]) indicate that any histone-binding protein (or other chemical factor) with an affinity in the range of the respective histone-DNA complex product can potentially facilitate nucleosome assembly in vitro. In vivo these histone-binding proteins may function in any of the aforementioned processes (I-IV) (Figure 3). Asf1 is an example of a histone chaperone with nucleosome assembly activity only under certain in vitro conditions [24, 81, 82] that does not directly assemble nucleosomes in a X. laevis extract system [42, 83]. Thus the ability to assemble nucleosomes in vitro may simply reflect an ability to bind histones tightly and avoid non-nucleosomal interactions.
The somewhat artificial scenario of the vitro reconstitution assay is further indicated by experiments showing that poly-glutamic acid or poly-aspartic acid successfully assembly nucleosomes [84]. Unlike Nap1, however, they do this by stabilizing the histone octamer even in the absence of DNA [84]. This alludes to a purpose for the acidic stretches often found in histone chaperones and highlights that in vitro nucleosome reconstitution can in fact be informative. In particular, it is useful to identify potential pathways of nucleosome formation and how they are biased by particular factors.
Another difficulty in applying in vitro mechanistic data in vivo relates to the specificity of a histone chaperone for particular histones. Nap1, for example, binds H2A-H2B and H3-H4 equally well in vitro [85]. Chz1 is also thought to be H2A.Z-H2B-specifc, and yet the co-structure fails to identify any H2A.Z-specific contacts [64]. It is thus more likely that specificity is achieved through direct competition or temporal and spatial regulation of histone chaperone activity in vivo. The fact that histone chaperones are often found in complex with other factors suggests that spatial and temporal regulation could be achieved by accessory subunits. Furthermore, it is still uncertain to what extent activities from biochemically independent, non-homologous histone chaperones are redundant in the cell.
3.3 Substrate and product remain obscure
The third major obstacle in identifying the mechanism of a histone chaperone is pinpointing the biological context of activity. The previously described prerequisites of nucleosome formation (I-VI) are examples of possible contexts (Figure 3). Layered upon these somewhat generic components of nucleosome assembly, there are also DNA-templated events such as replication, repair (of which there are many types) and transcription. Activities of histone chaperones may be regulated by specific contexts or DNA-templated events. CAF1, for example, readily associates with histones, but only exhibits nucleosome assembly during active DNA replication [45, 48]. It is unclear if this functional coupling results from specific association with the replication machinery, an allosteric activation mechanism or replication-specific features of the acceptor DNA or chromatin. Histone ptms and alternative nucleosome conformations have been hypothesized to influence histone chaperone activity [37]. To determine histone chaperone mechanism, the precise biological context of activity must to be defined.
Moreover, precisely how nucleosome structure is affected during DNA replication, repair and transcription is the subject of much debate and ongoing research (see [86] for a thorough review). The lack of evidence for a biochemical or genetic link between chaperoning of H3-H4 and H2A-H2B raises the question; are octameric, left-handed nucleosomes the universal substrate and product of histone chaperone-mediated chromatin assembly and disassembly? It will be difficult to determine histone chaperone mechanism as long as the constituents and features of the relevant forms of chromatin remain obscure. It is the thermodynamics of assembly and disassembly of these nucleosomal substrates and products, which allows histone chaperone mechanisms to be postulated. Overcoming this hurdle is a challenge not only restricted to those interested in histone chaperone mechanism, but DNA biology in general.
4.0 Models of Histone Chaperone Function
4.1 Preface
In spite of the many challenges in studying histone chaperones, several hypotheses regarding their mechanism once in the nucleus can be generated. These hypotheses deal with the following important questions:
How do histone chaperones prevent promiscuous histone interactions?
Do histone chaperones do more than just prevent histone aggregation?
Are nucleosome assembly and disassembly distinct processes?
How are histone chaperone pathways regulated?
