Abstract
pAp (3′-5′ phosphoadenosine phosphate) is a by-product of sulfur and lipid metabolism and has been shown to have strong inhibitory properties on RNA catabolism. In the present paper we report a new target of pAp, PARP-1 [poly(ADP-ribose) polymerase 1], a key enzyme in the detection of DNA single-strand breaks. We show that pAp can interact with PARP-1 and inhibit its poly(ADP-ribosyl)ation activity. In vitro, inhibition of PARP-1 was detectable at micromolar concentrations of pAp and altered both PARP-1 automodification and heteromodification of histones. Analysis of the kinetic parameters revealed that pAp acted as a mixed inhibitor that modulated both the Km and the Vmax of PARP-1. In addition, we showed that upon treatment with lithium, a very potent inhibitor of the enzyme responsible for pAp recycling, HeLa cells exhibited a reduced level of poly(ADP-ribosyl)ation in response to oxidative stress. From these results, we propose that pAp might be a physiological regulator of PARP-1 activity.
Keywords: bipolar disorder, lithium, 3′-5′ phosphoadenosine phosphate (pAp), poly(ADP-ribose) polymerase 1 (PARP-1)
Abbreviations: FBS, fetal bovine serum; NDK, nucleoside diphosphate kinase; pAp, 3′-5′ phosphoadenosine phosphate; PARP-1, poly(ADP-ribose) polymerase 1; PAPS, phosphoadenosine 5′ phosphosulfate; Sfn/Orn, small fragment nuclease/oligoribonuclease
INTRODUCTION
pAp (3′-5′ phosphoadenosine phosphate) is a ubiquitous 3′-phosphorylated nucleotide present in organisms from all domains of life [1]. In mammalian cells, pAp is produced primarily by the transfer of the sulfate group of PAPS (phosphoadenosine 5′ phosphosulfate) to various acceptor molecules [2]. This transfer is catalysed by a number of sulfotransferases and plays a crucial role in various biochemical pathways. For example, sulfonation is involved in the synthesis of sulfate-rich macromolecules such as heparan sulfate, the activation or inactivation of xenobiotics, the inactivation of hormones and catecholamines, and the elimination of end-products of catabolism [3]. Apart from sulfur metabolism, pAp is also generated from CoA during the transfer of the 4-phosphopantetheine group to ACP (acyl carrier protein) in fatty acid synthesis or to secondary metabolites such as peptide antibiotics, surfactin or polyketides [4].
pAp is recycled into AMP and Pi by pAp-phosphatases. pAp-phosphatases have been described in many organisms, including bacteria, yeast, plants and mammals [5–8], and belong to a protein family defined by a structurally conserved core domain. A characteristic feature of this protein family is that all members are competitively inhibited by sub-millimolar concentrations of lithium [9]. As pAp-phosphatase exhibits a very low inhibitory constant, Ki, for lithium [10], it has been proposed that pAp-phosphatase could be a molecular target of lithium toxicity. Indeed, overexpression of pAp-phosphatase can rescue lithium toxicity both in yeast and bacteria [11,12]. In addition, lithium is widely used in the treatment of bipolar disorder [13,14], and several authors have proposed that pAp-phosphatase could be one of the therapeutic targets of lithium [9,10]. This hypothesis was supported by the low Ki of lithium for human pAp-phosphatase [10] compatible within the range of therapeutic concentrations of lithium in the plasma of patients (0.8–1 mM). Importantly, the Ki of human pAp-phosphatase is ten times lower than that of GSK3β (glycogen synthase kinase 3β), another known target of lithium [15].
The inhibition of pAp-phosphatase by lithium is predicted to result in the accumulation of intracellular pAp, which in turn could affect various enzymes and biological processes within the cells. In fact, pAp inhibits in vitro several proteins, such as the different sulfotransferases that use PAPS as substrate [3], NDK (nucleoside diphosphate kinase) [16] and several nucleases involved in the degradation of RNA. Indeed, the ribonuclease Xrn1p, responsible in yeast for most cytoplasmic RNA turnover [17], and oligoribonucleases from Escherichia coli and humans are strongly inhibited by micromolar amounts of pAp [12,18].
In the present study, we have identified an additional target of pAp in human cells. Using pAp-affinity chromatography, we found that PARP-1 [poly(ADP-ribose) polymerase-1] could bind selectively to immobilized pAp. We further demonstrated that PARP-1 activity was strongly inhibited by micromolar concentrations of pAp. We also showed that lithium treatment of cells strongly reduced PARP-1 activity in response to oxidative stress. PARP-1 is a key enzyme in the detection of DNA single-strand breaks and is crucial for the recruitment of the DNA repair machinery to these lesions [19]. In the case of severe DNA damage, PARP-1 is also important in regulating the balance between DNA repair and cell death [20]. The inhibition of PARP-1 activity by pAp suggests a link between sulfur and lipid metabolism and the DNA repair machinery.
