Abstract
The present study was carried out to examine the effect of valproic acid (VPA), an important histone deacetylase inhibitor, on the in vitro development and expression of the epigenetic marker histone H3 lysine 9 (H3K9ac) in bovine somatic cell nuclear transfer (SCNT) embryos. We found that treatment with 4 mM VPA for 24 h could significantly improve the development of bovine SCNT embryos. Compared with the no-treatment group, the cleavage rate was higher (69.79±0.99% vs. 65.11±1.02%, p<0.05), as was the blastocyst rate (39.99±1.29% vs. 34.87±1.74%, p<0.05). Moreover, the rate of apoptosis (1.91±0.48% vs. 5.67±0.40%, p<0.05) in blastocysts was greatly reduced after VPA treatment. Valproic acid treatment also increased the immunofluorescent signal for H3K9ac in SCNT embryos in a pattern similar to that of in vitro fertilized (IVF) embryos. In conclusion, we demonstrated that VPA can significantly improve the in vitro developmental competence and enhance the nuclear reprogramming of bovine SCNT embryos.
Introduction
Since the first successful cloning of an animal by somatic cell nuclear transfer (SCNT) was performed in sheep (Wilmut et al., 1997), many other animals have also been cloned, such as cattle, goats, pigs, cats, rats, mules, horses, and dogs. However, the efficiency of SCNT was still low and remains an obstacle to potential applications in agriculture and regenerative medicine (Yang et al., 2007). Many approaches have been used to attempt to improve the efficiency of SCNT, including ameliorating ennucleation procedures (Kuhholzer et al., 2000; Costa-Borges et al., 2011), optimizing the oocyte activation time after reconstruction (Wakayama et al., 2003; Wakayama and Yanagimachi, 2001), chemical treatment of donor cells before SCNT (Enright et al., 2003), serial rounds of nuclear transfer (Wakayama et al., 2005), and aggregation of clones (Boiani et al., 2003). Analyses of cloned embryos or offspring revealed that abnormal epigenetic modifications, such as DNA methylation and histone modifications (Dean et al., 2001; Humpherys et al., 2002; Inoue et al., 2002; Kang et al., 2001; Ohgane et al., 2004; Santos et al., 2003; Suemizu et al., 2003; Wang et al., 2007), rather than genetic abnormalities, might be a key factor affecting the cloning efficiency. Epigenetic reprogramming, which involves modifications in chromatin-associated proteins and DNA, is an important feature of nuclear reprogramming in SCNT embryos. Dynamic interactions between DNA methylations and acetylations in the amino-terminal domains of core histones are thought to regulate DNA functions and control gene expression (Li et al., 2007; Wu et al., 2007).
Histone modification is an important epigenetic modification and includes acetylation, phosphorylation, methylation, and ubiquitination (Fischle et al., 2003). Numerous studies have suggested that elevated levels of histone acetylation in cloned embryos could improve the reprogramming efficiency. The global and local patterns of histone acetylation contribute considerably to nuclear reprogramming (Turner, 2000). Particular histone modifications can be used to predict transcription directly, and histone modifications induced by developmental or environmental cues, with potential coding roles, could be heritable from one cell generation to another, without inducing transcriptional change.
Histone modification seems to be closely involved in the complex changes in gene expression that drive early development (Azuara et al., 2006). For example, the acetylated form of histone H3 lysine 9 (H3K9ac) is associated with active chromatin configurations (Rice and Allis, 2001). Various methods have been used to regulate histone acetylation, such as trichostatin A (TSA) (Ding et al., 2008; Enright et al., 2003; Kishigami et al., 2006; Li et al., 2008), sodium butyrate (NaBu) (Das et al., 2010; Shi et al., 2003; Yang et al., 2007), Scriptaid (Van Thuan et al., 2009; Zhao et al., 2009, 2010), and valproic acid (VPA) (Miyoshi et al., 2010a). Valproic acid, a short-chain fatty acid that inhibits histone deacetylase (HDAC), has been used for decades in the treatment of epilepsy and is also effective as a mood stabilizer and in migraine headaches and schizophrenia. Valproic acid was recently demonstrated as a drug for inducing the reprogramming of differentiated cells. For example, Valproic acid enhances in vitro development and oct-3/4 expression of miniature pig somatic cell nuclear transfer embryos (Miyoshi et al., 2010a).Valproic acid has also been shown to induce reprogramming of mouse fibroblasts with only three of the four transcription factors that are usually needed (Oct-3/4, Sox2, c-Myc, and Klf4), with a considerably higher efficiency than TSA (Huangfu et al., 2008a) and valproic acid can improve the in vitro and full-term development of B6CBAF1 mouse SCNT embryos, at a similar level as TSA (Huangfu et al., 2008a). Recently, the use of miR367 and VPA can cooperate in a powerful way to reprogram somatic cells to pluripotency (Anokye-Danso et al., 2011). Moreover, valproic acid enabled reprogramming of primary human fibroblasts with only two factors, Oct4 and Sox2, without the need for c-Myc or Klf4 (Huangfu et al., 2008b). The superior beneficial effects of VPA on the in vitro reprogramming of somatic cell nuclei using transcription factors prompted us to investigate whether a similar trend would be observed in the in vitro reprogramming of bovine SCNT embryos.