4.2 Mechanistic implications of distinct histone-binding modes
Based on in vitro biochemistry, unbound or free H3-H4 and H2A-H2B units are presumed to be ‘sticky’ in the cell. The excess positive surface charges of free histones limit the formation of the histone octamer, as well as the (H3-H4)2 tetramer at physiological conditions [84]. As a consequence, normally protected, hydrophobic histone-histone interfaces become solvent-exposed outside of the nucleosomal octameric arrangement [23, 25, 87-89]. In this state of high potential energy, the histones readily engage in non-specific interactions, electrostatic or hydrophobic in nature, with nucleic acids, proteins or other cellular components [14, 28]. The putative detriment of these non-nucleosomal complexes to nucleosome formation has been illustrated by a study with Nap1 [14]. The histone chaperone activity of Nap1 can be attributed to its ability to inhibit H2A-H2B binding to DNA through contacts between Nap1 and H2A-H2B. The lack of structural or mutational data, however, means the molecular determinants of the Nap1-H2A-H2B complex are unknown. The available thermodynamic parameters therefore cannot be readily interpreted on a molecular level. Based on available structural and biochemical data, we propose that histone chaperones exhibit two alternative histone-binding modes (Figure 4). Both modes prevent promiscuous histone interactions. It remains to be seen if both modes can be employed by a single histone chaperone.
The first mode involves neutralizing excess positive charge by providing acidic residues to alleviate charge repulsion. This mode seems to provide little selectivity for H2A-H2B or H3-H4 (and in some cases the linker histones), and alludes to a purpose of the large negatively charged surfaces found in numerous histone chaperones. The acidic residues may be structured or unstructured in the apo-histone chaperone, and may form either tight or transient interactions with the histones. Phosphorylation of non-acidic residues will also contribute to this mode, as observed for NPM [90, 91]. The involvement of ptms that regulate chaperone-histone affinity might be crucial to ensure chaperone activity occurs in the applicable biological context. While counteracting electrostatic repulsion favors self-association of histone hetero-dimers into (H3-H4)2 tetramers and potentially histone octamers, steric constraints imposed by chaperone architecture would confer additional specificity. For example, the symmetric geometry of Nap1 and Vps75 facilitates binding of an obligate (H3-H4)2 tetramer [92]. The mechanistic benefits of electrostatic shielding stem from the histone chaperones ability to directly compete with non-specific electrostatic interactions, particularly with DNA. Under favorable thermodynamic conditions, pre-formed (H3-H4)2 tetramers, H2A-H2B dimers or histone octamers can be ‘presented’ to the recipient DNA or other histone chaperones.
The second, fundamentally different mode is to block histone hydrophobic surfaces. This mode is evident in the co-structures of Asf1-H3-H4 (Figure 4B), HJURP-cenH3-H4 (Figure 4C), Scm3-cenH3-H4 and Chz1-H2A.Z-H2B [64, 72, 74, 78, 79]. This mode is likely to be more specific as surface-exposed amino acids are divergent between H2A-H2B and H3-H4. In fact, subtle variations in the hydrophobic surfaces of the H3-H4 dimer may form the basis of the histone variant specificity observed for a number of histone chaperones. Histone chaperones that employ this mode are expected to retain histone-binding at high ionic strength. Indeed, Asf1, DAXX, HJURP and Scm3 histone complexes are stable at high salt, with hydrophobic contacts outweighing salt bridges and hydrogen bonds [44, 60, 74, 78, 79]. While the most immediate effect of this mode counteracts aggregation via hydrophobic histone surfaces, the chaperone can also prevent nucleosome formation through steric occlusion. As described previously, Asf1 and HJURP/Scm3 prevent (H3-H4)2 tetramer formation and occlude part of the DNA-binding site of the H3-H4 dimer (Figure 4) [72, 74, 78, 79]. Promiscuous histone interactions are thus efficiently prevented, despite retaining some of the excess positive charge in the chaperone-histone complex.
The use of diverse and somewhat complementary histone-binding modes reinforces the variety of mechanisms employed by histone chaperones to accomplish the seemingly simple task of preventing aggregation. This diversity, however, sets the stage for the formation of multi-chaperone complexes. Histones may be passed between chaperones within these complexes before eventually being made available for nucleosome assembly. Other components of these complexes may be integral in fine-tuning histone-binding and release onto DNA or other pre-nucleosomal complexes. This resolves the conundrum of histone chaperones essentially rendering the histones inert for any (specific or non-specific) interactions with other histones or DNA.
