Abstract
Background
In Thomas' formalism for modeling gene regulatory networks (GRNs), branching time, where a state can have more than one possible future, plays a prominent role. By representing a certain degree of unpredictability, branching time can model several important phenomena, such as (a) asynchrony, (b) incompletely specified behavior, and (c) interaction with the environment. Introducing more than one possible future for a state, however, creates a difficulty for ordinary simulators, because infinitely many paths may appear, limiting ordinary simulators to statistical conclusions. Model checkers for branching time, by contrast, are able to prove properties in the presence of infinitely many paths.
Results
We have developed Antelope ("Analysis of Networks through TEmporal-LOgic sPEcifications", http://turing.iimas.unam.mx:8080/AntelopeWEB/), a model checker for analyzing and constructing Boolean GRNs. Currently, software systems for Boolean GRNs use branching time almost exclusively for asynchrony. Antelope, by contrast, also uses branching time for incompletely specified behavior and environment interaction. We show the usefulness of modeling these two phenomena in the development of a Boolean GRN of the Arabidopsis thaliana root stem cell niche.
There are two obstacles to a direct approach when applying model checking to Boolean GRN analysis. First, ordinary model checkers normally only verify whether or not a given set of model states has a given property. In comparison, a model checker for Boolean GRNs is preferable if it reports the set of states having a desired property. Second, for efficiency, the expressiveness of many model checkers is limited, resulting in the inability to express some interesting properties of Boolean GRNs.
Antelope tries to overcome these two drawbacks: Apart from reporting the set of all states having a given property, our model checker can express, at the expense of efficiency, some properties that ordinary model checkers (e.g., NuSMV) cannot. This additional expressiveness is achieved by employing a logic extending the standard Computation-Tree Logic (CTL) with hybrid-logic operators.
Conclusions
We illustrate the advantages of Antelope when (a) modeling incomplete networks and environment interaction, (b) exhibiting the set of all states having a given property, and (c) representing Boolean GRN properties with hybrid CTL.
Background
Gene regulatory network models
A major challenge in current biology is relating spatio-temporal gene expression patterns to phenotypic traits of an organism. These patterns result partly from complex regulatory interactions sustained principally by genes and encoded proteins. The complexity of such interactions exceeds the human capacity for analysis. Thus, mathematical and computational models of gene regulatory networks (GRNs) are indispensable tools for tackling the problem of mapping the genotype into the phenotype. These models have been fruitfully applied in numerous biological systems (e.g., [1-4]).
Within the various kinds of GRN models [5], Boolean GRNs are especially valuable for their simplicity and for nonetheless having a rich behavior yielding meaningful biological information [6,7]. Examples where Boolean GRNs have been successfully used are: the segment polarity gene network of Drosophila melanogaster [4,8], the flower organ determination GRN of Arabidopsis thaliana [9], the mammalian cell cycle [10], and the yeast cell cycle [11,12].
In a Boolean GRN, each gene has only two possible activation values: active (1) or inactive (0); intermediate expression levels are neglected. A network state at time t is a vector containing the activation values of all the genes in the GRN at time t. In addition, time is viewed as proceeding in discrete steps. The value of every gene X at time t + 1 is specified by a Boolean function of the values of its regulators at time t.
Branching time
Boolean GRNs are closely related to the formalism developed by Thomas and his collaborators [13-15]. Thus, computer systems for Boolean GRNs are often influenced by Thomas' formalism, which employs GRN models with branching time, allowing states with more than one immediate future [[13], p. 33]. A network state with more than one immediate future represents the fact that the next state of the regulatory system modeled by such a GRN can be any one of several states. Hence, the next state of the modeled system is only partially determined. Let us then say that there is an indetermination in the network. This indetermination in the system's behavior reflects a certain degree of unpredictability that can be identified with several important phenomena.
Asynchrony
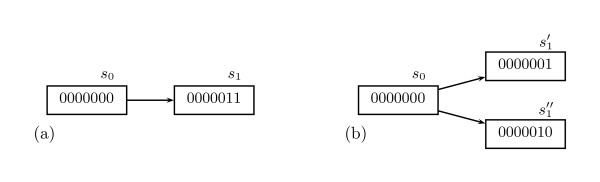
One such phenomenon is asynchrony [[13], p. 33]. Experiments for inferring gene interaction do not normally establish the length of time between state changes. Hence, when such experiments indicate the change in value of two genes, say, it is preferable to model such a situation with a single state having two successors, one for each change, as illustrated in Figure 1. The reasons are that we do not know the relative values of both delays in real biological systems [[13], p. 44] and that complete synchrony might be practically impossible [[13], pp. 33, 55].
Figure 1.
A fragment of the state-transition graph of a Boolean GRN exemplifying asynchrony. Assume that the behavior of a network specifies a simultaneous transition of the value of the two rightmost genes from 0 to 1 (panel (a)). If we exclude the possibility of simultaneous changes, it might be more realistic to model such a phenomenon with an indetermination (panel (b)).
Many computer systems based on, or inspired by, Thomas' formalism (such as BooleanNet [16], BoolNet [17], GINsim [18-20], GNBox [21,22], SMBioNet [23,24], and SQUAD [25-27]) employ asynchronous models. Thomas' formalism, however, incorporates two additional phenomena with indeterminations, that are typically excluded in such systems.
Incompletely specified behavior
One such additional phenomenon is incompletely specified behavior [[13], p. 24]. This behavior may emerge, first, from a "synthetic" approach [[13], pp. 60-67], where we are interested in all Boolean GRNs having certain properties (e.g., a certain set of steady states) regardless of other properties. The tables specifying the network behavior would then have outputs whose value "does not matter" [[13], p. 24]. Second, lack of some of the experimental information of a regulatory system also emerges as incompletely specified behavior. In this case, the behavior tables would have outputs whose value we do not know.
Interaction with the environment
Another phenomenon usually neglected in computer systems for GRN analysis and that can be modeled with branching time is that of interaction with the environment. Assume that the next state of a regulatory system depends on the temperature: If the temperature is low, the system's next state will be one, but if the temperature is high, the system's next state will be different. Another example is the unpredictability of radiation-induced apoptosis [28]. In this case, for the same degree of radiation some cells will initiate apoptosis while others will not. Thomas and D'Ari reflect such an unpredictability with an "input variable" [[13], pp. 33-35] of an unknown value. This phenomenon can be readily incorporated with indeterminations.
Simulators
Boolean GRNs are sometimes studied with simulators (e.g., Atalia [9], BooleanNet [16], and BoolNet [17]). A simulator attempts to replicate the behavior of a system by performing state changes in the same order as they occur in the system being modeled. Hence, network paths are traversed forward from one state to the next. In the presence of a state with more than one successor, such a straightforward approach must be complemented with additional mechanisms. Two of such mechanisms are: (a) a random device (randomly selecting one successor) and (b) backtracking (systematically selecting one successor after another by remembering which successors of each state have already been selected) coupled with a cycle-detection mechanism.
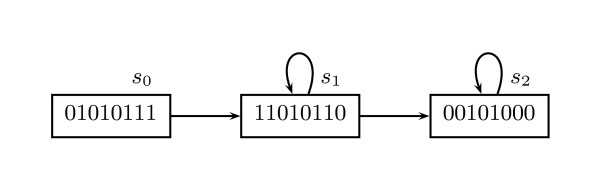
A random device, on the one hand, allows for only drawing statistical conclusions. The reason is that in the presence of a state with more than one successor, the number of paths may be infinite [6], as depicted in Figure 2. Backtracking and cycle detection, on the other hand, can be inefficient (taking, in the worst case, an exponential amount of time in the size of the network [[29], p. 82]).
Figure 2.