The objective of the present experiment was (1) to determine the effect of VPA treatment on the in vitro development of bovine SCNT embryos; and (2) to assess the effects of VPA treatment on the expression of the epigenetic marker H3K9ac.
Materials and Methods
Unless otherwise indicated, all reagents were purchased from Sigma (St. Louis, MO, USA). Procedures were approved by the Animal Care and Use Committee of Northwest A&F University and performed in accordance with animal welfare and ethics standards.
Oocyte collection and maturation
Bovine ovaries were obtained from slaughterhouses, sampled in sterile 0.9% NaCl at 15–20°C (Wang et al., 2011) and transported to the laboratory within 4 h. Bovine cumulus–oocyte complexes were collected from follicles that were 2–8 mm in diameter and cultured in TCM-199 (Gibco, Grand Island, NY, USA) with 10% (v/v) FBS, 1 μg/mL 17β-estradiol and 0.075 IU/mL human menopausal gonadotropin, then incubated at 38.5°C in a humidified incubator of 5% CO2 in air for 20 h.
Donor cell preparation
Fibroblasts were obtained from the ear skin of a 7-day-old fetus. Fetal tissues were minced, digested, and cultured in Dulbecco's Modified Eagle's medium (DMEM, Gibco) containing 10% FBS, 1 mM sodium pyruvate, 100 IU/mL penicillin and 100 mg/mL streptomycin under 5% CO2 in air at 38.5°C. Isolated cells were washed and cultured in DMEM, which was replaced by fresh medium every 24 h. Once fibroblasts reached 90% confluence, they were trypsinized and reconstituted at a concentration of 1×106 cells/mL. A fibroblast fetal cell line derived from a fetus between passages 2 and 5 was used for SCNT.
Embryo production by SCNT
After maturation, cumulus cells were denuded by treatment with 0.1% bovine testicular hyaluronidase in PBS. Matured oocytes with a polar body were selected for SCNT in PBS supplemented with 7.5 μg/mL cytochalasin B and 10% FBS. Oocytes were then enucleated by aspirating the first polar body and a small amount of surrounding cytoplasm using a beveled glass pipette with 20-μm internal diameters. A donor cell was injected into the perivitelline space of the recipient cytoplasm, then fused using two closely spaced electrical pulses of 35 V for 10 μsec. The reconstructed embryos were kept in mSOF [supplemented with 8 mg/mL fatty acid-free bovine serum albumin (BSA)], 1% MEM nonessential amino acid solution and 2% BME essential amino acid solution containing 5 μg/mL cytochalasin B for 2 h before activation. The fused embryos were then activated in 5 μM ionomycin for 3 min followed by 4-h exposure to 1.9 mM 6-dimethylaminopurine in mSOF.
In vitro fertilization
Matured oocytes were rinsed three times in Fert-Talp medium supplemented with 30 mg/mL heparin, 1.65 mg/mL hypotaurine, 0.27 mg/mL epinephrine, and 4.5 mg/mL penicillamine. About 30–35 oocytes were transferred into droplets of fertilization medium covered with mineral oil that had been equilibrated for 8 h at 38.5°C in 5% CO2 in air. A thawed semen pellet was then centrifuged (1000×g for 10 min) and spermatozoa were resuspended in fertilization medium to a concentration of 1×106 cells per mL. Sperm suspension solution was added to the fertilization droplets. The oocytes and sperm were incubated for at least 8 h.
Postactivation treatment and embryo culture
The SCNT and IVF embryos were rinsed three times in mSOF and then treated with various concentrations of VPA in mSOF for different durations. After treatment the embryos were washed three times again, transferred into mSOF medium under mineral oil, and cultured for 7 days at 38.5°C in a humidified incubator containing 5% CO2 in air. Cleavage and blastocyst formation rates were evaluated on days 2 (24 h) and 7 (144 h), respectively, with the day of SCNT or IVF designated day 0.
