Abstract
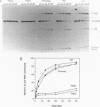
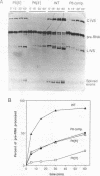
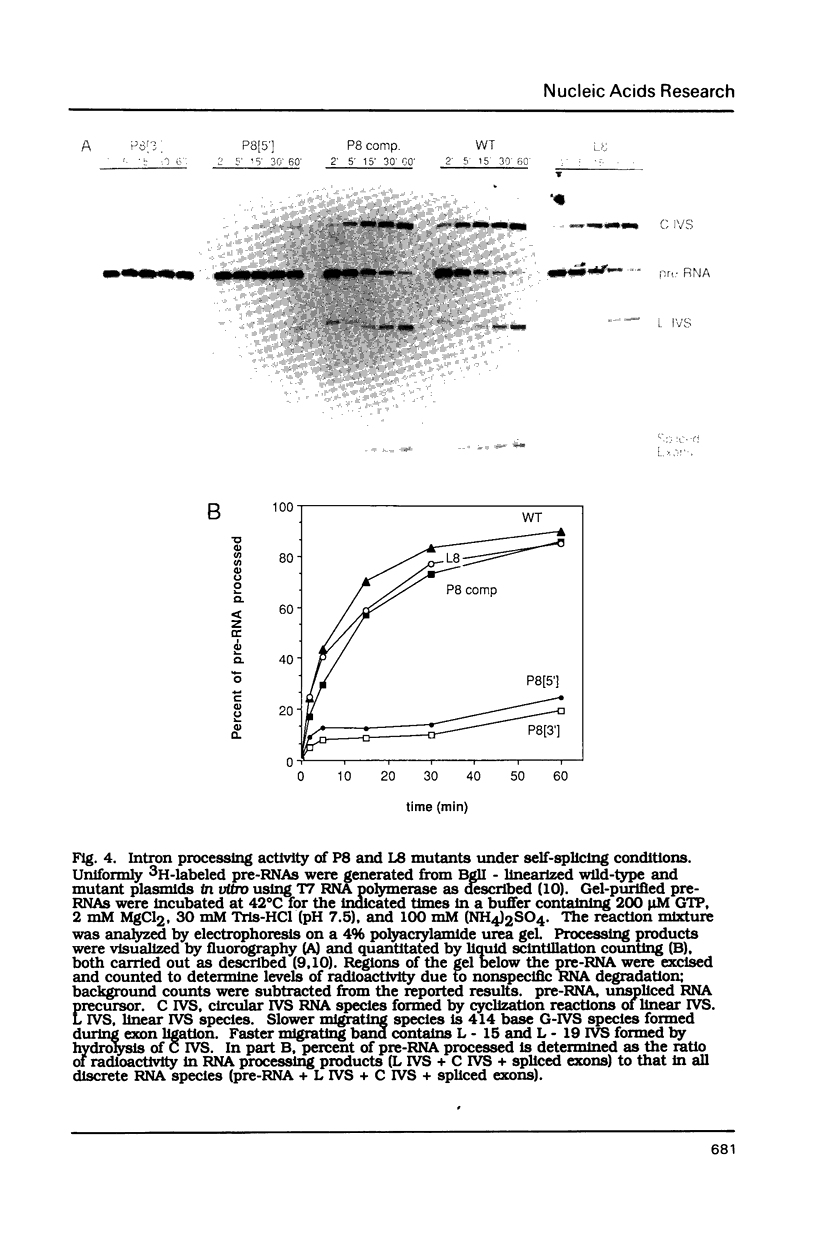
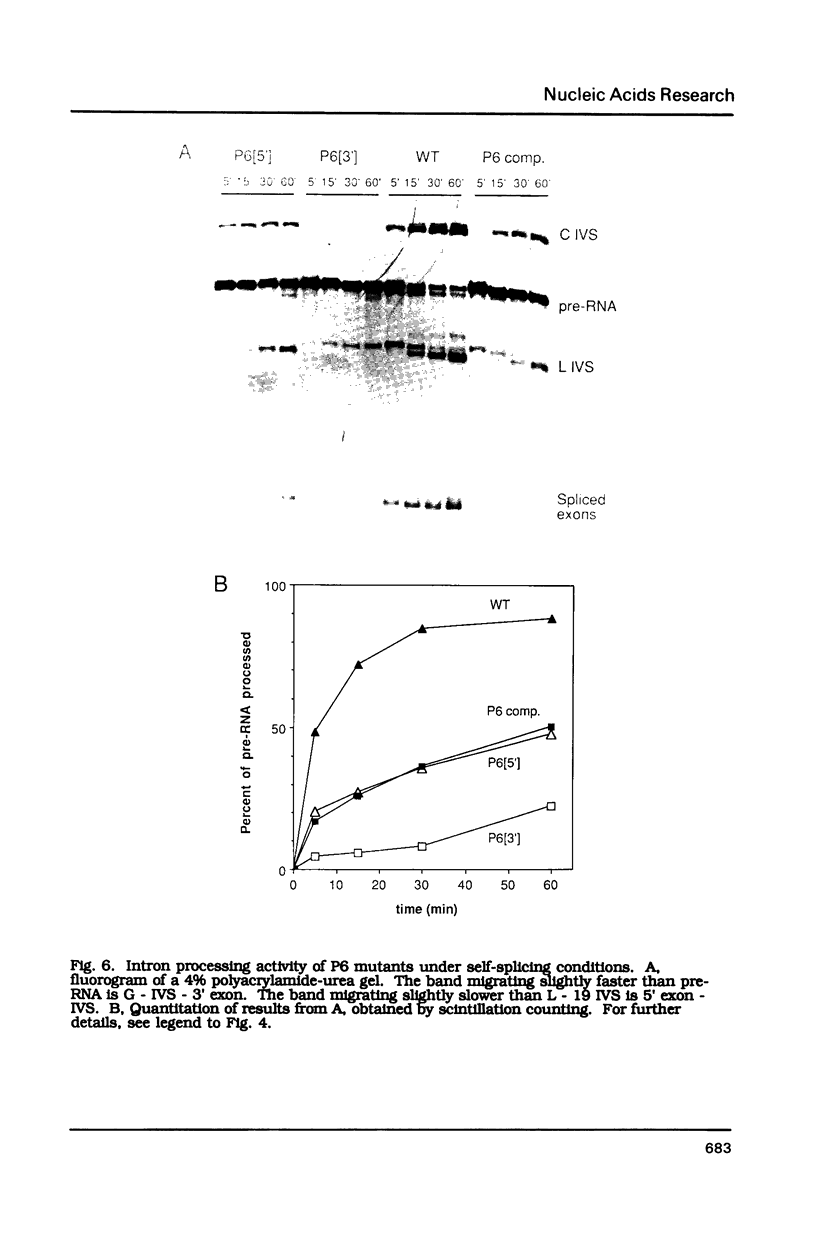
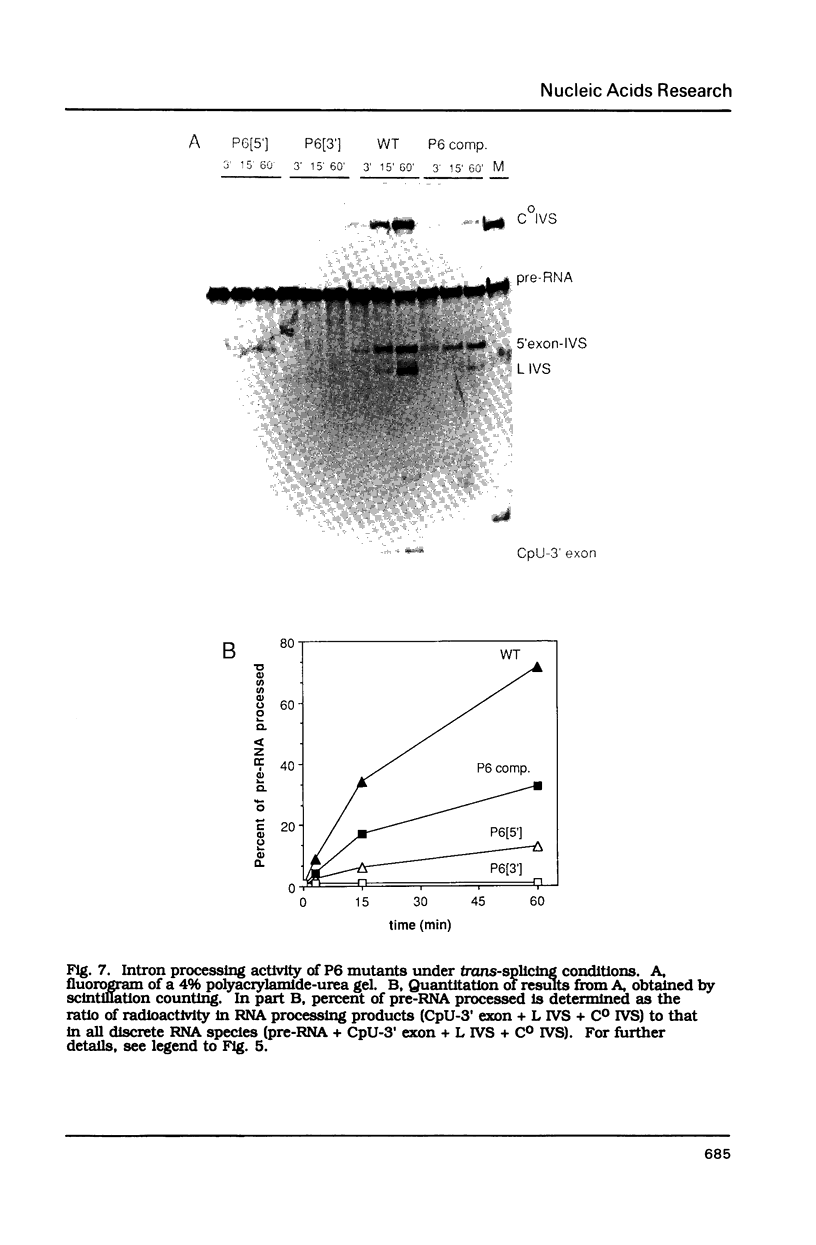
Compensatory mutations have been constructed which demonstrate that P8 and P6, two of nine proposed base-pairing interactions characteristic of group I introns, exist within the folded structure of the Tetrahymena thermophila rRNA intervening sequence, and that these secondary structure elements are important for splicing in E. coli and self-splicing in vitro. Two-base mutations in the 5' and 3' segments of P8 are predicted to disrupt P8 and a strong splicing-defective phenotype is observed in each case. A compensatory four-base mutation in P8 is predicted to restore pairing, and results in the restoration of splicing activity to nearly wild type levels. Thus, we conclude that P8 exists and is essential for splicing. In contrast to the strong phenotypes generally exhibited by mutations which disrupt RNA secondary structure, a two-base mutation in L8, the loop between P8[5'] and P8[3'], results in only a slight decrease in splicing activity. We also tested P6, a pairing which is proposed to consist of only two base-pairs in this intron. A two-base mutation in P6[3'] reduces splicing activity to a greater extent than does a two-base mutation in P6[5']. Comparison of the activities of these mutants and a compensatory P6 four-base mutant support the existence of P6, and suggest that the P6 pairing may be particularly important in the exon ligation step of splicing.
