Abstract
CBSs (cystathionine β-synthases) are eukaryotic PLP (pyridoxal 5 *-phosphate)-dependent proteins that maintain cellular homocysteine homoeostasis and produce cystathionine and hydrogen sulfide. In the present study, we describe a novel structural arrangement of the CBS enzyme encoded by the cbs-1 gene of the nematode Caenorhabditis elegans. The CBS-1 protein contains a unique tandem repeat of two evolutionarily conserved catalytic regions in a single polypeptide chain. These repeats include a catalytically active C-terminal module containing a PLP-binding site and a less conserved N-terminal module that is unable to bind the PLP cofactor and cannot catalyse CBS reactions, as demonstrated by analysis of truncated variants and active-site mutant proteins. In contrast with other metazoan enzymes, CBS-1 lacks the haem and regulatory Bateman domain essential for activation by AdoMet (S-adenosylmethionine) and only forms monomers. We determined the tissue and subcellular distribution of CBS-1 and showed that cbs-1 knockdown by RNA interference leads to delayed development and to an approximately 10-fold elevation of homocysteine concentrations in nematode extracts. The present study provides the first insight into the metabolism of sulfur amino acids and hydrogen sulfide in C. elegans and shows that nematode CBSs possess a structural feature that is unique among CBS proteins.
Keywords: cystathionine β-synthase (CBS), Caenorhabditis elegans, domain architecture, homocysteine, hydrogen sulfide, knockdown
Abbreviations: AdoMet, S-adenosylmethionine; BN, blue native; BS3, bis(sulfosuccinimidyl) suberate; CBS, cystathionine β-synthase; CGL, cystathionine γ-lyase; DTT, dithiothreitol; EST, expressed sequence tag; GFP, green fluorescent protein; LC–MS/MS, liquid chromatography–tandem MS; PLP, pyridoxal 5*-phosphate; RNAi, RNA interference; RT, reverse transcription; SEC, size-exclusion chromatography; UTR, untranslated region; WT, wild-type
INTRODUCTION
Methionine and cysteine are sulfur amino acids that play important roles in many biochemical reactions. Methionine, an essential amino acid, can be irreversibly converted into cysteine in a series of reactions. Methionine is first converted into AdoMet (S-adenosylmethionine), which serves as a methyl donor in various transmethylation reactions. A product of these transmethylations, S-adenosylhomocysteine, is further converted into homocysteine, which is a key intermediate in the metabolism of sulfur amino acids. In animal tissues, homocysteine is universally remethylated to methionine by methionine synthase using methyltetrahydrofolate as the methyl donor. In addition, a number of tissues can convert homocysteine into cystathionine and further to cysteine via the transsulfuration pathway through two PLP (pyridoxal 5*-phosphate)-dependent enzymes, CBS (cystathionine β-synthase) and CGL (cystathionine γ-lyase) [1].
CBS is a cytosolic enzyme that catalyses the formation of cystathionine with the release of water or hydrogen sulfide, depending on whether homocysteine is condensed with serine or cysteine. The human and rodent CBSs that have been characterized are tetrameric enzymes and each of their ~63 kDa polypeptide chains contains three different domains. The N-terminal domain binds haem, the presence of which has been suggested to increase CBS activity during the oxidation of the intracellular environment [1a,2]. Other studies suggest that haem may play a structural role that is necessary for the correct folding of the CBS protein [3,4]. The middle portion of the polypeptide chain forms the catalytic domain and is well conserved among the fold-type II PLP-dependent proteins [5]. The C-terminal domain possesses two defined CBS domains, a hydrophobic domain, CBS1, and a less conserved domain, CBS2, that are together referred to as the Bateman domain. Together, these two CBS domains bind AdoMet, an allosteric activator of mammalian CBS [6]. The C-terminal domain of mammalian CBS is also thought to be responsible for multimerization of the enzyme into homotetramers and higher oligomeric forms [6,7]. The C-terminal autoinhibitory domain of mammalian CBS can be removed by in vitro or in vivo proteolytic processing, yielding a ~45 kDa truncated form (45CBS) that forms dimers and is more active than the full-length enzyme [6,8,9].
The canonical domain architecture of mammalian CBSs is not conserved across phyla. The CBS enzymes of Saccharomyces cerevisiae, Trypanosoma cruzi and Drosophila melanogaster have been experimentally characterized; the N-terminal haem-binding domain is absent in yeast and protozoan CBS, in contrast with its presence in Drosophila [10–12], whereas the catalytic domain is conserved in the CBS enzymes of all three of these species. The C-terminal portion exhibits the highest degree of variability. The yeast and Drosophila CBS proteins contain the Bateman domain, but lack a response to AdoMet. Interestingly, although the C-terminal portion of the yeast CBS inhibits the activity of the enzyme and supports the formation of tetramers and octamers [13], Drosophila CBS forms only dimers [12]. In contrast, the protozoan CBS does not contain the Bateman domain and is not activated by AdoMet. Although its C-terminus is shortened, the protozoan CBS is still able to form tetramers [11]. The phylogenetic variability in the domain architecture of CBSs suggests that the activity of these enzymes is regulated differently in evolutionarily distant organisms.
In the present study, we characterized the structural and functional properties of the CBS in Caenorhabditis elegans, a well-established model organism used to study human diseases. We first identified a transcriptionally active gene encoding CBS in C. elegans, we then determined its pattern of expression and characterized the enzymatic and structural properties of the encoded protein. Finally, we determined the phenotypic effects of cbs-1 inhibition using RNA-mediated interference. These data describe novel structural features that are unique among CBS enzymes and provide the first insight into the metabolism of sulfur amino acids and hydrogen sulfide in C. elegans.
EXPERIMENTAL
C. elegans strains
The WT (wild-type) C. elegans Bristol strain N2 was obtained from the C. elegans Stock Center (University of Minnesota, Minneapolis, MN, U.S.A.), and the RB839 strain carrying the F54A3.4 (ok666) allele was provided by the C. elegans Gene Knockout Consortium (Oklahoma Medical Research Foundation, Oklahoma City, OK, U.S.A.). Worm cultures were maintained as described previously [14].
Bioinformatics
BLASTp searches were performed by online BLAST software using the C. elegans protein database (release WS215). Protein domain modelling was performed by Swiss-model (automatic modelling mode) using the crystal structure of human 45CBS (PDB code 1JBQ, chain A) as a template [15]. PDB structures were subsequently evaluated in the Prosa program [16] and visualized in Swiss-PDBViewer 4.0.4 [17]. Phylogenetic trees were constructed in the online portal system Mobyle [18]. Multiple alignments of amino acid sequences were performed using ClustalW2 online software with default parameters [19]. Conserved regions were also separated for further analysis by ClustalW2. For phylogenetic analysis, alignment was bootstrapped 100 times and analysed by the maximal likelihood method using the PHYML 3.0 program [20]. Bootstrap output trees were analysed by the PHYLIP 3.67 CONSENSE program; the final tree shape was visualized in the Dendroscope program [21].
PCR amplification and DNA sequencing
Nematode cDNA was prepared by RT (reverse transcription) using isolated total RNA from mixed stages of N2 worms and a RT kit with an oligo(dT) primer (Promega). Open reading frames of ZC373.1 and F54A3.4 were amplified by PCR using either cDNA prepared by RT–PCR or a C. elegans cDNA library (Invitrogen) as the template (a list of the primers is given in Supplementary Table S1 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). PCR products were cloned into the pCR4-TOPO vector (Invitrogen), and the authenticity of the DNA sequence was verified by dideoxy sequencing using an ABI PRISM 3100-Avant sequencer (Applied Biosystems).
GFP (green fluorescent protein) reporter assay
To determine the expression pattern of cbs-1, we generated a translational fusion vector using the PCR fusion technique described previously [22]. The 1.8 kb of 5′ upstream sequence and the entire coding region of ZC373.1 were amplified by PCR using primers A and B (Supplementary Table S1), and genomic C. elegans DNA as a template. The vector pPD95.75 was used as a template for amplification of the GFP-coding sequence using primers C and D (Supplementary Table S1). The two PCR products were mixed and used as a template for PCR fusion using nested primers E and F (Supplementary Table S1). The 6.8-kb PCR product was injected into C. elegans hermaphrodite gonads together with the plasmid pRF4 as a phenotypic marker for injection. Transgenic animals were separated, and the F2 progeny were screened for the GFP signal. An Olympus BX60 microscope and a Nikon Eclipse E800 with C1 confocal module and 488 nm laser and differential interference contrast optics were used for specimen examination.