Immunodetection of H3K9ac and counting of nuclei in blastocysts
For immunodetection of H3K9ac, embryos were rinsed in PBS, fixed for 20 min in 4% paraformaldehyde, and then transferred to PBS supplemented with 3% BSA and 0.5% Triton X-100. Next, samples were blocked in 2% BSA in PBS overnight at 4°C, then exposed to primary antibodies (rabbit polyclonal antibody against histone H3K9ac, Abcam, Hong Kong, diluted to 1:500) overnight at 4°C. Samples were washed three times for 10 min each in 0.2% PVA-PBS and incubated for 2 h at room temperature in the presence of 1:500 diluted secondary antibodies (goat antirabbit IgG, Beyotime, Nantong, China). Samples were rinsed three times for 10 min each in 0.2% PVA-PBS. Specimens were examined by epifluorescence using a Nikon eclipse Ti-S microscope (Nikon, Tokyo, Japan). All images were captured using Nikon DS-Ri1 digital camera and saved in TIFF format.
Average opitical intensity was measured using Image-Pro Plus 6.0. Images were converted to grayscale and inverted. After correcting optical density (average cytoplasmic intensity were measured for normalization to background). All individual nuclei of embryos at the two-cell, four-cell, eight-cell, and blastocyst stages, and 30 nuclei per blastocyst were outlined, integrated optical density (IOD) and area were measured, the average normalized fluorescence intensity for a single embryo was represented by “sum IOD/sum area.” In order to minimize the difference among embryos in the same batch and among batches, all images were captured using the same settings in the same environment, and normalized to their own cytoplasmic background before measurement. Experiments were performed in three replicates per stage with 10 embryos per replicate for each embryo type. The level of histone acetylation/histone methylation of embryos was represented by mean value of embryos±SD.
For counting, blastocysts were collected 7 days (144 h) after activation and fixed in 4% paraformaldehyde for 30 min at room temperature and washed three times in 0.2% PVA-PBS. Blastocysts were permeabilized in 0.5% Triton X-100 for 30 min. TUNEL analysis was performed using the fluorescein in the DeadEnd™ Fluorometric TUNEL System (Promega, Madison, WI, USA) following the manufacturer's recommendations. After the TUNEL reaction, embryos were rinsed three times in 0.2% PVA-PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI) for 3 min, and washed three times and examined by epifluorescence using a Nikon eclipse Ti-S microscope. All images were captured using Nikon DS-Ri1 digital camera and saved in TIFF format.
Experimental design and statistical analysis
Experiment 1
SCNT embryos were treated with various concentrations of VPA (0, 2, 4, or 10 mM) for 24 h after activation. Cleavage rates (24 h), blastocyst rates (144 h), and hatching rates (192 h) were recorded to assess their in vitro developmental capacity.
Experiment 2
SCNT embryos were treated with 4 mM VPA for different durations (0, 12, 24, or 48 h) after activation. Cleavage rates (24 h), blastocyst rates (144 h), and hatching rates (192 h) were recorded to assess their in vitro developmental capacity.
Experiment 3
SCNT embryos treated with or without 4 mM VPA for 24 h were collected at the blastocyst stage for counting and TUNEL assay.
Experiment 4
SCNT embryos treated with or without 4 mM VPA for 24 h and IVF embryos were collected at the two-cell, four-cell, eight-cell, and blastocyst stages for detection of the level of H3K9 histone acetylation.
Experiments were repeated at least three times. Data were analyzed by one-way ANOVA and Tukey's LSD test using SPSS 13.0 software, and p<0.05 was regarded as statistically significant.
Results
Experiment 1: Effect of different concentrations of VPA on the development of bovine SCNT embryos in vitro
There were differences in the cleavage rates (58.22±2.08% vs. 74.11±2.82%, p<0.05) of embryos among the different concentrations of VPA (Table 1). The group treated with 4 mM VPA achieved the highest cleavage rate (74.11±2.82%). After treatment with 4 mM VPA, the blastocyst formation rate (38.69±1.58%) and hatching rate (55.31±3.72%) were also higher than those treated with 0, 2, or 10 mM VPA.
Table 1.
