Abstract
Introduction
The innate immune system depends on molecules collectively known as pattern recognition receptors (PRRs) to survey the extracellular space and the cytoplasm for the presence of dangerous pathogens, pathogen-derived molecules, or even self-derived molecular danger signals, which arise from tissue damage. Absent in melanoma 2 (AIM2) is a newly discovered PRR involved in the sensing of dangerous cytosolic DNA produced by infection with DNA viruses.
Discussion
Remarkably, recent studies in AIM2-deficient mice showed that AIM2 is uniquely involved in sensing infection with the intracellular bacteria Francisella tularensis and subsequently triggering caspase-1-mediated pro-inflammatory cytokine production and macrophage cell death, which activate other components of the immune system and eliminate the infected macrophages. Here, we provide an overview of our current understanding of the role of AIM2 in innate immunity against F. tularensis in particular, and how infection of macrophages with this pathogen is thought to activate AIM2.
Keywords: Inflammasome, caspase-1, AIM2, innate immunity, inflammation, Francisella tularensis
Pattern Recognition Receptors in Innate Immunity
The innate immune system is the first line of defense against invading organisms and their associated substances. Molecular components of this system are evolutionarily conserved and are found in fungi, plant, and animal cells. All animal cells contain at least one or more of receptors collectively called pathogen or pattern recognition receptors (PRRs) [1, 2]. These receptors are specialized in sensing highly conserved molecular components of the invading organisms known as pathogen-associated molecular patterns (PAMPs), and in some cases, they also sense endogenous damage-associated molecular patterns (DAMPs) generated by tissue damage. PRRs are found in the cytoplasmic or endosomal membranes, or in the cell cytosol.
The Toll-Like Receptors
The best characterized among the PRRs is the Toll-like receptor (TLR) family, which to date contains 11 human and 13 mouse family members, each with distinct ligand specificity. TLRs are membrane-spanning proteins localized on the cytoplasmic or endosomal membranes [3], where they recognize specific extracellular microbial ligands, such as bacterial and viral nucleic acids and proteins, bacterial and fungal cell wall components and lipoproteins, and in some instances host-derived ligands, such as DNA, through their extracellular leucine-rich repeat (LRR) motif [3–5]. When engaged by their ligands, TLRs trigger a set of downstream signal transduction pathways via recruiting one or more Toll/interleukin-1 receptor homology (TIR) domain-containing adaptor molecules such as myeloid differentiation primary response gene 88 (MyD88), MyD88-adaptor-like (MAL/TIRAP), TIR-domain-containing adaptor protein-inducing interferon-β (TRIF/TICAM1), and TRIF-related adaptor molecule (TRAM/TICAM2) [1, 4, 6]. These various TLR-mediated signal transduction pathways ultimately lead to the activation of several transcriptional factors including nuclear factor-kappa B (NF- κB), interferon regulatory factors such as IRF-3 and IRF-7, and other more general transcription factors. These transcriptional events culminate in the induction of pro-inflammatory cytokines including type I interferons IFNα and IFNβ, and interleukins, which activate macrophage and dendritic cells and enhance the cytotoxic lymphocyte response to the infected cells [1, 6, 7].
The Retinol-Inducible Gene-I (RIG-I)-Like Receptor (RLR) and Nucleotide-Binding Oligomerization Domain (Nod)-Like Receptor (NLR) Proteins
Infection with intracellular pathogens also inadvertently leads to the release of their PAMPs, including nucleic acids, into the cytosol of infected cells where specialized PRRs detect their presence [2]. These PRRs includes retinol-inducible gene-I (RIG-I)-like receptor (RLR) families and nucleotide-binding oligomerization domain (Nod)-like receptor (NLR). Unlike the membrane-spanning TLRs, RLRs and NLRs are localized in the cytoplasm. As their localization indicates, these receptors function to survey the cytoplasm for the presence of PAMPs and tissue damage-derived molecules [2]. For example, RIG-I and melanoma differentiation-associated gene 5 (MDA5) recognize viral double-stranded RNA (dsRNA) through their C-terminal RNA helicase domain and subsequently signal via the adaptor protein MAVS (mitochondrial antiviral signaling protein, also known as IPS-1, Cardif and VISA) [2, 8], and Tank binding kinase 1 to activate the transcriptional pro-inflammatory NF- κB and the antiviral type I interferon response pathways. Intriguingly, recent results demonstrated that RIG-I can also signal through the adaptor ASC (apoptosis-associated speck-like protein containing a CARD) to trigger the non-transcriptional caspase-1-dependent inflammasome activation pathway in response to infection with RNA viruses [9].