Bacterial expression and protein purification
Initially, recombinant CBS-1 was expressed as a fusion protein with an N-terminal GST tag and further purified by affinity chromatography to 75% purity (see Supplementary Figure S4, lane 6) according to a previously described procedure for human CBS [23]. The contaminating polypeptide with the highest abundance, a 40-kDa fragment that represented approximately 20% of the total protein, was identified as the N-terminal portion of CBS-1 (residues 1–375) by peptide mass fingerprinting using MS detection (results not shown). This N-terminal fragment was observed with similar abundance even when the purification procedure was modified to limit proteolytic cleavage of the recombinant protein (the modification involved performing affinity chromatography at 4°C and increasing the concentration of protease inhibitors in the bacterial crude extract). To overcome this obstacle that was not previously reported for other CBS orthologues, we constructed a new vector that produced double-tagged CBS-1 with a cleavable N-terminal GST tag and a C-terminal His tag. The open reading frame of the ZC373.1 gene encoding CBS-1 was amplified by PCR using a C. elegans cDNA library as the template. PCR was performed with Taq polymerase using primers P and R (Supplementary Table S1). The 2.1-kb DNA fragment obtained by digestion of the PCR product with BamHI and XhoI was cloned into the BamHI- and XhoI-digested pGEX-6p-1 vector. Express Competent Escherichia coli cells (New England Biolabs) were transformed with the plasmid that encodes double-tagged CBS-1 (GST–CBS–1–His6) and cultured in the presence of 100 μM IPTG (isopropyl β-D-thiogalactopyranoside) at 18°C for 24 h. The GST–CBS-1 fusion protein was purified according to the purification protocol for human CBS described previously [24] with the following modifications: after cleavage by the PreScission protease (GE Healthcare), recombinant CBS-1 was loaded on to a Ni-Sepharose column that had been equilibrated with IMAC buffer [20 mM phosphate (pH 7.5), containing 0.5 M NaCl, 20 mM imidazole and 1 mM DTT (dithiothreitol)]. The column was washed with IMAC buffer containing 50 mM imidazole. CBS-1 was then eluted with IMAC buffer containing 75 mM imidazole. The protein enrichment procedure yielded approximately 1 mg of CBS-1 per litre of bacterial culture. The purity of isolated CBS-1 was analysed by SDS/PAGE [pre-cast 3–8% gradient gel (Invitrogen)] with Coomassie Brilliant Blue staining. The protein concentration was determined using Bradford reagent (Sigma–Aldrich) with BSA as the standard. The absorption spectrum of CBS-1 was recorded using a UV–visible spectrophotometer (Shimadzu UV-2550) at room temperature (25°C).
SEC (size-exclusion chromatography)
SEC was performed on an HPLC platform (Shimadzu LC-10A system). Recombinant purified CBS-1 was loaded on to a Bio-Sil SEC HPLC column (catalogue number 125-0060, Bio-Rad Laboratories) that had been previously equilibrated with buffer containing 50 mM Tris/HCl (pH 8.0), 1 mM DTT and 100 mM NaCl. The analysis was performed at a flow rate of 1.0 ml/min at 25°C; the elution profile was obtained by measurement of the absorbance at 280 nm. Calibration was performed using ferritin, aldolase, conalbumin (GE Healthcare), BSA (Thermo Fisher Scientific) and human 45CBS produced in E. coli and purified as described previously [24].
Native PAGE, BN (blue native)-PAGE and chemical cross-linking
Native electrophoresis was performed on 8% polyacrylamide gels using the Laemmli buffer system without SDS [25]. Per lane, 5 μg of CBS-1 and of the standards (BSA and human 45CBS) were loaded. BN electrophoresis was performed as described previously [26] with the High Molecular Weight Calibration kit for electrophoresis (GE Healthcare) and rabbit aldolase as the protein marker. Chemical cross-linking was performed using three different concentrations of BS3 [bis(sulfosuccinimidyl) suberate]; the molar ratios of CBS-1 (0.5 mg/ml) to the cross-linker were 1:10, 1:50 and 1:100. Cross-linked proteins were analysed using precast 3–8% gradient polyacrylamide gels. As a positive control for efficient cross-linking, we used dimeric human 45CBS reacted with BS3 at a protein/cross-linker molar ratio of 1:10. All of the proteins analysed by electrophoretic techniques were stained with EZ Blue Gel reagent (Sigma–Aldrich).
Pulse proteolysis
Pulse proteolysis of CBS-1 in a urea gradient was performed with thermolysin as described previously for human CBS [27].
Fluorescence-based thermal-shift assay
Protein samples (0.5 mg/ml) were dissolved in 20 mM Tris/HCl (pH 8.0), and 5×Sypro Orange dye (Bio-Rad Laboratories). Using the real-time PCR Detection System CFX96 Touch (Bio-Rad Laboratories), the proteins were incubated in a thermal gradient from 25°C to 70°C at increments of 0.5°C and with 1-min-hold intervals. The degree of protein unfolding was monitored by a FRET (fluorescence resonance energy transfer) channel that captured the spectral properties of Sypro Orange unfolded protein complexes (excitation wavelength≈470 nm and emission wavelength≈570 nm). The data were analysed by CFX Manager software, and the melting temperatures were determined using the first derivative spectra.
CD and fluorescence spectroscopy
The CD spectra of CBS-1 protein variants [0.5 mg/ml in 50 mM phosphate buffer (pH 7.5)] were recorded using a Jasco J-810 chiroptic spectrometer. The intrinsic fluorescence of CBS proteins in 50 mM Tris/HCl (pH 8.0), was measured in the same buffer using a PerkinElmer LS55 fluorescence spectrometer. The excitation wavelength for tryptophan was 298 nm (slit width of 5 nm) with an emission signal scanned from 300 to 700 nm (slit width of 5 nm).
Determination of substrate specificity
All enzyme assays were performed at 25°C with an incubation time of 10 min to ensure a linear increase in cystathionine or cysteine production. The reaction mixtures (50 μl) contained 1 μg/ml purified recombinant CBS-1, 10 mM tested substrates in the combinations shown in the Results section, 1 mM PLP, 1 mM DTT, 1 mg/ml BSA and 150 mM Tris/HCl (pH 7.0). The reactions were stopped by the addition of 25 μl of 1 M trichloracetic acid, and the reaction products were determined by HPLC [28] or LC–MS/MS (liquid chromatography–tandem MS) analysis [29] with the modifications described below.
Temperature and pH optima and kinetic analysis
We measured cystathionine production using LC–MS/MS analysis [29] with the following modifications: assays were performed in 100 mM Bis/Tris buffer with 2 μg/ml purified recombinant CBS-1 and unlabelled serine as the substrate. The temperature optimum for CBS-1 activity was determined in 5°C temperature intervals from 5°C to 80°C at pH 8.0, and the pH optimum of CBS-1 was determined at 25°C in 0.5 pH unit intervals using 100 mM Bis/Tris buffer at pH 6–10. Kinetic analyses at different concentrations of serine or homocysteine were performed at 25°C and pH 8.0, and the data were evaluated by non-linear data fitting using software Origin 8 (OriginLab). All measurements were repeated four times and the results are shown as means±S.D.
Site-directed mutagenesis and preparation of CBS-1 protein variants
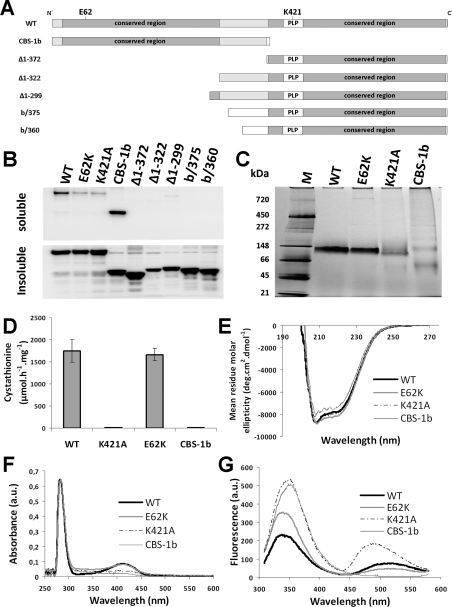
We prepared and analysed a series of mutant CBS-1 enzymes that included two missense variants of full-length CBS-1 (E62K and K421A) and six truncated CBS-1 variants (CBS-1b, Δ1–372, Δ1–322, Δ1–299, b/360 and b/375) (Figure 4A). All CBS-1 variants were cloned into the pGEX vector, which produces GST-tagged proteins. The sequences of primers used for cloning and site-directed mutagenesis are shown in Supplementary Table S1. Proteins were expressed and purified according to the procedure developed for CBS-1. The yields of purified mutant proteins were slightly lower, typically approximately 0.5 mg per litre of bacterial culture. The mutant proteins were analysed by UV–visible spectroscopy, CD and fluorescence spectroscopy and by BN-PAGE as described above for CBS-1. The catalytic activities for the reaction of serine with homocysteine were assessed in 100 mM Tris/HCl (pH 8.5) at 25°C.
Figure 4. Structural and enzymatic analysis of recombinant CBS-1 variants.
