Abstract
Background
Tuberculosis (TB) is a serious health problem in Tibet where Tibetans are the major ethnic group. Although genotyping of Mycobacterium tuberculosis (M. tuberculosis) isolates is a valuable tool for TB control, our knowledge of population structure of M. tuberculosis circulating in Tibet is limited.
Methodology/Principal Findings
In our study, a total of 576 M. tuberculosis isolates from Tibetans in Tibet, China, were analyzed via spoligotyping and 24-locus MIRU-VNTR. The Beijing genotype was the most prevalent family (90.63%, n = 522). Shared-type (ST) 1 was the most dominant genotype (88.89%, n = 512). We found that there was no association between the Beijing genotype and sex, age and treatment status. In this sample collection, 7 of the 24 MIRU-VNTR loci were highly or moderately discriminative according to their Hunter-Gaston discriminatory index. An informative set of 12 loci had similar discriminatory power with 24 loci set.
Conclusions/Significance
The population structure of M. tuberculosis isolates in Tibetans is homogeneous and dominated by Beijing genotype. The analysis of 24-locus MIRU-VNTR data might be useful to select appropriate VNTR loci for the genotyping of M. tuberculosis.
Introduction
Tuberculosis (TB) remains a major health problem in China. A 2000 national TB epidemiology survey conducted in China reported the average prevalence of TB amounts to 367 per 100,000 (0.0036%), with an estimated 4.5 million active pulmonary TB patients and 1.5 million new infections a year [1]. The prevalence rate of TB in western China was higher than central and eastern regions of the country. Furthermore, the increase of multi-drug resistant (MDR) TB in China inhibits the cure/treatment of the disease.
Tibet Autonomous Region (Tibet) is located in the Qinghai-Tibet Plateau of western China and Tibetans account for more than 90% of this population. Based on the 1990 national TB epidemiology survey in China, the prevalence rate of TB in Tibet (1203.06/100,000) was higher than anywhere else in China. In 2005, 4291 active pulmonary TB cases were reported in Tibet, posing a serious threat to the public health in Tibet [2].
Molecular typing of M. tuberculosis strains has proven to be a valuable tool for TB control in terms of tracking transmission chain, detecting suspected outbreaks, and identifying successful clones [3]. During the last few years, several PCR-based methods have been developed including spoligotyping and mycobacterial interspersed repetitive unit-variable number tandem repeat typing (MIRU-VNTR). Spoligotyping is a rapid and convenient typing method that is useful for the recognition of M. tuberculosis complex lineages on the basis of the presence or the absence of some specific spacer sequences in the direct repeat region of the mycobacterial genome [4], [5]. In addition, the database SITVIT2 (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo) has been developed for M. tuberculosis complex lineage identification by utilizing spoligotype signature matching [5]. Nevertheless, spoligotyping is not informative for Beijing genotype strains because almost all strains in this genotype share an identical spoligotype [6], [7]. The MIRU-VNTR method has proven to be faster and easier to perform and has been considered a good alternative to the gold standard method IS6110-RFLP [8], [9], [10], [11]. The 24-locus MIRU-VNTR method has been proposed to be the reference in standard typing and several reports have shown its appropriateness for population-based studies of TB transmission [10], [12], .
Our study aimed to examine the strain diversity and the prevalence of Beijing genotype strains in Tibet. The power of the 24-locus MIRU-VNTR scheme to differentiate the Beijing genotype strains from Tibet was also investigated.
Results
Description of isolates
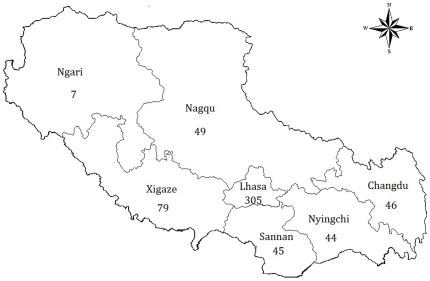
A total of 576 M. tuberculosis isolates from 329 male and 247 female Tibetan TB patients with a median age of 34 (range 8 to 85 years) were included in the study. These isolates comprised of 305 isolates from the Lhasa region, 79 from the Xigaze region, 49 from the Nagqu region, 46 from the Changdu region, 45 from the Sannan region, 44 from the Nyingchi region and 8 from the Ngari region (Figure 1). New cases consisted of 317 patients, and the remaining 259 patients were retreatment patients.
