Abstract
HIV-1 drug resistance monitoring in resource-poor settings is crucial due to limited drug alternatives. Recent reports of the increased prevalence of CXCR4 usage in subtype C infections may have implications for CCR5 antagonists in therapy. We investigated the prevalence of drug resistance mutations and CXCR4 coreceptor utilization of viruses from HIV-1 subtype C-infected children. Fifty-one children with virological failure during highly active antiretroviral therapy (HAART) and 43 HAART-naive children were recruited. Drug resistance genotyping and coreceptor utilization assessment by phenotypic and genotypic methods were performed. At least one significant drug resistance mutation was present in 85.4% of HAART-failing children. Thymidine analogue mutations (TAMs) were detected in 58.5% of HAART-failing children and 39.0% had ≥3 TAMs. CXCR4 (X4) or dual (R5X4)/mixed (R5, X4) (D/M)-tropic viruses were found in 54.3% of HAART-failing and 9.4% of HAART-naive children (p<0.0001); however, the HAART-failing children were significantly older (p<0.0001). In multivariate logistic regression, significant predictors of CXCR4 usage included antiretroviral treatment, older age, and lower percent CD4+ T cell counts. The majority of genotypic prediction tools had low sensitivity (≤65.0%) and high specificity (≥87.5%) for predicting CXCR4 usage. Extensive drug resistance, including the high percentage of TAMs found, may compromise future drug choices for children, highlighting the need for improved treatment monitoring and adherence counseling. Additionally, the increased prevalence of X4/D/M viruses in HAART-failing children suggests limited use of CCR5 antagonists in salvage therapy. Enhanced genotypic prediction tools are needed as current tools are not sensitive enough for predicting CXCR4 usage.
Introduction
In 2007, approximately 2 million children under 15 years were living with the human immunodeficiency virus type 1 (HIV-1) worldwide with 280,000 of these from South Africa, of whom only 32,000 were receiving highly active antiretroviral therapy (HAART).1 Drug resistance is a major hindrance to the successful clinical management of HIV-1 infection and, particularly in children, may be exacerbated by poor adherence, drug toxicity, and inadequate metabolism of drugs leading to the incomplete suppression of viral replication.2
Maraviroc, a recently approved CCR5 antagonist,3 is a plausible alternative for those failing current HAART regimens. However, there is inadequate information to guide the use of CCR5 antagonists either as part of first-line or salvage therapy for HIV-1 subtype C (HIV-1C), the predominant subtype in the epidemic. Viral isolates are classified as R5 viruses if they use the CCR5 coreceptor exclusively, X4 viruses if they use CXCR4 exclusively, or R5X4 (dual) viruses if they are able to use both coreceptors.4 CXCR4-using viruses usually emerge in a large proportion of subtype B infections during the advanced phase of disease in both adults and children and have been associated with a rapid CD4+ T cell decline and progression to AIDS.5,6 Earlier studies of HIV-1C viruses have demonstrated a predominance of R5 isolates in all stages of disease including advanced disease, with minimal switch to CXCR4 tropism in both adults and children.7,8 The predominance of R5 viruses among HIV-1C isolates may imply that CCR5 inhibitors such as maraviroc may be more efficacious against this viral subtype. However, recent reports have found about 30% of CXCR4-using viruses in untreated and treated HIV-1C-infected adults from South Africa, Malawi, and Zimbabwe, suggesting a shift in viral properties.9–11
This study investigated drug resistance and coreceptor usage among children initiating and failing HAART treatment in South Africa, where HIV-1C is the most common subtype. Factors were explored for associations with drug resistance patterns and viral tropism in these children.
Materials and Methods
Patient cohort
A total of 94 HAART-failing (n=51) and HAART-naive (n=43) children were recruited from King Edward VIII hospital in Durban, KwaZulu-Natal, South Africa between August 2008 and January 2010. Eleven samples had viral loads below the limit of detection of the Amplicor assay (<400 copies/ml) and two samples were of poor quality, leaving 41 HAART-failing and 40 HAART-naive children for analysis. Children were defined as having failed treatment according to clinical, immunological, and virological criteria outlined in the 2005 South African guidelines for the management of HIV-infected children.12 HAART-naive children exposed to antiretroviral therapy for the prevention of mother-to-child transmission (PMTCT) were included in the study.
CD4% and HIV-1 RNA load
CD4 cell counts and percentages (CD4%) were enumerated with the FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using the Multitest kit (CD4/CD3/CD8/CD45). HIV-1 plasma viral loads were quantified using the COBAS Amplicor Monitor test, version 1.5 (Roche Diagnostics).
HIV-1 resistance genotyping
HIV-1 genotyping was performed for all 41 HAART-failing children and a subset of 13 HAART-naive children, six of whom had received single-dose nevirapine (sdNVP) for PMTCT, using the HIV-1 ViroSeq Genotyping System v2.0 (Celera Diagnostics, Alameda, CA). Sequences were submitted to the Stanford drug resistance database (http://hivdb.stanford.edu/) for interpretation of drug resistance mutations.