(A) Illustration of the CBS-1 variants expressed in E. coli. (B) Detection of CBS-1 variants in E. coli lysate after expression using soluble and insoluble fractions separated by centrifugation. (C) BN-PAGE of purified recombinant CBS-1 variants shows the monomeric status of the WT, K421A and E62K proteins. The N-terminal domain exhibits monomeric and oligomeric forms. Molecular mass markers are shown in kDa on the left-hand side. (D) CBS activity of the purified CBS-1 variants; K421A and CBS-1b have no CBS activity. Results are means±S.D. (E) CD spectra at far-UV show a helical secondary structure for all of the purified CBS-1 variants. (F) The UV–visible spectrum of purified recombinant CBS-1 variants of equal concentration shows peaks in the 280 and 412 nm region, indicating light absorption by aromatic amino acids and PLP respectively. Soret peaks typical for haem are not present. (G) Emission spectrum after excitation of the tryptophan residues at 298 nm of purified recombinant CBS-1 variants of equal concentration.
RNA-mediated interference
The cbs-1-specific sequence (~350 bp in length) was prepared by PCR amplification of a C. elegans cDNA library primers G and H (Supplementary Table S1) and cloned into the pCR4-TOPO vector. Single-stranded RNAs were prepared from linearized DNA by in vitro transcription using T3 DNA-dependent RNA polymerase (construct DNA digested by NotI) and T7 DNA-dependent RNA polymerase (construct DNA digested by SalI). The sense and antisense single-stranded RNAs were mixed and incubated at 68°C for 10 min, followed by incubation at 37°C for 30 min. The double-stranded RNA was further purified by phenol/chloroform extraction and precipitated by ethanol; the RNA pellet was diluted in water to an approximate concentration of 2 μg of RNA/μl. The double-stranded RNA was injected into the gonads of young adult hermaphrodite worms as described previously [30]. The embryos of microinjected animals were synchronized in 9–12 h intervals. Nematodes were grown at 16°C on nematode growth medium plates and fed with E. coli strain OP50. After RNAi (RNA interference), the nematodes were seeded in 1× PBS buffer on 2% agarose and screened by their phenotype.
Determination of CBS-1 antigen levels and measurement of enzymatic activity in C. elegans extracts
Worms were grown at 16°C as described above and collected 7 days after embryo microinjection as a mixed population of all larval stages. Worm lysates were prepared by sonication of worm pellets resuspended in 1 vol. of 100 mM PBS containing protease inhibitor cocktails for prokaryotic (P8465, Sigma–Aldrich) and eukaryotic (P8340, Sigma–Aldrich) cells. Crude extracts were centrifuged for 1 h at 4°C and 20000 g and the supernatants were used for the determination of CBS-1 levels and CBS activity. Western blotting was used to examine CBS-1 antigen levels after RNAi. The samples were submitted to SDS/PAGE (pre-cast 3–8% gradient gel), and protein immunodetection was performed by Western blot analysis using custom-made rabbit polyclonal anti-CBS-1 antibody prepared against purified recombinant CBS-1 (Exbio Praha). Actin, which was detected using a rabbit anti-actin antibody (Abcam), was used for the normalization of protein loading. The signal levels of CBS-1 and actin were determined by chemiluminescence (Pierce) by employing the ChemiGenius station and Gene Tools software for semi-quantification [31]. The enzyme assay was performed according to a previously described protocol [29] with the modification that the reaction mixture was incubated at 16°C for 30 min.
Measurement of metabolites in C. elegans extracts
Worm lysates were prepared by sonication of worm pellets that had been resuspended in 1 vol. of 100 mM PBS without protease inhibitors (Cole-Parmer GE130 Ultrasonic processor, amplitude 20 for 2 min with l s on/off pulses). The crude extracts were centrifuged at 20000 g for 1 h at 4°C, and the supernatants were used for HPLC aminothiol determination as described previously [28]. The cystathionine concentration was determined by LC–MS/MS using the EZ:faast kit for amino acid analysis (Phenomenex) [29]. The concentrations of all metabolites measured were normalized to the amount of protein present in the sample.
RESULTS
CBS in C. elegans is encoded by ZC373.1
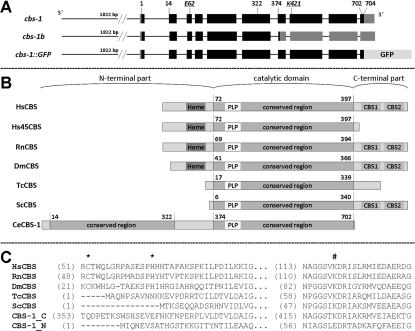
We used a BLASTp search as an in silico approach to identify genes that encode a CBS in C. elegans. Using the query sequences of three enzymes of the CBS family (human CBS, trypanosomal CBS and bacterial cysteine synthases), we identified ten genes with predicted amino acid sequences that are homologous with the catalytic domains of the known CBSs; these genes are annotated in WormBase (http://www.wormbase.org/) as ‘CBS and related proteins’. Alignment of the predicted amino acid sequences of the C. elegans genes ZC373.1 and F54A3.4 revealed the highest homology with human CBS (UniProt entry P35520). These predicted proteins exhibited 54% sequence identity with the human protein, whereas the other eight predicted proteins showed lower homology, with 21–44% sequence identity (Supplementary Table S2 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). The BLASTp searches using all ten nematode CBS homologues as the query sequences against the UniProt database, together with phylogenetic analysis, indicated that only ZC373.1 and F54A3.4 are homologous with CBS, whereas the remaining eight amino acid sequences are homologous with other proteins within the family of fold-type II PLP-dependent enzymes (Supplementary Figure S1 at http://www.BiochemJ.org/bj/443/bj4430535add.htm).
An annotation in the WormBase database shows that the ZC373.1 gene is trans-spliced to SL1 and contains ten exons, including 23 bp of the 5′-UTR (untranslated region) and 149 bp of the 3′-UTR followed by a polyadenylation sequence. The F54A3.4 gene is predicted to contain either eight exons without any 5′- or 3′-UTRs (http://www.wormbase.org/) or only seven exons terminated by a 77 bp 3′-UTR sequence (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/). To determine whether the ZC373.1 and F54A3.4 genes are transcribed and spliced into the predicted full-length mRNAs, we analysed their coding regions by RT–PCR and by sequencing of PCR products. We found two differently spliced variants of the ZC373.1; one sequence is identical with the WormBase annotation (cbs-1), and the other is a new ZC373.1 splice variant (cbs-1b) containing a 5-bp shortening of exon 7 in its 5′-terminus that leads to a frame-shift with a premature stop codon at amino acid residue 377 (Figure 1A and Supplementary Figure S2 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). In contrast, we were unable to amplify either of the two hypothetical full-length F54A3.4 mRNAs using several PCR conditions, various primers and various cDNA templates.
Figure 1. Organization of the cbs-1 gene and domain architecture of CBS in various organisms.
(A) Gene organization. The top diagram shows the organization of gene ZC373.1 (cbs-1) encoding CBS-1. The numbers indicate the codon position encoding the appropriate amino acid. Exons are indicated as black boxes, and the 5′- and 3′-UTR sequences are indicated as grey boxes. The middle diagram shows the novel splice ZC373.1 variant cbs-1b. The bottom diagram shows the translational fusion construct used in the GFP reporter assay. The length of the entire promoter used in the cbs-1–GFP construct is indicated by the number of base pairs of the 5′-upstream sequence. (B) Domain organization of various CBSs. The published structures of different CBSs were analysed for the presence of a haem-binding site (marked Heme), conserved catalytic regions with a PLP-binding site (marked PLP) and Bateman domain composed of two CBS domains (CBS1 and CBS2). The primary structures are aligned by the PLP-binding lysine residue; the numbers indicate the first and the last amino acid residues of conserved domains in the protein sequence. The aligned proteins are HsCBS (Homo sapiens CBS, UniProt entry P35520), Hs45CBS (truncated human CBS with 1–413 residues), RnCBS (Rattus norvegicus CBS, UniProt entry P32232), DmCBS (D. melanogaster CBS, UniProt entry Q9VRD9), TcCBS (T. cruzi CBS, UniProt entry Q9BH24), ScCBS (S. cerevisiae CBS, UniProt entry P32582) and CBS-1 (C. elegans CBS, UniProt entry Q23264). (C) Amino acid alignment of haem- and PLP-binding sites in various CBSs with separated N- and C-terminal conserved regions of CBS-1. The N-terminal region of CBS-1 does not contain the lysine residue that binds PLP. The cysteine and histidine residues that bind haem are indicated by asterisks, and the PLP-binding lysine residues are indicated by #.
Because we did not succeed in detecting the F54A3.4 mRNA by RT–PCR, we used additional approaches to examine the possible role of this gene in C. elegans. In silico analysis of the GenBank® database revealed three ESTs (expressed sequence tags) of F54A3.4: CK587466.1, CB389123.1 and FN902238.1; however, only FN902238.1 has been mapped to the sense strand of the F54A3.4 region (http://www.ncbi.nlm.nih.gov/nucleotide/). Furthermore, the proteomic database PeptideAtlas did not contain any peptide matches to the hypothetical protein F54A3.4 (http://www.peptideatlas.org/) [32]. Moreover, expression analysis using translational fusion proteins F54A3.4–GFP and ZC373.1–GFP (cbs-1–GFP) (see below) showed that the GFP signals reflecting the expression pattern of the appropriate genes were observed only in worms carrying ZC373.1–GFP, in contrast with the expression patterns observed in several worms carrying F54A3.4–GFP. Finally, F54A3.4 does not appear to have functional significance in C. elegans because the mutant strain RB839, which carries a deletion of F54A3.4, showed CBS activity and homocysteine concentrations indistinguishable from those of the WT strain (results not shown), and did not exhibit abnormal behavioural or a developmental phenotype (results not shown).