Effect of Different Concentrations of VPA Treatment on the Development of Bovine SCNT Embryos In Vitro
| |
|
|
No. (mean %±SEM) of embryos developed to |
|
|
|---|---|---|---|---|---|
| Concentration of VPA (mM) | Repeat times | No. of embryos cultured | ≥Two-cell | Blastocyst | No. of hatching blastocysts (mean %±SEM) |
| 0 | 4 | 422 | 278 (65.89±1.99)a | 136 (32.22±1.72) | 58 (42.57±0.99) |
| 2 | 4 | 426 | 290 (68.06±0.82)a,b | 138 (32.46±1.62) | 58 (41.90±3.01) |
| 4 | 4 | 430 | 318 (74.11±2.82)b,c | 166 (38.69±1.58)a | 92 (55.31±3.72)a |
| 10 | 4 | 416 | 242 (58.22±2.08)d | 120 (28.86±0.91) | 52 (43.66±3.57) |
Different superscripts in the same column show significant differences: (p<0.05).
Cleavage percentage: number of embryos cleaved/number of embryos cultured.
Blastocyst percentage: number of blastocysts/number of embryos cultured.
Hatching percentage: number of hatching blastocysts/number of blastocysts.
Experiment 2: Effect of different durations of VPA treatment on development
Difference in cleavage rates (60.27±3.62% vs. 65.91±1.94%, p<0.05) was observed among the treatment groups (Table 2). However, the blastocyst formation rates (38.46±3.52%) of the group treated with VPA for 24 h were higher than those of other groups. The hatching blastocyst rate of the group treated with VPA for 48 h was 47.33±3.31%, which was greater than that of the other groups.
Table 2.
Effect of Different Durations of VPA Treatment on the Development of Bovine SCNT Embryos In Vitro
| |
|
|
No. (mean %±SEM) of embryos developed to |
|
|
|---|---|---|---|---|---|
| Duration of VPA (h) | Repeat times | No. of embryos cultured | ≥Two-cell | Blastocyst | No. of hatching blastocysts (mean %±SEM) |
| 0 | 4 | 508 | 312 (61.47±2.37) | 160 (31.54±2.00)a | 68 (42.17±3.49)ab |
| 12 | 4 | 576 | 352 (61.14±2.10) | 204 (35.35±4.01)ab | 60 (30.53±4.30)a |
| 24 | 4 | 552 | 364 (65.91±1.94)a | 212 (38.46±3.52)b | 76 (35.51±2.59)ab |
| 48 | 4 | 564 | 340 (60.27±3.62) | 180 (32.00±4.01)ab | 84 (47.33±3.31)b |
Different superscripts in the same column show significant differences: (p<0.05).
Cleavage percentage: number of embryos cleaved/number of embryos cultured.
Blastocyst percentage: number of blastocysts/number of embryos cultured.
Hatching percentage: number of hatching blastocysts/number of blastocysts.
Experiment 3: Effect of VPA treatment on development
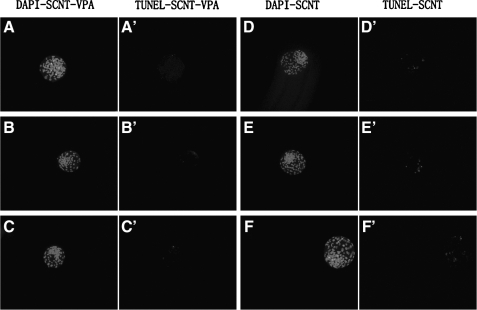
The cleavage rates (65.11±1.02% vs. 69.79±0.99%, p<0.05) and the blastocyst formation rates (34.87±1.74% vs. 39.99±1.29%, p<0.05) were higher following VPA treatment than the untreated SCNT groups. The apoptosis rates (1.91±0.48% vs. 5.67±0.40%, p<0.05) of embryos were decreased in the treated group (Fig. 1). However, the cell number (105±3.58 vs. 106±2.27) showed no observable differences (Table 3).
FIG. 1.
TUNEL assay of blastocysts. Each sample was counterstained with DAPI to visualize DNA.
Table 3.
Effect of VPA Treatment on the Development of Bovine SCNT Embryos In Vitro
| |
|
|
No. (mean %±SEM) of embryos developed to |
|
|
|
|---|---|---|---|---|---|---|
| VPA treatment | Repeat times | No. of embryos cultured | ≥Two-cell | Blastocyst | No. of cells per blastocyst | No. (mean %±SEM) of apoptosis cells per blastocyst |
| SCNT-VPA | 4 | 536 | 374 (69.79±0.99)a | 214 (39.99±1.29)a | 105±3.58 | 2 (1.91±0.48)a |
| SCNT (control) | 4 | 522 | 340 (65.11±1.02) | 182 (34.87±1.74) | 106±2.27 | 6 (5.67±0.40) |
Different superscripts in the same column show significant differences: (p<0.05).