MATERIALS AND METHODS
pAp-affinity chromatography on nuclear extracts
pAp-affinity chromatography was performed as described previously [12]. HeLa cells were grown in spinner flasks in DMEM (Dulbecco's modified Eagle's medium) containing 7% FBS (fetal bovine serum). Nuclear extracts were prepared from 108 cells as described by Dignam et al. [21]. Extracts were incubated with cyanogen-bromide-activated agarose beads coupled to pAp (pAp-agarose beads; Sigma–Aldrich). Cyanogen-bromide-activated agarose beads blocked with glycine were used as a control for non-specific binding. Nuclear extract was added to blocked agarose beads and rotated for 2 h at 4°C to clear the extract from proteins binding non-specifically to agarose. Supernatant was divided equally and added to washed pAp-agarose beads (pAp-binding fraction) or blocked agarose beads (control). After incubation at 4°C for 1.5 h, the beads were washed extensively with wash buffer [50 mM Hepes (pH 7.5), 10 mM CaCl2, 50 mM KCl and 0.5 M NaCl] (ten times in 800 μl). Elution was performed by adding hot SDS sample buffer followed by incubation at 65°C for 15 min. Aliquots (20 μl) of each sample were analysed by SDS/PAGE (12% gels).
Comparative binding of PARP-1 to AMP- or pAp-agarose
Samples containing 1 μg of partially purified PARP-1 (Trevigen) and 100 μg of BSA in 50 μl of binding buffer [5 mM MgCl2, 100 mM NaCl and 50 mM Tris/HCl (pH 8.0)] were incubated in the presence or absence of 3 mM pAp for 20 min at 4°C, then mixed with 5 μl of AMP-agarose or pAp-agarose resin and incubated for 1 h at 4°C in Durapore filter units (Millipore). Unbound material was removed by centrifugation at 3000 g for 1 min, after which the beads were washed by adding successively 50 μl and 25 μl of binding buffer. These different fractions were pooled into the ‘unbound fraction’. Elution of bound proteins was performed by adding a 50 μl aliquot of hot (70°C) SDS sample buffer (NuPage, Invitrogen). After 10 min of incubation at 70°C, the eluate was collected by centrifugation at 3000 g for 1.5 min. The elution procedure was repeated and the two eluates were combined and reapplied to the beads. After 5 min incubation at 95°C, the eluate was collected as described above. A final rinse of the beads was performed by adding 25 μl of SDS sample buffer at 95°C and centrifuging for 3 min at 5000 g. All eluates were pooled into the ‘bound fraction’. Aliquots (10 μl) of unbound or bound fraction were analysed by SDS/PAGE followed by detection of PARP-1 by Western blot analysis using monoclonal anti-PARP-1 antibodies (clone C2-10; BD Pharmingen).
In vitro assays of PARP-1 activity
The activity assay was adapted from a previously published protocol used to measure PARP-1 automodification [22]. Samples (50 μl) containing 50 mM Tris/HCl (pH 7.8), 4 mM MgCl2, 200 μM DTT (dithiothreitol), 0.01% Tween 20 and 100 ng of purified recombinant human PARP-1 (Alexis Biochemicals) were pre-incubated for 10 min at room temperature (20°C) in the presence or absence of pAp. Poly(ADP-ribosyl)ation was initiated by adding 50 μl of a solution containing 300 ng of DNase-I treated salmon sperm DNA (Sigma), 800 μM NAD+ (Sigma) (400 μM final) and 3.7×103 Bq of 32P-radiolabelled NAD+ (PerkinElmer). Reactions were performed at room temperature and terminated by the addition of ice-cold 3-aminobenzamide (Sigma) to a final concentration of 7 mM. Reaction mixtures were immediately filtered on a nitrocellulose membrane using a slot-blot device. [32P]NAD+ was detected using a storage phosphor screen and quantified on a Typhoon® reader using ImageQuant® software.
PARP-1 heteromodification activity on histones was measured using the HT Universal Colorimetric PARP Assay kit with Histone-Coated Strip wells from Trevigen® according to the manufacturer's instructions, except that reactions were stopped after 45 min instead of 1 h.
In vivo assay of PARP-1 activity
HeLa cells were grown on coverslips in RPMI 1640 medium containing 10% FBS, supplemented with lithium chloride from 0 to 5 mM. Oxidative stress was applied to the cells by washing with PBS, and exposing cells to PBS containing 800 μM H2O2. After 6 min of stress, cells were immediately fixed by putting coverslips in methanol/acetone at −20°C. Following this, immunodetection of poly(ADP-ribose) was performed according to the method of Amé et al. [22]. Acquisition of images was realized with an epifluorescence microscope and quantification of the fluorescence signal of at least 100 randomly chosen nuclei was performed with ImageJ software (NIH). The amount of PARP-1 and its integrity in cells treated or not with lithium were monitored by Western blot analysis using a polyclonal antibody from Pharmigen at a dilution of 1:600.