On the basis of the findings listed above, F54A3.4 appears to be a pseudogene and was not further examined in the present study. All of the data above strongly indicate that the C. elegans genome contains only one expressed orthologue of the human CBS gene, i.e. ZC373.1. In accordance with the recommended nomenclature, this gene was named cbs-1.
CBS-1 is a cytoplasmic enzyme that is expressed in the hypodermis and intestine, and in muscle cells
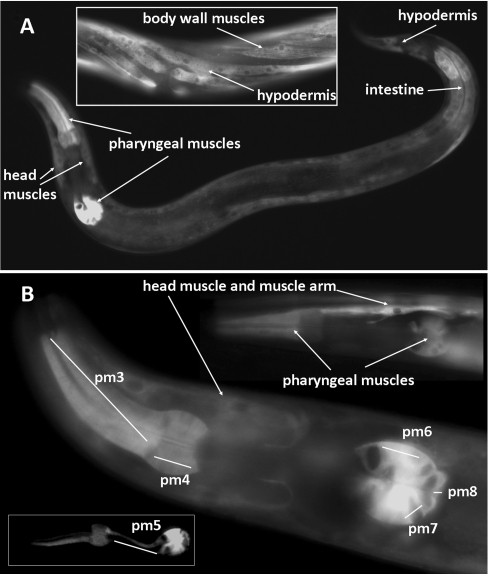
To determine the expression pattern and subcellular localization of cbs-1, we constructed the translational vector cbs-1–GFP, which contains the promoter and the entire CBS-1 sequence tagged at the C-terminus with GFP (Figure 1A). In worms expressing cbs-1–GFP, the GFP signal was observed in the hypodermis, intestine, body-wall muscle cells and pharyngeal muscles pm3, pm4, pm5, pm6, pm7 and pm8 in all larval stages as well as in adults (Figure 2). Our data using a translational reporter showed a similar expression pattern, as did previous transcriptional screens, and a novel expression of cbs-1 in pharyngeal muscles. We did not observe a GFP signal in embryos, although previous transcriptional screens and peptide mapping studies have reported expression of cbs-1 in this developmental stage [32,33]. The observed GFP signal was distributed diffusely within cells and spared the nucleus, suggesting that CBS-1 is localized in the cytoplasm. These data provide the first reported insight into the tissue and subcellular localization of nematode CBS-1 at the protein level and indicate which nematode tissues can metabolize homocysteine to cystathionine and/or cysteine to hydrogen sulfide.
Figure 2. Expression pattern of cbs-1 in worms.
The images show transgenic worms that carry the translational fusion vector cbs-1–GFP. (A) L4 larval stage showing the distribution of the GFP signal in the pharyngeal muscles, intestine, hypodermis and muscle cells. The middle part of the adult body (inset) shows the GFP signal in the body wall muscles and hypodermis. (B) Head of the worm showing GFP signal in pharyngeal muscles and a head muscle cell with its muscle arm; the GFP signal is distributed in the pharyngeal muscles pm3, pm4, pm6, pm7 and pm8. Some worms also exhibited a GFP signal in pm5 (inset).
CBS-1 is a haem-independent protein that lacks activation by AdoMet
To experimentally characterize the structural and enzymatic properties of CBS-1, cbs-1 cDNA was expressed in E. coli. Recombinant CBS-1 (704 residues of native CBS-1 with five additional amino acids at the N-terminus, six additional histidine residues at the C-terminus and a size of ~78 kDa) was purified to greater than 95% purity (Supplementary Figure S4 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). The UV–visible absorption spectrophotometry of the purified recombinant CBS-1 showed a peak at 412 nm, confirming the presence of covalently bound PLP that forms an internal aldimine, but it did not reveal a Soret band associated with a haem moiety (Figure 4F). This analysis confirmed that, in contrast with other characterized metazoan CBS enzymes, the nematode enzyme is a haem-independent CBS.
We next tested four reactions that have been described for previously characterized CBSs: (i) cystathionine-synthesizing activity that produces cystathionine and water from homocysteine and serine; (ii) formation of cystathionine and hydrogen sulfide from homocysteine and cysteine; (iii) cysteine synthase activity that produces cysteine from O-acetylserine and hydrogen sulfide; and (iv) serine sulfhydrylase activity in which cysteine is synthesized from serine and hydrogen sulfide. CBS-1 exhibited high enzymatic activity for synthesis of cystathionine from homocysteine utilizing either serine or cysteine and considerably lower cysteine synthase and serine sulfhydrase activities for synthesis of cysteine. The specific activities of CBS for the production of cystathionine from serine and cysteine were ~1500 μmol·h−1·mg−1 and ~300 μmol·h−1·mg−1 respectively, and its specific cysteine synthase and serine sulfhydrase activities were ~5 μmol·h−1·mg−1 and ~30 μmol·h−1·mg−1 respectively. None of these activities were stimulated by 1 mM AdoMet (results not shown), which is consistent with the absence of a Bateman domain in CBS-1 (see below). These data show that the nematode CBS-1 enzyme exhibits typical CBS activity and that it is not activated by AdoMet.
CBS-1 has a unique structural arrangement
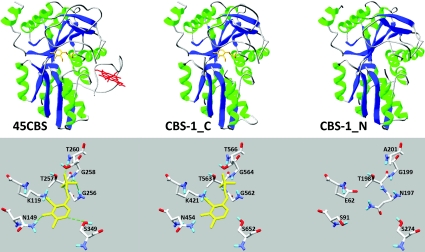
Alignment of the predicted amino acid sequence of CBS-1 with the sequences of previously characterized human, rat, Drosophila, trypanosome and yeast CBS enzymes revealed that the C. elegans enzyme possesses unique and novel domain architecture. In contrast with other CBSs, C. elegans CBS-1 lacks both the haem-binding N-terminus and the entire C-terminus found in other species (Figures 1B and 1C). Moreover, amino acid alignment together with protein modelling revealed that a single polypeptide chain of CBS-1 contains a unique tandem arrangement of two conserved CBS cores that belong to a family of fold-type II PLP-dependent proteins (Figure 1B and 3). Phylogenetic analysis of these two CBS-1 modules revealed that, in contrast with the C-terminal module, the N-terminal module has a lower homology with other CBS enzymes and does not belong to any of the fold-type II PLP-dependent protein families tested (Supplementary Figure S3 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). Furthermore, the critical PLP-binding lysine residue in the N-terminal module is replaced by a glutamic acid residue (Figures 1C and 3). Analysis of the PLP-binding site using homology modelling with the structure of 45CBS as the template revealed that the fully conserved glycine residue (Gly256 in human 45CBS) in the N-terminal module is replaced by a bulky asparagine residue (Asn197) that may sterically affect PLP binding (Figure 3). Thus the in silico data strongly suggest that the N-terminal module of nematode CBS-1 cannot bind the PLP essential for the catalytic activity of the enzyme.
Figure 3. Computationally modelled CBS-1 domains.
The images show the fold and PLP-binding site of human 45CBS, C-terminal module of CBS-1 (CBS-1_C) and N-terminal module of CBS-1 (CBS-1_N). The crystal structure of the human enzyme shows hydrogen bonds between amino acid residues and PLP, as indicated by broken green lines. Computational modelling of the individual CBS-1 modules revealed that both modules belong to the family of fold-type II PLP-dependent proteins and that the N-terminal module cannot bind PLP due to the absence of lysine and glycine residues in the consensus PLP-binding pocket.
The catalytic activity of CBS-1 is mediated only by the C-terminal module
To confirm the hypothesis that the catalytic function of CBS-1 is mediated only by its C-terminal module, we generated individual CBS-1 modules in E. coli. While the CBS-1b variant, which lacks the C-terminal module of CBS-1, was highly soluble after expression in E. coli, all of the cloned CBS-1 variants without the N-terminal module showed substantially decreased solubility (Figure 4B) that prevented successful purification of proteins containing only the C-terminal module. However, we purified and characterized the CBS-1b and used UV–visible and fluorescence spectrometry to determine that it does not bind PLP; we also found that CBS-1b had virtually undetectable catalytic activity (Figures 4D, 4F and 4G and Table 1). These observations demonstrate that the PLP-binding site essential for catalytic activity is not located in the N-terminal module of the CBS-1 protein. Although CD spectrometry showed that the CBS-1b has a helical secondary structure similar to that of the WT CBS-1 protein (Figure 4E), BN-PAGE revealed that purified CBS-1b precipitates and may form higher-order oligomers (Figure 4C). These data indicate that the presence of both CBS-1 modules in one subunit is necessary for the maintenance of the global structural stability of CBS-1 and/or for proper folding of the recombinant protein.