Cleavage percentage: number of embryos cleaved/number of embryos cultured.
Blastocyst percentage: number of blastocysts/number of embryos cultured.
Apoptosis percentage: number of apoptosis cells/number of cells per blastocyst.
Experiment 4: Effect of VPA treatment on H3K9 acetylation
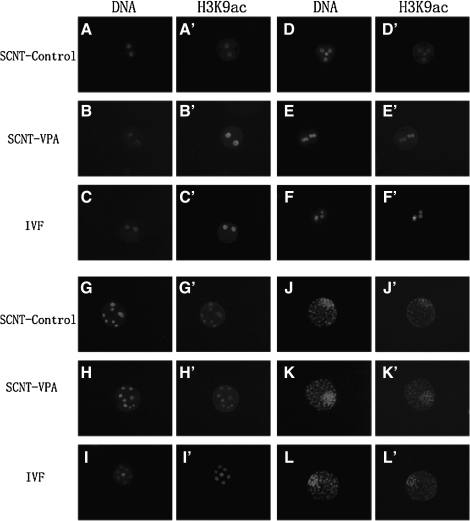
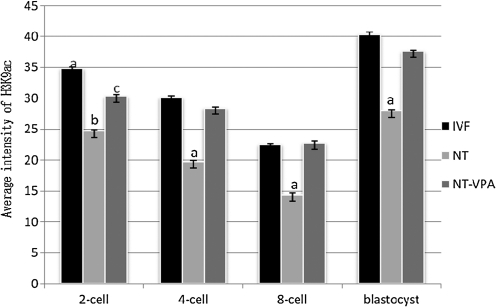
The intensity of H3K9ac staining at the two-cell, four-cell, eight-cell, and blastocyst stages is shown in Figure 2. From the two-cell to the eight-cell stage, the intensity of H3K9ac staining was decreased in all the treated groups, but the blastocyst stage showed the highest intensity of all the stages (Fig. 3). The intensity of staining in the IVF group was highest at every stage and the treatment group was next highest. After treatment with VPA the intensity of H3K9ac was significantly higher than that of the untreated group and was elevated to a similar level to that of the IVF counterparts.
FIG. 2.
Acetylation of histone H3 on lysine 9 (H3K9 acetylation) in two-cell stage (A–C′), four-cell stage (D–F′), eight-cell stage (G–I′), and blastocyst stage (J–L′) bovine embryos treated without (control) or with (VPA) and IVF.
FIG. 3.
Fluorescent intensity for H3K9ac in VPA-treated SCNT embryos, untreated SCNT embryos, and untreated IVF embryos. Average optical intensity was measured using Image-Pro Plus 6.0. Different superscripts in the same stage (two-cell, four-cell, eight-cell, and blastocyst) show statistically significant differences: (p<0.05).
Discussion
In the present study, we investigated the effect of VPA, an HDAC inhibitor, on the in vitro development of bovine SCNT embryos. After treatment with 4 mM VPA for 24 h, the cleavage rate and blastocyst rate were improved and the epigenetic marker H3K9ac was also elevated to a level similar to that of IVF counterparts. Here we have demonstrated that VPA can improve the in vitro development potential of bovine SCNT embryos and enhance nuclear reprogramming.
Previous studies have suggested that incomplete reprogramming was the main cause of low SCNT efficiency. Abnormal epigenetic modifications, such as DNA methylation and histone modification, occurring following SCNT (Dean et al., 2001; Kang et al., 2001; Ohgane et al., 2004; Santos et al., 2003) were likely associated with the low success of cloning. TSA, a widely used HDAC inhibitor, has been shown to enhance the development competence of SCNT embryos in mouse, pig, rabbit, and cattle. However, others reported that TSA has no effect on the in vitro development of rabbit SCNT embryos. In addition, although embryos treated with or without TSA could develop to term in rabbits, the offspring from the embryos treated with TSA died within 1 h to 19 days (Meng et al., 2009). Bovine SCNT embryos treated with 50 ng/mL TSA resulted in significantly lower blastocyst development (9.9%) than control groups (20%) (Wu et al., 2008). Higher TSA concentrations or long treatment times resulted in a significant reduction in the success of cloning. An overdose of TSA might also cause developmental defects after implantation (Svensson et al., 1998). Considering the harmful effects of TSA, we evaluated the effect of another HDAC inhibitor, valproic acid, on the in vitro developmental potential of bovine SCNT embryos. Valproic acid has been demonstrated to promote reprogramming of differentiated cells and induce pluripotent stem cells with three transcription factors, or even only two, Oct4 and Sox2 (Huangfu et al., 2008a, 2008b). After SCNT, valproic acid can induce acetylation of core histones, change the structure in chromatin, and enhance DNA demethylation (Kishigami et al., 2006) to improve the efficiency of reprogramming. The effect of VPA on reprogramming may be due to the collective effects of upregulation of embryonic stem cell specific genes and downregulation of embryonic fibroblast specific genes (Huangfu et al., 2008a). Valproic acid enhances the ability of miniature pig SCNT embryos to develop into blastocysts and maintains their ability to express Oct-3/4 (Miyoshi et al., 2010a). Moreover, it did not harm embryo development (Ono et al., 2010). We therefore investigated the effect of VPA on the in vitro developmental potential of SCNT embryos.