4.3 Thermodynamic and kinetic components of histone chaperone activity
Potential mechanisms of histone chaperones can be illustrated using simplified reaction diagrams such as those shown in figure 5. These diagrams are based on thermodynamic constants of complexes involving histone chaperones, histones and/or DNA. As described in section 4.2, free histones readily engage in electrostatic interactions with DNA. These interactions can be specific and associated with nucleosome formation (Figure 5, right) or so-called non-specific resulting in irreversible precipitation in vitro (Figure 5, left). The specific complexes may be (H3-H4)2 tetramer bound to DNA (tetrasome), tetrasome plus one copy of H2A-H2B dimer (hexasome), or the nucleosome. In the absence of a histone chaperone, the predominant reaction product will be the non-specific complexes because the specific complexes involve only a small subset of all potential histone-DNA interactions (Figure 5A).
Fig 5.
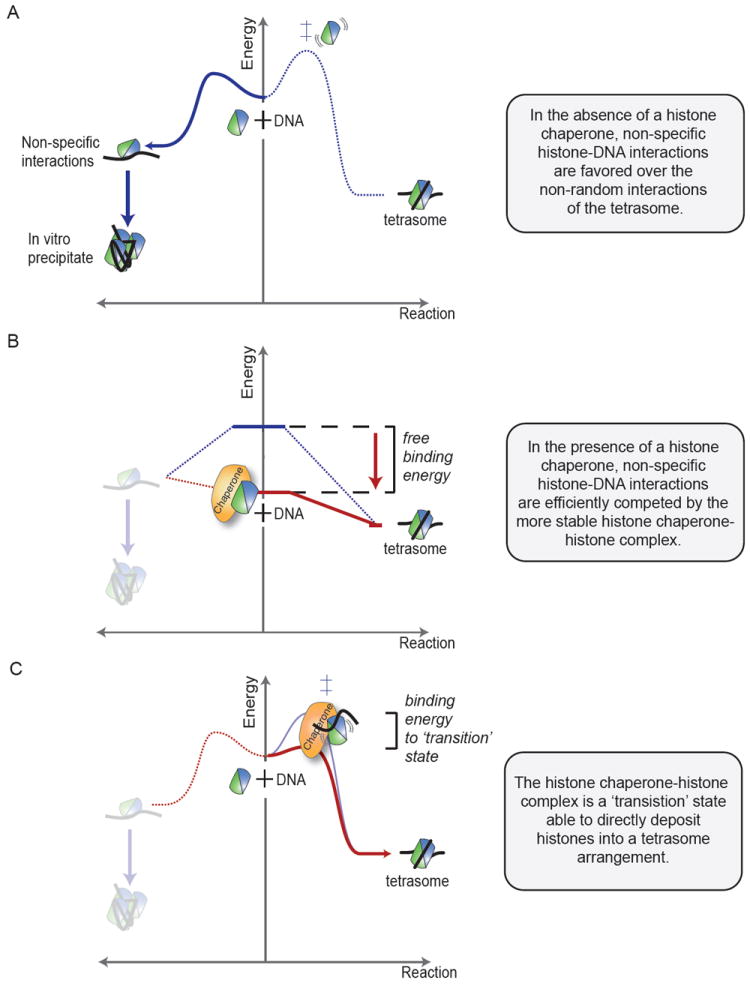
Models of histone chaperone function illustrated as free-energy reaction diagrams. The change in free-energy is plotted along the reaction coordinate as the substrate (DNA and histones, middle) becomes the product (precipitate, left, or tetrasome, right). Analogous to a standard chemical reaction, substrates are postulated to go through a ‘transition state’ of highest free-energy (‡) before forming the product. The transition state represents a kinetic barrier for the reaction. A shows that in the absence of histone chaperones, non-specific interactions of histones (here H3-H4) with DNA are favored (left) over the tetrasome formation (right). Tetrasome formation requires histones and DNA to encounter in a non-random orientation. B shows that a Nap1-type chaperone binds histones with a free-energy similar to the final tetrasome product. In the chaperone-bound state, non-specific interaction with DNA (left) is costly, and thus the tetrasome formation (right) is favored. C shows an alternative mode of chaperone action. The chaperone binds to form a ‘transition state’ that is competent to directly deposit histones onto DNA. This state might represent a chaperone-histone-DNA trimeric complex, or a conformationally activated histone moiety that is primed to form nucleosomal DNA contacts (right).