Table 1. Enzymatic and structural properties of purified CBS-1 variants.
ND, not detected, Trp, tryptophan.
| Protein | WT | K421A | E62K | CBS-1b |
|---|---|---|---|---|
| Catalytic activity (μmol·h−1·mg−1) | 1742±259 | 0.41±0.03 | 1663±144 | 0.27±0.05 |
| Absorption ratio 280/412 nm | 7.6 | 16.1 | 7.9 | 29.2 |
| PLP absorption maximum (nm) | 412 | 403 and 418 | 412 | ND |
| Trp relative fluorescence | 233 | 534 | 354 | 505 |
| Trp fluorescence wavelength maximum (nm) | 338 | 350 | 339 | 352 |
| Relative delayed fluorescence | 76 | 181 | 48 | ND |
Because analysis of the isolated N-terminal module indicated possible disruption of its native structure, we also generated and purified the CBS-1 mutants K421A, which abolishes a canonical PLP-binding site in the C-terminal module, and E62K, which creates a putative PLP-binding site in the N-terminal module. We observed altered fluorescence-based tryptophan spectra of the mutant proteins; their relative fluorescence showed different quenching of the tryptophan emission, and the existence of a wavelength maximum shift from ~340 to ~350 nm indicates higher accessibility of the tryptophan residues to the polar solvent in the K421A mutant (Figure 4G and Table 1). In contrast, both mutants retained oligomeric status identical with the WT as determined by BN-PAGE (Figure 4C), and CD measurements showed that the protein's secondary structure is not affected by either the K421A or E62K mutation (Figure 4E). The catalytic activity, PLP saturation and fluorescent properties of the E62K mutant were similar to those of WT (Figures 4D, 4F and 4G), supporting the idea that the structural properties of the N-terminal module do not permit PLP binding even if the canonical lysine residue is present. In contrast, the K421A mutant binds significantly less of the PLP cofactor, as determined by UV–visible absorption spectroscopy. The mutant enzyme's residual affinity for PLP probably results from the formation of an external aldimine; this affinity is manifested in its UV–visible spectrum by the presence of two bands with maxima at 403 and 418 nm and by the lack of the sharp maximum at 412 nm that is typical for internal aldimines (Figure 4F). The formation of an external aldimine in K421A was confirmed by fluorescence spectroscopy and, when excited at 298 nm, the emission spectrum of the mutant protein revealed a significantly higher extent of delayed fluorescence of Schiff bases for K421A in comparison with WT, E62K and CBS-1b (Figure 4G). Enhanced delayed fluorescence due to formation of external aldimines in the active site of the mutant enzyme has also been reported for the bacterial O-acetylsulfhydrylase mutant K42A [34]. Taken together, these experiments provide additional evidence that the catalytic activity of C. elegans CBS-1 is mediated only by the C-terminal module and that its N-terminal module cannot bind PLP cofactor either as an internal or as an external aldimine.
Analysis of the quaternary structure of nematode CBS-1 suggests a monomeric status of CBS-1
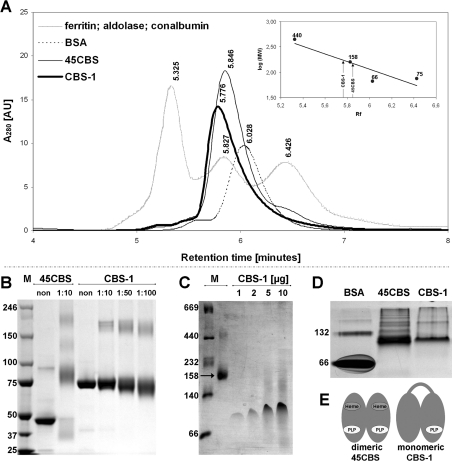
We analysed the quaternary structure of recombinant nematode CBS-1 to determine whether CBS-1 exists as a monomer with a structural arrangement similar to human 45CBS (the C-terminally truncated human CBS that lacks a Bateman domain and forms dimers of 90 kDa [15]), or whether CBS-1 forms dimers or higher-order oligomers. We first performed SEC using the standard proteins ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa) and BSA (66 kDa). To control for possible differences in the Stokes radii of the standard proteins and the CBS-related proteins that may influence their retention on the column, we analysed human 45CBS in parallel. Nematode CBS-1 exhibited a tailing peak with a retention time of 5.776 min (Figure 5A); on the basis of the calibration curve, the apparent native molecular mass of the protein was determined to be ~170 kDa. However, SEC of human 45CBS indicated a native molecular mass of approximately 150 kDa, suggesting that calibration with standard proteins may result in overestimation of the molecular mass of CBS proteins. According to the molecular mass markers used, SEC yielded ambiguous results compatible with both a monomeric and dimeric structure of CBS-1.
Figure 5. Determination of the quaternary structure of CBS-1.
(A) SEC. The bold solid curve represents the elution profile of purified recombinant CBS-1, which has a retention time of 5.776 min and an estimated molecular mass of 168 kDa. The thin solid curve represents human 45CBS, which has a retention time of 5.846 min and an estimated molecular mass of 148 kDa. The dashed curve represents BSA with a retention time of 6.028. The grey (dotted) curve represents molecular standards eluted at the following retention times: ferritin (440 kDa), 5.325 min; aldolase (158 kDa), 5.827 min; and conalbumin (70 kDa), 6.426 min. AU, absorbance unit. (B) Cross-linking. Purified CBS-1 and human 45CBS were cross-linked with BS3 in appropriate molar ratios of protein/modifier, as indicated in the Figure, and subjected to SDS/PAGE. In contrast with human 45CBS, which forms dimers, the mobility of CBS-1 does not change after cross-linking. (C) BN-PAGE. CBS-1, with a molecular mass of 78 kDa (four different amounts of loaded protein), migrates between molecular mass markers of 66 kDa and 140 kDa. The molecular protein mass markers include thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), BSA (66 kDa) and aldolase (158 kDa). (D) Native PAGE. CBS-1, with a molecular mass of 78 kDa, migrates between molecular mass markers of 66 kDa and 132 kDa, similar to a ~90 kDa dimer of human 45CBS. In (B–D) the molecular mass is given in kDa on the left-hand side. M, marker. (E) Schematic diagram of the hypothetical quaternary structure of CBS-1 and comparison of its structure with that of human 45CBS.
We next used additional techniques including native electrophoresis, BN electrophoresis and chemical cross-linking followed by SDS/PAGE to determine the most likely quaternary structure of CBS-1. These three techniques congruently showed that the 78 kDa nematode CBS-1 exists predominantly as a monomer. The evidence, which is shown in Figures 5(B)–5(D), is as follows: (i) in native PAGE, nematode CBS-1 migrates similarly to the 90 kDa marker of dimeric human 45CBS and between fractions containing monomeric and dimeric BSA respectively (66 kDa and 132 kDa); (ii) on BN electrophoresis, CBS-1 migrates between molecular mass markers of 66 and 140 kDa; and (ii) chemical cross-linking of CBS-1 did not result in changes in protein migration, suggesting modification of amino acid side chains within a single polypeptide chain, whereas human 45CBS readily formed a cross-linked dimeric product with a molecular mass of ~100 kDa. On the basis of these results, we propose that, in contrast with CBS enzymes from other species, recombinant nematode CBS-1 does not form oligomeric structures in vitro. Because the conserved catalytic regions of CBS-1 are homologous with each other, we hypothesize that they form an internal interface similar to that formed by subunit dimerization of human 45CBS (Figure 5E).
CBS-1 is more sensitive to denaturation and is more active than human 45CBS
We hypothesized that the above-described differences in the oligomeric assembly of nematode CBS-1 and human 45CBS might result in differences in the energetics of the two proteins. To explore this hypothesis, we used a fluorescence-based thermal-shift assay and pulse proteolysis in a urea gradient. Both approaches revealed significantly lower stability of CBS-1 compared with the human 45CBS; the melting point of CBS-1 was 10°C lower than that of human 45CBS, and the resistance of CBS-1 to urea-induced unfolding decreased by ~2.8 M (Table 2 and Supplementary Figure S5 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). These data show that the nematode CBS-1 is less energetically stable than the human 45CBS; this finding may be due to a lower energy of the interdomain interface or a higher structural flexibility of the worm CBS-1.