Full text
PDF


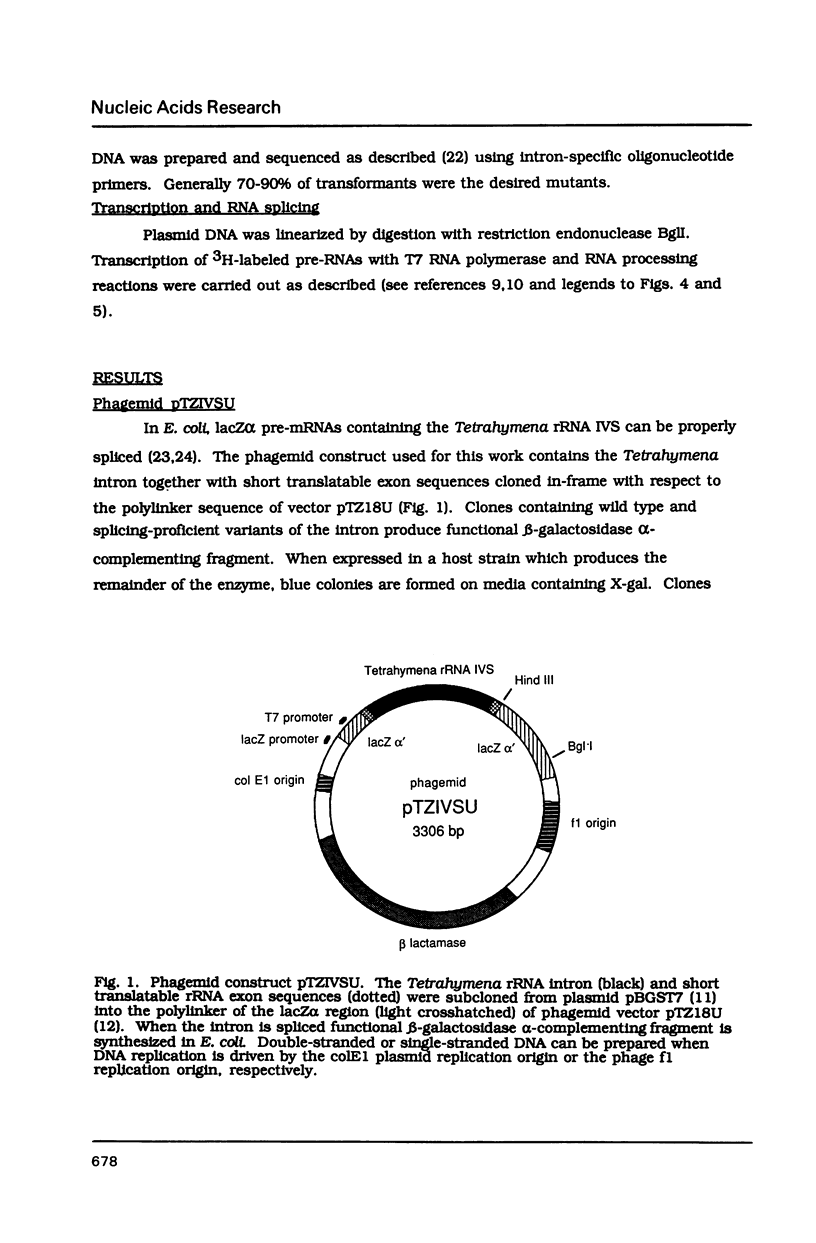
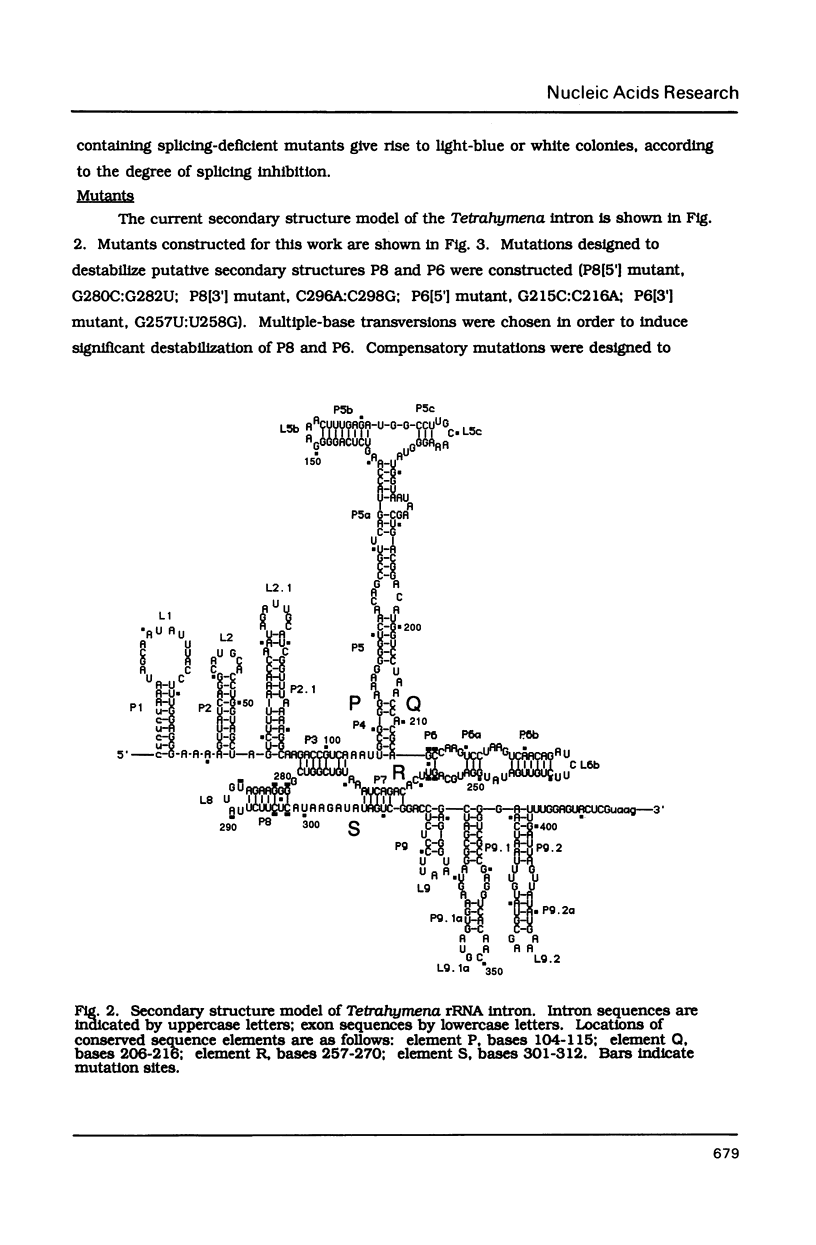







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Belfort M., Chandry P. S., Pedersen-Lane J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb Symp Quant Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Irvine K. D., Kaneko K. J., Kerker B. J., Oettgen A. B., Tierney W. M., Williamson C. L., Zaug A. J., Cech T. R. Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell. 1986 Apr 25;45(2):167–176. doi: 10.1016/0092-8674(86)90380-6. [DOI] [PubMed] [Google Scholar]
- Burke J. M. Molecular genetics of group I introns: RNA structures and protein factors required for splicing--a review. Gene. 1988 Dec 20;73(2):273–294. doi: 10.1016/0378-1119(88)90493-3. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Holl J., Rödel G., Schweyen R. J. Suppressor mutations identify box9 as a central nucleotide sequence in the highly ordered structure of intron RNA in yeast mitochondria. EMBO J. 1985 Aug;4(8):2081–2085. doi: 10.1002/j.1460-2075.1985.tb03895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Mierendorf R. C., Pfeffer D. Direct sequencing of denatured plasmid DNA. Methods Enzymol. 1987;152:556–562. doi: 10.1016/0076-6879(87)52061-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Perea J., Jacq C. Role of the 5' hairpin structure in the splicing accuracy of the fourth intron of the yeast cob-box