Phenotypic characterization of HIV-1 tropism
HIV-1 coreceptor usage was determined from the plasma of children with viral loads greater than 1000 copies/ml using the enhanced sensitivity Trofile coreceptor tropism assay (Monogram Biosciences Inc., South San Francisco, CA).13–15
Genotypic prediction of HIV-1 tropism
RNA isolation, cDNA synthesis, and first round polymerase chain reaction (PCR) were carried out using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) with primers OFM19 and VIF1 as previously described16 for amplification of the 3-kb env gene. Second round PCR was carried out using Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes, Finland) with primers ENV1Adir and ENVM.17 Second round cycling conditions were at 98°C for 30 s, followed by 35 cycles at 98°C for 10 s, 65°C for 30 s, and 72°C for 4 min, followed by a final extension at 72°C for 10 min. Amplified products were gel purified using the QIAquick Gel Extraction kit (Qiagen) and cloned and sequenced as previously described with the ABI Prism 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA).17
Sequences were assessed for typical features of X4 viruses, including a V3 net charge above +4.5 and basic amino acids at positions 11 and/or 25 (11/25 rule), both of which are predictive of CXCR4 usage.18,19 Other features include an increased V3 length >35 amino acids and a more variable crown motif.18
Coreceptor genotypic prediction tools, included calculation of the V3 net charge; the 11/25 rule; C-PSSMsinsi (http://indra.mullins.microbiol.washington.edu/webpssm/)20; geno2pheno[coreceptor] (http://coreceptor.bioinf.mpi-inf.mpg.de/)21; a combination of the first four criteria where the majority prediction was considered as the final genotype prediction; and the following tools: C4.5, C4.5 with positions 8 and 12 only, PART, and SVM available at http://genomiac2.ucsd.edu:8080/wetcat/v3.html. These were all assessed for reliability against the gold standard phenotypic Trofile assay results.
Phylogenetic analysis
Phylogenetic trees were constructed from the pol and env sequences to determine the HIV-1 subtype. Sequences were aligned with subtype reference strains from the Los Alamos HIV-1 database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) using MEGA v4.0.22 Maximum likelihood trees were constructed using PhyML23 with the General Time Reversible plus Gamma model determined by FindModel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) and viewed using FigTreev1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). Intersubtype recombination was assessed using the recombination identification program (RIP) (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html) and SimPlot v3.5.1.24 Mean genetic distances were calculated with the Kimura two-parameter model in MEGA v4.0. GenBank accession numbers are HM623494 to HM623611.
Statistical analysis
Associations between baseline variables and coreceptor usage, having drug-resistant virus (≥1 major drug resistance mutation) and ≥3 thymidine analogue mutations (TAMs), were explored. Continuous and categorical variables were tested using either the unpaired Student's t-test or Mann-Whitney U test (as appropriate) and Fisher's exact test, respectively. Baseline predictors of CXCR4 usage were further explored using multivariate logistic regression. Baseline variables included age, gender, current and nadir CD4%, log HIV-1 viral load, duration of treatment, WHO stage, and sdNVP for PMTCT. Sensitivity and specificity for predicting CXCR4 usage were calculated. Statistical analyses were performed using GraphPad Prism v5.01 and SAS v9.1. p values <0.05 were considered to be statistically significant.
Ethical approval
The Biomedical Research Ethics Committee of the University of KwaZulu-Natal approved this study and informed consent was obtained from guardians of participants.
Results
Patient characteristics
Demographic and clinical data are summarized in Table 1. The median age of HAART-failing children (7.9 years) was significantly higher than for HAART-naive children (0.9 years; p<0.0001). The median nadir CD4% was significantly lower in HAART-failing (9.0%) than in HAART-naive children (14.0%; p=0.008). There was no significant difference between the two groups in terms of median current CD4%. Viral load was significantly lower in HAART-failing children compared to HAART-naive children (4.9 versus 5.9 log10 copies/ml, respectively; p<0.0001).
Table 1.