Table 2. Stability and enzymatic properties of CBS-1 compared with human 45CBS.
cm, concentration of urea at which a fraction of folded proteins comprises 50% of the entire protein population.
| Protein | Nematode CBS-1 | Human 45CBS |
|---|---|---|
| Oligomeric status | Monomer | Dimer |
| Michaelis constant Km (mM) | ||
| Serine | 5.57±0.68 | 2.20±0.46† |
| Homocysteine | 4.29±0.97 | 0.33±0.07† |
| Turnover number kcat (s−1) | ||
| Serine | 48.12±2.95 | 13.81±0.88† |
| Homocysteine | 43.31±4.33 | 10.88±0.72† |
| Catalytic efficiency kcat/Km (mM−1·s−1) | ||
| Serine | ~8.5 | ~6 |
| Homocysteine | ~10 | 26.97±5.87† |
| Characteristics of protein denaturation | ||
| Midpoint of urea concentration cm (M) | 1.21±0.02 | 4.08±0.07* |
| Melting point Tm (°C) | 41 | 51 |
We considered the possibility that the observed structural and energetic differences between the nematode CBS-1 and human 45CBS result in different catalytic properties. We determined the temperature and pH optima and the kinetic parameters for the major CBS reaction, which produces cystathionine from serine and homocysteine. The CBS-1 protein exhibited the highest activity at pH 8.5 and 30°C (Supplementary Figure S6 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). These conditions are in accordance with the results of the thermal stability assay (see above). We speculate that the lower temperature optimum of CBS-1 compared with the human enzymes (37°C) may reflect the lower body temperature of nematodes living in the soil. We also found that the affinity of CBS-1 for homocysteine is lower than that of 45CBS (Table 2); however, we observed inhibition of CBS activity at 7.5 and 10 mM homocysteine and this inhibition prevented the activity from increasing to more than ~1500 μmol·h−1·mg−1 (Supplementary Figure S6). Inhibition by high concentrations of homocysteine has been previously reported for yeast and human CBS enzymes [13,35]. Taken together, these data show that the nematode CBS-1 subunit is approximately 4-fold more active compared with the human 45CBS subunit as expressed by the turnover number (Table 2).
CBS-1 mediates nematode development and maintains homocysteine homoeostasis
To explore the functional significance of cbs-1 in C. elegans, we silenced the cbs-1 gene by RNA-mediated interference and determined the phenotypic consequences of such silencing. To confirm the efficacy of cbs-1 RNAi, we measured the amounts of CBS-1 antigen and CBS activity in worm extracts of CBS-1-inactivated and WT worms. Western blot analysis using an anti-CBS-1 antibody showed that after RNAi treatment worms exhibited a CBS-1 level that was approximately 10% that of the control strain (Supplementary Figure S7A at http://www.BiochemJ.org/bj/443/bj4430535add.htm). Although the mean CBS activity of normal worms was 36.0 nmol·h−1·mg−1, cbs-1 RNAi animals exhibited a mean activity of only 5.4 nmol·h−1·mg−1, approximately ~15% of the control level (Supplementary Figure S7B). The results from both Western blot analysis and CBS activity measurement consistently confirmed that the RNAi experiments efficiently reduced the amount and activity of CBS-1.
RNAi resulted in a developmental delay phenotype in 97% of the worms (515 out of 530 individuals tested). These animals reached the egg-laying adult stage no earlier than the 9th day after embryo hatching, in contrast with control worms, which reached the same stage on the 5th day of development. The affected larvae had a shorter body length than the controls (Supplementary Figure S8 at http://www.BiochemJ.org/bj/443/bj4430535add.htm). After RNAi of larvae, the most severe abnormalities were observed in the tissues that express the highest amount of CBS-1 (i.e. gut and pharynx; see the data above on the translational cbs-1–GFP vector). The gut cells of these animals showed reduced pigment granule birefringence under Nomarski optical microscopy (Supplementary Figure 8), and the anterior bulb of the pharynx exhibited abnormal morphology, with a balloon-like appearance and enlarged diameter (Supplementary Figure 8). These data show that CBS-1 is essential for normal development in nematodes.
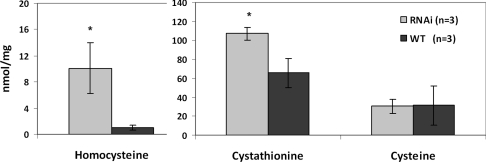
We determined metabolic flux through the trans-sulfuration pathway by measuring homocysteine, cystathionine and cysteine concentrations in worm homogenates. To eliminate possible differences in metabolic fluxes in worms at various stages of development, the worms were collected at the latest larval developmental stage (L4). The homocysteine and cystathionine levels in C. elegans extracts were ~10× and ~1.6× higher in the knockdown strain than in the controls, whereas cysteine concentrations did not differ between the two strains (Figure 6). The observation of elevated homocysteine levels in CBS-1-knockdown worms strongly supports an important role of this enzyme in maintaining homocysteine homoeostasis in C. elegans.
Figure 6. Metabolite levels in crude extracts of CBS-1-knockdown animals.
The concentration of metabolites (in nanomoles per mg of protein) from RNAi and control experiments respectively. Homocysteine and cystathionine concentrations in CBS-1-deficient worms are significantly higher (10.1 and 107.1 nmol/mg of protein respectively) than in control worms (1.0 and 65.6 nmol/mg of protein respectively). Results are means±S.D. from three independent experiments. *P<0.05, as determined by Student's t test.
DISCUSSION
Evidence that CBS in C. elegans is encoded by cbs-1
In the present paper, we identified the cbs-1 gene in C. elegans, which encodes an enzyme with cystathionine-synthesizing activity and is important for normal development of the nematode. Using a BLASTp search against a C. elegans protein database, we identified two nematode genes, ZC373.1 and F54A3.4, that are highly homologous with CBS genes in other species. However, several lines of evidence demonstrate that only ZC373.1 encodes a CBS, whereas F54A3.4 is probably a pseudogene. The gene denoted F54A3.4 has not been detected by RT–PCR, was not found by in silico searches in the appropriate EST and proteomic databases, and its partial deletion did not elicit biochemical or morphological phenotypic abnormalities. In contrast, ZC373.1 mRNA has been detected by RT–PCR, its ESTs and peptides were annotated in appropriate databases, the enzyme was shown to be expressed endogenously in its entire length, and its knockdown resulted in severe biochemical and phenotypic consequences. Most importantly, the purified CBS-1 enzyme exhibited enzymatic properties consistent with previously characterized CBS enzymes from other species.
The unique domain architecture of nematode CBS enzymes
In silico analysis of the CBS-1 protein sequence showed that the CBS-1 of C. elegans possesses a unique multi-domain architecture that has not been reported previously for any other CBS. The unusual structure of CBS-1 includes the lack of a haem-binding region, the lack of a Bateman domain and the tandem arrangement of two conserved catalytic regions of which only the C-terminal region is catalytically active. Such a domain arrangement of predicted CBS enzymes in fully sequenced organisms has been found only in organisms from the nematode phylum, showing an evolutionarily divergent arrangement of the CBS protein in this phylum. Interestingly, the nematode Loa loa possesses a PLP-binding site in both CBS modules (Supplementary Figure S9 at http://www.BiochemJ.org/bj/443/bj4430535add.htm), suggesting that the unusual and unique structure of CBS enzymes in nematodes probably originates from a duplication of the conserved catalytic region in a common ancestor followed by mutations abolishing PLP binding in the N-terminal module.
To our knowledge there is no evidence, except of nematode CBS proteins, regarding fold-type II PLP-dependent proteins lacking a PLP-binding site. Thus the function, if any, of the N-terminal module in the nematode CBS-1 protein remains unclear. Several pieces of experimental evidence obtained in the present study clearly show that this module does not have canonical catalytic function. We speculate that mutations in this portion of the nematode CBS enzyme may have permitted the acquisition of novel structural and functional properties, such as changes in protein stability and folding, protein–protein interactions, or regulation of enzyme activity. Studies of truncated variants suggest that the N-terminal module may be important for proper folding and subsequent stability of CBS-1 (see above). Because CBS-1 forms a monomer, it is probable that its N-terminal and C-terminal modules interact to form a structure similar to that of the human 45CBS dimer. However, the proposed interdomain interaction cannot be sufficiently supported by the computational modelling procedures using previously solved crystal structures of CBS proteins, and thus it requires further study of the three-dimensional spatial arrangement of CBS-1 at atomic resolution. The N-terminal module of CBS-1 may also have a regulatory role. The existence of tandem duplicated conserved modules of which only one is catalytically active in a single polypeptide is similar to the well-known case of tyrosine protein kinases [JAKs (Janus kinases)]. In tandem with a catalytically active kinase domain, these kinases have a catalytically inactive pseudo-kinase domain that has been implicated in the regulation of their activity [36]. Alternatively, the N-terminal module of CBS-1 may also play a role in protein–protein interactions, such as the interactions with the SUMOylation enzyme apparatus or huntingtin that have been described for human and rodent CBS-1 orthologues respectively [37,38].
More intriguingly, expression of the spliced variant cbs-1b shows that the N-terminal module of CBS-1 can be produced in vivo without the catalytic C-terminal module. This finding suggests that the non-catalytic module may play a role in additional biological processes independent of the catalytic module. However, it should be noted that misspliced variants with premature stop codons are commonly targeted by a cellular RNA nonsense-mediated decay mechanism [39]; therefore, the existence of a separate nematode N-terminal domain in vivo should be investigated in future studies.