The NLR family on the other hand contains 16 family members of which many are still without a clear function. NLRs contain important functional and regulatory domains including N-terminal caspase recruitment domain (CARD), pyrin domain (PYD), or baculovirus inhibitor of apoptosis repeat (BIR), followed by central NACHT domain and C-terminal LRR motifs [10, 11]. The first identified NLR members, NOD1 and NOD2, have been shown to activate NF-κB transcriptional pathway, similar to TLR or RLR, in response to the presence of bacterially derived cell wall components such as MDP in the cytoplasm [2]. Subsequent studies revealed that a subset of NLR proteins such as NLRP1 (Nalp1), NLRP3 (Nalp3, cryopyrin), and NLRC4 (Ipaf) are involved in the production of the mature IL-1β and IL-18 pro-inflammatory cytokines and the induction of pyroptotic cell death through the activation of the cysteine protease caspase-1 [11]. Although it is still not clear how these NLR proteins recognize microbial pathogen- or danger-derived molecular signals, it is now believed that these NLRs assemble “inflammasome” complexes with or without the adaptor protein ASC [11]. These complexes serve as molecular platforms to recruit and activate pro-caspase-1 to produce the active caspase-1, which processes pro-IL-1β and pro-IL-18 into their corresponding mature cytokines, IL-1β and IL-18. The active caspase-1 can also cleave other cellular proteins leading to a distinct form of cell death termed pyroptosis [12, 13].
Innate Immunity to Cytosolic DNA
Although considerable progress in our understanding of the pathways involved in sensing and responding to extracellular PAMPs and cytosolic dsRNA has been made over the last decade, the pathways involved in recognition and innate immunity to cytosolic DNA have only been recently discovered. Like dsRNA, cytosolic double-stranded DNA (dsDNA) can also activate both the transcriptional NF-κB and type I interferon pathways (in most cell types), as well as a non-transcriptional caspase-1 activation pathway (in macrophages and monocytes; reviewed in [14]).
Recent studies identified molecular mechanisms by which cytoplasmic DNA is recognized by innate immune cells leading to the activation of these two distinct pathways [15–20]. A putative receptor for cytoplasmic DNA called DAI (DNA-dependent activator of IFN-regulatory factors, also known as Z-DNA binding protein 1) has been suggested to be involved in the induction of type I interferons in response to cytosolic DNA [21, 22]. However, studies in DAI-deficient cells and mice showed that these mice have normal responses to cytoplasmic DNA [23], indicating that this factor is redundant or might be required for sensing modified DNA produced by a specific intracellular pathogen. More recently DNA-dependent RNA polymerase III was shown to be an important mediator of type I interferon response to cytoplasmic DNA [19, 20]. RNA polymerase III apparently recognizes cytosolic dsDNA and converts it to dsRNA, which is in turn recognized by RIG-I leading to the activation of the transcriptional NF-κB and type I interferon response pathways.
Studies from our lab and others demonstrated that cytoplasmic dsDNA is also recognized by the interferon-inducible protein, absent in melanoma 2 (AIM2) [15–18], which binds to the dsDNA through its HIN-200 domain, and assembles a large inflammasome complex with it [15]. In the following sections, we focus on the role of the AIM2 inflammasome in the innate immune responses to microbial and non-microbial danger signals and the mechanisms involved in the regulation of its activity.
The AIM2 Inflammasome
The term inflammasome collectively refers to oligomeric molecular platforms or assemblies that recruit and activate caspase-1. These assemblies are formed in the cytoplasm of innate immune cells like macrophages and dendritic cells when these cells are exposed to danger signals [11]. The basic structure of the inflammasome consists of a “danger-sensing initiator” component, a “pro-caspase-1-activating” component, and the “effector” component, which is the mature caspase-1 itself. In the case of the AIM2 inflammasome, the AIM2 protein serves as the initiator component, which recognizes the cytosolic DNA ligand through its DNA binding HIN-200 domain [15–18], whereas the ASC protein acts as the pro-caspase-1-activating component of this inflammasome [24].