Patient Demographic and Clinical Characteristics
| Characteristics | Children failing HAART (n=41) | Children naive to HAART (n=40) | p value |
|---|---|---|---|
| Age, median years (IQR) | 7.9 (4.8–10.4) | 0.9 (0.5–2.8) | <0.0001a |
| Black race | 41 (100.0) | 39 (97.5) | 0.49b |
| Male gender | 24 (58.5) | 18 (45.0) | 0.27b |
| Nadir CD4%, median (IQR) | 9.0 (3.1–13.5) (n=33) | 14.0 (7.5–22.0) (n=37) | 0.008a |
| Current CD4 cell count, median cells/mm3 (IQR) | 532.0 (212.0–1121.0) | 386.0 (227.0–943.5) (n=37) | 0.45a |
| Current CD4%, median (IQR) | 18.0 (9.0–24.0) | 14.0 (7.5–22.0) (n=37) | 0.47a |
| Current plasma HIV-1 viral load, median log10 copies/ml (IQR) | 4.9 (4.4–5.4) | 5.9 (5.6–6.8) | <0.0001a |
| Current WHO stage | (n=40) | ||
| I | 1 (2.5) | 0 (0.0) | |
| II | 15 (37.5) | 1 (2.5) | |
| III | 18 (45.0) | 20 (50.0) | |
| IV | 6 (15.0) | 19 (47.5) | 0.003b |
| Current drug regimen | |||
| d4T, 3TC, EFV | 25 (61.0) | ||
| d4T, 3TC, LPV/r • | 6 (14.6) | ||
| d4T, DDI, EFV * | 1 (2.4) | ||
| AZT, 3TC, NVP | 3 (7.3) | ||
| AZT, 3TC, EFV○ | 3 (7.3) | ||
| AZT, DDI, EFV ♦ | 1 (2.4) | ||
| AZT, DDI, LPV/r ▪ | 1 (2.4) | ||
| d4T, ABC, LPV/r * | 1 (2.4) | ||
| Duration of HAART prior to study recruitment, median months (IQR) | 28.6 (19.7–37.5) (n=38) | ||
| History of single-dose NVP for PMTCT | 10 (26.3) (n=38) | 18 (47.4) (n=38) | 0.09b |
Data are no. (%) of children unless otherwise indicated. For cases in which the data are incomplete, the n value is indicated. ABC, abacavir; AZT/ZDV, azidothymidine; DDI, didanosine; d4T, stavudine; EFV, efavirenz; HAART, highly active antiretroviral therapy; IQR, interquartile range; LPV/r, lopinavir boosted with ritonavir; NVP, nevirapine; PMTCT, prevention of mother-to-child transmission; 3TC, lamivudine; WHO, World Health Organization.
Prior treatment is indicated with italicized drug/s changed • d4T, 3TC, ritonavir (n=1); *unknown;○d4T, 3TC, EFV (n=1) and AZT, 3TC, NVP (n=1); ♦ d4T, 3TC, kaletra; ▪ d4T, 3TC, EFV.
Statistical tests: aMann–Whitney U test and bFisher's exact test (for WHO stage analysis, stages I, II, and III were grouped together).
Of the children failing treatment, 33 (80.5%) were receiving two nucleoside reverse transcriptase inhibitors (NRTIs) plus one nonnucleoside reverse transcriptase inhibitor (NNRTI), while eight (19.5%) children were receiving two NRTIs plus one protease inhibitor (PI). The median duration on HAART prior to study recruitment was 28.6 months.
HIV-1 drug resistance
Only a subset of HAART-naive children was genotyped for drug resistance due to limited sample volume, since the median age of these children was below 1 year old. Drug-associated mutations were found in 4 of 13 (30.8%) HAART-naive patients and included T74S (PI mutation) in one patient, both L10V (PI mutation) and T69N (NRTI mutation) in one patient, L10V in another patient, and E138G (NNRTI mutation) in one patient. None of these are included in the World Health Organization (WHO) list for surveillance of drug resistance.25 Among HAART-failing children, 85.4% had ≥1 significant drug resistance mutation to one drug class and 80.5% to two drug classes. One patient had ≥1 drug resistance mutation to all drug classes; however, this patient was not on a PI-inclusive regimen. The only PI mutation detected in this patient, L33F, is classified as a major mutation using the Stanford drug resistance database but has recently being changed from a major to a minor mutation in the International AIDS Society-USA drug resistance mutation list.26 Only one of eight children on a PI-inclusive regimen had major protease mutations (M46I, I54V, L76V, and V82A) detected and this was the only patient on ritonavir alone (with d4T and 3TC) prior to switching to lopinavir/ritonavir (LPV/r). The minor protease mutation, T74S, found in HIV-1C-untreated persons, was detected in six (14.6%) HAART-failing PI-naive children and may be important in this context as it has been found to restore the fitness of multiresistant HIV-1C viruses.27
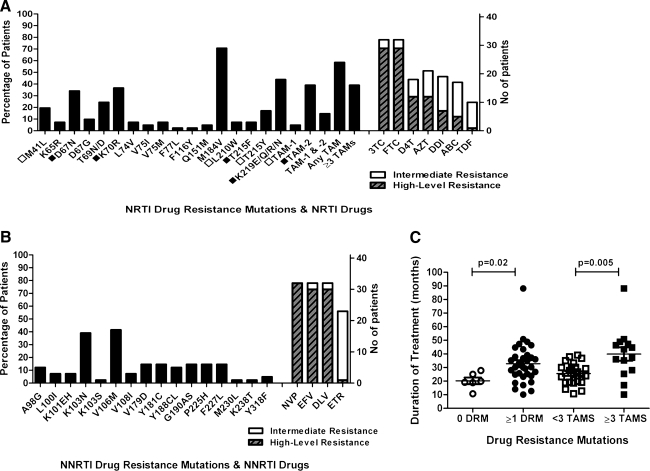
The prevalence of drug resistance mutations and the levels of NRTI and NNRTI resistance are represented in Fig. 1A and B, respectively. The most common NRTI mutations were M184V (70.7%), K219E/Q/R/N (43.9%), K70R (36.6%), and D67N (34.1%). Notably, 7.3% of children had K65R, which may impact the inclusion of tenofovir (TDF) in future treatment regimens and 7.3% had a deletion at reverse transcriptase codon 69 (not shown). The most common NNRTI mutations were V106M (41.5%) and K103N (39%). A high proportion of children had ≥1 TAM (58.5%), with 39.0% having ≥3 TAMs. Both TAM resistance pathways were observed in this study, with the TAM-2 pathway predominating over the TAM-1 pathway (39.0% and 4.9%; respectively) and 14.6% of children having mutations from both pathways.