Figure 1. Map of Tibet showing the distribution of M. tuberculosis included in the present study (the number indicate the absolute number of isolates per region).
Spoligotyping
In terms of spoligotyping, we recognized a total of 22 distinct spoligotypes among the 576 isolates (Figure 2). Comparison of the spoligotyping results with the SITVIT2 database and application of the published rules for definition of the Beijing lineage (hybridized to at least three of the spacers 35 to 43 in the genomic direct-repeat region and showed an absence of hybridization to spacers 1 to 34) [7], permitted us to assign 567 isolates to 3 known spoligotype lineages, whereas 9 isolates could not be matched to any, and are, thus referred to as ‘new’ (Figure 2).
Figure 2. Spoligotypes of the 576 M. tuberculosis isolates.
From left to right: 1) UPGMA dendrogram generated by 22 spoligotypes. 2) spoligotying patterns. 3) Spoligo-International-Type number. 4) Genetic lineage according to SITVIT2 database. 5) Strain numbers.
Clustering analysis revealed that 563 isolates were grouped into 9 clusters containing 2 to 512 isolates, while the other 13 isolates inhibited unique spoligotypes. The largest spoligotype lineage was Beijing genotype(90.63%, 522 isolates), most (512 strains) of which belonged to the classical Beijing genotype with a pattern that depicted the absence of the first 34 spacer oligonucleotides and the presence of spacers 35 to 43 [7], [15]. The next most common lineage was the ill-defined T lineage with 27 strains (4.69%), followed by CAS lineage with 18 strains (3.13%). The high rate of clustering (96.18%) was due to the lower discriminatory power of spoligotyping for the Beijing genotype.
Furthermore, there were no statistical significant associations between the Beijing lineage and age (P>.05), sex (P>.05), and treatment history (P>.05) (Table 1) in our study.
Table 1. Statistic analysis between Beijing genotype and sex, age, and treatment status.
| No. | No. of Beijing genotype | OR | P | 95%CI | |
| All patients | 576 | 522 | |||
| Sex | |||||
| Male | 329 | 298 | 1 | ||
| Female | 247 | 224 | 0.983 | 1.000 | 0.558–1.731 |
| Age | |||||
| 0–19 | 57 | 49 | 1 | ||
| 20–39 | 340 | 312 | 0.561 | 0.211 | 0.242–1.300 |
| 40–59 | 143 | 129 | 0.678 | 0.457 | 0.268–1.716 |
| 60– | 36 | 32 | 0.781 | 0.763 | 0.217–2.809 |
| TB treatment history | |||||
| New patient | 317 | 287 | 1 | ||
| Treatment previously | 259 | 235 | 0.980 | 1.000 | 0.558–1.723 |
24-locus MIRU-VNTR
When using the 24-locus MIRU-VNTR method to typing the 576 isolates, the full set of results was obtained for 517 isolates. For 59 isolates, no PCR products were obtained at one or more loci. These observations remained consistent even after repeated testing. These findings might result from the chromosomal deletion, nucleotide polymorphism in the sequences complementary to PCR primers [16], or decreased quality of DNA. In this study, these cases whereby no PCR products were obtained were excluded from the cluster analysis.
In total, 24-locus MIRU-VNTR method differentiated 247 genotypes among the 517 isolates (Table S1). A total of 229 isolates had unique patterns and the remaining 228 formed 62 clusters (2 to 37 isolates per cluster). The allelic diversity of each MIRU-VNTR locus for 517 isolates was estimated by using the Hunter-Gason discriminatory index (HGDI) (Table 2). The discriminatory power for 2 loci (QUB11b and MIRU31) exceeded 0.6 and these were regarded as highly discriminatory [17]. Five loci (QUB26, Mtub21, QUB4156, MIRU26 and MIRU20) showed moderately discrimination (0.3≤h≤0.6). Other loci were found to be less polymorphic, with HGDI within the range of 0 to 0.3. Locus MIRU24 was monomorphic.