A histone chaperone may promote specific complex formation by lowering the free energy of the histones (Figure 5B). Free energy is lowered by the formation of a chaperone-histone complex that is thermodynamically favored over non-specific histone-DNA complexes. This mechanism has been proposed for Nap1 [14, 32], which binds either H3-H4 or H2A-H2B with nano-molar affinity in vitro [76]. Through these interactions, Nap1 assembles nucleosomes via tetrasome and hexasome intermediates (Figure 2) [32]. The two key measurements are the affinities of H2A-H2B for DNA (non-specific), and H2A-H2B for the tetrasome (specific) [14]. The absolute numbers vary depending on the DNA sequence, with the well-positioned 601 sequence [77] measuring 44 ± 5 nM and 13 ± 3 nM correspondingly [14]. These numbers fit well with the Nap1 affinity for H2A-H2B of 7.8 ± 0.4 nM insofar as H2A-H2B will prefer to bind Nap1 over DNA (7.8 < 44 nM), but equally likes being bound to Nap1 or tetrasome (7.8 ~ 13 nM). When generalized to other histone chaperones with nano-molar histone affinities, it seems that chaperones may make histones available for nucleosome formation (Figure 5B, right) by disfavoring non-specific histone-DNA complexes (Figure 5B, left). Unfortunately, as little is known about the kinetics of chaperone-histone complexes, it is unclear if the substrates of nucleosome assembly are the chaperone-histone complexes or the small proportion of free histones in dynamic equilibrium.
Histone chaperones may also guide nucleosome assembly in a more directed fashion by presenting the histones in the ‘correct’ orientation (Figure 5C). In contrast to the case above (Figure 5B), the chaperone does not necessarily lower the free energy of the histones. Instead, it is involved in a transient chaperone-histone-DNA complex that is resolved by DNA wrapping and chaperone dissociation. By increasing the likelihood of specific interactions between histone and DNA, the chaperone skews the reaction kinetics towards a specific histone-DNA complex (ie. tetrasome, hexasome or nucleosome) (Figure 5C, right). The formation of a transient ternary complex causes an apparent decrease in activation energy to be surmounted for nucleosome formation, favoring it over non-specific histone-DNA interactions (Figure 5C, left). While such ternary complexes have not been observed directly, as expected by their transient nature, they can be inferred in a few in vitro cases. Asf1, for example, facilitates formation of a disome [(H3-H4)1-DNA] opposed to the tetrasome formed in the absence of chaperone [24]. Similarly, for the reverse, nucleosome disassembly reaction, chaperone-nucleosome complexes may occur for FACT [52] and Nap1 [93]. It should also be noted that a transient, ternary complex may also be involved in preventing non-specific histone-DNA interactions. If histone chaperones do directly deposit histones, how is the transition made kinetically and thermodynamically more favorable? A possible answer is that histone chaperones exploit the conformational flexibility of the histone fold. Co-structures involving p46/p55 and Scm3 suggest that the histone fold can be distorted upon chaperone binding [68, 75, 79]. These conformational changes may represent high-energy ‘transition’ states resolved by nucleosome formation upon encounter of DNA.
4.4 Nucleosome assembly versus disassembly
One might wonder if nucleosome disassembly is a direct reversal of a given assembly pathway. Chaperone-histone affinities range from micromolar to nanomolar and are thus equivalent of the nanomolar free energy of a nucleosome [14]. As such, it has been speculated that histone chaperones can directly facilitate histone eviction. This notion is supported by excess Nap1 removing H2A-H2B dimers from a preformed nucleosome in vitro [93]. As discussed previously, affinities of H2A-H2B for tetrasome and Nap1 are matched to allow assembly reactions to be driven in both directions. The actual direction will depend on the abundance of Nap1, histones and DNA. FACT has also been shown to be competent to remove H2A-H2B from a nucleosome in the context of transcription [70]. It is important to note however that the energy required to evict H2A-H2B (and in fact H3-H4) depends on nucleosome conformation and stability, with histone ptms, DNA sequence and buffer all being influential variables [37]. These variables most likely contribute to the likelihood of histone chaperone-mediated nucleosome disassembly in vivo.
