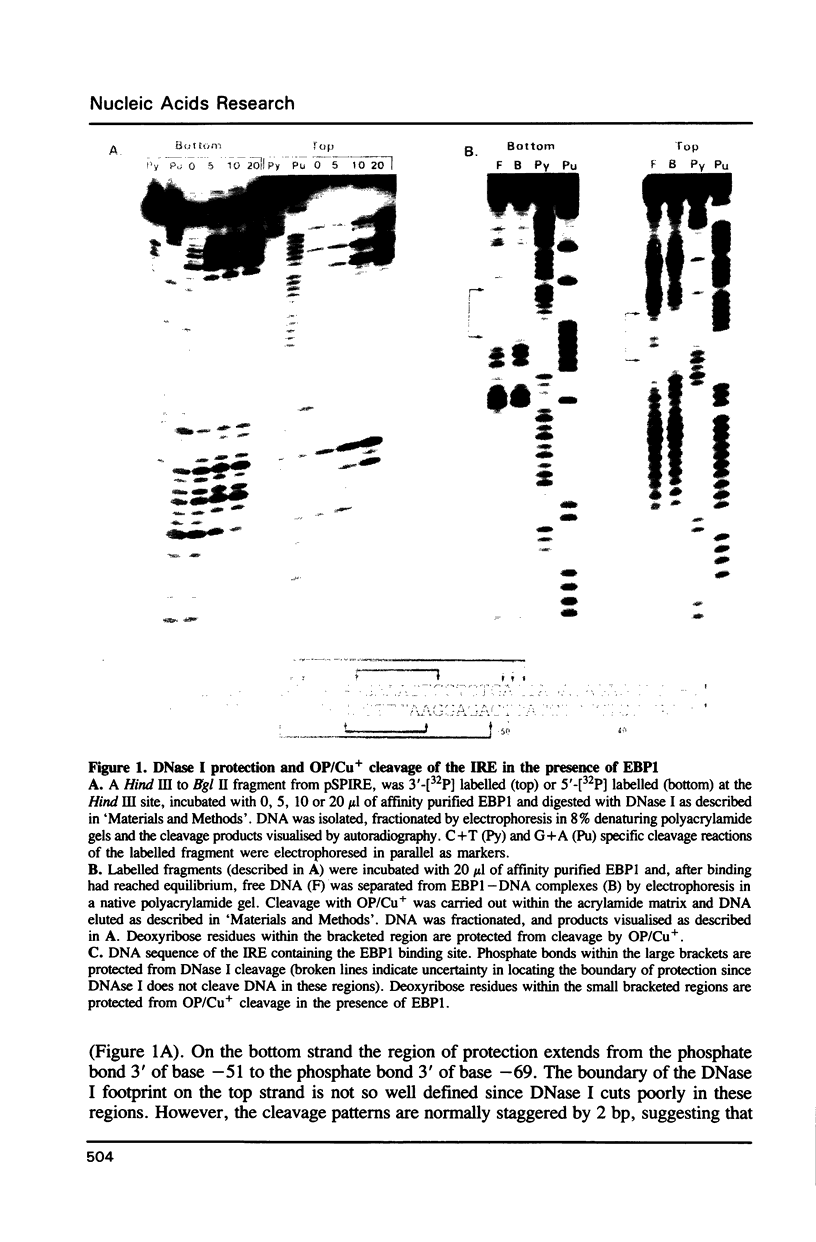
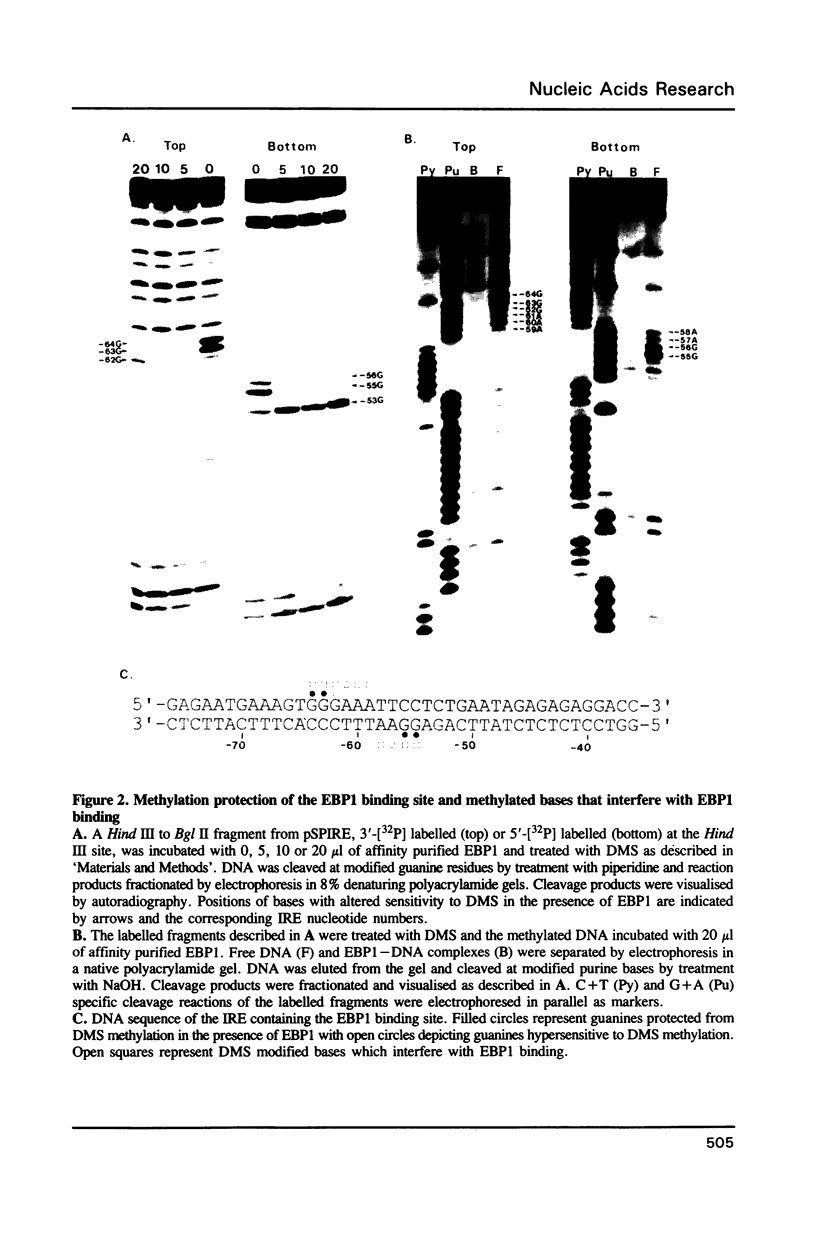
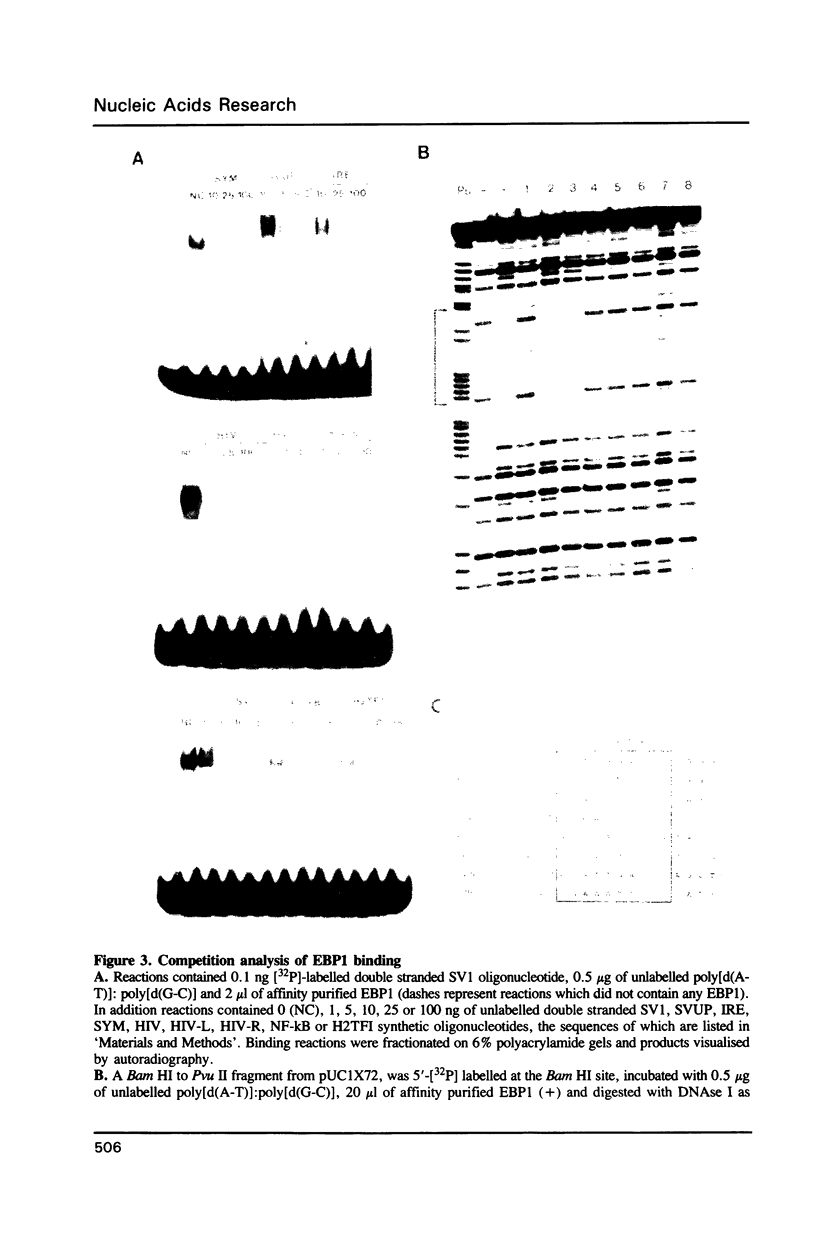
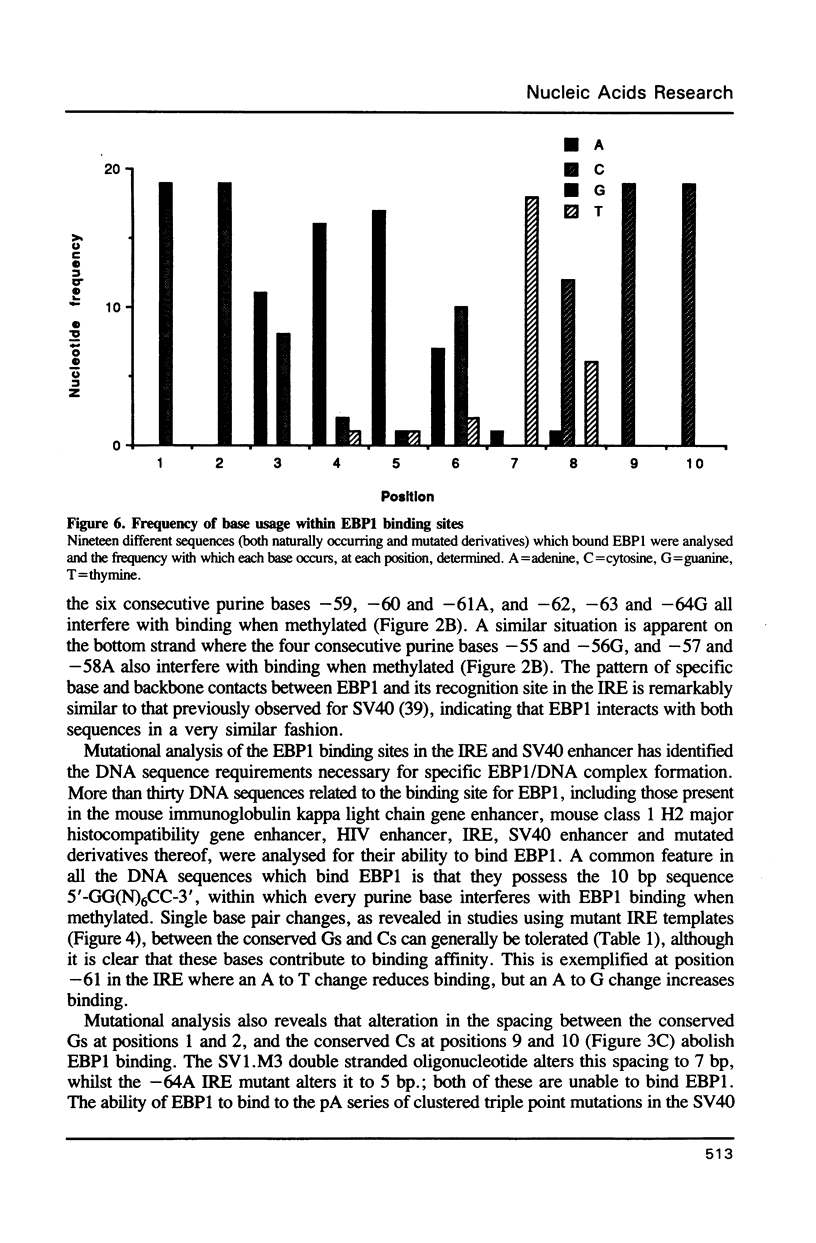
Abstract
Short DNA sequence motifs have been identified in viral and cellular enhancers which represent the binding sites for a variety of trans- acting factors. One such HeLa cell factor, EBP1, has been purified and shown to bind to sequences in the SV40 enhancer. The PRDII element in the human beta-interferon gene regulatory element (IRE) shows strong sequence similarity to the EBP1 binding site in the SV40 enhancer. We demonstrate here that EBP1 binds to its sites in the SV40 enhancer and IRE in a similar manner, making base specific contacts over one complete turn of the DNA double helix. Mutational analysis of the EBP1 sites in the IRE and SV40 enhancer has identified the DNA sequence requirements necessary for specific EBP1/DNA complex formation. In addition, 34 DNA sequences related to the EBP1 binding site were analysed for their ability to bind EBP1. Sequences constituting high affinity binding sites possess the sequence 5'-GG(N)6CC-3'. Single base pair changes in the region between the conserved Gs and Cs can generally be tolerated although it is clear that these intervening bases contribute to binding affinity. Mutations in the recognition site which could lead to gross structural changes in the DNA abolish EBP1 binding.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baeuerle P. A., Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988 Apr 22;53(2):211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Sharp P. A. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2Kb class I major histocompatibility gene. Mol Cell Biol. 1987 Jan;7(1):305–313. doi: 10.1128/mcb.7.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Clark L., Pollock R. M., Hay R. T. Identification and purification of EBP1: a HeLa cell protein that binds to a region overlapping the 'core' of the SV40 enhancer. Genes Dev. 1988 Aug;2(8):991–1002. doi: 10.1101/gad.2.8.991. [DOI] [PubMed] [Google Scholar]
- Cochran M. D., Weissmann C. Modular structure of the beta-globin and the TK promoters. EMBO J. 1984 Nov;3(11):2453–2459. doi: 10.1002/j.1460-2075.1984.tb02155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I., Fromental C., Augereau P., Wildeman A., Zenke M., Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986 Oct 9;323(6088):544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
- Davidson I., Xiao J. H., Rosales R., Staub A., Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988 Sep 23;54(7):931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromental C., Kanno M., Nomiyama H., Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988 Sep 23;54(7):943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Wilson R. N., Weinberg R. A. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986 Dec;6(12):4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Maniatis T. Overlapping positive and negative regulatory domains of the human beta-interferon gene. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1447–1451. doi: 10.1073/pnas.85.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Zinn K., Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985 Jun;41(2):509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Israël A., Kimura A., Kieran M., Yano O., Kanellopoulos J., Le Bail O., Kourilsky P. A common positive trans-acting factor binds to enhancer sequences in the promoters of mouse H-2 and beta 2-microglobulin genes. Proc Natl Acad Sci U S A. 1987 May;84(9):2653–2657. doi: 10.1073/pnas.84.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Haslinger A., Karin M., Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987 Jan 22;325(6102):368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Tilly K., Maniatis T. Fine structure genetic analysis of a beta-globin promoter. Science. 1986 May 2;232(4750):613–618. doi: 10.1126/science.3457470. [DOI] [PubMed] [Google Scholar]
- Nabel G., Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987 Apr 16;326(6114):711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Nomiyama H., Fromental C., Xiao J. H., Chambon P. Cell-specific activity of the constituent elements of the simian virus 40 enhancer. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7881–7885. doi: 10.1073/pnas.84.22.7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mutul J., Macchi M., Wasylyk B. Mutational analysis of the contribution of sequence motifs within the IgH enhancer to tissue specific transcriptional activation. Nucleic Acids Res. 1988 Jul 11;16(13):6085–6096. doi: 10.1093/nar/16.13.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986 Dec 26;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Serfling E., Lübbe A., Dorsch-Häsler K., Schaffner W. Metal-dependent SV40 viruses containing inducible enhancers from the upstream