RESULTS
PARP-1 binds to pAp-agarose
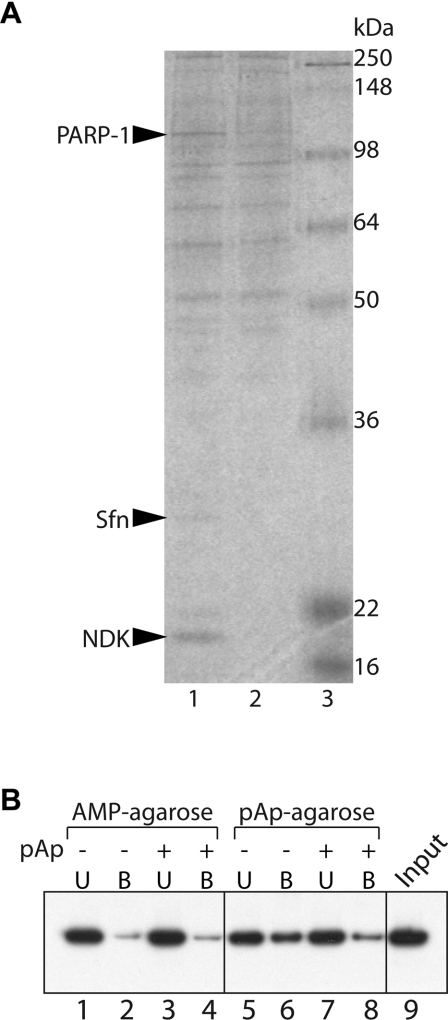
To identify potential targets of pAp in human cells, we performed pAp-affinity chromatography using nuclear extracts from HeLa cells. This approach has been successfully employed to identify the pAp-binding protein Sfn/Orn (small fragment nuclease/oligoribonuclease) from cytoplasmic extracts of HeLa cells [12]. pAp-affinity chromatography was performed in the presence of CaCl2, an efficient inhibitor of pAp-phosphatase activity. As shown in Figure 1(A), three proteins were specifically enriched in the pAp-agarose-bound fraction as compared with protein bound to control beads. Analysis of these proteins by in-gel trypsin digestion and LC-MS/MS (tandem MS) analysis revealed that the two lower bands of 20 and 30 kDa corresponded to NDK and Sfn/Orn respectively. These two proteins had previously been identified in cytoplasmic extracts [12] and are known to be inhibited by pAp [12,16]. The upper band of 110 kDa was identified as PARP-1. The identification was obtained with three peptides covering 3.8% of the total mass: VVDRDSEEAEIIRK, HPDVEVDGFSELR and TLGDFAAEYAK. The overall score of the identification, using ThermoFinnigan Bioworks software, was 50.
Figure 1. Physical interactions between pAp and PARP.
(A) SDS/PAGE of pAp-affinity chromatography from HeLa cell nuclear extracts. The gel was stained with Colloidal Blue. Lane 1, pAp-binding fraction; lane 2, binding on control agarose beads; lane 3, molecular mass markers. (B) PARP-1-affinity chromatography with AMP-agarose or pAp-agarose. Partially purified PARP-1 (1 μg) was incubated with AMP-agarose (lanes 1–4) or pAp-agarose (lanes 5–8) in the presence (+) or absence (−) of 3 mM pAp. Detection of PARP-1 in the bound (B) or unbound (U) fractions was performed by Western blot analysis. Lane 9, PARP-1 input.
Specificity of the interaction between pAp and PARP-1
To confirm the interaction between PARP-1 and pAp, we performed pAp-affinity chromatography using a recombinant enzyme produced in E. coli. As a control for binding specificity, we also tested the ability of PARP-1 to bind to AMP-agarose beads. As shown in Figure 1(B), only limited amounts of PARP-1 bound to AMP-agarose (lane 2). This binding was not affected by the addition of free pAp (lane 4) and probably corresponded to the non-specific adsorption of PARP-1 to agarose beads. In contrast, significant amounts of PARP-1 bound to pAp-agarose beads (lane 6) and, more importantly, this binding was significantly prevented by the presence of 3 mM pAp (lane 8). These results suggested that PARP-1 could interact specifically with pAp-agarose.
Inhibition of PARP-1 by adenine nucleotides
The interaction between PARP-1 and pAp prompted us to explore whether pAp might affect the catalytic activity of PARP-1. PARP-1 is composed of three domains: a DNA-binding domain, an automodification domain and a catalytic domain [23] responsible for the poly(ADP-ribosyl)ation activity. The DNA-binding domain senses DNA breaks and subsequently activates PARP-1 activity [19]. Poly(ADP-ribosyl)ation is a post-translational modification of proteins (reviewed in [24]), which occurs by successive addition of ADP-ribose using NAD+ as a substrate. Numerous proteins have been shown to be poly(ADP-ribosyl)ated by PARP-1, including PARP-1 itself [25,26].