A fragment of the state-transition graph of a Boolean GRN showing the appearance of infinitely many paths. Infinitely many paths appear in this Boolean GRN because of one state (s1) having more than one future and occurring in a cycle. Some paths are: (s0s1s2 ...), (s0s1s1s2 ...), (s0s1s1s1s2 ...), ... A simulator using a random device traverses the model forward, state by state, following a single path of the state graph, limiting the use of such a tool to drawing only statistical conclusions about all paths in models such as this one. Model checkers, by contrast, can prove precise properties, even in the presence of infinitely many paths resulting from states having more than one future.
There are two important approaches for circumventing these difficulties. One of these techniques is an elaboration of backtracking so as to increase its efficiency by requiring certain constraints to be satisfied as the network is traversed [30]. The work by Corblin et al. [21,22] uses this approach. Another relevant method is model checking.
Model checking
Model checking [31,32] is a collection of techniques for automatically verifying properties especially of discrete systems. The main ideas of model checking appeared 30 years ago [31,32]. At present, numerous model-checking tools exist. Model checking is routinely used, mainly for hardware verification, but also for software verification [33], and was distinguished with the A. M. Turing award in 2007. Model checking has been advocated for analyzing biological systems with increasing interest [6,24,34-43].
A model checker normally has as input (1) a "Kripke structure" representing a discrete system (comprising a finite number of states), (2) a distinguished "initial" state (or set of states) in the Kripke structure, and (3) a "temporal-logic" formula expressing a desirable property, that may or may not hold (i.e., be true) at a state. The output of the model checker is either a confirmation or a denial that the formula holds at the initial state(s) (given by the user as part of the input).
In a Kripke structure time is branching, so that there may be more than one possible future of a given state. The introduction of branching time may produce infinitely many forward traversals (see Figure 2). Model checkers, however, unlike simulators randomly selecting a successor state, can systematically analyze such infinitely many possibilities [6]. Intuitively, this is often done by traversing the Kripke structure in reverse and accumulating the set of all states at which a subformula holds. Model checking amounts, thus, to performing an exhaustive search (in the presence of branching time). Such a search plays the role of a mathematical proof establishing a property for infinitely many paths.
Programming vs. formula writing
By being based on properties formalized in temporal logic, model checkers have another advantage over simulators. The decision of whether or not a state satisfies a property of interest is programmed in the simulator itself. Therefore, if an unforeseen property appears during the usage of a Boolean GRN simulator, such a property must be incorporated in the simulator by modifying program code. This renders simulators rigid: either the user's needs are anticipated or reprogramming must be done.
Compared with simulators, model checkers exhibit the benefit of having replaced programming with temporal-logic formula writing. Instead of having to modify the computer program of a simulator, many new queries can be dealt with by writing new temporal-logic formulas (as long as the queries can be expressed in the selected logic), which (unlike large programs and their modifications) are concise and self-contained.
Organization of this paper
In the Implementation section, we first illustrate both Computation-Tree Logic (CTL) [31] and its hybrid extension, Hybrid CTL (which we based on [44,45]), chosen to be able to express interesting properties for Boolean GRN analysis and construction. The term "hybrid" here means a combination of propositional modal logic with classical predicate logic, and should not to be confused with hybrid model checking, combining discrete with continuous variables. The Implementation section subsequently covers the model-checking algorithms and some implementation details. Next we show, in the Results section, the use of the Antelope model checker in the presence of indeterminations either caused by environment interaction or by an incompletely specified behavior. Finally, the Discussion section reviews other similar software systems, compares Antelope with such systems, and outlines features planned for the future.
Implementation
This section first covers the temporal logics used by Antelope. After explaining CTL, we turn our attention to its hybrid extension. Next, we cover the model-checking algorithms, as well as additional implementation issues.
Computation-Tree Logic
We now give a short account of CTL and refer the reader to additional file 1 of this paper for a gentle introduction and to additional file 2 for a formal definition of CTL. More thorough treatments can be found in [46-50].
Boolean and temporal operators
Formulas in CTL can have Boolean operators, such as not and or. In addition, such formulas can have "temporal operators", allowing us to refer to formulas holding in the future of a particular state. In this case, we must indicate whether we mean some future or all futures. Hence, it is possible to refer either (1) to some path starting in the present with the "modality" E, or (2) to all paths starting in the present with the modality A. Similarly, it is possible to refer (a) to the immediate future with the modality X, (b) to any state in the present or any point in the future with the modality F, or (c) to all states in the present and in the future with the modality G. Table 1 summarizes these modalities.
Table 1.
CTL modalities
| modality | meaning |
|---|---|
| E | some path (i.e., there Exists a path) |
| A | All paths |
| X | neXt state (i.e., immediate future) |
| F | any state either in the present or in the Future |
| G | all states in the present and in the future (Global) |
A temporal operator is composed of a modality in the upper part together with a modality in the lower part of this table, which results in six temporal operators. (Often more temporal operators are included in CTL [49].) For example, a formula asserting that there exists a path such that in the present or in the future g0 does not hold (i.e., g0 is inactive) and g1 does hold (i.e., g1 is active) would be: "EF((not g0) and g1)". Hence, assuming that there is a single state s in which g0 does not hold and g1 does hold, this formula can be used to obtain the basin of attraction of such a state, with a model checker computing all states at which a given formula holds. The formula "AX ((not g0) and g1)" holds at all states from which it is necessary to reach s in one step, i.e., states which have s as their only next state. The formula "EX ((not g0) and g1)" holds at all states from which it is possible to reach s in one step, i.e., states which have s as a next state (and possibly other next states because of indeterminations). Other CTL formulas can characterize, for instance, whether or not it is necessary to go through a state s1 to reach another state s2. See [51] for a list of CTL formulas specifying various biological properties.
Some properties not expressible in CTL
There do not exist, however, CTL formulas for characterizing steady states (i.e., a formula holding exactly at the set of all steady states of an arbitrary Boolean GRN) [51], or oscillations. This motivates the use of a more expressive logic than CTL. Antelope provides a "hybrid" extension of CTL.
Hybrid Computation-Tree Logic
This subsection is devoted to Hybrid CTL. We refer the reader to additional file 1 of this paper for a gentle introduction and to additional file 2 for a formal definition of Hybrid CTL. Deeper treatments of hybrid logics are in [52,53].
State variables
The main idea behind the hybrid extension of a temporal logic consists in the addition of variables allowing us to refer to states (i.e., state variables). The downarrow binder "↓σ" sets the state variable σ to the current state of evaluation. The formula "↓σ.AX σ", for example, characterizes the set of states which have themselves as their only next state. Hence, Hybrid CTL allows us to characterize the set of steady states. Moreover, by employing branching time, we are able to distinguish between two kinds of steady state. When a state has only one transition from and to itself, following Thomas and D'Ari [13], we will call it a stable steady state. When a state has, in addition to a self-loop, other transitions going to other states, following [13], we will call it an unstable steady state (named "stationary" state in [51]). Hybrid CTL formulas for calculating both these sets of states are: "↓σ.AX σ", for the set of stable steady states, and "↓σ.EXσ", for the union of the sets of stable and unstable steady states.
Other formulas
Attractors of various sizes and oscillations
The notion of a steady state can be generalized in an attractor, possibly involving more than one state. A steady state would then be a one-state attractor. A formula characterizing attractors of any size would be: "↓σ.EX EF σ".
Another interesting formula would be "↓σ.EX((not σ) and EX σ)", which holds at states belonging to a size-two attractor. Oscillations, where a gene is alternatively active and inactive, can also be characterized in Hybrid CTL: Additional file 1 explains a formula for the basin of attraction of possible oscillations. We refer the reader to the Antelope web site http://turing.iimas.unam.mx:8080/AntelopeWEB/ for more formulas.
Algorithms
CTL
Antelope uses a standard "labeling" algorithm [46] for ordinary CTL formulas. Labeling algorithms for model checking are so called because we can think of each state as being labeled with the subformulas holding at that state.