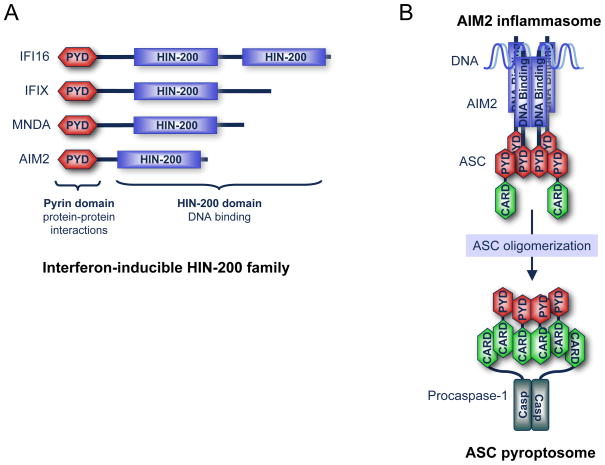
AIM2 was identified in a functional screen for putative melanoma tumorigenicity suppressor genes (reviewed in [25]). AIM2 belongs to the hematopoietic interferon-inducible HIN-200 protein family, of which all family members have a similar structure comprised of an N-terminal PYD and a C-terminal HIN-200 domain (also called IFI-200 domain) containing two oligonucleotide/oligosaccharide-binding folds (Fig. 1a). Four human and five mouse HIN-200 family members were cloned and characterized [25]. Although all HIN-200 family members can bind to DNA, only AIM2 is able to interact with ASC through PYD–PYD interactions and is primarily localized in the cytoplasm [15, 16]. Our results show that cytoplasmic DNA interacts with the HIN-200 domain of AIM2 and causes it to form large oligomers visible by confocal microscopy [15]. The fully assembled oligomeric AIM2/DNA complex then recruits the adaptor protein ASC through PYD–PYD interactions leading to its oligomerization into a structure we coined “the ASC pyroptosome” [13] (Fig. 1b). Subsequently, the ASC pyroptosome recruits pro-caspase-1 through protein–protein interactions between the CARDs of ASC and those of pro-caspase-1 molecules, leading to auto-activation of caspase-1 by the induced proximity mechanism [26]. Activated caspase-1 then induces the maturation of the IL-18 and IL-1β cytokines, as well as pyroptotic cell death [13, 15].
Fig. 1.
A schematic representation of the interferon-inducible HIN-200 family and the AIM2 inflammasome. a Human interferon-inducible HIN-200 family, IFI16, IFIX, MNDA, and AIM2. PYD: pyrin domain involved in protein–protein interactions. HIN-200 domain is a DNA binding domain. b The oligomerized AIM2–DNA complex forms the AIM2 inflammasome, which serves as a molecular platform to recruit ASC through protein–protein interactions between the PYDs of the AIM2 molecules and those of ASC molecules. This causes the ASC molecules to form a large supramolecular cluster called the pyroptosome. Subsequently, the ASC pyroptosome recruits pro-caspase-1 through protein-–protein interactions between the CARDs of the ASC pyroptosome and those of the monomeric pro-caspase-1 molecules. These interactions result in activation of pro-caspase-1 into the active caspase-1 protease.
Role of AIM2 in the Pro-inflammatory Response to Transfected or Virally Produced Cytosolic DNA
Despite these recent advances in our understanding of the mechanism by which cytosolic DNA is recognized by AIM2 leading to caspase-1-dependent production of the pro-inflammatory cytokines IL-1β and IL-18 and cell death, we still know little about the exact role of the AIM2 signaling pathway in the pro-inflammatory response to and innate immunity against intracellular microbial and viral pathogens in vivo, and whether there exist other redundant AIM2-independent pathways that might fulfill this purpose. Most of our current understanding of AIM2 was based on in vitro experiments using AIM2-targeting small interfering RNA (siRNA) strategies and transfection and reconstitution experiments in 293T cells. The siRNA strategies have the inherent disadvantages that they do not achieve complete removal of the AIM2 protein from macrophages and could also lead to activation of type I interferon response. This might lead to altered innate immune responses to microbial and viral pathogens and potentially complicate the interpretation of any results obtained with siRNA knockdown experiments. To overcome these limitations, we recently generated AIM2-deficient mice to study pathogen–host interactions and to provide us with a continuous source of innate immune cells and tissues to define the precise role of the AIM2 inflammasome in the host pro-inflammatory response to and innate immune defense against viral and microbial pathogens in vivo and in vitro [27].