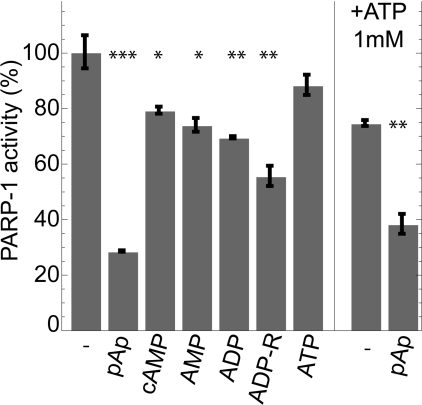
We first assessed the effect of pAp on PARP-1 automodification, namely its ability to auto-poly(ADP-ribosyl)ate in vitro. For this purpose, we measured the incorporation of [32P]NAD+ into poly(ADP-ribose) as described in the Materials and methods, using a substrate concentration of 400 μM in the presence or absence of pAp. In the absence of pAp, 1 μg of PARP-1 incorporated more than 14 pmol of NAD+ per s. This activity is in agreement with the previously reported PARP-1 activity following activation by cleaved DNA [27,28]. We found that in the presence of 50 μM pAp, poly(ADP-ribosyl)ation activity was reduced by more than 70%. To determine whether this inhibitory effect was specific for pAp, we tested PARP-1 activity in the presence of various adenine derivatives. As shown in Figure 2, pAp was by far the strongest inhibitor of PARP-1. With the exception of ADP-ribose, other adenine derivatives only inhibited PARP-1 to a limited extent. These results are consistent with a previous study by Niedergang et al. [29], who characterized the effect of various nucleotides on PARP-1 activity. These authors found that, among the nucleotides tested, 50 μM ADP-ribose inhibited approximately 40% of the PARP-1 activity, in good agreement with the results of the present study (Figure 2). The results of the present study indicate that pAp was the strongest inhibitor among all adenine derivatives tested thus far.
Figure 2. Inhibition of PARP-1 activity by various adenine derivatives.
PARP-1 activity was measured as described in the Materials and methods section, in the presence of 400 μM [32P]NAD+ with an incubation time of 7 min. Inhibition of PARP-1 activity was measured in the presence of 50 μM of the indicated adenine nucleotides. The two assays shown on the right-hand side were performed in the presence of 1 mM ATP. Values are means±S.D. for duplicate experiments, and are expressed as the percentage of activity without adenine nucleotides. 100% corresponds to 14 pmol of incorporated ADP-ribose per s. *P<0.05; **P<0.01; ***P<0.001, as compared with the control (with or without 1 mM ATP respectively) calculated using Student's t test.
We also tested the effect of pAp on PARP-1 activity in the presence of ATP as several studies have previously shown that millimolar concentrations of ATP inhibit PARP-1 activity [28,29]. As shown in Figure 2, PARP-1 activity was reduced by ~25% in the presence of 1 mM ATP. However, under these conditions, pAp, at a concentration of 50 μM, still exhibited a very significant inhibitory effect on PARP-1 activity.
pAp-mediated PARP-1 inhibition
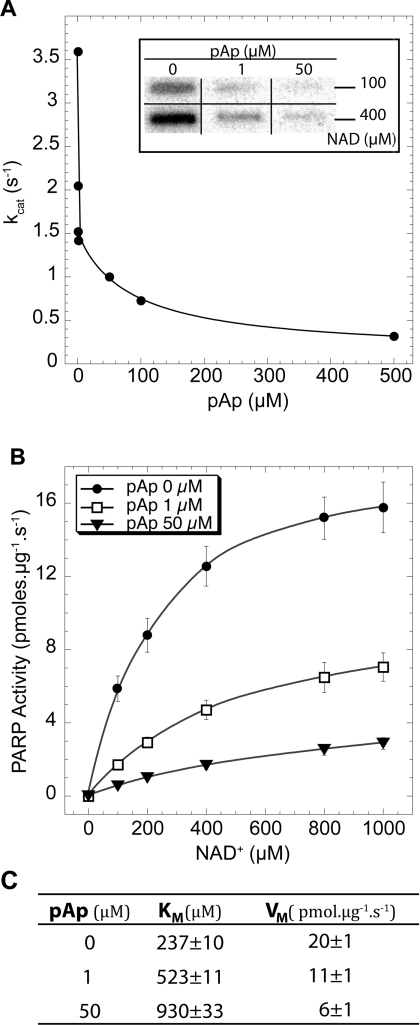
Figure 3(A) shows PARP-1 activity, measured in the presence of a fixed substrate concentration of 400 μM [32P]NAD+, as a function of the pAp concentration. As can be seen, pAp inhibited the reaction rate of PARP-1 in a dose-dependent manner, with an apparent inhibitory constant, Ki, in the micromolar range (0.5–1 μM).
Figure 3. Kinetic parameters of pAp inhibition of PARP-1 activity.
(A) The kcat, defined as mol of NAD+ incorporated/mol of PARP-1 per s, is expressed as a function of the pAp concentration. PARP-1 activity was measured as described in Figure 2. Inset: PhosphorImager autoradiography of poly(ADP-ribosyl)ated PARP-1 blotted on to a nitrocellulose membrane. (B) Activity of PARP-1, expressed as pmol of NAD+ incorporated/μg of PARP-1 per s, as a function of the NAD+ concentration, and in the presence of the pAp concentrations indicated. For each data point ADP-ribose incorporation was measured at 2, 5, 7 and 10 min. The activity was then determined from the slope of the ADP-ribose incorporated as a function of time. (C) Kinetic parameters of PARP-1 deduced from the results shown in (B).