In contrast to H2A-H2B, the (H3-H4)2 tetramer has higher dissociation energy and more potential to be kinetically trapped. In the context of the nucleosome, kinetically trapped simply means that the DNA does not visit an ‘unwrapped’ conformation for long enough to allow the histones to dissociate. The relevance of the nucleosome kinetic trap is indicated by excess Asf1 failing to disassemble nucleosomes or tetrasomes at physiological salt [24]. Thermodynamics alone would predict that Asf1 would disassembly nucleosomes, given its nanomolar affinity that is comparable to the tetrasome energy [24]. Interestingly, thermal unwinding of the DNA still does not allow Asf1 to compete for H3-H4 dimer, likely because at least half of the tetrasome DNA would need to be unwrapped to uncover its binding site [24]. In light of nucleosomes being kinetically trapped, it seems probable that histone chaperones alone cannot directly disassemble nucleosomes in vivo. However, they may facilitate disassembly alongside ATP-dependent remodelers, as hydrolysis of two to three ATP molecules would theroretically provide sufficient energy to completely dissociate all histone-DNA interactions within the nucleosome [94]. Furthermore, mechanical stress imposed by DNA-templated processes such as the positive super-coils induced by traveling RNA polymerase, might destabilize nucleosomes sufficiently to allow direct disassembly [95]. Therefore, the dynamic environment in vivo will alleviate some of the kinetic barriers observed in vitro and allow a direct competition of histone chaperones and DNA for histones. Nucleosome density could therefore be controlled by mass action of histone chaperones with high histone affinity.
The acidic stretches found in many histone chaperones may also function in nucleosome disassembly. That is, they may initiate the unwrapping of the nucleosomal DNA (or stabilize it in an ‘open’ state) to allow ATP-dependent chromatin remodeling activity or histone eviction [93]. This is consistent with acidic stretches also being found in other chromatin modifying enzymes, such as the histone methyltransferase SET1, as well as ATP-dependent remodelers such as SWR1 and ATRX. An accessory function accounts for the fact that these acidic stretches are sometimes dispensable and not necessarily conserved.
4.5 Channeling histones along a histone chaperone pathway
The regulation of histone chaperone activity is also at the core of their in vivo mechanism. How do histone chaperones ensure that the thermodynamic flow of histones is regulated in an ordered fashion? This is particularly important as there is evidence that chromatin assembly pathways compete for histone supply [60]. A theme emerging from recent literature is the regulation of histone hand-offs by histone ptms. For example, the acetylation of H4-lysine 5 and H4-lysine 12 is well-established and promotes histone association with nuclear import complexes [96]. First discovered in yeast and later in mammals, H3 is further acetylated at lysine 56 before chromatin assembly [97-99]. The histone chaperone Asf1 is a required cofactor in the acetylation of H3 at lysine 56 [100, 101]. The multiple acetylation of H3-H4 in turn facilitates nuclear import and may enhance H3 affinity for CAF1 thereby promoting chromatin assembly [98, 100, 102, 103]. Following incorporation, H3-lysine 56 is deacetylated, potentially creating a more stable nucleosome [14]. A series of histone hand-offs regulated by histone ptms can thus be delineated. Such a series would fine-tune histone chaperone activity in the context of chromatin homeostasis, essentially creating a series of checkpoints that keep the system ordered [102-106].
