Abstract
Fluconazole resistance is an important type of resistance in Candida because in most countries, fluconazole is the drug of choice for vulvovaginal candidiasis. Candida species resist fluconazole by various mechanisms but there is paucity of data on these in our environment. Such mechanisms include among others, over-expression of the ERG11 gene, which codes for synthesis of the target enzymes in the fungus. The aim of this study was to screen Candida spp. resistant to fluconazole for the expression of ERG11 gene. Fluconazole susceptibility test was performed on 28 clinical strains of Candida species previously obtained from students of a School of Nursing in Lagos, Nigeria. They were identified by API Candida, CHROMagar candida and germ tube test. Using 25 mcg discs, fluconazole susceptibility was determined by the disc diffusion method and results were interpreted in accordance with the Clinical Laboratory Standard Institute (CLSI) criteria; sensitive (S), resistant (R) and susceptible dose dependent (SDD). The R and SDD isolates were subsequently evaluated for the presence of ERG11 gene. Of the 28 clinical isolates, 14 were identified as C. albicans and six as C. tropicalis. The remaining isolates were identified as C. glabrata (2), C. famata (2) C. kefyr (2) one each of C. parapsilosis and C. guilliermondii respectively. In this study, 18 were susceptible (S) to fluconazole, eight were SDD and two were resistant to the antifungal agent. Out of the 14 C. albicans isolates, 12 were susceptible, one showed high level resistance and similar number showed susceptible dose dependence. ERG11 was detected in three susceptible dose dependent Candida species. This analysis demonstrates that susceptible dose dependence should not be overlooked as it may be associated with the presence of ERG11 gene and resistance to fluconazole. There is a need to consider routine antifungal susceptibility testing for Candida species causing vulvovaginitis.
Keywords: Fluconazole, ERG11 gene, Candida species, antifungal susceptibility
Introduction
Candida is a yeast fungus found as part of the muco-cutaneous flora of humans. There are approximately 200 species, among which are Candida albicans, glabrata, tropicalis, stellatoidea, parapsilosis, catemilata, ciferri, guilliermondii, haemulonii, kefyr and krusei [1]. Candida albicans is the most common species and is found in the vagina of 20-50% of healthy and asymptomatic women [1]. Non-albicans such as C. glabrata and C. tropicalis are also found in vaginal specimens [2]. Although Candida species occur as normal vaginal flora, opportunistic conditions such as diabetes, pregnancy and other immune depressants in the host enable them to proliferate and cause infection. About 75% of adult women experience at least one episode of vulvovaginal candidiasis during their lifetime among which approximately 40-50% would experience further episodes and 5% will develop recurrence, with at least three symptomatic episodes in one year [2-4]. The number of episodes tends to be more in women who are sexually active, pregnant, immunocompromised or on contraceptive pills [5-8].
A number of antifungal agents especially azoles are available to treat vulvovaginal candidiasis. However, fluconazole is a triazole introduced in the 1990’s for the treatment of infections due to Candida and other opportunistic yeasts [9]. Currently, it is recommended in various guidelines as the first drug of choice because it is less toxic and can be taken as a single oral dose [9,10]. Ergosterol is an essential component of the fungal plasma membrane which maintains the integrity of fungal cell wall. Lanosterol 14α-demethylase enzyme is a key enzyme in the synthesis of ergosterol. This enzyme is the target of fluconazole which it inhibits with a resulting impairment of ergosterol synthesis and depletion of ergosterol in the fungal cell membrane.
Production of lanosterol 14α-demethylase is encoded by the ERG11 gene. Over-expression of ER11 gene results in production of a large amount of lanosterol 14α-demethylase and this favours continuous synthesis of ergosterol and maintenance of the integrity of the cell wall which enables Candida to resist fluconazole [11-14]. This type of resistance has been associated with widespread and continuous usage of fluconazole as prophylaxis [11]. In addition, mutations in ERG11 gene exist and contribute to resistance in about 65% of fluconazole-resistant Candida species [11,15]. Genetic variation due to mutation in ERG11 gene among strains of Candida species is responsible for significant differences observed in their susceptibilities to fluconazole and itraconazole [16].