Table 2. HGDI of the 24 MIRU-VNTR loci for the whole sample and for the Beijing genotype isolates.
| ETRA | ETRB | ETRC | MIRU2 | MIRU4 | MIRU10 | MIRU16 | MIRU20 | MIRU23 | MIRU24 | MIRU26 | MIRU27 | |
| Whole sample | 0.169 | 0.109 | 0.110 | 0.003 | 0.097 | 0.193 | 0.222 | 0.413 | 0.031 | 0.000 | 0.482 | 0.078 |
| Beijing genotype | 0.089 | 0.030 | 0.049 | 0.000 | 0.082 | 0.030 | 0.171 | 0.434 | 0.034 | 0.000 | 0.430 | 0.056 |
Determination of a minimal set of MIRU-VNTR loci for differentiating Beijing genotype strains
To identify a minimal set of MIRU-VNTR loci for differentiating Beijing genotype strains in Tibet, the allelic diversity of each MIRU-VNTR locus were calculated separately (Table 2). When comparing the allelic diversity among all the isolates and Beijing genotype strains, 21 of the 24 MIRU-VNTR loci showed lower allelic diversity among Beijing genotype strains and were consistent with the close genetic relationships of those strains. The 3 loci that showed more allelic diversity among Beijing genotype strains were MIRU20, MIRU23 and Mtub29. Two loci (MIRU2 and MIRU24) were conserved among all Beijing genotype strains (h = 0.000).
Based on the allelic diversity of each MIRU-VNTR locus among Beijing genotype strains, the cumulative HGDI of the MIRU-VNTR locus combination was calculated and compared (Table 3). The cumulative HGDI of 22-locus VNTR was equal to that of the 24-locus MIRU-VNTR. The top 12 MIRU-VNTR loci combination appears to provide an HGDI close to that of the 24-locus MIRU-VNTR method.
Table 3. The cumulative HGDI with successive addition of each MIRU-VNTR locus.
| Locus combination | VNTR alias | VNTR locus | No. of patterns | No. of clusters | No. of clustered isolates | No. of isolates in each cluster | Clustering rate (%) | HGDI (cumulative) |
| 1 | VNTR2163 | QUB11b | ||||||
| 2 | VNTR3192 | MIRU31 | 24 | 17 | 466 | 2–135 | 94.9 | 0.8590 |
| 3 | VNTR4052 | QUB26 | 69 | 35 | 439 | 2–71 | 85.4 | 0.9332 |
| 4 | VNTR4156 | QUB4156 | 96 | 44 | 421 | 2–65 | 79.7 | 0.9506 |
| 5 | VNTR1955 | Mtub21 | 119 | 50 | 404 | 2–64 | 74.8 | 0.9582 |
| 6 | VNTR2059 | MIRU20 | 125 | 50 | 398 | 2–63 | 73.57 | 0.9598 |
| 7 | VNTR2996 | MIRU26 | 158 | 57 | 372 | 2–52 | 66.59 | 0.9713 |
| 8 | VNTR0424 | Mtub04 | 175 | 62 | 360 | 2–48 | 63.00 | 0.9751 |
| 9 | VNTR0802 | MIRU40 | 189 | 60 | 344 | 2–48 | 60.04 | 0.9784 |
| 10 | VNTR3690 | Mtub39 | 196 | 60 | 337 | 2–44 | 58.56 | 0.9805 |
| 11 | VNTR1644 | MIRU16 | 207 | 62 | 328 | 2–41 | 56.24 | 0.9830 |
| 12 | VNTR4348 | MIRU39 | 222 | 61 | 312 | 2–39 | 53.06 | 0.9850 |
| 13 | VNTR2165 | ETR A | 228 | 63 | 308 | 2–39 | 51.80 | 0.9855 |
| 14 | VNTR0580 | MIRU4 | 233 | 63 | 303 | 2–38 | 50.74 | 0.9860 |
| 15 | VNTR3007 | MIRU27 | 235 | 64 | 302 | 2–38 | 50.32 | 0.9863 |
| 16 | VNTR0577 | ETR C | 239 | 62 | 296 | 2–37 | 49.47 | 0.9867 |
| 17 | VNTR2401 | Mtub30 | 241 | 61 | 293 | 2–37 | 49.05 | 0.9869 |
| 18 | VNTR2531 | MIRU23 | 242 | 61 | 292 | 2–37 | 48.84 | 0.9870 |
| 19 | VNTR3171 | Mtub34 | 244 | 61 | 290 | 2–37 | 48.41 | 0.9872 |
| 20 | VNTR2461 | ETR-B | 245 | 62 | 290 | 2–37 | 48.20 | 0.9875 |
| 21 | VNTR0960 | MIRU10 | 246 | 62 | 289 | 2–37 | 47.99 | 0.9876 |
| 22 | VNTR2347 | Mtub29 | 247 | 62 | 288 | 2–37 | 47.78 | 0.9877 |
| 23 | VNTR0154 | MIRU2 | 247 | 62 | 288 | 2–37 | 47.78 | 0.9877 |