gene. EMBO J. 1985 Dec 1;4(12):3281–3288. doi: 10.1002/j.1460-2075.1985.tb04078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. V., Cech T. R. Coupling of Tetrahymena ribosomal RNA splicing to beta-galactosidase expression in Escherichia coli. Science. 1985 May 10;228(4700):719–722. doi: 10.1126/science.2986286. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Ray J. A., Edwards S. W., Scazzocchio C., Davies R. W. The Tetrahymena rRNA intron self-splices in E. coli: in vivo evidence for the importance of key base-paired regions of RNA for RNA enzyme function. Cell. 1985 Feb;40(2):371–380. doi: 10.1016/0092-8674(85)90151-5. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Holl J., Schweyen R. J., Rödel G., Kaudewitz F. Processing of yeast mitochondrial RNA: involvement of intramolecular hybrids in splicing of cob intron 4 RNA by mutation and reversion. Cell. 1983 May;33(1):195–202. doi: 10.1016/0092-8674(83)90348-3. [DOI] [PubMed] [Google Scholar]
- Williamson C. L., Tierney W. M., Kerker B. J., Burke J. M. Site-directed mutagenesis of core sequence elements 9R', 9L, 9R, and 2 in self-splicing Tetrahymena pre-rRNA. J Biol Chem. 1987 Oct 25;262(30):14672–14682. [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. Oligomerization of intervening sequence RNA molecules in the absence of proteins. Science. 1985 Sep 13;229(4718):1060–1064. doi: 10.1126/science.2412290. [DOI] [PubMed] [Google Scholar]