Say that the formula given by the user is φ. The labeling algorithm starts by considering the simplest subformulas of φ, that is, the names of the genes. For each gene g, labeling all states at which the formula "g" holds is easy, as that information is already present in the Kripke structure.
Next, the labeling algorithm proceeds to more complex subformulas, until φ is reached, by treating the operator of each such subformula by cases. For instance, if the subformula is of the form "ψ1 and ψ2", then the labeling algorithm computes the set of states at which such a subformula holds as the intersection of the set of states at which ψ1 holds with the set of states at which ψ2 holds. All Boolean operators can be treated by combining set operations, like union, intersection, and set difference.
The labeling algorithm treats some temporal operators, such as AX, by using equivalences. For example, "AX ψ" is equivalent to "not EX not ψ". The rest of the temporal operators, however, must be dealt with explicitly. For all such primitive operators the labeling algorithm traverses the Kripke structure in reverse. Take for instance "EX ψ", which holds if there Exists a neXt state at which ψ holds. Given the set of states at which ψ holds, the labeling algorithm treats "EX ψ" by obtaining all states which have an immediate successor in such a set, i.e., all the predecessors of the states in such a set. The labeling algorithm processes operators such as EG by repetitively traversing the Kripke structure in reverse.
Hybrid CTL
The labeling algorithm is efficient (taking polynomial time in the size of the Kripke structure). The additional expressiveness of hybrid operators, such as "↓" comes at a price, however. Given a CTL formula φ, the computation of the set of states at which a formula of the form "↓σ.φ" holds involves calling the labeling algorithm with φ once for each state. The decrease in efficiency is even more if the "↓" operator appears nested. Antelope, however, treats certain patterns in special ways, requiring less time than a direct approach.
More implementation issues
Antelope is a symbolic model checker [54], representing state sets by Reduced, Ordered Binary-Decision Diagrams (BDDs) [55]. (In particular, Antelope employs JavaBDD [56], which in turn uses BuDDy [57].)
Representation of a set of states
A BDD is a representation of a Boolean function. Thus, to use a BDD for representing a set of states in a Kripke structure we must view such a set as a Boolean function. This is possible if each row of the truth table of the Boolean function corresponds to an element which may or may not belong to such a set. The value of such a function will be 1 at exactly those states belonging to the set.
Representation of a set of transitions
In addition to representing sets of states, BDDs are used for representing the set of transitions of Kripke structures. In this case, the Boolean function has twice as many variables as there are genes. The reason is that each transition (corresponding to a row in the truth table of such a function) has both a source and a terminating state. BDDs are often surprisingly concise, allowing the verification of many large Kripke structures, with more than 1020 states [54]. We refer the reader to [49] for a detailed description of BDDs and their use in symbolic model checking.
Optimizations
Apart from the use of BDDs, Antelope has several "optimizations" (i.e., special treatment of particular patterns so as to increase the efficiency). For example, a straightforward formula characterizing the states with more than one successor has the pattern "↓σ.EX ↓τ.φ". If evaluated as described in the Algorithms section, this formula would call the labeling algorithm a number of times proportional to the square of the number of states (O(|S|2), where |S| is the number of states). To find the set of states with more than one successor, however, it is not necessary to visit all states for each state of the Kripke structure. It suffices to be able to enumerate the successors of each state. Antelope treats the formula for characterizing the states with more than one successor as a special case so that the CTL model-checking algorithm is called with φ as input a number of times linear in the size of the Kripke structure (O(|S| + |R|), where |R| is the number of transitions).
Another optimization is that of the operator EY (for "Exists Yesterday"), which is the converse of EX. Although this operator need not be primitive, Antelope does treat it as primitive by simply traversing the transitions forward. This operator allows the user to view Antelope as a kind of simulator.
Additional file 3 has a table comparing the verification times for a few models with respect to some properties of increasing complexity.
Input formats
Antelope accepts two formats for describing the Boolean GRN: tables and equations. In both cases, the values of a gene (at the current time step) are specified as a Boolean relation which depends on the values of (some) genes (at the previous time step). A table can be viewed as an extension of an ordinary truth table, where stars are allowed on the right-hand side, denoting indeterminations. Sometimes, however, it may be more convenient to use a logical formula instead of a truth table. Hence, Antelope accepts equations, each of which is of the form:
where the left-hand side represents the value of the gene X at the current time step, and the right-hand side is an arbitrary Boolean function (defined employing the usual Boolean operators, such as conjunction, disjunction, or negation) on the values of genes at the previous time step. To be able to represent indeterminations, we need two equations with the same left-hand side. We refer the reader to the Antelope user's manual, which appears in additional file 4, and in the URL http://turing.iimas.unam.mx:8080/AntelopeWEB/.
Results
We now exemplify the use of Antelope for analyzing Boolean variants of the A. thaliana root stem cell niche GRN. Stem cells or initials are undifferentiated cells from which particular cell types of the organisms are generated; the microenvironment in which stem cells are located is called the stem cell niche.
Anatomically, stem cell niches are conformed by two different cell types, the stem cells themselves, and another cell or group of cells sometimes generically called organizer cells [58]. The organizer cells maintain the stem cells in the undifferentiated state through short-range signals. Understanding how the different cells conforming stem cell niches are specified, as well as how the balance between cell division and cell differentiation is maintained in the niches, is central for understanding the development, growth and regeneration processes occurring in plants and animals. In particular, plant stem cell niches constitute valuable model systems for studying regenerative and plastic developmental processes, as these organisms grow new organs and structures throughout their life [58,59].
We focus on the root stem cell niche of A. thaliana, that is located near the root tip and is well characterized at the anatomical and molecular level (see the recent review in [60]). This niche is conformed by the so-called quiescent center (QC), which is in turn conformed by the organizer cells of the root SCN, and is surrounded by four different stem cell types [59]. Each of these four types of stem cell will give rise to a different cell lineage: vascular, cortex/endodermal, epidermal, and columella/root-cap cells. However, in this contribution two of the stem cell types (epidermal and root-cap cells) are considered as only one since the available experimental evidence is not enough to distinguish between them at the gene expression level (see more details in [61]), leaving only four types of initial cells (QC, vascular, cortex/endodermal (CEI), and epidermal/root-cap (CEpI) initials).
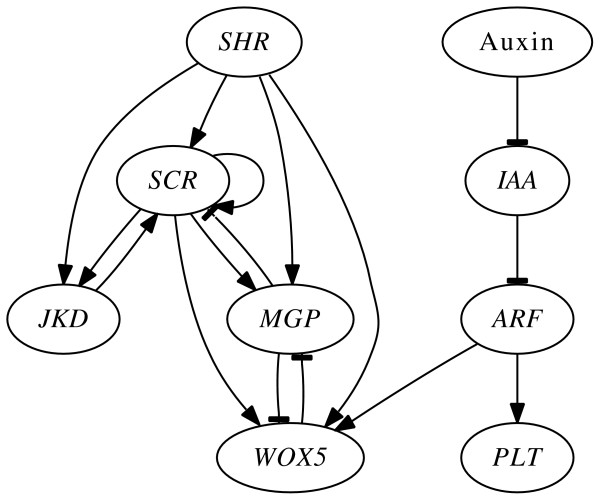
Besides being thoroughly characterized at the anatomical level, the root stem cell niche of A. thaliana has been relatively well described from a molecular and genetic perspective. Indeed, some of the molecular components that are necessary to establish and maintain the root SCN cellular patterning have been recently uncovered. Among these components are the genes SHORT-ROOT (SHR) and its target gene SCARECROW (SCR), the immediately downstream genes of the dimer SHR/SCR, and other genes that interact with them. Another set of relevant genes includes the PLETHORA (PLT) genes, which have been proposed to be key components of the molecular readout of the plant hormone auxin. Finally, the QC specific gene WUSCHEL RELATED HOMEOBOX5 (WOX5) is fundamental for root SCN organization [60,62-64]; see the graphical representation of the interactions between these genes in Figure 3. Moreover, the expression patterns of these genes and the localization of their corresponding proteins have been described. Thus, it is possible to postulate a gene expression profile that characterizes each of the SCN cell types mentioned above according to the Table 2.