The interpretive breakpoints for Candida species tested against fluconazole are based on the analysis of treatment outcomes of infections [17,18]. In addition to the traditional categories of “susceptible” and “resistant”, the interpretive breakpoint includes a novel category, “susceptible-dose dependent” [17,18]. The SDD category encompasses MICs of 16 and 32 μg/ml (or a width of clearance zone of 15-18 mm as determined by the disc diffusion method. Isolates for which fluconazole MICs were above 64 μg/ml (≤ 14 mm) were termed resistant (R), whereas those inhibited at a lower concentration of 8 μg/ml (> 19 mm) were labeled susceptible (S). This study was conducted to evaluate the susceptibility of clinical isolates of Candida to fluconazole and to investigate isolates with resistance and SDD susceptibilities for the expression of ERG11 genes.
Materials and methods
Study design
Fluconazole susceptibility testing was carried out on clinical isolates of Candida spp. obtained from some young women. The resistant isolates and susceptible dose dependent species were subjected to polymerase chain reaction using primers that identify ERG11 gene expression.
Study population
A total of 28 clinical isolates of Candida were obtained from high vaginal swabs collected from students of the school of nursing of the Lagos University Teaching Hospital, Lagos Nigeria between May and August, 2008. All patients gave written informed consent to be recruited for this study, which was approved by the Research and Ethics Committee of the College of Medicine, University of Lagos, Proc. No. CM/COM/8/VOL.XIX and Lagos University Teaching Hospital (LUTH) Idi-Araba Proc. No. ADM/DCST/221/VOL.10. The samples were collected using sterile swab sticks and cultured on sabouraud dextrose agar at 37°C for 24 hours. Isolates were characterized using API Candida kit (Biomerieux, France) CHROM agar Candida (Biolmerieux, France) and germ tube formation. The pure isolates were stored in 10% glycerol in Brain-Heart infusion broth at - 80°C.
Antifungal drug susceptibility testing
Suspension of inoculums was prepared in 5 ml of sterile saline (0.85%) and the turbidity adjusted to 0.5 McFarland standards. Within 15 minutes of adjusting the turbidity, each isolate was plated onto a dried surface of a sterile Mueller- Hinton + Glucose + Methylene Blue agar plate respectively using a sterile cotton swab. Antimicrobial disks containing 25 μg of fluconazole were dispensed onto the surface of the inoculated agar plate. Each disk was pressed down to ensure its complete contact with the agar surface. The plates were incubated at 37°C and examined after 24 hours of incubation. The zones of inhibition were measured in millimeter and the results were interpreted using validated CLSI interpretive breakpoints for in vitro susceptibility testing of fluconazole [17]. Candida species were reported as susceptible S (zone diameter ≥ 19 mm); susceptible dose dependent SDD, (15 to 18 mm) and resistant R (≤ 14 mm).
DNA extraction
Yeast strains were cultured on Sabouraud Dextrose Agar (SDA) at 37°C for 24 hours. The ERG11 genes of all fluconazole resistant (< 14 mm zones) and susceptible doses dependent isolates (15-18 mm zones) were amplified using PCR. DNA was extracted from yeast isolates using a method previously described [19] and slightly modified in this study. Briefly, isolates were harvested from culture media into 30 ml sterile de-ionized water and washed at 6,000 rpm for 15 min and washed once in distilled water. The cell pellets were resuspended in 700 μl of lysis buffer (10 mM Tris-HCl [pH 8.0], 250 mM EDTA [pH 8.0], 0.5% Triton X-100 [vol/ vol]) supplemented with 3 mg/ml of lyticase enzyme and incubated at 37°C overnight. The spheroplasts were subsequently lysed by incubating the sample with 3 mg/ml of proteinase K at 55°C for 2 h, after which the proteinase K was inactivated at 95°C for 10 min. An equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added, the tube was mixed by inversion and centrifuged at 12,000 rpm for 15 minutes. The aqueous layer in the eppendorf tube was transferred to a fresh tube, and an equal volume of ice-cold isopropanol was added, the DNA was precipitated by centrifugation at 12,000 rpm for 15 min, while the pellet formed was washed twice with 70% ethanol. Air-dried pellets were re-suspended in 200 μl of TE buffer and stored at - 20°C prior to use. DNA concentrations were estimated against known concentrations of lambda DNA using Nanodrop spectrophotometer (ND 1000). Candida DNA was diluted to a working concentration of 10 ng/μl.