When 4 mM VPA was applied to SCNT embryos for 24 h, the cleavage rate was significantly higher than in control groups and the morphological characteristics were also better. In contrast to our results, there were no significant differences in the cleavage rates in miniature pig (Miyoshi et al., 2010a). The response to VPA treatment is probably characteristic of the species; in bovine SCNT embryos, at least, it increases the competence of reprogramming. The morphological characteristics at early stages would be useful for predicting the developmental fate of embryos (Holm et al., 1998), and rapid cleavage at the early stages has been regarded as a sign of greater developmental competence (Van Soom et al., 1992). This suggests that the positive effect of hyperacetylation on nuclear reprogramming occurred at an early stage of embryo development. The blastocyst rate following treatment was also higher than in the control groups, which was in accordance with other reports that Scriptaid and TSA treatment both improved the blastocyst rate in pigs (Zhao et al., 2010) and that rates of blastocyst treated with either TSA or VPA was clearly superior to those of untreated SCNT embryos (Costa-Borges et al., 2010). This indicates that VPA could significantly increase the efficiency of bovine SCNT in vitro. Furthermore, the rate of apoptosis in cloned embryos was higher in the control group, and in VPA-treated embryos it was low. The low apoptosis rate indicated that VPA treatment could generate high-quality blastocysts. In addition, the apoptosis rate was reduced after treatment with m-carboxycinnamic acid bishydroxamide (CBHA) (Dai et al., 2010). The total number of cells in blastocysts treated with VPA was not significantly different from that in blastocysts cultured without VPA treatment, which indicates that VPA does not affect cell proliferation in the bovine SCNT embryos. Similarly, the total cell number in blastocysts was reported to show no significant differences between the treated and untreated groups in miniature pig (Miyoshi et al., 2010b; Zhao et al., 2009).
We also tested the level of the fluorescence signal for H3K9ac. The signal was higher in the VPA-treatment group than in the no-treatment group, which indicates that HDACs were effectively inhibited by VPA treatment. Similar results have been reported by others: the acetylation of H3K9, H3K14, H4K5, H4K12, and H4K16 was increased after HDAC inhibitor treatment of SCNT embryos derived from various cell types (Iager et al., 2008; Li et al., 2008; Rybouchkin et al., 2006; Shi et al., 2008; Wang et al., 2007). After treatment with VPA, we found that the fluorescence signal for H3K9ac was increased to a level similar to that of the IVF counterparts, and the increase was especially at great an early stage of embryonic development. This suggests that increased histone acetylation could facilitate nuclear reprogramming by maintaining higher order chromatin structure, regulating DNA functions, and inducing transcription and replication (Cosgrove et al., 2004; Jenuwein and Allis 2001). After treatment, the high level of histone acetylation might result in the formation of transcriptionally permissive euchromatic regions and facilitate the reprogramming of genes that regulate embryonic development. Although the potential mechanism of how VPA treatment improves the efficiency of cloning remains unknown, we suggest that VPA-induced hyperacetylation of the histones changes the chromatin structure after nuclear transfer. Such hyperacetylation is an important part of nuclear reprogramming.
In summary, our study demonstrates that VPA modifies the early morphological characteristics of embryonic development and significantly increases the blastocyst rate. The level of H3K9ac was also elevated by VPA treatment to a similar level to that of IVF. Valproic acid could be used to improve the in vitro developmental competence and enhance the nuclear reprogramming of bovine SCNT embryos.