Figure 3.
The interaction diagram of the GRN underlying cell type determination in the root stem cell niche of the model plant A. thaliana. The abbreviated names of the genes are inside ellipses and the edges correspond to the regulatory interactions. Auxin is a morphogene. The genes are: Auxin/INDOLE-3-ACETIC ACID (Aux/IAA), AUXIN RESPONSE FACTOR (ARF), JACKDAW (JKD), MAGPIE (MGP), PLETHORA (PLT), SCARECROW (SCR), SHORTROOT (SHR), and WUSCHEL-RELATED HOMEBOX5 (WOX5). Ordinary arrow heads denote activation; T-bar arrow heads denote inhibition.
Table 2.
Expected expression profiles for the cells conforming the A. thaliana root stem cell niche
| Cell type | PLT | Auxin | ARF | Aux/IAA | SHR | SCR | JKD | MGP | WOX5 |
|---|---|---|---|---|---|---|---|---|---|
| QC | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| Vascular | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| CEI | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| CepI | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
In order to define the rules for a Boolean GRN model for this system, we considered all the genes that have been reported to play a relevant role in the specification of the root stem cells and gathered the available experimental information for the regulation of their expression [60,61]; see Figure 3. These data included mostly molecular genetics experiments, such as experiments with plants containing a mutant allele of a gene. The resulting rules can be summarized in the following logical statements (uploaded in Antelope under the name 'Root gene regulatory network''):
// SHR; without regulators
// Auxin; without regulators
PLT: = ARF;
AUXINS: = AUXINS;
IAA: = ~ AUXINS;
ARF: = ~ IAA;
SHR: = SHR;
SCR: = SHR & SCR & (JKD | ~MGP);
JKD: = SHR & SCR;
MGP: = SHR & SCR &~ WOX;
WOX: = ARF & SHR & SCR & (~ MGP | WOX);
As has been proposed for other systems (e.g., [1,9]), we expected the stable steady states of our GRN model to correspond to the gene expression profiles characterizing the different stem cells within the root niche of A. thaliana (table above). Thus, from our knowledge of the system, we expected four stable steady states. The expected steady states are indeed obtained after postulating a mutual negative interaction between WOX5 and MGP, which gives rise to a new testable prediction [61].
Using this GRN model, we first illustrate the use of indeterminations representing incomplete experimental data. Next, we use indeterminations for modeling the influence of unpredictable external signals.
Experimental gap
Steady states and SCARECROW
While developing the truth tables for this GRN, we detected an experimental gap. We know that SCARECROW (SCR), a target gene of the dimer SHORTROOT (SHR)/SCR [62,63], either loses or diminishes its own expression in the JACKDAW single mutant (jkd) in the stem cell niche [64]. The same is true for SCR-dependent quiescent-center marker QC25 [65]. The MAGPIE mutant (mgp), by contrast, has no visible phenotype. Finally, the mgp jkd double mutant recovers the SCR expression [64] (but see [66] for different results).
Based on this information, we established the truth table for SCR, which appears in Table 3. Observe the indetermination, reflecting the fact that activity could or could not be lost in a jkd background. Antelope produced three stable steady states, but four unstable steady states (see the Hybrid Computation-Tree Logic subsection for definitions of stable and unstable steady states). Hence, removing the indetermination in the above table may recover the four expected stable steady states. We performed the jkd loss-of-function simulation in our models to distinguish which of the two possibilities (i.e., no SCR transcription in jkd or SCR transcription in jkd) recovered the expected states. Interestingly, following the GRN state transitions backwards, using the EX operator, we noted that if SCR is unable to be expressed in jkd, then neither the WUSCHEL-RELATED HOMEBOX5 (WOX5) (another quiescent-center marker, dependent on SCR [60]) expression nor the SCR expression disappeared at the quiescent-center.
Table 3.
Truth table for SCR
| SHR | SCR | JKD | MGP | SCR' |
| 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 1 | 0 |
| 0 | 0 | 1 | 0 | 0 |
| 0 | 0 | 1 | 1 | 0 |
| 0 | 1 | 0 | 0 | 0 |
| 0 | 1 | 0 | 1 | 0 |
| 0 | 1 | 1 | 0 | 0 |
| 0 | 1 | 1 | 1 | 0 |
| 1 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 1 | 0 |
| 1 | 0 | 1 | 0 | 0 |
| 1 | 0 | 1 | 1 | 0 |
| 1 | 1 | 0 | 0 | 1 |
| 1 | 1 | 0 | 1 | * |
| 1 | 1 | 1 | 0 | 1 |
| 1 | 1 | 1 | 1 | 1 |
Furthermore, our jkd mutant does cause a loss of the cortex-endodermis initials attractor, contrary to what is observed in experimental jkd mutants [64], suggesting that jkd only diminishes SCR expression. Again, following the GRN transitions backwards for the case in which jkd loss-of-function does not lose SCR expression, we found that the system was able to recover the jkd loss-of-function mutant. Based on the result found with the system including indeterminations, we replaced the star by a 1 in the table for SCR. Once the indetermination was so removed, we obtained four stable steady states.
External signals
FAS and SCR
Let us now exemplify Antelope as used for modeling the effect of external signals that affect one or more GRN nodes. The root stem cell niche of A. thaliana is affected by several external signals, such as genes and molecules from modules involved in other processes in the organism. For example, Kaya and collaborators [67] reported that FASCIATA1 (FAS1) and FASCIATA2 (FAS2), hereafter collectively called FAS, affect SCR expression. In the fas mutant, SCR expression is deregulated and can be either expressed or not expressed in almost any cell of the root stem cell niche. Similarly, Inagaki and collaborators [68] reported the TECHBI (TEB) mutants also affecting SCR expression. Again, when TEB is mutated, SCR may or may not be expressed through the endodermal layer, the cortex-endodermis initial cells, and the quiescent center.
We incorporated FAS by adding a variable FAS to the truth table for SCR. For FAS = 1, the truth table obtained in the "Experimental gap" subsection was used. For FAS = 0, by contrast, all the right-hand sides of the new truth table had indeterminations. In the case of TEB, we only used indeterminations for the right-hand side of the SCR table where the output was 1 for the teb mutant. We found that under these conditions the original four attractors were preserved in both cases. We also found that in the fas mutant, SCR could be expressed in any of the four original attractors, while in the teb mutant SCR could or could not be expressed either in the quiescent center or in the cortex-endodermis attractor. It is worth noting that in both cases the basins of attraction changed. For instance, consider the states that without any indetermination originally led to the cortex-endodermis attractor. Such states could now lead to vascular initials due to SCR indeterminations, as expected given the experimental evidence. It is also important to note that even though SCR expression is clearly affected in real roots, cells may not switch among cell types. However, the results derived from modeling the GRN using Antelope are consistent with data currently available and demonstrate the utility of this tool when we deal with networks in which the truth tables for some genes are not completely known. Figures 4 and 5 show screenshots of this analysis.
Figure 4.