The AIM2-deficient mice develop normally and have no phenotypic abnormalities indicating that AIM2 is not critical for physiological homeostasis. Interestingly, bone marrow-derived macrophages from these mice were found to be defective in caspase-1 activation and induction of pyroptotic cell death in response to cationic liposome-delivered cytosolic DNA from different sources including the synthetic DNA poly(dA:dT). In contrast, these cells had normal pro-inflammatory (caspase-1 activation and IL-1β processing) and cell death responses to NLRP1-, NLRP3-, and NLRC4-specific stimuli, indicating that AIM2 is specifically involved in the recognition and pro-inflammatory and cell death responses to cytosolic DNA.
Vaccinia virus is a member of the poxviruses, which are the largest known DNA viruses, distinguishable from other viruses by their ability to replicate their DNA entirely in the cytoplasm of infected cells. Vaccinia viral DNA is therefore not sequestered from the cytoplasm and can potentially be detected by AIM2. Consistent with this, AIM2-deficient macrophages were defective in caspase-1 activation in response to infection with Vaccinia virus [27]. However, we observed a small amount of caspase-1 activation by Vaccinia virus in AIM2-deficient macrophages if the cells were infected for longer periods of time (≥24 h; unpublished observation), perhaps through activation of the NLRP3 inflammasome. This indicates that Vaccinia virus activation of the pro-inflammatory response is primarily AIM2-dependent, but could still elicit a relatively small pro-inflammatory response in the absence of AIM2 through activation of other inflammasomes. Whether AIM2 is involved in the recognition of and the pro-inflammatory responses to other DNA viruses such as adenovirus is not yet clear. Work from Jurg Tschopp’s lab demonstrated that adenoviruses are recognized primarily by the NLRP3 inflammasome [28]. Although adenovirus is a DNA virus, its DNA might not be sensed by the AIM2 inflammasome. Adenoviral particles attach to their cell surface receptors (CAR or CD46 depending on serotype) and are internalized within an endosome by endocytosis via clathrin-coated pits. Subsequently, they escape the endosome and are transported to the nuclear pore complex, which facilitates the transportation of the viral DNA into the nucleus, where gene expression, viral replication, and assembly occur [29]. Therefore, unlike Vaccinia, the DNA of adenoviruses is most likely sequestered from the cytoplasm at all stages of viral entry, transportation, and replication and thus cannot be detected by AIM2. As a result, adenovirus infection is expected to be primarily sensed by the NLRP3 inflammasome as demonstrated before [28], which is activated by many cellular danger signals, particularly those that could lead to endosomal/lysosomal destabilization.
Taken together, studies in AIM2-deficient macrophages provided definitive genetic proof that the AIM2 inflammasome is indeed critical for recognition of cytosolic DNA whether delivered into the cytoplasm artificially by cationic liposome-mediated transfection, or infection with Vaccinia virus. It is thus expected that AIM2 might also play a critical role in the pro-inflammatory response to other viruses which produce cytosolic DNA during their life cycle (e.g., herpes simplex virus-1 “HSV-1” and perhaps single-stranded RNA retroviruses, which reverse transcribe their RNA into double-stranded DNA in the cytoplasm). Because activation of the AIM2 inflammasome not only leads to generation of pro-inflammatory cytokines, but also to killing and elimination of the infected cells, we propose that AIM2 is likely critical for efficient clearance of viral infection and improving survival of animals specifically infected with cytosolic DNA viruses, and possibly other viruses in general, which produces cytoplasmic DNA during their replication in infected macrophages.
Role of AIM2 in the Pro-inflammatory Response to Francisella tularensis Infection
F. tularensis is a facultative highly infectious intracellular gram-negative bacteria and the causative agent of tularemia, a serious infectious disease in humans and animals with high mortality rates. Because F. tularensis is a highly infectious pathogen, it represents a major concern to the public as a possible bioterrorism agent. Understanding the molecular determinants of its pathogenesis and virulence and the cellular innate immune pathways that recognize and eliminate this pathogen is thus of clear health interest, especially for the development of novel antimicrobial strategies and vaccines targeting this pathogen.