| 24 | VNTR2387 | MIRU24 | 247 | 62 | 288 | 2–37 | 47.78 | 0.9877 |
Discussion
Our results demonstrated that the population structure of M. tuberculosis isolates in Tibet appears to be very homogeneous, as only 3 spoligotype lineages were obtained for the 576 isolates, with 90.63% of the isolates belonging to the Beijing genotype. The predominance of the Beijing genotype in China is well documented in other studies. It is prevalent in Beijing (80 to 92.6%), Tianjin (91.7%), Heilongjiang (89.5%), Jilin (89.9%), and Shanghai (89%), but less prevalent in Guangxi (55.3%), Fujian (54.5%), and Guangdong (25%) [15], [18], [19], [20], [21]. Hence, Tibet is one of the regions where the proportion of the Beijing genotype is the highest. Owing to the special geographic and living habit, the predominance of a narrow range of genotypes might imply their long-standing presence in Tibet and maybe linked with limited contact with other populations.
Our results showed that there was no association between the prevalence of Beijing genotype and sex, age, and treatment status. This indicated that maybe there were other factors that contributed to the spread of Beijing genotype strains. Demographic factors may be responsible for the dominance of Beijing genotype strains based on a co-evolution between the host and the pathogen [22], [23]. This estimation for the correlation between the Beijing genotype and sex, age, and treatment status were, perhaps, biased by a smaller sample size of non-Beijing genotype strains compared to Beijing genotype strains.
We found that the spoligotyping method could not effective distinguish Beijing genotype strains. Therefore in our study, these strains were further subjected to the newly proposed 24-locus MIRU-VNTR method and we found that the allelic diversity of the VNTR loci varied significantly at each locus. Among the 24 loci we investigated, QUB11b and MIRU31 were highly discriminative (h≥0.6), QUB4156, Mtub21, MIRU20 and MIRU26 were moderately discriminative, and other loci were poorly discriminative. When scrutinizing the 24-locus scheme, we found that MIRU24 and MIRU2 remained monomorphic in every published setting in China, Japan and Russia [20], [24], [25]. Locus MIRU24 remained monomorphic, which mirrorsi the previous observation that this locus is phylogenetically conserved.
When comparing the HGDI of this locus set with the VNTR loci reported in other areas (Table 4) [20], [24], [25], [26], [27], [28], [29], [30], we found that the allelic diversity of these loci were different from that described in other reports related to analyses of Beijing genotype strains. Many lowly polymorphic loci (MIRU31 and MIRU20) from Beijing, Shanghai, Wuhan, Hong Kong, and Russia may be able to discriminate Beijing genotype strains in Tibet. Most of the VNTR loci showed higher discriminatory power for Japan than China and Russia. A generally lower discriminatory power of VNTR loci for Russia may be considered a reflection of the recent clonal expansion of the Beijing genotype strains in that country. Although MIRU-VNTR loci showed a variation in the ability to differentiate Beijing genotype strains from different geographical areas, this may be attributed to the dissimilarities in the population structure of the circulating M. tuberculosis strains in distinct geographic areas.