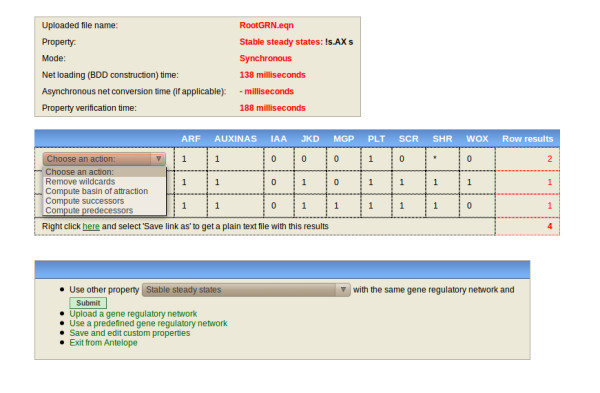
Screenshot of Antelope showing the stable steady states for the stem cell niche GRN without indeterminations. The upper frame displays the name of the file being analyzed, the analysis performed (with the Hybrid CTL formula), and the mode by which the property was checked (synchronous or asynchronous). The middle frame displays the analysis results. The bottom frame displays new actions that can be done. The stable steady states correspond to the root SCN cellular types.
Figure 5.
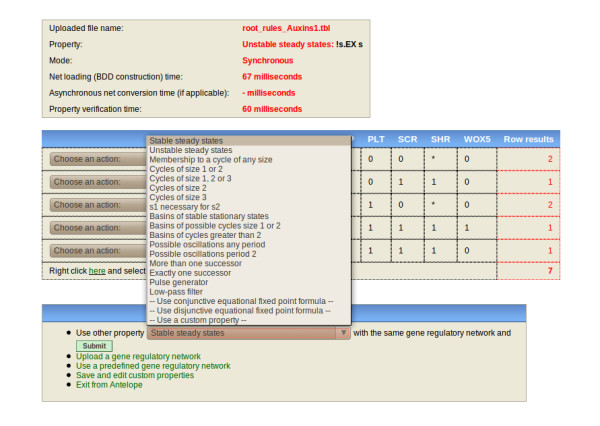
Screenshot of Antelope showing the results for unstable steady state search for the stem cell niche GRN with an indetermination in the SCR logical rule. This indetermination represents a mutation in FAS. As observed, SCR activity is present even in the absence of SHR, which is indispensable for SCR activity. It is important to note that some of the changes can only be observed analyzing the basins of attraction. For instance, the steady states for QC and CEI (two different cell types) are not possible without SCR presence; hence, a change of one steady state for another is only observable through their basins of attraction.
Other properties
These two analyses were based on indeterminations, stable and unstable steady states, and basins of attraction of such states. When designing and analyzing larger GRNs, more complex state attributes, such as global properties or conditional reachability may be useful.
For example, all the states occurring in either one-state or two-state attractors (which may be either stable or unstable) satisfy the formula "↓σ.EX EX σ". The formula "EF(↓σ.EX EX σ)", in turn, can be used to calculate the basins of attraction of all such attractors. Hence, the formula "not(EF (↓σ.EX EX σ))" would characterize the complement of all such basins of attraction. This is equivalent to the set of all states in the basins of attraction of attractors with more than two states. Similarly, the set of states occurring in exactly two-state attractors can be calculated with the formula "↓σ.EX ((not σ) and EX σ)". These global properties cannot be expressed by CTL formulas.
Conditional reachability can be expressed with the EU operator (for "Exists Until"), a generalization of EF. Whereas "EFs2" holds at all states from which it is possible to reach state s2, "E[φ U s2]" holds at all states from which it is possible to reach s2 by going only through states at which the formula φ holds. For instance, the set of states from which it is possible to reach s2 without going through s1 corresponds to the formula "E[(not s1) U s2]". By contrast, the set of states from which it is possible to reach s2 only by going through s1 at least once is the complement of the previous set of states with respect to the basin of attraction of s2: "not(E[(not s1) U s2]) and EF s2". In formulas having such schemata, we would need to name states. Such a naming is possible in CTL by identifying a state with the conjunction of its nonnegated active genes and its negated inactive genes. Antelope, by contrast, provides more concise ways of referring to a state, with a number which, if written in binary, follows the lexicographic order of the names of the genes. We refer the reader to the Antelope user's manual and site.
Discussion
Other related systems
We now describe other systems relevant for us. For brevity, we have to exclude certain works: First, we leave out Boolean GRN simulators, such as Atalia [9], BooleanNet [16], and BoolNet [17]. Second, we omit research based on structures other than Kripke structures; examples are: a work utilizing the LTL (Linear-time Temporal Logic) model checker of the Maude system [34], works using reactive modules with the Mocha model checker [42,43], and those employing probabilistic model checking with PRISM [35,37,38]. We start with systems based on Thomas' formalism and proceed with systems using continuous approaches.
GNBox
GNBox [21,22] applies constraint logic programming techniques [30] to Thomas' formalism [13]. Such a formalism establishes a search space resulting from states possibly having more than one successor. A straightforward implementation of a logic programming language (without constraints) typically traverses a search space following a depth-first, top-down discipline in the same way as an ordinary simulator. Unlike a simulator employing a random device, however, such an implementation utilizes backtracking. Observe that a depth-first, top-down discipline together with backtracking can take an exponential amount of time in the size of the model [[29], p. 82]. Constraint logic programming languages, nevertheless, use constraints to efficiently traverse the search space. In particular, GNBox expresses constraints as a Boolean satisfiability (SAT) problem that is turned over to a dedicated SAT solver. This approach is able to model many possible GRNs, thereby pruning the search space and eliminating the need for performing numerous simulations. By expressing desired properties as constraints, GNBox can find parameter values of GRNs represented in Thomas' framework.
GINsim
GINsim [18-20] also uses a variant of Thomas' formalism. As in such a formalism, networks in GINsim have indeterminations representing asynchrony. GINsim computes the state transition graph of the GRN (presumably with forward traversal together with backtracking because of the indeterminations) before proceeding to analyze a trajectory selected by the user. GINsim can also classify circuits in the interaction diagram (i.e., can identify "functional" circuits) and can compute the set of all (stable) steady states of GRNs which do not have indeterminations using MDDs, a multi-value generalization of BDDs. Finally, GINsim can find the strongly connected components of the state-transition graph or the interaction graph.
SMBioNet and Mateus et al.'s system
SMBioNet [23,24] employs a variant of Thomas' formalism as well. The input is an interaction diagram of the GRN under study, together with desired properties expressed as CTL formulas. The output is a set of all the models conforming to the given interaction diagram and which also satisfy the given formulas. Candidate models are generated by instantiating parameters and then tested with a model checker.
Another system also using both Thomas' formalism and temporal logic is that by Mateus et al. [39]. Inequalities over the parameters of the model are obtained from the interaction diagram. These inequalities are augmented with LTL formulas specifying desirable properties of the model. The model is traversed forward and paths that do not satisfy the constraints are eliminated, so that only paths satisfying the constraints are retained.
SQUAD
SQUAD [25-27] combines a continuous model, employing ordinary differential equations, with a Boolean model of the network. The user provides the interaction diagram of the network, from which SQUAD obtains a continuous model. To find steady states of the continuous model, SQUAD first converts such a model into an approximate Boolean asynchronous model. (Thomas' formalism is not used because such a formalism has "proved to scale badly for large networks" [26].) In the Boolean model, SQUAD then computes, using BDDs and a random device, the set of states probably belonging to attractors of any size and occurring in attractors without indeterminations (called "steady" states in [26,27]). Next, SQUAD repetitively uses such states as initial states in a continuous simulator to search for steady states in the continuous model. Perturbations may be introduced to confirm that such steady states are stable and to identify the effect of specific genes.
GNA
GNA [40,69-72] is based on piecewise-linear differential equations. Unlike other systems using this formalism, the user need not specify precise values of parameters. Instead, less precise intervals are employed. States are qualitative and represent ranges of concentrations of proteins, so that simulations are also qualitative. In addition, GNA computes a discrete abstraction [73] of the continuous model, that can be verified with standard model checkers (NuSMV and CADP). The user in this case can express simple properties in CTL. For more complex properties, the GNA group has developed its own logic, called Computation Tree Regular Logic [74]. This logic extends CTL with regular expressions and fairness operators, allowing the expression of properties such as multistability and oscillations. Finally, GNA has a formula editor, guiding the user in writing new formulas.