FIG. 1.
(A) Frequency of selected drug resistance mutations and levels of resistance in HAART-failing children to the NRTIs and to (B) NNRTIs. In (C) associations between duration of treatment and either having ≥1 significant drug resistance mutation compared to none or having ≥3 TAMs compared to <3 TAMs are shown. In Fig. 1A open squares indicate TAM-1 pathway mutations (M41L, L210W, and T215Y); closed squares indicate TAM-2 pathway mutations (D67N, K70R, T215F, and K219E/Q/R/N). HAART, highly active antiretroviral therapy; ABC, abacavir; AZT/ZDV, azidothymidine; DDI, didanosine; DLV, delavirdine; DRM, drug resistance mutation; D4T, stavudine; EFV, efavirenz; ETV, etravirine; FTC, emtricitabine; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NVP, nevirapine; TAMs, thymidine analogue mutations; TDF, tenofovir; 3TC, lamivudine.
High-level resistance was observed in 70.7% of HAART-failing children to both lamivudine (3TC) and emtricitabine (FTC), in 29.3% of children to stavudine (d4T) and azidothymidine (AZT), in 17.1% to didanosine (DDI), in 12.2% to abacavir (ABC), and in 2.4% to TDF (Fig. 1A). Intermediate resistance to TDF was observed in 22.0% of children and the combined level of intermediate and high-level resistance to ABC, d4T, DDI, and AZT was between 41.5% and 51.2% (Fig. 1A). Seventy-eight percent and 73.2% of children had high-level resistance to NVP and both delavirdine (DLV) and efavirenz (EFV), respectively (Fig. 1B). High-level resistance to etravirine (ETR) was found in 2.4% of patients, while 53.7% had intermediate resistance to ETR (Fig. 1B).
The only significant associations with drug-resistant virus or the accumulation of ≥3 TAMs were longer duration of treatment (p=0.02 and p=0.005; respectively) (Fig. 1C).
Some NNRTI resistance mutations were found to persist for long periods. One patient who switched from d4T/3TC/EFV to AZT/DDI/LPV/r 2 years ago still had K101EH, V106M, and V179D. A child who had only received d4T/3TC/LPV/r, except for sdNVP for PMTCT 3.7 years previously, had Y181C.
Viral tropism
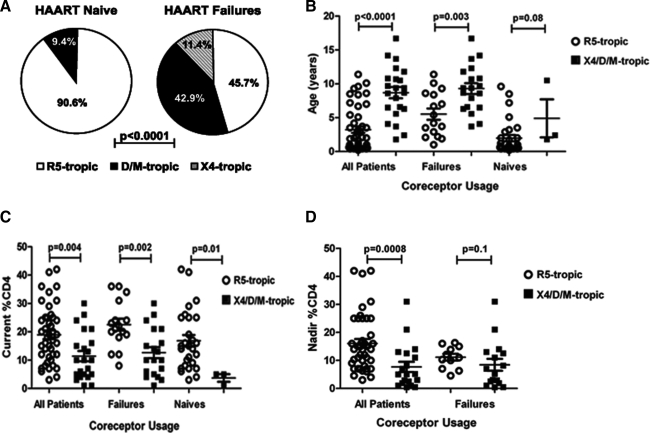
Phenotypic characterization was successful with 35 of 40 HAART-failing and 32 of 39 HAART-naive children. Nonreportable results were likely due to inadequate plasma volumes. HAART-failing children had a significantly higher proportion of X4/D/M viruses compared to the HAART-naive children (54.3% versus 9.4%, respectively; p<0.0001) (Fig. 2A).
FIG. 2.
(A) Comparison of coreceptor usage in HAART-failing and HAART-naive children, (B) associations between coreceptor usage and age, (C) current CD4%, and (D) nadir CD4% of all and of the HAART-failing children alone.
Older children had significantly more X4/D/M viruses compared to younger children (all children: p<0.0001 and HAART-failing children: p=0.003) (Fig. 2B). There was also a trend for children with X4/D/M viruses to be older among the HAART-naive children (p=0.08) (Fig. 2B). Children with X4/D/M viruses had significantly lower current CD4% compared to children with R5-only viruses and this was observed in all children, in the HAART-failing and HAART-naive children (p=0.004, p=0.002, and p=0.01, respectively) (Fig. 2C). Similarly, a lower nadir CD4% was significantly associated with X4/D/M viruses in all children (p=0.0008). However, this was not evident only in the HAART-failing children (p=0.1) (Fig. 2D). In a multivariate logistic regression, HAART treatment, older age, and lower current CD4% were found to be significant predictors of CXCR4 usage (Table 2). Nadir CD4% was excluded from this analysis due to incomplete data. Among HAART-failing children, there was no association found between tropism and the presence of ≥3 TAMs (p=1.0, data not shown).