Table 4. Allelic diversity of different MIRU-VNTR in Beijing genotype strains from different locations.
| VNTR alias | Tibet, China(this study) | Beijing, China | Heilongjiang China | Shanghai, China | Wuhan, China | Hong Kong,China | Hong Kong, China | Kobe, Japan | St. Petersburg, Russia |
| QUB11b | 0.694 | 0.651 | 0.704 | 0.655 | 0.669 | 0.618 | 0.772 | 0.205 | |
| MIRU31 | 0.617 | 0.169 | 0.395 | 0.246 | 0.23 | 0.156 | 0.200 | 0.322 | 0.160 |
| QUB26 | 0.525 | 0.518 | 0.607 | 0.595 | 0.314 | 0.299 | 0.741 | 0.636 | |
| QUB4156 | 0.519 | 0.395 | 0.182 | 0.492 | 0.167 | 0.611 | 0.082 | ||
| Mtub21 | 0.491 | 0.556 | 0.396 | 0.523 | 0.393 | 0.330 | |||
| MIRU20 | 0.440 | 0.014 | 0.061 | 0.25 | 0.022 | 0.120 | |||
| MIRU26 | 0.429 | 0.353 | 0.596 | 0.612 | 0.60 | 0.200 | 0.383 | 0.520 | |
| Mtub04 | 0.224 | 0.306 | 0.391 | 0.297 | 0.459 | 0 | |||
| MIRU40 | 0.221 | 0.194 | 0.292 | 0.147 | 0.23 | 0.196 | 0.327 | 0.122 | |
| Mtub39 | 0.166 | 0.171 | 0.174 | 0.061 | 0.186 | 0 | |||
| MIRU16 | 0.158 | 0.068 | 0.200 | 0.242 | 0.55 | 0.058 | 0.310 | 0.082 | |
| MIRU39 | 0.147 | 0.119 | 0.290 | 0.286 | 0.03 | 0.320 | 0.221 | 0 | |
| ETR A | 0.090 | 0.232 | 0.238 | 0.031 | 0.188 | 0.201 | 0.147 | 0.158 | |
| MIRU4 | 0.066 | 0.120 | 0.212 | 0.061 | 0.08 | 0.072 | 0.019 | 0.086 | 0 |
| MIRU27 | 0.058 | 0.014 | 0.031 | 0.08 | 0.115 | 0 | |||
| ETR C | 0.054 | 0.094 | 0.057 | 0.165 | 0.022 | 0.042 | |||
| Mtub30 | 0.033 | 0.068 | 0.133 | 0.091 | 0.403 | 0.042 | |||
| MIRU23 | 0.033 | 0.014 | 0.061 | 0.25 | 0.176 | 0 | |||
| Mtub34 | 0.029 | 0.014 | 0.089 | 0.065 | 0 | ||||
| ETR B | 0.029 | 0.014 | 0 | 0.064 | 0 | 0 | 0 | ||
| MIRU10 | 0.025 | 0.144 | 0.154 | 0.195 | 0.52 | 0.377 | 0.419 | 0.082 | |
| Mtub29 | 0.013 | 0.119 | 0.123 | 0.061 | 0.043 | 0.087 | |||
| MIRU2 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| MIRU24 | 0 | 0 | 0 | 0 | 0 | 0 |
The 24-locus MIRU-VNTR scheme has some technical limitations: in China, MIRU-VNTR can be only performed manually and, consequently, was very time-consuming and tedious for this study. Some reports demonstrated that combination of 24-locus MIRU-VNTR and spoligotyping could improve the discriminatory power of M. tuberculosis strains. In addition, not all 24 loci are required for genotyping M. tuberculosis strains in any given situation because the number of loci required depends on the population structure of M. tuberculosis. In this study, the top 12 loci also demonstrated a high discriminatory power among Beijing genotype strains. A 12-locus VNTR typing method proposed in Japan is reported to be appropriate for discrimination of Beijing strains [31], but the discriminatory power of these loci sets require further study in different settings for other areas.
In conclusion, Beijing genotype strains appeared to be widely disseminated across Tibet. The analysis of MIRU-VNTR data might be useful to select appropriate VNTR loci for the genotyping of M. tuberculosis.
Materials and Methods
Ethics statement
The study was approved by the Ethics Committee of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. All patients in the study signed informed consent form.Clinical isolates and DNA samples.
We randomly collected a total of 590 M. tuberculosis isolates between 2006 and 2010 from 590 Tibetan patients at seven different regional Center of Disease Prevent and Control (CDC) in Tibet (Lhasa, Xigaze, Nyingchi, Sannan, Nagqu, Changdu, and Ngari). Chromosomal DNA was extracted by boiling a loopful of colonies from L-J slants in 400 µL of 10 mM tris-HCl and 1 mM EDTA (pH8.0) buffer for 10 minutes. The suspension was centrifuged at 12000 rpm for 10 minutes, and the supernatant was stored at −20°C until further use [32]. Fourteen isolates were excluded because of sample contamination and/or mixed infection as detected by double alleles in two or more MIRU-VNTR loci [10], [33]. A final sample of 576 isolates was retained for analysis.