F. tularensis primarily infects and replicates inside macrophages, and by doing so, it manages to avoid early detection by the immune system. It enters macrophages through the endocytic pathway by phagocytosis. Within 1 h after entry into the early endosomes/phagosome, it rapidly secretes bacterial proteins that disrupt the phagosomal membrane and facilitate its escape into the cytosol where it replicates freely from 4 to 24 h after infection [30]. The escape of F. tularensis from the early endosomes triggers potent type I interferon production as well as activation of the caspase-1 and cell death pathways (reviewed in [31]). The induction of type I interferon production by F. tularensis escape from the endosome appears to be dependent on IRF3 signaling and is independent of signaling by plasma membrane or endosomal TLRs, or cytosolic RIG-I/MDA5 or Nod1/Nod2 [31, 32]. Since this response is triggered by the presence of DNA in the cytosol, we believe that endosomal lysis of F. tularensis shortly after infection of macrophages and the subsequent disruption and escape of live F. tularensis from the phagosomes provides a source of DNA and the means for delivering this DNA for cytosolic recognition by different cytosolic nucleic acid sensors including AIM2 (see below). Consistent with this mechanism, we found that macrophages from AIM2-deficient mice are remarkably defective in their ability to induce caspase-1 activation and cell death in response to infection with F. tularensis [27]. However, these macrophages have no obvious defect in type I interferon production after F. tularensis infection, indicating that AIM2 is not a critical signaling component in the interferon production pathway.
Although macrophages from AIM2-deficient mice have normal caspase-1-dependent pro-inflammatory and cell death responses to NLRC4-, NLRP1-, and NLRP3-activating stimuli, they are clearly defective in their ability to mount similar responses against F. tularensis infection. It is thus clear that F. tularensis have developed mechanisms to evade detection by or prevent activation of the well-known host cell inflammasomes NLRP1, NLRP3, or NLRC4 and probably other as yet uncharacterized inflammasomes. Consequently, the new genetic data from the AIM2-deficient macrophages indicate that the AIM2 inflammasome is uniquely required for sensing of and activation of the caspase-1 dependent pro-inflammatory and cell death responses against F. tularensis infection.
Mechanism of Activation of AIM2 by F. tularensis
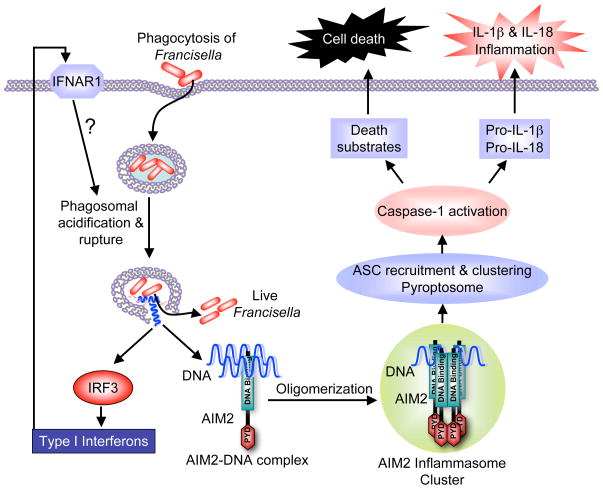
As discussed above, AIM2 is activated by cytosolic DNA through a mechanism involving binding of AIM2 to the DNA and subsequent oligomerization of the DNA-bound AIM2 into a large oligomeric molecular platform that activates caspase-1 with the help of ASC. However, since F. tularensis DNA is encapsulated inside its cell wall, the cell wall must be destroyed to release the DNA into the cytoplasm, in order for AIM2 to sense F. tularensis infection. The precise mechanism by which this is achieved is not yet clear but we speculate that the cell wall of some phagocytosed F. tularensis bacteria is lysed when the phagosome is transiently acidified, shortly after it enters into the phagosomes, and eventually when the phagosomal membrane is degraded (within 1–2 h after bacterial phagocytosis), the DNA from the lysed bacteria escapes together with the live bacteria into the cytoplasm (Fig. 2). Supporting this hypothesis, inhibition of killing and degradation of F. tularensis in the phagosome using the phagosomal acidification inhibitors bafilomycin or NH4Cl completely inhibited these processes. Because phagosomal acidification inhibitors prevent killing and degradation of phagocytosed bacteria [33, 34] but do not prevent phagosomal permeabilization and escape of F. tularensis into the cytosol [35], these observations strongly suggest that phagosomal acidification is a critical event essential for providing a source for cytosolic DNA that subsequently activates both the AIM2 inflammasome and production of type I interferons.