Table 2.
Multivariate Logistic Regression Analysis for Predictors of CXCR4 Usage
| Variable | OR (95% CI) | p value |
|---|---|---|
| HAART treatment | 18.22 (1.58; 209.60) | 0.0199 |
| Age | 1.49 (1.06; 2.09) | 0.0221 |
| Current CD4% | 0.82 (0.71; 0.95) | 0.0075 |
| Current plasma HIV-1 viral load, log10 copies/ml | 1.86 (0.50; 6.99) | 0.3566 |
| WHO Stage (IV versus I to III) | 0.08 (0.01; 1.22) | 0.0686 |
A total of 64 children were included in this analysis. OR, odds ratio; CI, confidence interval; HAART, highly active antiretroviral therapy; WHO, World Health Organisation.
The env gene was successfully cloned in 40 of 41 HAART-failing and 24 of 24 HAART-naive children who had samples available. Phylogenetic analysis of the env region indicated that the viral isolates cloned were subtype C, except for two that clustered with subtype A and two confirmed intersubtype recombinants, B/C and A/C. All viral isolates with drug resistance data were subtype C in the pol region including these four nonpure C isolates. Phylogenetic analysis of the pol region did not indicate any clustering of samples based on exposure to antiretrovirals (ARVs). Additionally, the two groups (failing and naive) were 99.3% similar, with no significant differences between them. The mean genetic distance between the groups was 2.2% [standard error (SE)±0.2%] and within these groups were 2.3% and 2.0% (SE±0.2%) for the failing and naive children, respectively. The protease diversity between groups was 1.2% (SE±0.3%) and similarly 1.2% within these groups with standard errors of 0.3% and 0.4% for the failing and naïve children, respectively.
The V3 loop is an important determinant of coreceptor usage. Based on the phenotype, all X4 viruses, 10 (58.8%) D/M viruses, and eight (22.9%) R5 viruses had V3 net charges above +4.5. Basic amino acids at positions 11 and/or 25 of the V3 loop were found in two (50.0%) X4, four (23.5%) D/M, and two (5.7%) R5 viruses. All the R5 and 16 (94.1%) D/M viruses had V3 loops of 34 to 35 amino acids in length while three (75%) X4 viruses had lengths between 36 and 37 amino acids due to insertions of two amino acids before the crown motif. The crown motif was GPGQ in all R5 viruses as typically seen in HIV-1C sequences, whereas GPG(K/R) or GRG(H/Q) was seen in all four X4 viruses and seven (41.2%) D/M viruses.
The sensitivity and specificity for predicting CXCR4 usage in pure HIV-1C sequences using several genotypic prediction tools are shown in Table 3. Overall C-PSSMsinsi performed the best for predicting CXCR4 usage with a sensitivity and specificity of 75.0% and 87.5%, respectively. The remaining genotypic tools were poorly sensitive (≤65.0%) at predicting CXCR4 usage. In contrast, the specificities of these tools were all high with values ≥87.5% except for the net V3 charge rule with 78.1%.
Table 3.
Evaluation of Several Genotypic Tools for the Prediction of CXCR4 Usage
| |
Prediction of CXCR4 usagea |
|||
|---|---|---|---|---|
| Genotypic tool | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
| 11/25 charge rule | 30.0 | 96.9 | 83.0 | 74.0 |
| Net V3 charge rule | 65.0 | 78.1 | 59.0 | 82.0 |
| C-PSSMsinsi | 75.0 | 87.5 | 75.0 | 88.0 |
| Geno2pheno[coreceptor]b | 60.0 | 87.5 | 70.0 | 82.0 |
| Combined rulesc | 63.2 | 100.0 | 100.0 | 85.0 |
| C4.5 | 25.0 | 100.0 | 100.0 | 73.0 |
| C4.5 positions 8–12 | 25.0 | 100.0 | 100.0 | 73.0 |
| PART | 30.0 | 100.0 | 100.0 | 75.0 |
| SVMwetcat | 40.0 | 96.9 | 86.0 | 77.0 |
A total of 52 pure subtype C isolates with both phenotypic and genotypic data were included in this analysis.
A false-positive rate of 10% was used.
A combination of the first four genotypic tools was used where the majority prediction was considered as the final genotype prediction (n=47). PPV, positive predictive value; NPV, negative predictive value.