5.0 Outlook
Since their discovery, the unusual and unique biochemistry of the histone proteins has continued to astound and amaze. A wealth of advances over the last decade has transformed them from static to dynamic modulators of virtually all processes that act or depend on DNA. Closely associated both figuratively and literally, with the histones, is a network of histone chaperones. Through structural and functional diversity, the histone chaperones facilitate nucleosome formation. They are involved in all facets of histone biology, from folding in the cytoplasm to nuclear import, storage and deposition into chromatin. The precise mechanisms of histone chaperone function, however, have remained elusive. Figure 6 highlights key questions relevant to histone chaperone mechanism. The lack of chaperone-histone co-structures, difficulties interpreting in vitro results in an in vivo context, and a poor understanding of the biologically relevant chaperone substrate and product, have hindered mechanism determination. Even with these limitations, we can postulate several models of histone chaperone mechanism by considering the thermodynamics of nucleosome assembly and disassembly. These models relate to how histone chaperones bind to their substrates, prevent promiscuous histone interactions and balance nucleosome assembly and disassembly, the fate of chaperone-bound histones, and the importance of histone hand-offs between different histone chaperones. Testing these hypotheses will be the seed for future studies aimed at determining histone chaperone mechanism.
Fig. 6.
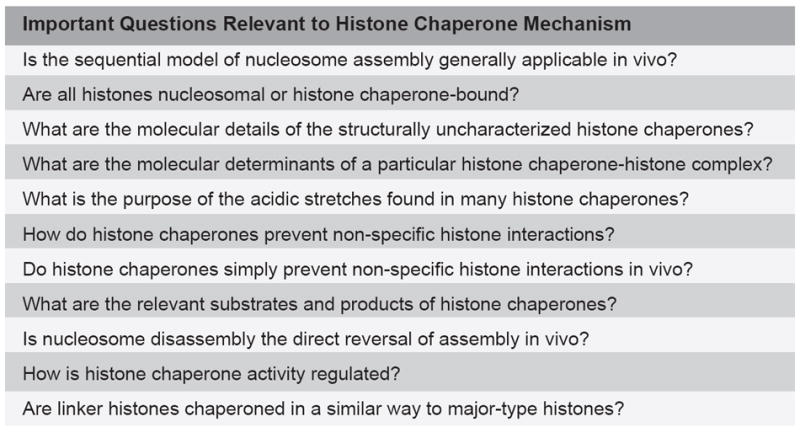
Important questions relevant to histone chaperone mechanism.
Highlights.
The unique biochemistry of histones causes a requirement for histone chaperones
Histone chaperones are histone-binding proteins that facilitate nucleosome assembly
Histones chaperones are structurally, functionally and mechanistically diverse
Histone chaperones prevent non-specific histone interactions
Histone chaperones adopt diverse histone-binding modes
Acknowledgments
SJE is supported by a Boehringer Ingelheim Funds fellowship and the David Rockefeller Graduate Program. S.D. is supported by the Howard Hughes Medical Institute. We thank K. Luger and D. Allis for comments on the manuscript, and the Luger and Allis groups for helpful discussion.
Abbreviations
- ATRX
Alpha Thalassemia/mental retardation syndrome X-linked
- Asf1
Anti-silencing Function 1
- cenH3
centromeric H3
- Chz1
Chaperone for Htz1/H2A-H2B dimer 1
- CAF1
Chromatin Assembly Factor 1
- DAXX
Death-domain Associated protein
- FACT
Facilitates Chromatin Transcription
- HIRA
HIR histone cell cycle regulation defective homolog A
- HJURP
Holliday Junction Recognition Protein
- Nap1
Nuclear assembly protein 1
- NASP
Nuclear Autoantigenic Sperm Protein
- NPM
Nucleosplasmin
- ptms
post-translational modifications
- Rtt106
Regulator of Ty1 Transposition 106
- Rtt109
Regulator of Ty1 Transposition 109
- RbAp46/48
Retinoblastoma binding protein 7/4 46/48
- SET1
SET domain-containing 1
- SWR1
SWI/SNF-related protein
- SSRP1
Structure Specific Recognition Protein 1
- Scm3
Suppressor of chromosome mis-segregation
- SPT16
Suppressor of Ty 16 homolog
- Vps75
Vacuolar Protein Sorting 75
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Marino-Ramirez L, Hsu B, Baxevanis AD, Landsman D. The Histone Database: a comprehensive resource for histones and histone fold-containing proteins. Proteins. 2006;62:838–842. doi: 10.1002/prot.20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 4.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Current opinion in genetics & development. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marino-Ramirez L, Jordan IK, Landsman D. Multiple independent evolutionary solutions to core histone gene regulation. Genome Biol. 2006;7:R122. doi: 10.1186/gb-2006-7-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewen ME. Where the cell cycle and histones meet. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 8.Talbert PB, Henikoff S. Histone variants - ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 9.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 14.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosgrove MS. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev Proteomics. 2007;4:465–478. doi: 10.1586/14789450.4.4.465. [DOI] [PubMed] [Google Scholar]