DNA amplification
The reaction solution consisted of 2.5 ml of PCR reaction buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 10 ng of target genomic DNA, 200 mM of dNTPs, 1 mM of each primer, 0.5 U Taq polymerase, and sterilized distilled water to a final volume of 25 ml. PCR was carried out by using primers that span the entire ERG11 open reading frame: 5’-GTT GAA ACT GTC ATT GAT GG (forward) and 5’-TCA GAA CAC TGA ATC GAA AG (reverse) [13,15]. The PCR was performed in a 25 well thermocycler (Epperndorf, Germany). The system was programmed to 3 min of denaturation at 92°C and this was followed by 30 cycles, each consisting of 1 min of denaturation at 92°C, 1 min of annealing at 43°C, and 1 min of extension at 72°C. At the final cycle, an additional 10 min of incubation at 72°C was performed for complete extension. For each PCR run, a negative control was also included containing the reaction buffer, dNTPs, Taq polymerase without the target DNA. Reference strain C. parapsilosis ATCC 22019 was included in each run. The amplicon was detected by electrophoresis on a 2% agarose gel performed at 70 V for 2.5 h. The gel was stained with ethidium bromide (0.5 μg/ml) and photographed under UV light with a Photo documentation system (GenoSens 1500). A DNA ladder of 100 bp (Promega, USA) was used as molecular weight marker.
Results
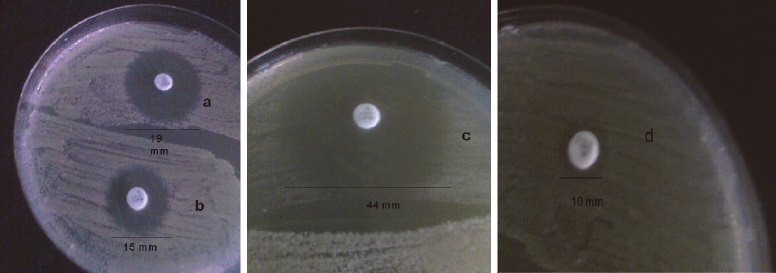
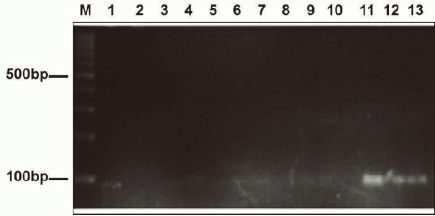
Out of the 28 Candida species investigated, 14 were characterized as C. albicans, six were C. tropicalis, two each were C. glabrata, C. famata, C. kefyr, one as C. guillermondii and C. parapsilosis respectively. The species distribution and in-vitro susceptibilities of the Candida isolates to fluconazole (Table 1). Antifungal susceptibility testing revealed that 18 Candida species were susceptible to fluconazole, two were resistant while eight were SDD. Of the 14 species of C. albicans tested, one was resistant, another one SDD while the remaining 12 were susceptible. Zones of inhibition showing the interpretive categories in accordance with the disc diffusion technique were presented on Figures 1A-D. ERG11 gene was amplified and detected in two resistant Candida isolates which included one C. albicans and one C. famata. The gene was also amplified in three SDD isolates comprising one each of C. albicans, C. parapsilosis and C. tropicalis respectively (Figure 2).
Table 1.