Fig. 2.
A schematic model depicting activation of the AIM2 inflammasome by F. tularensis infection. The bacteria enter macrophages through phagocytosis. Shortly after, the phagosome rapidly acidifies and ruptures releasing the phagocytosed bacteria and some bacterially derived DNA from lysed bacteria. This released DNA activates IRF3 by an as yet uncharacterized mechanism, which transcriptionally induces production of type I interferons. The newly synthesized type I interferons somehow enhance activation of the AIM2 inflammasome through an autocrine signaling pathway involving IFNAR1. The released DNA can also bind to the HIN-200 DNA-binding domain of AIM2. This interaction leads to oligomerization of AIM2 into a large AIM2 inflammasome cluster. The AIM2 inflammasome activates caspase-1 as described in Fig. 1, which in turn processes pro-IL-1β and pro-IL-18 to the active cytokines, IL-1β and IL-18, which induce inflammation. The activated caspase-1 also cleaves a number of cellular proteins leading to pyroptotic cell death. The nature of the caspase-1 substrates that lead to pyroptotic cell death, however, is not yet known.
Further support for the above mechanism is provided by the observations that mutants of F. tularensis subspecies novicida lacking the transcription factor MglA or genes within the F. tularensis pathogenicity island are unable to escape the phagosome [36–38], and therefore, are defective in their ability to stimulate type I interferon production or to induce caspase-1 activation and cell death. It will be interesting to find out in the future whether more virulent strains of F. tularensis such as F. tularensis subsp. tularensis, which infects humans, have developed means to prevent their lysis during their presence in the phagosomes, whereas less virulent strains, such as the FTT0748 and FTT0584 strains [31], which activate caspase-1 and cell death more efficiently than the wild-type strain F. tularensis novicida, have lost the activity of some of their protective genes, thus making them more susceptible to phagosomal lysis.
Role of Type I Interferon Signaling in F. tularensis-Induced Activation of the AIM2 Inflammasome
Previous work from Denise Monack’s group clearly showed that signaling by IFNβ is surprisingly required for efficient activation of caspase-1 and cell death in response to F. tularensis infection of macrophages [32]. Nevertheless, the mechanism by which IFNβ signaling through its IFNAR1 surface receptor enhances the caspase-1 and cell death responses to F. tularensis infection is not yet clear. Our results demonstrated that prior treatment of wild-type or IRF3-deficient macrophages with IFNβ accelerates the activation of the inflammasome by almost 2 h [27], but without affecting the expression levels of AIM2 proteins in these cells, indicating that IFNβ signaling does not modulate caspase-1 activation and cell death by modulating the expression level of AIM2. It is also unlikely that IFNβ signaling induces a factor that modulates the activity of AIM2 since IRF3-deficient macrophages have a normal caspase-1 activation response to transfected DNA as wild-type macrophages. Therefore, IFNβ signaling is most likely required upstream of cytosolic DNA release, possibly at the level of the phagosome, perhaps to enhance killing of F. tularensis and permeabilization of the phagosome. Future studies should clarify how type I interferon signaling potentiates the activity of the AIM2 inflammasome in response to F. tularensis infection.