Molecular typing methods
We performed spoligotyping according to a standard protocol as described by Kamerbeek et al [4]. We entered the results in an Excel spreadsheet in a 43-digit binary format representing the 43 spacers [5] and compared them to SITVIT2 database (an international spoligotype database at the Institute Pasteur de Guadeloupe), which is an updated version of the published SpolDB4 database [5].
In terms of the 24-locus MIRU-VNTR typing, we used PCR primers flanking each of the 24 loci that were described by Supply et al [10]. Each MIRU-VNTR locus was amplified individually in a 25-µL reaction volume in a 0.2-ml PCR tube. PCR products were analyzed by electrophoresis on a 2% agarose gel using 100 bp DNA ladder as size markers. The H37Rv strain was run as an additional control for accuracy. Sizing of the PCR fragments and assignment of the VNTR alleles were done using Bionumerics software version 5.0 (Applied Maths, Sint-Martens Laten, Belgium). Clusters were defined when 100% similarity was observed between patterns.
To minimize the risk of laboratory cross-contamination, DNA extraction and PCR amplification were conducted in separate rooms. The PCR laboratory has four disconnected rooms for preparation of the PCR mixtures, addition of the DNA, PCR amplification, and electrophoretic fractionation. Negative controls (sterile water) were included to control for cross contamination.
Analysis of genotyping
Bionumerics software version 5.0 and MIRU-VNTRplus (http://www.miru-vntrplus.org) were used to analyze genotyping data [34]. Clustering analysis was done using the unweighted pair group method with arithmetic averages (UPGMA). The Dice and categorical coefficients were used in spoligotyping and MIRU-VNTR, respectively. The Hunter-Gaston discriminatory index (HGDI) was used to evaluate the discriminatory power of the typing methods and the allelic diversity of the VNTR loci [35]. The clustering rate was defined as (nc - c)/n, whereby nc is the total number of clustered cases, c is the number of clusters, and n is the total number of cases in the sample [36].
The chi square test was used to assess association of Beijing genotye with sex, age, and treatment status by using SPSS 11.5 (SPSS Inc., Chicago, IL, USA). P<.05 was defined as statistically significant.
Supporting Information
24-locus MIRU-VNTR profile and spoligotyping profile of 517 isolates. The table provides the 24-locus MIRU-VNTR typing data and spoligotpying data in 517 isolates.
(XLSX)
Acknowledgments
We thank the staffs of the respective institutes in Tibet for their contribution to this study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the project “Transmission Mode of Tuberculosis” (2008ZX100/03-010-02) of National Key Program of Mega Infectious Disease and by the project of National Nature Science Funding in China (30860243). It was also funded by the project of State Key Laboratory for Infectious Disease Prevention and Control (2011SKLID2008).
References
- 1.National Technic Steering Group Of The Epidemiological Sampling Survey For T, Duanmu H. [Report on fourth national epidemiological sampling survey of tuberculosis]. Zhonghua Jie He He Hu Xi Za Zhi. 2002;25:3–7. [PubMed] [Google Scholar]
- 2.Tian R, Suolangzhaxi, He L. Analysis of tuberculosis patients registered in Tibet from 1994 to 2005. Tibet's Science & Techology. 2011;214:2. [Google Scholar]
- 3.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 4.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002;8:843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, et al. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol. 2004;42:4040–4049. doi: 10.1128/JCM.42.9.4040-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, et al. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 9.Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci U S A. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frothingham R, Meeker-O'Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144(Pt 5):1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 12.Maes M, Kremer K, van Soolingen D, Takiff H, de Waard JH. 24-locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis (Edinb) 2008;88:490–494. doi: 10.1016/j.tube.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Oelemann MC, Diel R, Vatin V, Haas W, Rusch-Gerdes S, et al. Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007;45:691–697. doi: 10.1128/JCM.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valcheva V, Mokrousov I, Narvskaya O, Rastogi N, Markova N. Utility of new 24-locus variable-number tandem-repeat typing for discriminating Mycobacterium tuberculosis clinical isolates collected in Bulgaria. J Clin Microbiol. 2008;46:3005–3011. doi: 10.1128/JCM.00437-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Affolabi D, Anyo G, Faihun F, Sanoussi N, Shamputa IC, et al. First molecular epidemiological study of tuberculosis in Benin. Int J Tuberc Lung Dis. 2009;13:317–322. [PubMed] [Google Scholar]