Acknowledgments
We thank Younan Wang for providing the Holstein cow ovaries. The present study was supported by grants from the National Key Project for Production of Transgenic Livestock, China (No. 2008ZX-08007-004).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Anokye-Danso F. Trivedi C.M. Juhr D., et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V. Perry P. Sauer S., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Boiani M. Eckardt S. Leu N.A., et al. Pluripotency deficit in clones overcome by clone–clone aggregation: epigenetic complementation? EMBO J. 2003;22:5304–5312. doi: 10.1093/emboj/cdg507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove M.S. Boeke J.D. Wolberger C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004;11:1037. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- Costa-Borges N. Santalo J. Ibanez E. Comparison between the effects of valproic acid and trichostatin A on the in vitro development, blastocyst quality, and full-term development of mouse somatic cell nuclear transfer embryos. Cell. Reprogram. 2010;12:437–446. doi: 10.1089/cell.2009.0108. [DOI] [PubMed] [Google Scholar]
- Costa-Borges N. Gonzalez S. Santal J., et al. Effect of the enucleation procedure on the reprogramming potential and developmental capacity of mouse cloned embryos treated with valproic acid. Reproduction. 2011;141:789. doi: 10.1530/REP-10-0455. [DOI] [PubMed] [Google Scholar]
- Dai X. Hao J. Hou X., et al. Somatic nucleus reprogramming is significantly improved by m-carboxycinnamic acid bishydroxamide, a histone deacetylase inhibitor. J. Biol. Chem. 2010;285:31002. doi: 10.1074/jbc.M110.136085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Z.C. Gupta M.K. Uhm S.J., et al. Increasing histone acetylation of cloned embryos, but not donor cells, by sodium butyrate improves their in vitro development in pigs. Cell. Reprogram. 2010;12:95–104. doi: 10.1089/cell.2009.0068. [DOI] [PubMed] [Google Scholar]
- Dean W. Santos F. Stojkovic M., et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 2001;98:13734. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. Wang Y. Zhang D., et al. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology. 2008;70:622–630. doi: 10.1016/j.theriogenology.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Enright B. Kubota C. Yang X., et al. Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol. Reprod. 2003;69:896. doi: 10.1095/biolreprod.103.017954. [DOI] [PubMed] [Google Scholar]
- Fischle W. Wang Y. Allis C.D. Histone and chromatin cross-talk. Curr. Opin. Cell. Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Holm P. Shukri N. Vajta G., et al. Developmental kinetics of the first cell cycles of bovine in vitro produced embryos in relation to their in vitro viability and sex. Theriogenology. 1998;50:1285–1299. doi: 10.1016/s0093-691x(98)00227-1. [DOI] [PubMed] [Google Scholar]
- Huangfu D. Maehr R. Guo W., et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008a;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D. Osafune K. Maehr R., et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008b;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Humpherys D. Eggan K. Akutsu H., et al. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. USA. 2002;99:12889. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iager A.E. Ragina N.P. Ross P.J., et al. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells. 2008;10:371–380. doi: 10.1089/clo.2007.0002. [DOI] [PubMed] [Google Scholar]
- Inoue K. Kohda T. Lee J., et al. Faithful expression of imprinted genes in cloned mice. Science. 2002;295:297. doi: 10.1126/science.295.5553.297. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Allis C.D. Translating the histone code. Science. 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kang Y.K. Koo D.B. Park J.S., et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat. Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kishigami S. Mizutani E. Ohta H., et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Kuhholzer B. Baguisi A. Overstrom E. Long-term culture and characterization of goat primordial germ cells. Theriogenology. 2000;53:1071–1079. doi: 10.1016/S0093-691X(00)00253-3. [DOI] [PubMed] [Google Scholar]
- Li B. Carey M. Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li J. Svarcova O. Villemoes K., et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–808. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Meng Q. Polgar Z. Liu J., et al. Live birth of somatic cell-cloned rabbits following trichostatin A treatment and cotransfer of parthenogenetic embryos. Cloning Stem Cells. 2009;11:203–208. doi: 10.1089/clo.2008.0072. [DOI] [PubMed] [Google Scholar]
- Miyoshi K. Mori H. Mizobe Y., et al. Valproic acid enhances in vitro development and Oct-3/4 expression of miniature pig somatic cell nuclear transfer embryos. Cell. Reprogram. 2010a;12:67–74. doi: 10.1089/cell.2009.0032. [DOI] [PubMed] [Google Scholar]