AIM2 Is Critical for Innate Immunity Against F. tularensis
The in vitro studies in isolated macrophages clearly demonstrated the key role that AIM2 plays in the caspase-1 activation and cell death pathways in response to infection with F. tularensis. The importance of AIM2 in the innate immunity to this pathogen became even more obvious when AIM2-deficient mice were challenged subcutaneously with a sublethal dose of F. tularensis. Remarkably, all of these mice died within 72 h after infection compared to only 33% of the wild-type mice, because of increased bacterial burden in their tissues [27]. These findings thus highlight the critical nature of the AIM2 inflammasome in innate immunity against this pathogen and most likely future studies will identify other as yet uncharacterized intracellular pathogens that are also sensed by the AIM2 inflammasome. As a final note, it is worth mentioning that although the AIM2-deficient mice have no defect in mounting a strong type I interferon response against F. tularensis, this response appears to be insufficient on its own to protect these mice from the lethality of F. tularensis infection. We propose that since macrophages represent the primary replication niche for F. tularensis, early killing of the infected macrophages through the AIM2-dependent ASC/caspase-1 cell death pathway prevents the spread of the infection to vital organs and reduces bacterial burden allowing sufficient time for the adaptive immune system to clear the bacteria from the infected tissues.
Concluding Remarks
We have focused here on a newly identified innate immune pathway involved in caspase-1 activation and cell death in response to dangerous cytoplasmic DNA. This pathway is mediated by the interferon-inducible protein AIM2. Our studies with AIM2-deficient mice or macrophages derived from these mice clearly demonstrated that AIM2 is involved in sensing cytosolic pathogen and non-pathogen-derived DNA. AIM2 is particularly involved in sensing cytosolic DNA generated by infection with Vaccinia virus and the intracellular bacteria F. tularensis. The AIM2 inflammasome represents a crucial surveillance and early response component of the innate immune system, because it not only alerts different components of the immune system through the production of IL-18 and IL-1β when it detects dangerous cytoplasmic DNA, but also destroys the infected macrophages by inducing caspase-1-dependent pyroptotic cell death to limit the spread of infection. Considering that some human auto-immune diseases such as systemic lupus erythematosus are linked to dysregulated immunity to self-generated nucleic acids, future studies with AIM2-deficient mice should provide important information on the role of AIM2 in these nucleic acid-dependent diseases.
Acknowledgments
This work was supported by NIH grants AG14357 and AR055398 to ESA.
Footnotes
Recent studies from K. Fitzgerald [39], confirmed our findings that AIM2 is critical for innate immunity against F. tularensis and Vaccinia virus infection.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 3.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–25. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 5.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- 6.O’ Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 7.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–90. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 8.Moore CB, Ting JP. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008;28:735–9. doi: 10.1016/j.immuni.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–9. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 10.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–7. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 12.Brodsky IE, Monack D. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin Immunol. 2009;21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–30. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 18.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 19.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–91. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–72. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477–82. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–5. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 23.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–9. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–22. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 25.Choubey D, Duan X, Dickerson E, Ponomareva L, Panchanathan R, Shen H, et al. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30(6):371–80. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–41. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 27.Fernandes-Alnemri T, Yu J, Juliana C, Solorzano L, Kang K, Wu J, et al. The AIM2 inflammasome is critical for innate immunity against F. tularensis. Nature Immunol. 2010;11(5):385–93. doi: 10.1038/ni.1859. Epub 2010 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–7. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 29.Wu E, Nemerow GR. Virus yoga: the role of flexibility in virus host cell recognition. Trends Microbiol. 2004;12:162–9. doi: 10.1016/j.tim.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–83. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry T, Monack DM. Activation of the inflammasome upon Francisella tularensis infection: interplay of innate immune pathways and virulence factors. Cell Microbiol. 2007;9:2543–51. doi: 10.1111/j.1462-5822.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- 32.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–94. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankenberg T, Kirschnek S, Hacker H, Hacker G. Phagocytosis-induced apoptosis of macrophages is linked to uptake, killing and degradation of bacteria. Eur J Immunol. 2008;38:204–15. doi: 10.1002/eji.200737379. [DOI] [PubMed] [Google Scholar]
- 34.Bidani A, Reisner BS, Haque AK, Wen J, Helmer RE, Tuazon DM, et al. Bactericidal activity of alveolar macrophages is suppressed by V-ATPase inhibition. Lung. 2000;178:91–104. doi: 10.1007/s004080000012. [DOI] [PubMed] [Google Scholar]
- 35.Clemens DL, Lee BY, Horwitz MA. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect Immun. 2009;77:1757–73. doi: 10.1128/IAI.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005;7:969–79. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 37.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–9. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–59. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- 39.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. doi: 10.1038/ni.1864. Epub 2010 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]