We further tested the effect of pAp on the PARP-1 kinetic parameters. The initial rate of ADP-ribose incorporation by PARP-1 was measured in the presence of increasing NAD+ concentrations at various concentrations of pAp. As shown in Figure 3(B), the initial rate of poly(ADP-ribosyl)ation could be fitted using classical Michaelis–Menten kinetics. Figure 3(C) shows the kinetic parameters calculated from the typical experiment presented in Figure 2(B). Both Km and Vmax of PARP-1 were affected by the presence of pAp. From a series of three independent experiments, we found that the Km of PARP-1 for NAD+ varied from 200 to 250 μM in the absence of pAp, from 500 to 550 μM in the presence of 1 μM pAp, and from 900 to 950 μM in the presence of 50 μM pAp. The maximal velocity Vmax of PARP-1 was also reduced by a factor of 3–4 when pAp was present at 50 μM as compared with control conditions. These results suggest a mixed inhibition of PARP-1 by pAp.
pAp inhibitory effect on histone modification
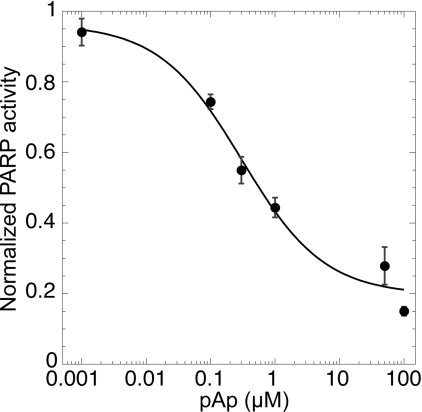
In addition to its automodification, PARP-1 also modifies various proteins, and in particular histone H1 [30], in order to recruit the DNA repair machinery. To test whether pAp could also inhibit this heteromodification, we measured the poly(ADP-ribosyl)ation of histone H1 in the presence of various concentrations of pAp. Figure 4 shows a dose-dependent inhibition of the PARP-1-catalysed histone modification by pAp. This inhibition was comparable with the pAp-mediated inhibition of PARP-1 automodification. We therefore propose that pAp is also likely to inhibit poly(ADP-ribosyl)ation of other PARP-1 targets.
Figure 4. pAp inhibition of histone poly(ADP-ribosyl)ation.
PARP-1 catalysed poly(ADP-ribosyl)ation of histone H1 was measured as described in the Materials and methods section in the presence of the indicated concentrations of pAp. The PARP-1 activity (arbitrary unit) is expressed as a percentage of the activity in the absence of pAp.
Effect of lithium treatment of cells on poly(ADP-ribosyl)ation activity
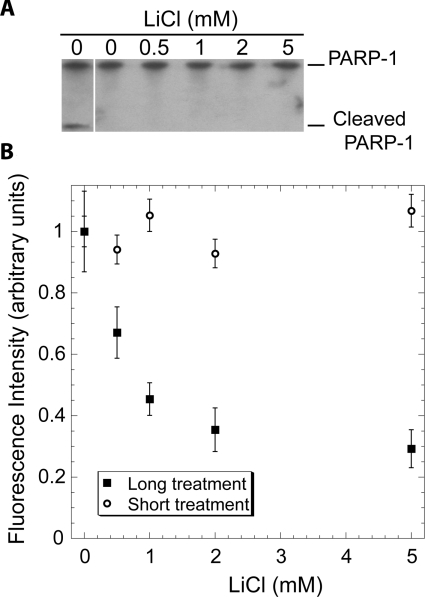
In order to evaluate whether the activity of PARP-1 might be regulated in vivo, as a response to changes in pAp levels, we quantified by immunofluorescence the poly(ADP-ribosyl)ation in response to oxidative stress in HeLa cells treated or not with lithium. As lithium is a very potent inhibitor of pAp-phosphatase, treatment with lithium should result in an increased concentration of pAp inside the cells. We verified by Western blot analysis that lithium treatment was not affecting the PARP-1 level in the cell (Figure 5A). This Western blot also shows that there was no detectable PARP-1 cleavage in cells treated with lithium, the caspase-induced cleavage of PARP-1 being known to inactivate the enzyme [31]. After lithium treatment, cells were exposed to oxidative stress using H2O2, and the resulting poly(ADP-ribosyl)ation was quantified by immunofluorescence. As shown in Figure 5(B), lithium treatment of HeLa cells resulted in a dose-dependent decrease of the poly(ADP-ribose) fluorescent signal intensity. This indicates that the HeLa cells treated with lithium exhibited a strongly reduced poly(ADP-ribosyl)ation activity in response to oxidative stress as compared with untreated cells.
Figure 5. Inhibition of poly(ADP-ribosyl)ation by lithium treatment.
(A) PARP-1 was detected by Western blot analysis in protein extracts from HeLa cells treated or not with lithium. The left-hand lane presents an extract from cells entering apoptosis as a positive control for PARP-1 cleavage. (B) HeLa cells were treated with the lithium concentrations indicated for 4 days (long) or 30 min (short) and were exposed to 800 μM H2O2 for 6 min. Poly(ADP-ribosyl)ation was then measured by immunofluorescence detection of poly(ADP-ribose) polymers as described in the Materials and methods section.