- Miyoshi K. Mori H. Mizobe Y., et al. Valproic acid enhances in vitro development and Oct-3/4 expression of miniature pig somatic cell nuclear transfer embryos. Cell. Reprogram. 2010b;12:67–74. doi: 10.1089/cell.2009.0032. [DOI] [PubMed] [Google Scholar]
- Ohgane J. Wakayama T. Senda S., et al. The Sall3 locus is an epigenetic hotspot of aberrant DNA methylation associated with placentomegaly of cloned mice. Genes Cells. 2004;9:253–260. doi: 10.1111/j.1356-9597.2004.00720.x. [DOI] [PubMed] [Google Scholar]
- Ono T. Li C. Mizutani E., et al. Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol Reprod. 2010;83:929–937. doi: 10.1095/biolreprod.110.085282. [DOI] [PubMed] [Google Scholar]
- Rice J.C. Allis C.D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell. Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Rybouchkin A. Kato Y. Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 2006;74:1083. doi: 10.1095/biolreprod.105.047456. [DOI] [PubMed] [Google Scholar]
- Santos F. Zakhartchenko V. Stojkovic M., et al. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr. Biol. 2003;13:1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Shi L.H. Miao Y.L. Ouyang Y.C., et al. Trichostatin A (TSA) improves the development of rabbit-rabbit intraspecies cloned embryos, but not rabbit-human interspecies cloned embryos. Dev. Dyn. 2008;237:640–648. doi: 10.1002/dvdy.21450. [DOI] [PubMed] [Google Scholar]
- Shi W. Hoeflich A. Flaswinkel H., et al. Induction of a senescent-like phenotype does not confer the ability of bovine immortal cells to support the development of nuclear transfer embryos. Biol. Reprod. 2003;69:301. doi: 10.1095/biolreprod.102.012112. [DOI] [PubMed] [Google Scholar]
- Suemizu H. Aiba K. Yoshikawa T., et al. Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev. Biol. 2003;253:36–53. doi: 10.1006/dbio.2002.0870. [DOI] [PubMed] [Google Scholar]
- Svensson K. Mattsson R. James T.C., et al. The paternal allele of the H19 gene is progressively silenced during early mouse development: the acetylation status of histones may be involved in the generation of variegated expression patterns. Development. 1998;125:61. doi: 10.1242/dev.125.1.61. [DOI] [PubMed] [Google Scholar]
- Turner B.M. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Van Soom A. Van Vlaenderen I. Mahmoudzadeh A., et al. Compaction rate of in vitro fertilized bovine embryos related to the interval from insemination to first cleavage. Theriogenology. 1992;38:905–919. doi: 10.1016/0093-691x(92)90165-n. [DOI] [PubMed] [Google Scholar]
- Van Thuan N. Bui H.T. Kim J.H., et al. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction. 2009;138:309. doi: 10.1530/REP-08-0299. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Cibelli J.B. Wakayama T. Effect of timing of the removal of oocyte chromosomes before or after injection of somatic nucleus on development of NT embryos. Cloning Stem Cells. 2003;5:181–189. doi: 10.1089/153623003769645848. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Mizutani E. Kishigami S., et al. Mice cloned by nuclear transfer from somatic and ntES cells derived from the same individuals. J. Reprod. Dev. 2005;1:765–772. doi: 10.1262/jrd.17061. [DOI] [PubMed] [Google Scholar]
- Wakayama T. Yanagimachi R. Effect of cytokinesis inhibitors, DMSO and the timing of oocyte activation on mouse cloning using cumulus cell nuclei. Reproduction. 2001;122:49. doi: 10.1530/rep.0.1220049. [DOI] [PubMed] [Google Scholar]
- Wang F. Kou Z. Zhang Y., et al. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Reprod. 2007;77:1007. doi: 10.1095/biolreprod.107.063149. [DOI] [PubMed] [Google Scholar]
- Wang Y. Zhao X. Su J., et al. Lowering storage temperature during ovary transport is beneficial to the developmental competence of bovine oocytes used for somatic cell nuclear transfer. Anim. Reprod. Sci. 2011;124:48–54. doi: 10.1016/j.anireprosci.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Schnieke A.E. McWhir J., et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wu J. Wang S.H. Potter D., et al. Diverse histone modifications on histone 3 lysine 9 and their relation to DNA methylation in specifying gene silencing. BMC Gnomics. 2007;8:131. doi: 10.1186/1471-2164-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Li Y. Li G.P., et al. Trichostatin A improved epigenetic modifications of transfected cells but did not improve subsequent cloned embryo development. Anim. Botechnol. 2008;19:211–224. doi: 10.1080/10495390802271482. [DOI] [PubMed] [Google Scholar]
- Yang X. Smith S.L. Tian X.C., et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Zhao J. Ross J.W. Hao Y., et al. Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol. Reprod. 2009;81:525. doi: 10.1095/biolreprod.109.077016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Hao Y. Ross J.W., et al. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell. Reprogram. 2010;12:75–83. doi: 10.1089/cell.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]