In vitro susceptibility pattern of Candida species to fluconazole
| Species | Number of isolates tested (%) | S (%) | SDD (%) | R (%) |
|---|---|---|---|---|
| C. albicans | 14(50) | 12(85.7) | 1(7.1) | 1(7.1 ) |
| C. tropicalis | 6(21.4) | 3(50) | 3(50) | - |
| C. glabrata | 2(7.14) | 1(50) | 1(50) | - |
| C. kefyr | 2(7.14) | 1(50) | 1(50) | - |
| C. famata | 2(7.14) | 1(50) | - | 1(50) |
| C. guilliermondii | 1(3.57) | - | 1(100) | - |
| C. parapsilosis | 1(3.57) | - | 1(100) | - |
| Total | 28(100) | 18(64.3) | 8(28.6) | 2(7.14) |
S: susceptible; SDD: susceptible dose dependent; R: resistant.
Figure 1.
Susceptibility pattern of Candida specie to of Fluconazole (25 μg) showing different sizes of inhibition zone. A. zone size of 19 mm (S); B. 15 mm (SDD); C. 44 mm (S); D. 10 mm (R).
Figure 2.
Agarose gel electrophoresis of the amplicon lane M: DNA marker, lane 2 (+) positive control, lane 3 (–) negative control and lanes 11, 12, 13 showing visible amplification of ERG11 gene with band size of 92 bp for SDD isolates; C. albicans, C. parapsilosis and C. tropicalis.
Discussion
Antifungal susceptibility testing of Candida is becoming recognized as a useful aid in optimizing the treatment of Candida infections as emergence of resistant strains continuously threatens fluconazole therapy. In this study, we provided data on fluconazole susceptibility of vaginal Candida species. Overall, most of the 28 isolates investigated were susceptible to fluconazole. C. albicans is generally reported to be susceptible to fluconazole [20-24]. Reports emanating from Brazil on MIC ranges of all isolates tested indicated susceptibility to fluconazole [20]. In Benin City Nigeria, 97.2% susceptibility was observed among isolates obtained from patients with genitourinary tract infections, but the study did not define the presence or absence of susceptible dose dependent strains [24]. Although most C. albicans studied were sensitive to fluconazole, only 67% were clearly sensitive while 22.2% were susceptible dose dependent. This is noteworthy because increasing the dosage of fluconazole for such susceptible dose dependent strains would likely yield good therapeutic results in-vivo [19-22]. C. tropicalis is usually susceptible to fluconazole, and in this study three (50%) were susceptible while three (50%) were susceptible dose dependent.
In recent times, most surveillance studies reported fluconazole resistance in both albicans and non-albicans species [21,22]. Our study showed that 57.1% of non-albicans species were susceptible dose dependent and one isolate of C. famata showed high level resistance. An overall resistance rate of 7.14% was noted; this result is similar to that of a previous study that observed resistance in 9.5% of Candida species obtained from HIV patients [25,26] and those with genitourinary tract infection [24] but a bit lower in another similar study [27]. Fluconazole is the first drug of choice for treating vaginitis in most parts of the world in accordance with recommended guidelines [9,10], however, due to the high cost it is not usually the first drug to be prescribed in Nigeria. It is usually prescribed after cotrimazole has failed to treat vaginitis, so a low resistance rate to it was expected. The level of resistance seen could be due to its widespread use as prophylactic, empiric and definitive therapy of candidal and other fungal infections in HIV/ AIDS persons [21-23,26,27] that are many in Nigeria [28]. We therefore, highlight the need for routine susceptibility testing before fluconazole is prescribed.
Fluconazole resistance in Candida species has been associated with over expression of ERG11 genes which encodes for lanosterol 14α-demethylase. Over-expression of ERG11 gene may occur due to genetic point mutation in ERG11 gene [11]. In this study, PCR amplified ERG11 gene in three of the isolates that were susceptible dose dependent. Constant exposure of Candida species to fluconazole has frequently been associated with resistance. Although ERG11 gene is present in all species of Candida, the primer used in this study was designed to detect point mutation within the ERG11 gene [13,15] apparently associated with resistance due to the exposure of these species to azoles hence the detection of ERG11 gene in some isolates and not in others. The detection rate of ERG11 gene observed in this study is similar to a previous finding that went further to determine the level of ERG11 expression [16]. Unfortunately, due to non availability of sequencing machine in our environment, it was not possible to determine the level of expression of the ERG11 genes.