As PARP-1 is responsible for more than 90% of the poly(ADP-ribosyl)ation activity inside the cells [32,33], this decrease is probably due to inhibition of PARP-1 activity. Importantly, the inhibition of PARP-1 activity was only seen after prolonged lithium treatment, as short-time (30 min) treatment of the cells (Figure 5B) had no effect on the poly(ADP-ribosyl)ation levels in response to oxidative stress. This indicates that lithium had no direct inhibitory effect on the PARP activity or on its activation by the oxidative stress signals in vivo. We also verified that, in vitro, lithium at concentrations up to 5 mM had no effect on the PARP-1-catalysed poly(ADP-ribosyl)ation of histone H1 (Supplementary Figure S1 at http://www.BiochemJ.org/bj/443/bj4430485add.htm). We conclude that the strong inhibition of PARP-1 activity in cells treated by lithium is indirect. We propose that, in cells exposed to lithium, the inhibition of pAp-phosphatase could result in progressive accumulation of pAp that, in turn, could bind to PARP-1 and directly inhibit its enzymatic activity.
DISCUSSION
Searching for potential targets of pAp in human cells, we found that PARP-1 from cell extracts bound specifically to pAp-agarose. Using purified PARP-1, we have demonstrated that this interaction was specific and resulted in a strong inhibition of PARP-1 activity. The half-maximum inhibition of enzymatic activity occurred at pAp concentrations below 1 μM. As adenine derivatives are known to slightly inhibit PARP-1 activity, we compared the effect of pAp with that of several other adenine nucleotides. We found that pAp was a more potent inhibitor than ADP-ribose, which was previously considered as the strongest inhibitor among adenine derivatives. We have also shown that both PARP-1 automodification and the heteromodification of histones were similarly affected by pAp. Moreover, characterization of PARP-1 kinetic parameters showed that pAp affected both the Km and Vmax, thus indicating a mixed inhibition.
In principle, pAp could target any of the three domains of PARP-1: the DNA-binding domain, the automodification domain or the catalytic domain [23]. As pAp inhibits both poly(ADP-ribosyl)ation of histones and PARP-1 automodification, it is unlikely that pAp would act on the automodification domain. On the basis of the homology of pAp with the NAD+ substrate, it is probable that pAp binds to the catalytic site of PARP-1. However, we cannot at present exclude that pAp could interfere with the activation of PARP-1 by damaged DNA.
As PARP-1 plays a key role in genome maintenance [24], the inhibition of PARP-1 by pAp may have important physiological consequences. PARP-1 activation by DNA single-strand breaks triggers: (i) in the case of limited DNA damage, the recruitment of repair factors to the DNA damage site [34], and (ii) in the case of massive DNA damage, the extensive incorporation of NAD+ into poly(ADP-ribose) polymers, which can ultimately lead to NAD+ depletion and cell death [35]. Given the major role of PARP-1 in regulating the balance between DNA repair and cell death, numerous inhibitors have been designed to be used as anticancer drugs [36,37] or to down-regulate inflammatory responses [38,39]. PARP-1 is also regulated by endogenous effectors, such as the previously described KIF4 (kinesin family member 4), which controls the activity-dependent survival of postmitotic neurons by regulating PARP-1 activity [40]. The results of the present study indicate that pAp could be another physiological modulator of PARP-1 activity.
In human cells, pAp is generated during sulfur and lipid metabolism and is recycled by the pAp-phosphatases. As these latter enzymes are potently inhibited by lithium, it is assumed that lithium treatment of cells could affect the intracellular levels of pAp, and, as a consequence, may ultimately alter the PARP-1 activity. Indeed, we show in the present study, for the first time, that treatment of HeLa cells by lithium inhibits PARP-1 activity in response to oxidative stress. At 1 mM lithium, which is the concentration of lithium found in the plasma of patients with bipolar disorder treated with lithium [41], PARP-1 activity was reduced by 50% as compared with untreated cells. We further showed that the PARP-1 inhibition by lithium treatment is indirect, as lithium had no effect in vitro on PARP-1 activity and, in vivo, the lithium-induced inhibition of PARP-1 activity was seen only under prolonged exposure of the cells to this salt. We propose that the inhibition of PARP-1 activity by lithium might indirectly result from the accumulation of intracellular pAp following the direct lithium inhibition of the pAp-phosphatases.
In human cells, pAp is a by-product of sulfur and lipid metabolism. The results of the present study suggest that, under conditions where pAp may accumulate inside the cells, this nucleotide could modulate the activity of PARP-1. However, to date, only very limited information on the in vivo concentration of pAp is available. The standard technique to measure the pAp concentration relies on HPLC separation of nucleotide, but cannot detect concentrations of pAp below 50 μM [42]. To our knowledge, variations in pAp concentrations have only been measured in yeast under conditions of elevated sulfur metabolic flux and when the cells were either depleted of pAp-phosphatase or treated with lithium, a known inhibitor of pAp-phosphatase [11,18]. In these situations, the pAp concentrations increased from undetectable levels to more than 1 mM. Consequently, these studies are difficult to extrapolate to human cells. A more recent study attempted to detect changes in pAp concentrations in brains of rats treated with lithium. The authors of this study could not detect any pAp using the HPLC-based technique described in the study [43].
The most precise measurement of pAp basal levels in various cell types was performed by Anderson et al. [44] using a bioluminescent enzymatic measurement of pAp on the basis of a sulfotransferase from Renilla reniformis. These authors found approximately 17 nmol of pAp per g of liver tissue from rabbits, an amount that would correspond to an intracellular pAp concentration of approximately 0.2–2 μM. The results of the present study suggest that PARP-1 can be inhibited by pAp at these concentrations. Given our Ki value in the micromolar range, potential variations of pAp concentration in the micromolar range could significantly affect the activity of PARP-1.