BIOCHAM
BIOCHAM [41] can analyze and simulate biochemical networks using Boolean, kinetic, and stochastic models. In addition, properties can be formalized in temporal logic (CTL or LTL with numerical constraints), so that a model checker can be used to validate such properties. BIOCHAM models a network of protein interactions as a set of biochemical reaction rules, such as A+B = > C. Indeterminations appear because such a rule, for instance, is translated into four transitions going out of the same state, resulting from the four combinations of either reactant A or reactant B being completely or incompletely consumed. In addition, BIOCHAM has a model-update module, repairing models that do not satisfy the formalized properties.
Comparison and planned features
On the one hand, compared with systems employing constraints, Antelope, by using BDDs, can compute large sets of states having a certain CTL property (e.g., a basin of attraction). On the other hand, compared with simulators, in addition to this benefit, Antelope can prove assertions about infinitely many paths, as opposed to only drawing statistical conclusions. It is interesting to observe, though, that some systems built around a simulator (e.g., GINsim and SQUAD) leave the simulation technique for BDDs when calculating steady states (or approximations to such states).
We also find differences between Antelope and other systems using model checking. For instance, SMBioNet, Mateus et al.'s system, GNA, and BIOCHAM perform model checking for verification, using a model checker to confirm or deny that a certain formula is satisfied. Antelope, by comparison, employs model checking for calculating sets of states.
A first clear limitation of Antelope when compared with systems based on Thomas' formalism (GNBox, GINsim, SMBioNet, and Mateus et al.'s system) is its being restricted to Boolean genes. We thus plan to extend Antelope with multi-valued genes. In this case, it would be interesting to try to incorporate into Antelope techniques using constraints, like those of GNBox, for determining parameter values.
Currently, Antelope's GRNs are only either completely synchronous or completely asynchronous. Another improvement would then be the possibility of representing partially asynchronous GRNs, as employed in [10]. Many of the systems we reviewed allow the user to draw the GRN, whereas currently Antelope only accepts textual formats for describing the GRN. Clearly, future versions of Antelope should also have such drawing capabilities. In addition, GNA, for instance, has a formula editor, which would be desirable in Antelope as well. By contrast, Antelope is a web application, requiring no installation of any local software from the user other than a standard web browser. Moreover, Antelope can also run locally, exhibiting advantages of both web and local applications.
We can mention two further additions requiring more substantial work. BIOCHAM has an update module, repairing faulty models. A similar update module would also enhance Antelope's features.
Another improvement, as with any model checker, would be the addition of more powerful methods for approaching the state-explosion problem. Currently, Antelope only has BDDs for representing large sets of states, but new techniques, such as CEGAR (Counterexample-guided abstraction refinement) [75] would enable Antelope to deal with larger GRNs.
Conclusions
Systems for analyzing and building Boolean GRNs employ branching time almost exclusively for representing asynchronous transitions. Thomas' work, however, represents two other important phenomena with branching time, namely incomplete specifications and environment interaction. A consequence of including these two other kinds of indetermination is that unstable steady states may appear. We have shown how having both stable and unstable steady states is useful for developing Boolean GRNs.
In addition, we reviewed and extended the advantages of model checking, as compared with simulation, in the presence of indeterminations. In particular, we observed that model checkers, unlike simulators randomly selecting a successor, can prove properties of a set of infinitely many paths. Another advantage we reviewed is that of handling new, unforeseen properties: While model checkers can often represent new properties with additional temporal-logic formulas, simulators require the incorporation of such properties in their program code.
We illustrated the advantages of two extensions to ordinary model checking. First, we noted that ordinary model checkers would only confirm or deny that all the states in a given set of states have a certain property. By contrast, we claimed that model checkers are more useful for reasoning about Boolean GRN when exhibiting the set of states that have a property of interest. Second, we observed that the logics (e.g., CTL and LTL) underlying many model checkers are not expressive enough for representing many interesting properties of Boolean GRNs. Antelope tries to overcome these two limitations by showing the set of states satisfying a given formula, and by employing a hybrid extension of CTL.
It is important to remark that model checkers for hybrid logics are both relevant and neglected. As pointed out in [76], "The implementation of model checkers for hybrid logics still remains a quite unexplored field of research". Other than Antelope, we only know of two hybrid model checkers [52,76]. These, however, employ a basic modal logic instead of CTL, and their implementations do not use BDDs. This makes Antelope the first symbolic model checker for Hybrid CTL (as far as we know) with which to experiment in the development of Boolean GRNs.
Availability and requirements
•Project name: Antelope
•Project home page: http://turing.iimas.unam.mx:8080/AntelopeWEB/
•Operating system(s): Platform independent
•Programming language: Java
•Other requirements: Any standard web browser
•License: GPL
•Any restrictions to use by non-academics: none other than those in GPL
Authors' contributions
GA did most of the web interface. JA participated in the design of, and wrote the code for, the previous version of Antelope's model checker. EA contributed to the design of the stem cell niche GRN from the literature data, used Antelope, and participated in writing the biology part of this paper, as well as the manual. MB also contributed to the design of the stem cell niche GRN from the literature data and wrote the rest of the biology part of this paper, as well as the manual. MC suggested using Hybrid CTL to overcome CTL limitations, participated in the design of Antelope, contributed to the presentation of these results, and wrote the formal definitions of CTL and Hybrid CTL (additional file 2). PG wrote the code for Antelope's model checker, connected the model checker with the web interface, embedded Antelope and Apache Tomcat in a single file, did the rest of the web interface, and added numerous features to Antelope. DAR participated in the design of Antelope and wrote the model-checking part of this paper. ERAB put forward the idea of testing Kauffman's hypothesis that Boolean GRNs can recover experimental gene expression profiles, and led the translation of actual data into the the stem cell niche GRN. All authors read and approved the final manuscript.
Supplementary Material
A gentle introduction to (Hybrid) Computation-Tree Logic. This additional file has gentle introductions to Computation-Tree Logic and Hybrid Computation-Tree Logic.
(Hybrid) Computation-Tree Logic. This additional file has formal definitions of Computation-Tree Logic and Hybrid Computation-Tree Logic.
Benchmarks. This additional file shows the execution time for several examples.
Antelope User's Manual. This additional file has the Antelope user's manual.
Contributor Information
Gustavo Arellano, Email: gustavo.arellano@metasoft.com.mx.
Julián Argil, Email: julian.argil@gmail.com.
Eugenio Azpeitia, Email: emazpeitia@gmail.com.
Mariana Benítez, Email: marianabk@gmail.com.
Miguel Carrillo, Email: miguel.mcb@gmail.com.
Pedro Góngora, Email: pedro.gongora@gmail.com.
David A Rosenblueth, Email: drosenbl@servidor.unam.mx.
Elena R Alvarez-Buylla, Email: eabuylla@gmail.com.
Acknowledgments
This paper owes much to Pablo Padilla-Longoria, who carefully read a previous version of this paper and subsequently had valuable discussions with us. We also thank Carlos Velarde, who patiently helped us with LATEX, Montserrat Alvarado, who helped us translating the first version of Antelope's manual and generating the figures, and who is in charge of turing.iimas.unam.mx, Gabriel Muñoz-Carrillo and Jorge Hernández, who helped us translating the first version of Antelope's windows and menus, Carlos Gershenson and Nathan Weinstein, who gave us useful suggestions, and Michael Dent and Michael Scott White, who proposed English changes. We gratefully acknowledge the facilities provided by the IIMAS, the Instituto de Ecología, and the Centro de Ciencias de la Complejidad. MB participated in this paper through the Centro de Ciencias de la Complejidad, by means of the "Red de Conacyt Complejidad, Ciencia y Sociedad". MB was also supported by the Czech Ministry of Education, Youth and Sports (grant LC06034). ERAB acknowledges the financial support from Conacyt grants 81433, 81542, and 90565, and PAPIIT grants IN210408, IN229009-3, and IN223607-3. DAR acknowledges the financial support from PAPIIT grant IN120509-3. Finally, we are grateful to the referees whose comments helped improve the previous version of this paper.