The ERG11 genes expression in the fluconazole resistant isolates is noteworthy especially since they were found in susceptible dose dependent isolates. This finding shows that the molecular mechanisms of fluconazole resistance in Nigerian isolates may likely include over-expression of ERG11 gene. It also demonstrates that susceptible dose dependence should not be overlooked as it may be associated with the expression of ERG11 gene.
Surveillance of the resistance mechanisms of fluconazole is a potentially valuable tool to track the emergence and spread of fluconazole-resistant Candida species and there is need to consider routine antifungal susceptibility testing for Candida species causing vulvuvaginitis in our environment.
References
- 1.Scott MD. Candida vulvovaginitis Aka: Vaginal Candidiasis, Note book. LLC. 2008:28–30. [Google Scholar]
- 2.Okungbowa F1, Isikhuemhen OS, Dede APO. The distribution frequency of Candida species in the genitor urinary tract among symptomatic individuals in Nigerian cities. Rev Iberoam Micol. 2003;20:60–63. [PubMed] [Google Scholar]
- 3.Sobel JD. Candidal vulvovaginitis. Clin Obstet Gynecol. 1993;36:153–212. doi: 10.1097/00003081-199303000-00021. [DOI] [PubMed] [Google Scholar]
- 4.Jombo GTA, Akpera MT, Hemba SH, Eyong KI. Symptomatic vulvovaginal candidiasis: knowledge, perceptions and treatment modalities among pregnant women of an urban settlement in west africa. Afri J Clin Exper Microbiol. 2011;12:32–37. [Google Scholar]
- 5.Jombo1 GTA, Opajobi SO, Egah DZ, Banwat EB. Denen Akaa P Symptomatic vulvovaginal candidiasis and genital colonization by Candida species in Nigeria. J Pub Health Epidemiol. 2010;2:147–151. [Google Scholar]
- 6.Jombo GTA, Egah DZ, Akosu JT, Bella IS. Beliefs and perceptions about acquired immunodeficiency syndrome (AIDS) of a Nigerian rural community. The implications for prevention policy and programs. Afr J of Clin Experl Microbiol. 2007;8:40–45. [Google Scholar]
- 7.Ogunfowokan AA, Ibrahim EM, Akintaju OM. Knowledge and management of vulvovaginal candidiasis in Nigeria. Afric J Mid Women’s Health. 2010;4:63–67. [Google Scholar]
- 8.Ohmit SE, Sobel JD, Schuman P, Duerr A, Mayer K, Rompalo A, Klein RS; HIV Epidemiology Research Study (HERS) Group. Longitudinal study of mucosal Candida species colonization and candidiasis among human immunodeficiency virus (HIV) seropositive and at risk seronegative women. J Infect Dis. 2003;188:118–127. doi: 10.1086/375746. [DOI] [PubMed] [Google Scholar]
- 9.Maertens JA. History of the development of azole derivatives. Clin Microbiol Infect. 2004;10:1–10. doi: 10.1111/j.1470-9465.2004.00841.x. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC) Sexually Transmitted Diseases Treatment Guidelines. 2002. http://www.cdc.gov/std/treatment/default.htm. [PubMed] [Google Scholar]
- 11.Kelly SL, Arnoldi A, Kelly DE. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans. 1993;21:1034–1038. doi: 10.1042/bst0211034. [DOI] [PubMed] [Google Scholar]
- 12.Manastır L, Ergon MC, Yucesoy M. Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses. 2009;54:99–104. doi: 10.1111/j.1439-0507.2009.01766.x. [DOI] [PubMed] [Google Scholar]
- 13.Perea S, Lopez-Ribot JL, Kirkpatrick WR, McAtee RK, Santillan RA, Martinez M, Calabrese D, Sanglard D, Patterson TF. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virusinfected patients. Antimicrob Agents Chemother. 2001;45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manastır L, Ergon MC, Yücesoy M. Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses. 2011;5:99–104. doi: 10.1111/j.1439-0507.2009.01766.x. [DOI] [PubMed] [Google Scholar]