- 16.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 17.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood C, Snijders A, Williamson J, Reynolds C, Baldwin J, Dickman M. Post-translational modifications of the linker histone variants and their association with cell mechanisms. FEBS J. 2009;276:3685–3697. doi: 10.1111/j.1742-4658.2009.07079.x. [DOI] [PubMed] [Google Scholar]
- 20.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 23.Banks DD, Gloss LM. Equilibrium folding of the core histones: the H3-H4 tetramer is less stable than the H2A-H2B dimer. Biochemistry. 2003;42:6827–6839. doi: 10.1021/bi026957r. [DOI] [PubMed] [Google Scholar]
- 24.Donham DC, II, Scorgie JK, Churchill ME. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxevanis AD, Godfrey JE, Moudrianakis EN. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 26.Thomas JO, Butler PJ. Characterization of the octamer of histones free in solution. J Mol Biol. 1977;116:769–781. doi: 10.1016/0022-2836(77)90270-4. [DOI] [PubMed] [Google Scholar]
- 27.Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 28.Stein A. DNA folding by histones: the kinetics of chromatin core particle reassembly and the interaction of nucleosomes with histones. J Mol Biol. 1979;130:103–134. doi: 10.1016/0022-2836(79)90421-2. [DOI] [PubMed] [Google Scholar]
- 29.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. II. Three transitions in nucleosomes resolved by salt-titration. J Mol Biol. 1987;196:399–411. doi: 10.1016/0022-2836(87)90700-5. [DOI] [PubMed] [Google Scholar]
- 30.Wilhelm FX, Wilhelm ML, Erard M, Duane MP. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978;5:505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazurkiewicz J, Kepert JF, Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J Biol Chem. 2006;281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 33.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worcel A, Han S, Wong ML. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Andrews AJ, Luger K. Nucleosome Structure(s) and Stability: Variations on a Theme. Annu Rev Biophys. 2010;40 doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 38.Stein A, Bina-Stein M, Simpson RT. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci USA. 1977;74:2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- 42.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 43.Osakabe A, Tachiwana H, Matsunaga T, Shiga T, Nozawa RS, Obuse C, Kurumizaka H. Nucleosome formation activity of human sNASP. J Biol Chem. 2010;285:11913–11921. doi: 10.1074/jbc.M109.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 46.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, Dekelver RC, Miller JC, Lee Y-L, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 49.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 50.Han J, Li Q, Mccullough L, Kettelkamp C, Formosa T, Zhang Z. Ubiquitylation of FACT by the Cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010;24:1485–1490. doi: 10.1101/gad.1887310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundu LR, Seki M, Watanabe N, Murofushi H, Furukohri A, Waga S, Score AJ, Blow JJ, Horikoshi M, Enomoto T, Tada S. Biphasic chromatin binding of histone chaperone FACT during eukaryotic chromatin DNA replication. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 53.Philpott A, Leno GH, Laskey RA. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- 54.Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- 55.Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shintomi K, Iwabuchi M, Saeki H, Ura K, Kishimoto T, Ohsumi K. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci U S A. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kepert JF, Mazurkiewicz J, Heuvelman GL, Toth KF, Rippe K. NAP1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J Biol Chem. 2005;280:34063–34072. doi: 10.1074/jbc.M507322200. [DOI] [PubMed] [Google Scholar]
- 58.Gadad SS, Senapati P, Syed SH, Rajan RE, Shandilya J, Swaminathan V, Chatterjee S, Colombo E, Dimitrov S, Pelicci PG, Ranga U, Kundu TK. The multifunctional protein nucleophosmin (NPM1) is a human linker histone H1 chaperone. Biochemistry. 2011;50:2780–2789. doi: 10.1021/bi101835j. [DOI] [PubMed] [Google Scholar]
- 59.Elsaesser SJ, Allis CD. HIRA and Daxx Constitute Two Independent Histone H3.3-Containing Predeposition Complexes. Cold Spring Harb Symp Quant Biol. 2010;75 doi: 10.1101/sqb.2010.75.008. [DOI] [PubMed] [Google Scholar]