region of metallothionein genes. EMBO J. 1985 Dec 30;4(13B):3851–3859. doi: 10.1002/j.1460-2075.1985.tb04157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuey D. J., Parker C. S. Binding of Drosophila heat-shock gene transcription factor to the hsp 70 promoter. Evidence for symmetric and dynamic interactions. J Biol Chem. 1986 Jun 15;261(17):7934–7940. [PubMed] [Google Scholar]
- Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog Nucleic Acid Res Mol Biol. 1975;15(0):219–284. [PubMed] [Google Scholar]
- Singh H., LeBowitz J. H., Baldwin A. S., Jr, Sharp P. A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988 Feb 12;52(3):415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5' element and c-fos 3' sequences. Cell. 1985 Oct;42(3):889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Zenke M., Schatz C., Wintzerith M., Grundström T., Matthes H., Takahashi K., Chambon P. Specific protein binding to the simian virus 40 enhancer in vitro. Mol Cell Biol. 1986 Jun;6(6):2098–2105. doi: 10.1128/mcb.6.6.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Macchi M., Rosales R., Vigneron M., Staub A., Chambon P. In vitro binding of several cell-specific and ubiquitous nuclear proteins to the GT-I motif of the SV40 enhancer. Genes Dev. 1987 Oct;1(8):794–807. doi: 10.1101/gad.1.8.794. [DOI] [PubMed] [Google Scholar]
- Yano O., Kanellopoulos J., Kieran M., Le Bail O., Israël A., Kourilsky P. Purification of KBF1, a common factor binding to both H-2 and beta 2-microglobulin enhancers. EMBO J. 1987 Nov;6(11):3317–3324. doi: 10.1002/j.1460-2075.1987.tb02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]