Possible biological roles of CBS-1 enzymatic activity in C. elegans
Because cbs-1 is expressed in a limited number of tissues, it is tempting to speculate on the role of this enzyme in the organs in which it is expressed. High expression of cbs-1 was observed throughout post-embryonic development in the intestine, which is characterized by secretion of digestive enzymes and high metabolic activity in C. elegans, such as the synthesis and storage of macromolecules and detoxification of xenobiotics [40]. Thus the expression of cbs-1 in C. elegans intestine may mirror the high expression of CBS in the mammalian liver, pancreas and small intestine, in which CBS plays an important role in homocysteine homoeostasis and/or in the provision of cysteine for glutathione production [41]. We hypothesize that the intestinal expression of cbs-1 in worms may serve similar purposes, namely removal of homocysteine or cysteine biosynthesis.
Expression of cbs-1 has also been observed in pharyngeal muscles and hypodermis. Because neither of these tissues shows high metabolic activity compared with the intestine, there are other possible explanations for CBS-1 activity in these tissues. Because both hypodermal cells and pharyngeal muscle cells secrete cuticle (http://www.wormatlas.org/), we propose that CBS-1 may provide cysteine, which is important for cuticle formation and its stabilization by disulfide bonds [42]. Another possible role for CBS-1 in muscle and hypodermis is the production of the neuromodulator and smooth muscle relaxant hydrogen sulfide [43]. The endogenous biosynthesis of H2S via CBS might serve for smooth muscle relaxation in the strongly innervated nematode pharynx or in regulating the expression of HIF-1 (hypoxia-inducible factor 1) target genes in the hypodermis [44]. Interestingly, although CBS is thought to be the main enzyme that produces hydrogen sulfide in the mammalian brain [45], we did not observe a GFP signal in neurons. This finding suggests that the endogenous production of hydrogen sulfide in C. elegans neurons is mediated by different enzymes than in other species or that the role of hydrogen sulfide in C. elegans neurons is negligible.
C. elegans as a model of CBS deficiency
Because many genes implicated in human diseases are well-conserved across phyla [46], C. elegans is considered by many investigators to be a suitable model for studying cellular and metabolic mechanisms in selected genetic disorders [47–49]. In addition to its low cost of maintenance and short generation time, other advantages of the C. elegans model include the possibility of observing cellular processes in vivo and of easily screening for the effects of novel therapies [50,51]. In the present study, we examined the morphological and biochemical effects of nematode CBS-1 deficiency. These effects may in part recapitulate the human disease homocystinuria, which results from CBS deficiency. Homocystinuria is characterized by increased tissue, plasma and urinary concentrations of homocysteine, and by decreased concentrations of cystathionine and cysteine [52,53]. Its clinical features include liver steatosis, connective tissue disorder, thromboembolism and various degrees of central nervous system involvement [53]. In our CBS-1–GFP localization study, CBS-1-knockdown worms exhibited abnormal morphology of several tissues that express the cbs-1 gene. Using light microscopy, we observed a reduced birefringent signal from pigment gut granules, which are considered to be lysosome-related organelles [54]. Although the function and composition of these granules has not been fully elucidated, the abnormal pattern of gut granules in CBS-1-knockdown animals may in part correspond to the liver steatosis observed in murine and human CBS deficiency. Furthermore, the observed abnormal pharyngeal morphology of CBS-1-deficient worms may possibly correspond to some of the neurological sequelae of human CBS deficiency. It appears that the CBS-knockdown nematodes produced in the present study may in part recapitulate some of the features of human homocystinuria due to CBS deficiency.
In the CBS-1-deficient nematodes produced in the present study, the amounts of CBS-1 antigen and enzyme activity decreased to ~10–15% those of WT worms. This degree of enzyme deficiency resulted in an approximately 10-fold increase in homocysteine concentrations in worm extracts compared with the WT strain, demonstrating an essential role for CBS-1 in maintaining homocysteine homoeostasis in C. elegans. Because exposure of worms to homocysteine in medium [55] leads to a similar developmental delay as the cbs-1 RNAi in the present study, it is conceivable that high tissue levels of homocysteine may be directly responsible for the developmental delay phenotype that we observed. Surprisingly, and in contrast with human patients with CBS deficiency [53], cystathionine levels in CBS-1-deficient worms were only slightly increased. However, a similar elevation in plasma cystathionine was reported for one murine model of CBS deficiency [56]. We hypothesize that elevated cystathionine in CBS-1-deficient worms may be caused by three possible mechanisms: (i) elevated homocysteine may inactivate CGL, as proposed previously for a murine model of CBS deficiency [56]; (ii) elevated homocysteine may lead to formation of cystathionine via condensation of cysteine and homocysteine by CGL [57]; and (iii) cystathionine may be synthesized by a hypothetical cystathionine γ-synthase in the reverse trans-sulfuration pathway using cysteine and O-succinylhomoserine. Moreover, the CBS-1-deficient worms observed in the present study did not exhibit cysteine depletion, which is a common feature of human CBS deficiency. We hypothesize that cysteine levels in deficient worms are maintained by sufficient cysteine intake from E. coli or by biosynthesis of cysteine via a hypothetical sulfur assimilation pathway because C. elegans possesses several bacterial and plant cysteine synthase homologues (see above, [58]).
Online data
AUTHOR CONTRIBUTION
Roman Vozdek designed and performed most of the experiments and wrote the first draft of the paper; Aleš Hnízda purified and further characterized recombinant CBS-1; Jakub Krijt measured aminothiols and cystathionine by HPLC and LC–MS/MS in the appropriate studies; Marta Kostrouchová co-ordinated the experiments with C. elegans; and Viktor Kožich co-ordinated the whole project. All authors have extensively revised various versions of the paper and approved its final version prior to submission.
ACKNOWLEDGEMENTS
We thank Dr A. Fire for vector pPD95.75, and Eva Zouharová, Mrs Kateřina Raková, Hana Prouzová, Jitka Honzíková and Professor Milan Kodíček for technical assistance and advice.
FUNDING
This work was supported by a Wellcome Trust International Senior Research Fellowship in Biomedical Science in Central Europe [number 070255/Z/03/Z], the Grant Agency of the Charles University in Prague [grant numbers 21709 and SVV262502], the Ministry of Education of the Czech Republic [grant number MSM0021620806] and the Czech Science Foundation [grant number 304/08/0970].
References
- 1.Finkelstein J. D. The metabolism of homocysteine: pathways and regulation. Eur. J. Pediatr. 1998;157:S40–S44. doi: 10.1007/pl00014300. [DOI] [PubMed] [Google Scholar]
- 1a.Taoka S., Ohja S., Shan X., Kruger W. D., Banerjee R. Evidence for heme-mediated redox regulation of human cystathionine beta-synthase activity. J. Biol. Chem. 1998;273:25179–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R., Zou C. G. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Arch. Biochem. Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 3.Cherney M. M., Pazicni S., Frank N., Marvin K. A., Kraus J. P., Burstyn J. N. Ferrous human cystathionine beta-synthase loses activity during enzyme assay due to a ligand switch process. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 4.Smith A. T., Majtan T., Freeman K. M., Su Y., Kraus J. P., Burstyn J. N. Cobalt cystathionine beta-synthase: a cobalt-substituted heme protein with a unique thiolate ligation motif. Inorg. Chem. 2011;50:4417–4427. doi: 10.1021/ic102586b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta P. K., Christen P. The molecular evolution of pyridoxal-5′phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 2000;74:129–184. doi: 10.1002/9780470123201.ch4. [DOI] [PubMed] [Google Scholar]
- 6.Kery V., Poneleit L., Kraus J. P. Trypsin cleavage of human cystathionine beta-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch. Biochem. Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 7.Taoka S., Widjaja L., Banerjee R. Assignment of enzymatic functions to specific regions of the PLP-dependent heme protein cystathionine beta-synthase. Biochemistry. 1999;38:13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 8.Skovby F., Kraus J. P., Rosenberg L. E. Biosynthesis and proteolytic activation of cystathionine beta-synthase in rat liver. J. Biol. Chem. 1984;259:588–593. [PubMed] [Google Scholar]
- 9.Zou C. G., Banerjee R. Tumor necrosis factor-α-induced targeted proteolysis of cystathionine beta-synthase modulates redox homeostasis. J. Biol. Chem. 2003;278:16802–16808. doi: 10.1074/jbc.M212376200. [DOI] [PubMed] [Google Scholar]
- 10.Jhee K. H., McPhie P., Miles E. W. Yeast cystathionine beta-synthase is a pyridoxal phosphate enzyme but, unlike the human enzyme, is not a heme protein. J. Biol. Chem. 2000;275:11541–11544. doi: 10.1074/jbc.c000056200. [DOI] [PubMed] [Google Scholar]