- 15.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. Application of realtime quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother. 2004;48:2124–2131. doi: 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Source Over expression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. 2010;38:536–545. doi: 10.1177/147323001003800216. [DOI] [PubMed] [Google Scholar]
- 17.Pfaller MA, Diekema DJ, Rex JH, Espinel-Ingroff A, Johnson EM, Andes D, Chaturvedi V, Ghannoum MA, Odds FC, Rinaldi MG, Sheehan DJ, Troke P, Walsh TJ, Warnock DW. Correlation of MIC with outcome for Candida species tested against voriconazole: analysis and proposal for interpretive breakpoints. J Clin Microbiol. 2006;44:819–826. doi: 10.1128/JCM.44.3.819-826.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rex JH, Pfaller MA, Walsh TJ, Chaturvedi V, Espinel- Ingroff A, Ghannoum MA, Gosey LL, Odds FC, Rinaldi MG, Sheehan DJ, Warnock DW. Antifungal susceptibility testing: practical aspects and current challenges. Clin Microbiol Rev. 2001;14:643–658. doi: 10.1128/CMR.14.4.643-658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura K, Kaneko T, Yamamoto Y. Lysis of yeast cells by enzymes of Arthrobacter luteus. Arch Biochem Biophys. 1971;145:402–404. doi: 10.1016/0003-9861(71)90053-1. [DOI] [PubMed] [Google Scholar]
- 20.Azevedo AC, Bizerra FC, da Matta DA, de Almeida LP, Rosas R, Colombo AL. In vitro susceptibility a large collection of Candida Strains against fluconazole and voriconazole by using the CLSI disk diffusion assay Mycopathologia. 2011;171:411–416. doi: 10.1007/s11046-010-9387-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Kang Y, Yokoyama K, Gonoi T, Mikami Y. Molecular Differentiation and Antifungal Susceptibility of Candida albicans isolated from Patients with Respiratory Infections in Guiyang Medical College hospital china. Jap J Med Mycol. 2009:50. doi: 10.3314/jjmm.50.175. 175-178. [DOI] [PubMed] [Google Scholar]
- 22.Clancy CJ, Yu VL, Morris AJ, Snydman DR, Nguyen MH. Fluconazole MIC and the fluconazole dose/MIC ratio correlate with therapeutic response among patients with candidemia. Antimicrob Agents Chemother. 2005;49:3171–3177. doi: 10.1128/AAC.49.8.3171-3177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother. 2005;49:668–679. doi: 10.1128/AAC.49.2.668-679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akortha EE, Nwaugo VO, Chikwe NO. Antifungal resistance among Candida species from patients with genitourinary tract infection isolated in Benin City, Edo state, Nigeria. Afri J Microbiol Res. 2009;3:694–699. [Google Scholar]
- 25.Omar JMH, Mecky INM, Mainen JM, Elison NMS, Ferdinand M, Frans HMM, Wim HH, Antonius JMM, André JAM, Paul EV. Species distribution and in vitro antifungal susceptibility of oral yeast isolates from Tanzanian HIV-infected patients with primary and recurrent oropharyngeal candidiasis. BMC Microbiol. 2008;8:135. doi: 10.1186/1471-2180-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enwuru CA, Ogunledun A, Idika N, Enwuru NV, Ogbonna F, Aniedobe M, Adeiga A. Fluconazole resistant opportunistic Oro-pharyngeal Candida and non-candida yeast-like isolates from HIV infected patients attending ARV clinics in Lagos, Nigeria. Afri Health Sci. 2008;8:142–148. [PMC free article] [PubMed] [Google Scholar]
- 27.Nweze EI, Ogbonnaya UL. Oral Candida isolates among HIV-infected subjects in Nigeria. J Microbiol Immunol Infect. 2011;44:172–177. doi: 10.1016/j.jmii.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 28.UNGASS Country Progress Report. United Nations General Assembly Special Session (UNGASS); 2010; Nigeria. [Google Scholar]