Discussion
HIV-1-infected children in resource-poor settings may be particularly vulnerable to drug resistance due to limited monitoring and salvage options. Viral tropism in children has not been well characterized, particularly for non-B subtypes, which form the bulk of the HIV-1 epidemic. Improved intervention strategies will result in entry inhibitor drugs needed as salvage or earlier therapy to effectively manage the growing numbers of children surviving early mortality.28
This study found that the majority of HAART-failing children (80.5%) had developed at least one significant mutation to two drug classes. Six children (14.6%) had no major drug resistance mutations detected suggesting that nonadherence may be a problem for children in this setting. The only patient with multiple resistance mutations to PIs had been given unboosted ritonavir, previously shown to be strongly predictive of virological failure.29 HAART-failing children had resistance profiles consistent with those reported previously in HIV-1C-infected adults and children, with M184V, V106M, and K103N being the most prevalent mutations.30–32 No V106A was found, confirming previous reports indicating that V106M was the most common substitution in HIV-1C.33 The higher proportion of TAMs detected here (39.0%) and in a Malawian study (44%)30 may be explained by a longer duration of treatment of 28.6 and 36.5 months, respectively, compared to two previous HIV-1C studies31,34 in which only 12–13% of adults had ≥3 TAMs, but the duration of treatment was considerably shorter (10.8 months).31 Supporting this is the significant association found between increased length of treatment and either having drug-resistant virus or ≥3 TAMs. Consistent with other studies,35,36 two children had persistent NNRTI mutations despite stopping treatment several years previously, which may have a deleterious impact on future treatment.
High-level resistance to the majority of NRTIs and NNRTIs ranged from 12.2% to 78% with only TDF and ETR having a low prevalence of high-level resistance (2.4%). Although TDF is not yet approved for use in children, there is still a concern regarding the faster development of K65R in HIV-1C leading to TDF resistance and cross-resistance to several other drugs. A similar percentage of K65R (7.3%) was detected compared to previous HIV-1C studies (2.5% to 19.1%) in adults and children.30–32,34 Overall, the levels and patterns of NRTI and NNRTI resistance and the extensive TAMs are disconcerting, probably resulting from accumulating resistance mutations due to children remaining on failing regimens.
Although an increased prevalence of X4 viruses has been reported in patients exposed to antiretroviral therapy compared to naive patients, this has largely been attributed to lower nadir and/or current CD4+ T cell counts.37,38 Here, antiretroviral treatment, older age, and lower current CD4% were all significant predictors of CXCR4-usage in a multivariate analysis. In this study, interpretation of tropism data was confounded by HAART-naive children being much younger than HAART-failing children. This reflects the implementation of updated treatment guidelines in KwaZulu-Natal at the end of 2008 with children under a year being treated. These factors may require further analysis in longitudinal studies. The higher prevalence of X4/D/M viruses in recent HIV-1C viruses has been reported in both treated and untreated patients9,10 implying that a tropism switch to CXCR4 usage likely is occurring due to a maturing of the HIV-1C epidemic. No associations were found between coreceptor usage and either having drug-resistant virus or an accumulation of TAMs.
With the introduction of maraviroc as either first-line or salvage therapy, improved methods for predicting coreceptor usage are desirable. V3 loop characterization confirmed previous findings in which X4 viruses had a more variable crown motif, longer length, and higher V3 net charges than R5 viruses.11,18 In contrast to other studies,11,17,18 these characteristics were not found in the majority of D/M viruses except for a V3 net charge above +4.5 in 58.8%. The efficiency of genotypic predictions based on V3 loop sequences was also investigated with C-PSSMsinsi having the highest sensitivity and relatively high specificity for predicting CXCR4 usage (75.0 and 87.5%, respectively). The other tools had high specificity but much poorer levels of sensitivity. In a recent study, a combination of the 11/25 and net charge rules, geno2pheno [coreceptor], C-PSSMsinsi, and B-PSSM, proved to be quite sensitive and specific (80.0–93.3%) at predicting CXCR4 usage in HIV-1C viruses.11 In contrast, in two studies predictions of both B and non-B subtypes yielded much poorer levels of sensitivity but a similarly high specificity level using similar tools.39,40 There is a clear discrepancy concerning the sensitivity of these available prediction methods and enhanced laboratory methods for the detection of minority X4 variants, which are mostly underestimated using bulk PCR, are likely to increase their sensitivity.40 Our poorer genotypic prediction of CXCR4 usage could be related to the D/M viruses in this study being mixtures of R5 and X4 viruses; as only one clone generated was sequenced, the minority species may have been missed.
In conclusion, HAART-failing children had a high prevalence of drug resistance mutations and prolonged treatment on failing regimens is almost certainly leading to limited options of effective drugs available for salvage therapy. More than half of the HAART-failing children had either X4/D/M viruses compared to the small percentage of the HAART-naive children. Significant predictors of CXCR4 usage included antiretroviral treatment, older age, and lower current CD4%. CCR5 antagonists such as maraviroc are likely to be more valuable as part of a first-line regimen. Furthermore, improved and more affordable methods for the prediction of coreceptor usage are required to aid rational clinical use of CCR5 antagonists, particularly in resource-limited settings.