References
- von Dassow G, Meir E, Munro E, Odell G. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla E. A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. The Plant Cell. 2004;16:2923–2939. doi: 10.1105/tpc.104.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biology. 2006;4(10):1732–1748. doi: 10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R. In: Complex Networks. Ben-Naim E, Frauenfelder H, Toroczkai Z, editor. Springer; 2004. Boolean modeling of genetic regulatory networks; pp. 459–481. [Lecture Notes in Physics Vol. 650] [Google Scholar]
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. Journal of Computational Biology. 2002;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- Fisher J, Henzinger TA. Executable cell biology. Nature Biotechnology. 2007;25(11):1239–1249. doi: 10.1038/nbt1356. [DOI] [PubMed] [Google Scholar]
- Bornholdt S. Boolean network models of cellular regulation: prospects and limitations. J R Soc Interface. 2008;5:S85–S94. doi: 10.1098/rsif.2008.0132.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/S0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Benítez M, Corvera-Poiré A, Candor AC, de Folter S, de Buen AG, Garay-Arroyo A, García-Ponce B, Jaimes-Miranda F, Pérez-Ruiz RV, Pineiro-Nelson A, Sánchez-Corrales YE. Flower development. The Arabidopsis Book. 2010;8:e0999. doi: 10.1199/tab.0127. [Doi:10.1199/tab.0999] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré A, Naldi A, Chaouiya C, Thieffry D. Dynamical analysis of a genetic Boolean model for the control of the mammalian cell cycle. Bioinformatics. 2006;22(14):e124–e131. doi: 10.1093/bioinformatics/btl210. [DOI] [PubMed] [Google Scholar]
- Li F, Long T, Lu Y, Ouyang Q, Tang C. The yeast cell-cycle network is robustly designed. Proc Natl Acad Sci USA. 2004;101(14):4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidich MI, Bornholdt S. Boolean network model predicts cell cycle sequence of fission yeast. PLoS One. 2008;3(2):e1672. doi: 10.1371/journal.pone.0001672. [Doi:10.1371/journal.pone.0001672] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, D'Ari R. Biological Feedback. CRC Press; 1990. [Google Scholar]
- Thomas R. Regulatory networks seen as asynchronous automata. J Theor Biol. 1991. pp. 1–23.
- Thomas R, Thieffry D, Kaufman M. Dynamical behaviour of biological regulatory networks--I. Biological role of feedback loops and practical use of the concept of the loop-characteristic state. Bull Math Biol. 1995;57(2):247–276. doi: 10.1007/BF02460618. [DOI] [PubMed] [Google Scholar]
- Albert I, Thakar J, Li S, Zhang R, Albert R. Boolean networks simulations for life scientists. Source Code Biol Med. 2008;3(16) doi: 10.1186/1751-0473-3-16. [Doi: 10.1186/1751-0473-3-16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssel C, Hopfensitz M, Kestler HA. BoolNet--an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics. 2010;26(10):1378–1380. doi: 10.1093/bioinformatics/btq124. [Applications Note] [DOI] [PubMed] [Google Scholar]
- Chaouiya C, Remy E, Mossé B, Thieffry D. Qualitative analysis of regulatory graphs: a computational tool based on a discrete formal framework. Positive Systems, LNCIS. 2003;294:119–126. [Google Scholar]
- Gonzalez AG, Naldia A, Sánchez L, Thieffry D, Chaouiya C. GINsim: A software suite for the qualitative modelling, simulation and analysis of regulatory networks. BioSystems. 2006;84:91–100. doi: 10.1016/j.biosystems.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Naldi A, Berenguier D, Fauré A, Lopez F, Thieffry D, Chaouiya C. Logical modelling of regulatory networks with GINsim 2.3. BioSystems. 2009;97:134–139. doi: 10.1016/j.biosystems.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Corblin F, Tripodi S, Fanchon E, Ropers D, Trilling L. A declarative constraint-based method for analyzing discrete genetic regulatory networks. BioSystems. 2009;98:91–104. doi: 10.1016/j.biosystems.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Corblin F, Fanchon E, Trilling L. Applications of a formal approach to decipher discrete genetic networks. BMC Bioinfomatics. 2010;11:385. doi: 10.1186/1471-2105-11-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernot G, Comet JP, Richard A, Guespin J. Application of formal methods to biological regulatory networks: extending Thomas' asynchronous logical approach with temporal logic. Journal of Theoretical Biology. 2004;229:339–347. doi: 10.1016/j.jtbi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Khalis Z, Comet JP, Richard A, Bernot G. The SMBioNet method for discovering models of gene regulatory networks. Genes, Genomes and Genomics. 2009;3:15–22. [Google Scholar]
- Mendoza L, Xenarios I. A method for the generation of standardized qualitative dynamical systems of regulatory networks. Theoretical Biology and Medical Modelling. 2006;3:13. doi: 10.1186/1742-4682-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cara AD, Garg A, Micheli BD, Xenarios I, Mendoza L. Dynamic simulation of regulatory networks using SQUAD. BMC Bioinformatics. 2007;8:462. doi: 10.1186/1471-2105-8-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Xenarios I, Mendoza L, DeMicheli G. An efficient method for dynamic analysis of gene regulatory networks and in silico gene perturbation experiments. Proc Research in Computational Molecular Biology. 2007. pp. 62–76. [Lecture Notes in Computer Science No. 4453]
- Feinendegen LE. Significance of basic and clinical research in radiation medicine: challenges for the future. British Institute of Radiology supplement. 2005;27:185–195. [Google Scholar]
- Poole DL, Mackworth AK. Artificial Intelligence. Foundations of Computational Agents. Cambridge University Press; 2010. [Google Scholar]
- Cohen J. Constraint logic programming languages. Communications of the ACM. 1990;33(7):52–68. doi: 10.1145/79204.79209. [DOI] [Google Scholar]
- Clarke EM, Emerson EA. Proc Workshop on Logics of Programs. IBM Watson Research Center; 1981. Design and synthesis of synchronization skeletons using branching time temporal logic; pp. 52–71. [Lecture Notes in Computer Science No. 131] [Google Scholar]
- Quielle JP, Sifakis J. Specification and verification of concurrent systems in CESAR. Proc 5th International Symposium on Programming. 1981. pp. 337–350.