In conclusion, we have found that pAp is a strong inhibitor of PARP-1. We have also shown that treatment of cells with lithium results in a significant inhibition of PARP-1 activity in response to oxidative stress. We propose that an increase in intracellular pAp concentrations, occuring either when sulfur or lipid metabolism is very active or during lithium treatment (inhibiting pAp-phosphatase), could modulate PARP-1 activity and thus change the cellular response to DNA damage. Our results provide new insight into the understanding of the molecular effect of lithium in the treatment of bipolar disorder.
Online data
AUTHOR CONTRIBUTION
The experimental results shown in Figure 1 were obtained by Undine Mechold. MS analyses were performed by Vasily Ogryzko. Data shown in Figures 2, 3, 4 and 5 and Supplementary Figure S1 were obtained by Elie Toledano. Elie Toledano wrote the first draft of the paper. Undine Mechold and Daniel Ladant edited the paper prior to submission. Antoine Danchin was involved in initial supervision of the project. Undine Mechold and Daniel Ladant supervised the project.
FUNDING
This work was supported by the Centre National de la Recherche Scientifique and Institut Pasteur. E.T. was supported by the Université Pierre et Marie Curie. V.O. was supported by Association pour la Recherche sur la Cancer [grant numbers 5950 and 7659].
ACKNOWLEDGEMENTS
We thank Dr J.C. Amé for stimulating discussions and helpful advice. We also thank Agnès Ullmann and Scot Ouellette for critical reading of the paper prior to submission.
References
- 1.Gregory J. D., Lipmann F. The transfer of sulfate among phenolic compounds with 3′,5′-diphosphoadenosine as coenzyme. J. Biol. Chem. 1957;229:1081–1090. [PubMed] [Google Scholar]
- 2.Robbins P. W., Lipmann F. Identification of enzymatically active sulfate as adenosine-3′-phosphate-5′-phosphosulfate. J. Am. Chem. Soc. 1956;78:2652–2653. [Google Scholar]
- 3.Klaassen C. D., Boles J. W. Sulfation and sulfotransferases 5: the importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997;11:404–418. doi: 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- 4.Reuter K., Mofid M. R., Marahiel M. A., Ficner R. Crystal structure of the surfactin synthetase-activating enzyme Sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 1999;18:6823–6831. doi: 10.1093/emboj/18.23.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuwald A. F., Krishnan B. R., Brikun I., Kulakauskas S., Suziedelis K., Tomcsanyi T., Leyh T. S., Berg D. E. CysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murguia J. R., Belles J. M., Serrano R. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 7.Gil-Mascarell R., López-Coronado J. M., Bellés J. M., Serrano R., Rodríguez P. L. The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 1999;17:373–383. doi: 10.1046/j.1365-313x.1999.00385.x. [DOI] [PubMed] [Google Scholar]
- 8.López-Coronado J. M., Belles J. M., Lesage F., Serrano R., Rodriguez P. L. A novel mammalian lithium-sensitive enzyme with a dual enzymatic activity, 3′-phosphoadenosine 5′-phosphate phosphatase and inositol-polyphosphate 1-phosphatase. J. Biol. Chem. 1999;274:16034–16039. doi: 10.1074/jbc.274.23.16034. [DOI] [PubMed] [Google Scholar]
- 9.York J. D., Ponder J. W., Majerus P. W. Definition of a metal-dependent/Li+-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5149–5153. doi: 10.1073/pnas.92.11.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yenush L., Bellés J. M., López-Coronado J. M., Gil-Mascarell R., Serrano R., Rodríguez P. L. A novel target of lithium therapy. FEBS Lett. 2000;467:321–325. doi: 10.1016/s0014-5793(00)01183-2. [DOI] [PubMed] [Google Scholar]
- 11.Spiegelberg B. D., dela Cruz J., Law T.-H., York J. D. Alteration of lithium pharmacology through manipulation of phosphoadenosine phosphate metabolism. J. Biol. Chem. 2005;280:5400–5405. doi: 10.1074/jbc.M407890200. [DOI] [PubMed] [Google Scholar]
- 12.Mechold U., Ogryzko V., Ngo S., Danchin A. Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 2006;34:2364–2373. doi: 10.1093/nar/gkl247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cade J. F. Lithium salts in the treatment of psychotic excitement. Med. J. Aust. 1949;2:349–352. doi: 10.1080/j.1440-1614.1999.06241.x. [DOI] [PubMed] [Google Scholar]
- 14.Fountoulakis K. N. The contemporary face of bipolar illness: complex diagnostic and therapeutic challenges. CNS Spectrum. 2008;13:763–779. doi: 10.1017/s1092852900013894. [DOI] [PubMed] [Google Scholar]
- 15.Klein P. S., Melton D. A. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider B., Xu Y. W., Janin J., Véron M., Deville-Bonne D. 3′-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity. J. Biol. Chem. 1998;273:28773–28778. doi: 10.1074/jbc.273.44.28773. [DOI] [PubMed] [Google Scholar]
- 17.Dichtl B., Stevens A., Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todeschini A.-L., Condon C., Bénard L. Sodium-induced GCN4 expression controls the accumulation of the 5′ to 3′ RNA degradation inhibitor, 3′-phosphoadenosine 5′-phosphate. J. Biol. Chem. 2006;281:3276–3282. doi: 10.1074/jbc.M511688200. [DOI] [PubMed] [Google Scholar]