Acknowledgments
We thank the study participants and staff involved in this study from King Edward VIII hospital. We are also very grateful for the assistance with CD4+ T cell counts and HIV-1 viral loads performed by the CAPRISA laboratory staff. We thank Dr. Johannes Viljoen and the Africa Centre Virology Laboratory for the use of the ABI Prism 3130xl Genetic Analyzer. We thank Jennifer Fisher and Wei Huang for assistance with V3 sequence analysis and members of the Monogram Biosciences clinical reference laboratory for assistance with Trofile assays.
Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) [U01 AI068632], the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institute of Mental Health (NIMH) [AI068632]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was supported by the Statistical and Data Analysis Center at Harvard School of Public Health under the National Institute of Allergy and Infectious Diseases cooperative agreement #5 U01 AI41110 with the Pediatric AIDS Clinical Trials Group (PACTG) and #1 U01 AI068616 with the IMPAACT Group. Support of the sites was provided by the National Institute of Allergy and Infectious Diseases (NIAID) and the NICHD International and Domestic Pediatric and Maternal HIV Clinical Trials Network funded by NICHD (contract number N01-DK-9-001/HHSN267200800001C).
Further support was received from the Hasso Plattner Foundation, the South African DST/NRF Research Chair Initiative, and by NIH Grant 5R44AT057068 to Monogram Biosciences.
Author's contributions: Concept and design: T.N., M.A., R.B., and H.C. Recruitment and clinical management of patients: M.A. and R.B. Laboratory analyses and interpretation of data: T.N.G., M.L.G., N.P., Y.L., E.D.A., J.D.R., and T.N. Statistical analyses: T.N.G. and A.G. Drafting of the manuscript: T.N.G. and T.N. Critical review of the manuscript: all authors.
Author Disclosure Statement
Y.L., E.D.A., and J.D.R. are employees of Monogram Biosciences, Inc.
References
- 1.UNAIDS: Report on the global AIDS epidemic. http://www.unaids.org/en/KnowledgeCentre/HIVData/GlobalReport/2008/2008_Global_report.asp. [Jul 1;2009 ]. http://www.unaids.org/en/KnowledgeCentre/HIVData/GlobalReport/2008/2008_Global_report.asp
- 2.Hamers RL. Derdelinckx I. van Vugt M. Stevens W. Rinke de Wit TF. Schuurman R. The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antivir Ther. 2008;13:625–639. [PubMed] [Google Scholar]
- 3.Dorr P. Westby M. Dobbs S. Griffin P. Irvine B. Macartney M, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger EA. Doms RW. Fenyo EM. Korber BT. Littman DR. Moore JP, et al. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 5.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarlatti G. Tresoldi E. Bjorndal A. Fredriksson R. Colognesi C. Deng HK, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 7.Choge I. Cilliers T. Walker P. Taylor N. Phoswa M. Meyers T, et al. Genotypic and phenotypic characterization of viral isolates from HIV-1 subtype C-infected children with slow and rapid disease progression. AIDS Res Hum Retroviruses. 2006;22:458–465. doi: 10.1089/aid.2006.22.458. [DOI] [PubMed] [Google Scholar]
- 8.Ndung'u T. Sepako E. McLane MF. Chand F. Bedi K. Gaseitsiwe S, et al. HIV-1 subtype C in vitro growth and coreceptor utilization. Virology. 2006;347:247–260. doi: 10.1016/j.virol.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 9.Connell BJ. Michler K. Capovilla A. Venter WD. Stevens WS. Papathanasopoulos MA. Emergence of X4 usage among HIV-1 subtype C: Evidence for an evolving epidemic in South Africa. AIDS. 2008;22:896–899. doi: 10.1097/QAD.0b013e3282f57f7a. [DOI] [PubMed] [Google Scholar]
- 10.Johnston ER. Zijenah LS. Mutetwa S. Kantor R. Kittinunvorakoon C. Katzenstein DA. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J Virol. 2003;77:7682–7688. doi: 10.1128/JVI.77.13.7682-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raymond S. Delobel P. Mavigner M. Ferradini L. Cazabat M. Souyris C, et al. Prediction of HIV type 1 subtype C tropism by genotypic algorithms built from subtype B viruses. J Acquir Immune Defic Syndr. 2010;53:167–175. doi: 10.1097/QAI.0b013e3181c8413b. [DOI] [PubMed] [Google Scholar]
- 12.DOH: Guidelines for the management of HIV-infected children. http://www.doh.gov.za/docs/factsheets/guidelines/hiv/part5.pdf. [Dec 8;2008 ]. http://www.doh.gov.za/docs/factsheets/guidelines/hiv/part5.pdf
- 13.Whitcomb JM. Huang W. Fransen S. Limoli K. Toma J. Wrin T, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinh L. Han D. Huang W. Wrin T. Larson J. Kiss L, et al. Technical validation of an enhanced sensitivity Trofile HIV co-receptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antiviral Ther. 2008;13:A128. [Google Scholar]
- 15.Reeves JD. Coakley E. Petropoulos CJ. Whitcomb JM. An enhanced-sensitivity Trofile™ HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: A review of analytical and clinical studies. J Viral Entry. 2009;3:94–102. [Google Scholar]