- 17.Mokrousov I, Narvskaya O, Limeschenko E, Vyazovaya A, Otten T, et al. Analysis of the allelic diversity of the mycobacterial interspersed repetitive units in Mycobacterium tuberculosis strains of the Beijing family: practical implications and evolutionary considerations. J Clin Microbiol. 2004;42:2438–2444. doi: 10.1128/JCM.42.6.2438-2444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai LQ, Li WM, Li L, Dai ZJ, Bai DP, et al. [Study on the genotype of Mycobacterium tuberculosis isolates from hospitals in Tianjin]. Zhonghua Liu Xing Bing Xue Za Zhi. 2007;28:785–788. [PubMed] [Google Scholar]
- 19.Li WM, Wang SM, Pei XY, Liu ZQ, Zhong Q, et al. [DNA fingerprinting of Mycobacterium tuberculosis strains from Beijing, Guangdong and Ningxia]. Zhonghua Liu Xing Bing Xue Za Zhi. 2003;24:381–384. [PubMed] [Google Scholar]
- 20.Jiao WW, Mokrousov I, Sun GZ, Guo YJ, Vyazovaya A, et al. Evaluation of new variable-number tandem-repeat systems for typing Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J Clin Microbiol. 2008;46:1045–1049. doi: 10.1128/JCM.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H, Liu Z, Lv B, Zhang Y, Liu J, et al. Spoligotypes of Mycobacterium tuberculosis from different Provinces of China. J Clin Microbiol. 2010;48:4102–4106. doi: 10.1128/JCM.00549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oota H, Kitano T, Jin F, Yuasa I, Wang L, et al. Extreme mtDNA homogeneity in continental Asian populations. Am J Phys Anthropol. 2002;118:146–153. doi: 10.1002/ajpa.10056. [DOI] [PubMed] [Google Scholar]
- 23.Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NT, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwamoto T, Yoshida S, Suzuki K, Tomita M, Fujiyama R, et al. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol Lett. 2007;270:67–74. doi: 10.1111/j.1574-6968.2007.00658.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Chen J, Shen X, Gui X, Mei J, et al. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282:22–31. doi: 10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Liu Y, Zhang CL, Ji BY, Zhang LZ, et al. Genotypes and characteristics of clustering and drug susceptibility of Mycobacterium tuberculosis isolates collected in Heilongjiang Province, China. J Clin Microbiol. 2011;49:1354–1362. doi: 10.1128/JCM.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H, Wang F, Xiao Y, Ren Y, Chao Y, et al. Utility of mycobacterial interspersed repetitive unit typing for differentiating Mycobacterium tuberculosis isolates in Wuhan, China. J Med Microbiol. 2007;56:1219–1223. doi: 10.1099/jmm.0.47005-0. [DOI] [PubMed] [Google Scholar]
- 28.Kam KM, Yip CW, Tse LW, Leung KL, Wong KL, et al. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol Lett. 2006;256:258–265. doi: 10.1111/j.1574-6968.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 29.Kremer K, Au BK, Yip PC, Skuce R, Supply P, et al. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005;43:314–320. doi: 10.1128/JCM.43.1.314-320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mokrousov I, Narvskaya O, Vyazovaya A, Millet J, Otten T, et al. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J Clin Microbiol. 2008;46:3576–3584. doi: 10.1128/JCM.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murase Y, Mitarai S, Sugawara I, Kato S, Maeda S. Promising loci of variable numbers of tandem repeats for typing Beijing family Mycobacterium tuberculosis. J Med Microbiol. 2008;57:873–880. doi: 10.1099/jmm.0.47564-0. [DOI] [PubMed] [Google Scholar]
- 32.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, et al. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir Res. 2006;7:99. doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
24-locus MIRU-VNTR profile and spoligotyping profile of 517 isolates. The table provides the 24-locus MIRU-VNTR typing data and spoligotpying data in 517 isolates.
(XLSX)