- 11.Nozaki T., Shigeta Y., Saito-Nakano Y., Imada M., Kruger W. D. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine beta-synthase and serine acetyltransferase from Trypanosoma. J. Biol. Chem. 2001;276:6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- 12.Koutmos M., Kabil O., Smith J. L., Banerjee R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine beta-synthase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jhee K. H., McPhie P., Miles E. W. Domain architecture of the heme-independent yeast cystathionine beta-synthase provides insights into mechanisms of catalysis and regulation. Biochemistry. 2000;39:10548–10556. doi: 10.1021/bi001020g. [DOI] [PubMed] [Google Scholar]
- 14.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meier M., Janosik M., Kery V., Kraus J. P., Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiederstein M., Sippl M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 18.Neron B., Menager H., Maufrais C., Joly N., Maupetit J., Letort S., Carrere S., Tuffery P., Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 21.Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinf. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulin T., Etchberger J. F., Hobert O. Reporter gene fusions. WormBook. 2006. pp. 1–23. doi/10.1895/wormbook.1.106.1. The C. elegans Research Community, ed. [DOI] [PMC free article] [PubMed]
- 23.Frank N., Kent J. O., Meier M., Kraus J. P. Purification and characterization of the wild type and truncated human cystathionine beta-synthase enzymes expressed in E. coli. Arch. Biochem. Biophys. 2008;470:64–72. doi: 10.1016/j.abb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janosik M., Meier M., Kery V., Oliveriusova J., Burkhard P., Kraus J. P. Crystallization and preliminary X-ray diffraction analysis of the active core of human recombinant cystathionine beta-synthase: an enzyme involved in vascular disease. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:289–291. doi: 10.1107/s0907444900017893. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Wittig I., Braun H. P., Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 27.Hnizda A., Spiwok V., Jurga V., Kozich V., Kodicek M., Kraus J. P. Cross-talk between the catalytic core and the regulatory domain in cystathionine beta-synthase: study by differential covalent labeling and computational modeling. Biochemistry. 2010;49:10526–10534. doi: 10.1021/bi101384m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maclean K. N., Sikora J., Kozich V., Jiang H., Greiner L. S., Kraus E., Krijt J., Crnic L. S., Allen R. H., Stabler S. P., et al. Cystathionine beta-synthase null homocystinuric mice fail to exhibit altered hemostasis or lowering of plasma homocysteine in response to betaine treatment. Mol. Genet. Metab. 2010;101:163–171. doi: 10.1016/j.ymgme.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krijt J., Kopecka J., Hnizda A., Moat S., Kluijtmans L. A., Mayne P., Kozich V. Determination of cystathionine beta-synthase activity in human plasma by LC–MS/MS: potential use in diagnosis of CBS deficiency. J. Inherited Metab. Dis. 2011;34:49–55. doi: 10.1007/s10545-010-9178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 31.Janosik M., Oliveriusova J., Janosikova B., Sokolova J., Kraus E., Kraus J. P., Kozich V. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am. J. Hum. Genet. 2001;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrimpf S. P., Weiss M., Reiter L., Ahrens C. H., Jovanovic M., Malmstrom J., Brunner E., Mohanty S., Lercher M. J., Hunziker P. E., et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009;7:e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKay S. J., Johnsen R., Khattra J., Asano J., Baillie D. L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 34.Rege V. D., Kredich N. M., Tai C. H., Karsten W. E., Schnackerz K. D., Cook P. F. A change in the internal aldimine lysine (K42) in O-acetylserine sulfhydrylase to alanine indicates its importance in transimination and as a general base catalyst. Biochemistry. 1996;35:13485–13493. doi: 10.1021/bi961517j. [DOI] [PubMed] [Google Scholar]
- 35.Belew M. S., Quazi F. I., Willmore W. G., Aitken S. M. Kinetic characterization of recombinant human cystathionine beta-synthase purified from E. coli. Protein Expression Purif. 2009;64:139–145. doi: 10.1016/j.pep.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Aringer M., Cheng A., Nelson J. W., Chen M., Sudarshan C., Zhou Y. J., O'Shea J. J. Janus kinases and their role in growth and disease. Life Sci. 1999;64:2173–2186. doi: 10.1016/s0024-3205(98)00538-4. [DOI] [PubMed] [Google Scholar]
- 37.Kabil O., Zhou Y., Banerjee R. Human cystathionine beta-synthase is a target for sumoylation. Biochemistry. 2006;45:13528–13536. doi: 10.1021/bi0615644. [DOI] [PubMed] [Google Scholar]
- 38.Boutell J. M., Wood J. D., Harper P. S., Jones A. L. Huntingtin interacts with cystathionine beta-synthase. Hum. Mol. Genet. 1998;7:371–378. doi: 10.1093/hmg/7.3.371. [DOI] [PubMed] [Google Scholar]
- 39.Zahler A. M. Alternative splicing in C. elegans. WormBook. 2005. pp. 1–13. doi/10.1895/wormbook.1.31.1. The C. elegans Research Community, ed. [DOI] [PMC free article] [PubMed]
- 40.McGhee J. D. The C. elegans intestine. WormBook. 2007. pp. 1–36. doi/10.1895/wormbook.1.133.1. The C. elegans Research Community, ed. [DOI] [PMC free article] [PubMed]
- 41.Finkelstein J. D. Pathways and regulation of homocysteine metabolism in mammals. Semin. Thromb. Hemostasis. 2000;26:219–225. doi: 10.1055/s-2000-8466. [DOI] [PubMed] [Google Scholar]
- 42.Page A. P., Johnstone I. L. The cuticle. WormBook. 2007. pp. 1–15. doi/10.1895/wormbook.1.138.1. The C. elegans Research Community, ed. [DOI] [PMC free article] [PubMed]
- 43.Kimura H. Hydrogen sulfide: its production, release and functions. Amino Acids. 2011;41:113–121. doi: 10.1007/s00726-010-0510-x. [DOI] [PubMed] [Google Scholar]
- 44.Budde M. W., Roth M. B. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel–Lindau tumor suppressor-1 in C. elegans. Mol. Biol. Cell. 2010;21:212–217. doi: 10.1091/mbc.E09-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe K., Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuwabara P. E., O'Neil N. The use of functional genomics in C. elegans for studying human development and disease. J. Inherited Metab. Dis. 2001;24:127–138. doi: 10.1023/a:1010306731764. [DOI] [PubMed] [Google Scholar]
- 47.Chandler R. J., Aswani V., Tsai M. S., Falk M., Wehrli N., Stabler S., Allen R., Sedensky M., Kazazian H. H., Venditti C. P. Propionyl-CoA and adenosylcobalamin metabolism in Caenorhabditis elegans: evidence for a role of methylmalonyl-CoA epimerase in intermediary metabolism. Mol. Genet. Metab. 2006;89:64–73. doi: 10.1016/j.ymgme.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvo A. C., Pey A. L., Ying M., Loer C. M., Martinez A. Anabolic function of phenylalanine hydroxylase in Caenorhabditis elegans. FASEB J. 2008;22:3046–3058. doi: 10.1096/fj.08-108522. [DOI] [PubMed] [Google Scholar]
- 49.Fisher A. L., Page K. E., Lithgow G. J., Nash L. The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: a Caenorhabditis elegans model of type I tyrosinemia. J. Biol. Chem. 2008;283:9127–9135. doi: 10.1074/jbc.M708341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Link E. M., Hardiman G., Sluder A. E., Johnson C. D., Liu L. X. Therapeutic target discovery using Caenorhabditis elegans. Pharmacogenomics. 2000;1:203–217. doi: 10.1517/14622416.1.2.203. [DOI] [PubMed] [Google Scholar]
- 51.Kaletta T., Hengartner M. O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 52.Kraus J. P., Janosik M., Kozich V., Mandell R., Shih V., Sperandeo M. P., Sebastio G., de Franchis R., Andria G., Kluijtmans L. A., et al. Cystathionine beta-synthase mutations in homocystinuria. Hum. Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 53.Kraus J. P., Kožich V. Cystathionine-β-synthase and its deficiency. In: Carmel R., Jacobsen D. W., editors. Homocysteine in Health and Disease. Cambridge: Cambridge University Press; 2001. pp. 223–243. [Google Scholar]
- 54.Hermann G. J., Schroeder L. K., Hieb C. A., Kershner A. M., Rabbitts B. M., Fonarev P., Grant B. D., Priess J. R. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khare S., Gomez T., Linster C. L., Clarke S. G. Defective responses to oxidative stress in protein l-isoaspartyl repair-deficient Caenorhabditis elegans. Mech. Ageing Dev. 2009;130:670–680. doi: 10.1016/j.mad.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maclean K. N., Sikora J., Kozich V., Jiang H., Greiner L. S., Kraus E., Krijt J., Overdier K. H., Collard R., Brodsky G. L., et al. A novel transgenic mouse model of CBS-deficient homocystinuria does not incur hepatic steatosis or fibrosis and exhibits a hypercoagulative phenotype that is ameliorated by betaine treatment. Mol. Genet. Metab. 2010;101:153–162. doi: 10.1016/j.ymgme.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh S., Padovani D., Leslie R. A., Chiku T., Banerjee R. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 2009;284:22457–22466. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budde M. W., Roth M. B. The response of Caenorhabditis elegans to hydrogen sulfide and hydrogen cyanide. Genetics. 2011;189:521–532. doi: 10.1534/genetics.111.129841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.