- 16.Salazar-Gonzalez JF. Bailes E. Pham KT. Salazar MG. Guffey MB. Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A. Page T. Moore PL. Allgaier RL. Hiramen K. Coovadia HM, et al. Functional and genetic analysis of coreceptor usage by dualtropic HIV-1 subtype C isolates. Virology. 2009;393:56–67. doi: 10.1016/j.virol.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coetzer M. Cilliers T. Ping LH. Swanstrom R. Morris L. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology. 2006;356:95–105. doi: 10.1016/j.virol.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Fouchier RA. Groenink M. Kootstra NA. Tersmette M. Huisman HG. Miedema F, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen MA. Coetzer M. van 't Wout AB. Morris L. Mullins JI. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J Virol. 2006;80:4698–4704. doi: 10.1128/JVI.80.10.4698-4704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sing T. Low AJ. Beerenwinkel N. Sander O. Cheung PK. Domingues FS, et al. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir Ther. 2007;12:1097–1106. [PubMed] [Google Scholar]
- 22.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 24.Lole KS. Bollinger RC. Paranjape RS. Gadkari D. Kulkarni SS. Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett DE. Camacho RJ. Otelea D. Kuritzkes DR. Fleury H. Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson VA. Brun-Vezinet F. Clotet B. Gunthard HF. Kuritzkes DR. Pillay D, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 27.Soares EA. Santos AF. Gonzalez LM. Lalonde MS. Tebit DM. Tanuri A, et al. Mutation T74S in HIV-1 subtype B and C proteases resensitizes them to ritonavir and indinavir and confers fitness advantage. J Antimicrob Chemother. 2009;64:938–944. doi: 10.1093/jac/dkp315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Violari A. Cotton MF. Gibb DM. Babiker AG. Steyn J. Madhi SA, et al. Early antiretroviral therapy and mortality among HIV-infected infants. N Engl J Med. 2008;359:2233–2244. doi: 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies MA. Wood R. Van Cutsem G. Giddy J. Eley B. Rabie H, et al. 5th IAS Conference on Pathogenesis. Treatment and Prevention; Cape Town: 2009. Virologic failure, second-line antiretroviral therapy (ART) in children in South Africa: The international epidemiologic databases to evaluate AIDS (leDEA) Southern Africa collaboration. [Google Scholar]
- 30.Hosseinipour MC. van Oosterhout JJ. Weigel R. Phiri S. Kamwendo D. Parkin N, et al. The public health approach to identify antiretroviral therapy failure: High-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23:1127–1134. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marconi VC. Sunpath H. Lu Z. Gordon M. Koranteng-Apeagyei K. Hampton J, et al. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008;46:1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallis CL. Erasmus L. Varughese S. Ndiweni D. Stevens WS. Emergence of drug resistance in HIV-1 subtype C infected children failing the South African national antiretroviral roll-out program. Pediatr Infect Dis J. 2009;28:1123–1125. doi: 10.1097/INF.0b013e3181af5a00. [DOI] [PubMed] [Google Scholar]
- 33.Kantor R. Katzenstein DA. Efron B. Carvalho AP. Wynhoven B. Cane P, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallis CL. Mellors JW. Venter WD. Sanne I. Stevens W. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J Acquir Immune Defic Syndr. 2010;53:480–484. doi: 10.1097/QAI.0b013e3181bc478b. [DOI] [PubMed] [Google Scholar]
- 35.Joly V. Descamps D. Peytavin G. Touati F. Mentre F. Duval X, et al. Evolution of human immunodeficiency virus type 1 (HIV-1) resistance mutations in nonnucleoside reverse transcriptase inhibitors (NNRTIs) in HIV-1-infected patients switched to antiretroviral therapy without NNRTIs. Antimicrob Agents Chemother. 2004;48:172–175. doi: 10.1128/AAC.48.1.172-175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinson NA. Morris L. Gray G. Moodley D. Pillay V. Cohen S, et al. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr. 2007;44:148–153. doi: 10.1097/QAI.0b013e31802b920e. [DOI] [PubMed] [Google Scholar]
- 37.Briz V. Poveda E. del Mar Gonzalez M. Martin-Carbonero L. Gonzalez-Gonzalez R. Soriano V. Impact of antiretroviral therapy on viral tropism in HIV-infected patients followed longitudinally for over 5 years. J Antimicrob Chemother. 2008;61:405–410. doi: 10.1093/jac/dkm469. [DOI] [PubMed] [Google Scholar]
- 38.Hunt PW. Harrigan PR. Huang W. Bates M. Williamson DW. McCune JM, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194:926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 39.Garrido C. Roulet V. Chueca N. Poveda E. Aguilera A. Skrabal K, et al. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J Clin Microbiol. 2008;46:887–891. doi: 10.1128/JCM.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low AJ. Dong W. Chan D. Sing T. Swanstrom R. Jensen M, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS. 2007;21:F17–24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]