- 60.Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 63.Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat Struct Mol Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 66.Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci USA. 2007;104:4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, Laue ED, Mackay JP. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. J Biol Chem. 2011;286:1196–1203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 71.Regnard C, Desbruyeres E, Huet JC, Beauvallet C, Pernollet JC, Edde B. Polyglutamylation of nucleosome assembly proteins. J Biol Chem. 2000;275:15969–15976. doi: 10.1074/jbc.M000045200. [DOI] [PubMed] [Google Scholar]
- 72.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agez M, Chen J, Guerois R, van Heijenoort C, Thuret J-Y, Mann C, Ochsenbein F. Structure of the histone chaperone ASF1 bound to the histone H3 C-terminal helix and functional insights. Structure. 2007;15:191–199. doi: 10.1016/j.str.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 75.Murzina NV, Pei X-Y, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008;22:1265–1268. doi: 10.1101/gad.1676208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Molecular Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu R-M. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011 doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes to cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 82.Umehara T, Chimura T, Ichikawa N, Horikoshi M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes to cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 83.Ray-Gallet, Quivy, Silljé, Nigg, Almouzni The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma. 2007;116:487–496. doi: 10.1007/s00412-007-0112-x. [DOI] [PubMed] [Google Scholar]
- 84.Stein A, Whitlock JP, Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci USA. 1979;76:5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. J Biol Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 87.Eickbush TH, Moudrianakis EN. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 88.Karantza V, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: pH and ionic strength effects on the stability of the (H3-H4)/(H3-H4)2 system. Biochemistry. 1996;35:2037–2046. doi: 10.1021/bi9518858. [DOI] [PubMed] [Google Scholar]
- 89.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cotten M, Sealy L, Chalkley R. Massive phosphorylation distinguishes Xenopus laevis nucleoplasmin isolated from oocytes or unfertilized eggs. Biochemistry. 1986;25:5063–5069. doi: 10.1021/bi00366a014. [DOI] [PubMed] [Google Scholar]
- 91.Taneva SG, Muñoz IG, Franco G, Falces J, Arregi I, Muga A, Montoya G, Urbaneja MA, Bañuelos S. Activation of nucleoplasmin, an oligomeric histone chaperone, challenges its stability. Biochemistry. 2008;47:13897–13906. doi: 10.1021/bi800975r. [DOI] [PubMed] [Google Scholar]
- 92.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Molecular Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 94.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 95.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Molecular Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 96.Alvarez F, Munoz F, Schilcher P, Imhof A, Almozuni G, Loyola A. Sequential establishment of marks on soluble histones H3 and H4. J Biol Chem. 2011 doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 98.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang Y, Holbert MA, Delgoshaie N, Wurtele H, Guillemette B, Meeth K, Yuan H, Drogaris P, Lee E-H, Durette C, Thibault P, Verreault A, Cole PA, Marmorstein R. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011;19:221–231. doi: 10.1016/j.str.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barman H, Takami Y, Nishijima H, Shibahara K, Sanematsu F, Nakayama T. Histone acetyltransferase-1 regulates integrity of cytosolic histone H3-H4 containing complex. Biochemical and Biophysical Research Communications. 2008;373:624–630. doi: 10.1016/j.bbrc.2008.06.100. [DOI] [PubMed] [Google Scholar]
- 103.Glowczewski L, Waterborg JH, Berman JG. Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus. Molecular and Cellular Biology. 2004;24:10180–10192. doi: 10.1128/MCB.24.23.10180-10192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rufiange A, Jacques P-E, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 105.Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 106.Sharp JA, Rizki G, Kaufman PD. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]