- Emerson EA. 25 Years of Model Checking. Springer; 2008. The beginning of model checking: a personal perspective; pp. 27–45. [Lecture Notes in Computer Science No. 5000, DOI: 10.1007/978-3-540-69850-0_2] [Google Scholar]
- Eker S, Knapp M, Laderoute K, Lincoln P, Meseguer J, Sonmez K. Pathway logic: symbolic analysis of biological signaling. Proc Pacific Symposium on Biocomputing. 2002. pp. 400–412. [PubMed]
- Calder M, Vyshemirsky V, Gilbert D, Orton R. Analysis of signalling pathways using the PRISM model checker. Proc Computational Methods in Systems Biology. 2005. pp. 179–190. [Lecture Notes in Computer Science No. 4416]
- Ahmad J, Bernot G, Comet JP, Lime D, Roux O. Hybrid modelling and dynamical analysis of gene regulatory networks with delays. Complexus. 2006;3:231–251. doi: 10.1159/000110010. [DOI] [Google Scholar]
- Heath J, Kwiatkowska M, Norman G, Parker D, Tymchyshyn O. Probabilistic model checking of complex biological pathways. Theoretical Computer Science. 2008;391(3):239–257. doi: 10.1016/j.tcs.2007.11.013. [DOI] [Google Scholar]
- Ciocchetta F, Gilmore S, Guerriero ML, Hillston J. Integrated simulation and model-checking for the analysis of biochemical systems. Electronic Notes in Theoretical Computer Science. 2009;232:17–38. [Google Scholar]
- Mateus D, Gallois JP, Comet JP, Gall PL. Symbolic modeling of genetic regulatory networks. Journal of Bioinformatics and Computational Biology. 2007;5(2b):627–640. doi: 10.1142/S0219720007002850. [DOI] [PubMed] [Google Scholar]
- Batt G, Ropers D, de Jong H, Geiselmann J, Mateescu R, Page M, Schneider D. Validation of qualitative models of genetic regulatory networks by model checking: analysis of the nutritional stress response in Escherichia coli. Bioinformatics. 2005;21(Suppl 1):i19–i28. doi: 10.1093/bioinformatics/bti1048. [DOI] [PubMed] [Google Scholar]
- Calzone L, Fages F, Soliman S. BIOCHAM: An environment for modeling biological systems and formalizing experimental knowledge. Bioinformatics. 2006;22(14):1805–1807. doi: 10.1093/bioinformatics/btl172. [DOI] [PubMed] [Google Scholar]
- Li C, Nagasaki M, Ueno K, Miyano S. Simulation-based model checking approach to cell fate specification during Caenorhabditis elegans vulval development by hybrid functional Petri net with extension. BMC Systems Biology. 2009;3(42) doi: 10.1186/1752-0509-3-42. [Doi:10.1186/1752-0509-3-42] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J, Piterman N, Hajnal A, Henzinger TA. Predictive modeling of signaling crosstalk during C. elegans vulval development. PLoS Computational Biology. 2007;3(5):e92. doi: 10.1371/journal.pcbi.0030092. [Doi:10.1371/journal.pcbi.0030092] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior A. Past, Present and Future. Clarendon. 1967.
- Blackburn P, Seligman J. Hybrid languages. Journal of Logic, Language and Information. 1995;4:251–272. doi: 10.1007/BF01049415. [DOI] [Google Scholar]
- Clarke EM, Emerson EA, Sistla AP. Automatic verification of finite-state concurrent systems using temporal logic specifications. ACM Transactions of Programming Languages and Systems. 1986;8(2):244–263. doi: 10.1145/5397.5399. [DOI] [Google Scholar]
- Clarke EM, Grumberg O, Peled DA. Model Checking. MIT Press; 1999. [Google Scholar]
- Bérard B, Bidoit M, Finkel A, Laroussinie F, Petit A, Petrucci L, Schnoebelen P, McKenzie P. Systems and Software Verification. Model-Checking Techniques and Tools. Springer; 2001. [Google Scholar]
- Huth MRA, Ryan MD. Logic in Computer Science. Modelling and reasoning about systems. 2. Cambridge University Press; 2004. [Google Scholar]
- Baier C, Katoen JP. Principles of Model Checking. MIT Press; 2008. [Google Scholar]
- Chabrier-Rivier N, Chiaverini M, Danos V, Fages F, Schächter V. Modeling and querying biomolecular interaction networks. Theoretical Computer Science. 2004;325:25–44. doi: 10.1016/j.tcs.2004.03.063. [DOI] [Google Scholar]
- Franceschet M, de Rijke M. Model checking hybrid logics (with an application to semistructured data) Journal of Applied Logic. 2006;4(3):279–304. doi: 10.1016/j.jal.2005.06.010. [DOI] [Google Scholar]
- Areces C, ten Cate B. In: Handbook of Modal Logics. Blackburn P, Wolter F, van Benthem J, editor. Elsevier; 2006. Hybrid logics; pp. 821–868. [Google Scholar]
- Burch J, Clarke E, McMillan K, Dill DL, Hwang LJ. Symbolic model checking: 1020 states and beyond. Information and Computation. 1992;98:142–170. doi: 10.1016/0890-5401(92)90017-A. [DOI] [Google Scholar]
- Bryant RE. Graph-based algorithms for Boolean function manipulation. IEEE Transactions on Computers. 1986;C-35(8):1035–1044. [Google Scholar]
- Whaley J. JavaBDD 1.0b2. 2007. http://javabdd.sourceforge.net/ http://javabdd.sourceforge.net/
- Lind-Nielsen J. BuDDy 2.4. 2004. http://sourceforge.net/projects/buddy/ http://sourceforge.net/projects/buddy/
- Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8(5):345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446(7137):811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Azpeitia E, Benítez M, Vega I, Villarreal C, Alvarez-Buylla ER. Single-cell and coupled GRN models of cell patterning in the Arabidopsis thaliana root stem cell niche. BMC Syst Biol. 2010;4(135) doi: 10.1186/1752-0509-4-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M, Vernoux T, Busch W, Cui H, Wang J, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann J, Scheres B, Benfey P. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4(5):e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque M, Vernoux T, Jung J, Paquette A, Gallagher K, Wang J, Blilou I, Scheres B, Benfey P. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316(5823):421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21(17):2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17(3):354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Kaimi R, Colasanti J, Kozaki A. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol Biol. 2011. in press . [DOI] [PubMed]
- Kaya H, Shibahara K, Taoka K, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Suzuki T, Ohto M, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18(4):879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H, Geiselmann J, Hernández C, Page M. Genetic Network Analyzer: qualitative simulation of genetic regulatory networks. Bioinformatics. 2003;19(3):336–344. doi: 10.1093/bioinformatics/btf851. [DOI] [PubMed] [Google Scholar]
- Batt G, Ropers D, de Jong H, Geiselmann J, Mateescu R, Page M, Schneider D. Analysis and verification of qualitative models of genetic regulatory networks: a model-checking approach. IJCAI. 2005. pp. 370–375. [DOI] [PubMed]
- Ropers D, de Jong H, Page M, Schneider D, Geiselmann J. Qualitative simulation of the carbon starvation response in Escherichia coli. BioSystems. 2006;84:124–152. doi: 10.1016/j.biosystems.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Monteiro PT, Dumas E, Besson B, Mateescu R, Page M, Freitas AT, de Jong H. A service-oriented architecture for integrating the modeling and formal verification of genetic regulatory networks. http://www.biomedcentral.com/1471-2105/10/450. BMC Bioinformatics. 2009;10(450) doi: 10.1186/1471-2105-10-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt G, de Jong H, Page M, Geiselmann J. Symbolic reachability analysis of genetic regulatory networks using discrete abstractions. Automatica. 2008;44:982–989. doi: 10.1016/j.automatica.2007.08.004. [DOI] [Google Scholar]
- Mateescu R, Monteiro PT, Dumas E, de Jong H. In: Proc Automated Technology for Verification and Analysis (ATVA) Cha SS, Choi JY, Kim M, Lee I, Viswanathan M, editor. 2008. Computation Tree Regular Logic for genetic regulatory networks; pp. 48–63. [Lecture Notes in Computer Science No. 5311, Seoul, Korea] [Google Scholar]
- Clarke E, Grumberg O, Jha S, Lu Y, Veith H. Counterexample-guided abstraction refinement for symbolic model checking. Journal of the ACM. 2003;50(5):752–794. doi: 10.1145/876638.876643. [DOI] [Google Scholar]
- Mosca A, Manzoni L, Codecasa D. HyLMoC a model checker for hybrid logic. Proc 24th Italian Congress on Computational Logic (CILC-09) 2009.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A gentle introduction to (Hybrid) Computation-Tree Logic. This additional file has gentle introductions to Computation-Tree Logic and Hybrid Computation-Tree Logic.
(Hybrid) Computation-Tree Logic. This additional file has formal definitions of Computation-Tree Logic and Hybrid Computation-Tree Logic.
Benchmarks. This additional file shows the execution time for several examples.
Antelope User's Manual. This additional file has the Antelope user's manual.