- 19.Ménissier-de Murcia J., Molinete M., Gradwohl G., Simonin F., de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J. Mol. Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- 20.Dantzer F., Amé J., Schreiber V., Nakamura J., Ménissier-de Murcia J., de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amé J.-C., Hakmé A., Quenet D., Fouquerel E., Dantzer F., Schreiber V. Detection of the nuclear poly(ADP-ribose)-metabolizing enzymes and activities in response to DNA damage. In: Walker J. M., editor. The Nucleus. Humana Press; 2009. pp. 267–283. [DOI] [PubMed] [Google Scholar]
- 23.Murcia G., Schreiber V., Molinete M., Saulier B., Poch O., Masson M., Niedergang C., Murcia J. M. Structure and function of poly(ADP-ribose) polymerase. Mol. Cell. Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- 24.D'Amours D., Desnoyers S., Da Silva I., Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 25.Desmarais Y., Ménard L., Lagueux J., Poirier G. G. Enzymological properties of poly(ADP-ribose)polymerase: characterization of automodification sites and NADase activity. Biochim. Biophys. Acta. 1991;1078:179–186. doi: 10.1016/0167-4838(91)99007-f. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza-Alvarez H., Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J. Biol. Chem. 1993;268:22575–22580. [PubMed] [Google Scholar]
- 27.Kristensen T., Holtlund J. Purification of poly(ADP-ribose) polymerase from Ehrlich ascites tumor cells by chromatography on DNA-agarose. Eur. J. Biochem. 1976;70:441–446. doi: 10.1111/j.1432-1033.1976.tb11035.x. [DOI] [PubMed] [Google Scholar]
- 28.Kun E., Kirsten E., Mendeleyev J., Ordahl C. P. Regulation of the enzymatic catalysis of poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, Histones H1 and H3, and ATP. Biochemistry. 2004;43:210–216. doi: 10.1021/bi0301791. [DOI] [PubMed] [Google Scholar]
- 29.Niedergang C., Okazaki H., Mandel P. Properties of purified calf thymus poly(adenosine diphosphate ribose) polymerase. Eur. J. Biochem. 1979;102:43–57. doi: 10.1111/j.1432-1033.1979.tb06261.x. [DOI] [PubMed] [Google Scholar]
- 30.Althaus F. R. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J. Cell Sci. 1992;102:663–670. doi: 10.1242/jcs.102.4.663. [DOI] [PubMed] [Google Scholar]
- 31.Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., Poirier G. G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 32.Althaus F. R., Richter C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol. Biol. Biochem. Biophys. 1987;37:1–237. [PubMed] [Google Scholar]
- 33.Shieh W. M., Amé J.-C., Wilson M. V., Wang Z.-Q., Koh D. W., Jacobson M. K., Jacobson E. L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 34.Dantzer F., Schreiber V., Niedergang C., Trucco C., Flatter E., Rubia G. D. L., Oliver J., Rolli V., Ménissier-de Murcia J., de Murcia G. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin P. M., Lewis P. J., Davies M. I., Skidmore C. J., Shall S. The effect of gamma radiation and neocarzinostatin of NAD and ATP levels in mouse leukaemia cells. Biochim. Biophys. Acta. 1978;543:576–582. doi: 10.1016/0304-4165(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 36.Delaney C. A., Wang L.-Z., Kyle S., White A. W., Calvert A. H., Curtin N. J., Durkacz B. W., Hostomsky Z., Newell D. R. Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin. Cancer Res. 2000;6:2860–2867. [PubMed] [Google Scholar]
- 37.Muñoz-Gámez J. A., Martín-Oliva D., Aguilar-Quesada R., Cañuelo A., Nuñez M. I., Valenzuela M. T., Ruiz de Almodóvar J. M., De Murcia G., Oliver F. J. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem. J. 2005;386:119–125. doi: 10.1042/BJ20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kauppinen T. M., Swanson R. A. The role of poly(ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;145:1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 39.Jagtap P., Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discovery. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 40.Midorikawa R., Takei Y., Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 41.DSM-IV. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 42.Murguia J. R., Belles J. M., Serrano R. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 1996;271:29029–29033. doi: 10.1074/jbc.271.46.29029. [DOI] [PubMed] [Google Scholar]
- 43.Shaltiel G., Deutsch J., Rapoport S., Basselin M., Belmaker R., Agam G. Is phosphoadenosine phosphate phosphatase a target of lithium's therapeutic effect? J. Neural Transm. 2009;116:1543–1549. doi: 10.1007/s00702-009-0298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson J. M., Hori K., Cormier M. J., Marlene A. D. Methods Enzymol. Vol. 57. Academic Press; 1978. A bioluminescence assay for PAP (3′, 5′-diphosphoadenosine) and PAPS (3′-phosphoadenylyl sulfate) pp. 244–257. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.