Abstract
Escherichia coli G3/10 is a component of the probiotic drug Symbioflor 2. In an in vitro assay with human intestinal epithelial cells, E. coli G3/10 is capable of suppressing adherence of enteropathogenic E. coli E2348/69. In this study, we demonstrate that a completely novel class II microcin, produced by probiotic E. coli G3/10, is responsible for this behavior. We named this antibacterial peptide microcin S (MccS). Microcin S is coded on a 50.6 kb megaplasmid of E. coli G3/10, which we have completely sequenced and annotated. The microcin S operon is about 4.7 kb in size and is comprised of four genes. Subcloning of the genes and gene fragments followed by gene expression experiments enabled us to functionally characterize all members of this operon, and to clearly identify the nucleotide sequences encoding the microcin itself (mcsS), its transport apparatus and the gene mcsI conferring self immunity against microcin S. Overexpression of cloned mcsI antagonizes MccS activity, thus protecting indicator strain E. coli E2348/69 in the in vitro adherence assay. Moreover, growth of E. coli transformed with a plasmid containing mcsS under control of an araC PBAD activator-promoter is inhibited upon mcsS induction. Our data provide further mechanistic insight into the probiotic behavior of E. coli G3/10.
Introduction
Microcins are ribosomally synthesized antimicrobial peptides with a low molecular mass. Produced by enterobacteria, mostly Escherichia coli, microcin synthesis is sharply activated under stress conditions such as limitation of nutrients [1]–[4]. Microcins exert potent antibacterial activity against closely related species, which offers a highly competitive advantage in the intestinal microflora [5]. Microcin-producers are resistant to the microcin they produce, which is mediated by at least one resistance-conferring gene located within one gene cluster. Most of the 14 known microcins are plasmid-encoded [5]–[10], but chromosomally-encoded antibacterial peptides have also been described [11]. The probiotic strain E. coli Nissle 1917 (EcN) is known to produce microcins M and H47 [12]. In this study we show that probiotic E. coli G3/10 produces a novel microcin, that we named microcin S (MccS). E. coli G3/10 is one of six E. coli genomotypes present in the probiotic drug Symbioflor 2 (DSM17252). the product has been successfully used for the treatment of functional gastrointestinal disorders, in particular irritable bowel syndrome in adults and children [13], [14]. Probiotics are defined as living microorganisms, which upon ingestion in certain numbers, exert health benefits beyond inherent basic nutrition [15]. The mechanisms that enable a strain to serve as a probiotic are poorly understood. Nevertheless, the antimicrobial activity of microcins could positively influence the stability of the intestinal microflora. Given its extensive clinical safety record, a microcin-producing strain containing no virulence factors can clearly fulfill the definition of a probiotic. In contrast to enterobacterial microcins, food-borne lactic acid bacteria produce lanthionine-containing peptide antibiotics. The so-called lantibiotics of gram-positive bacteria are already used for food preservation [16]. The use as an antitumor agent [17] or as an alternative to classical antibiotics in infectious diseases [18], [19] are two further applications where bacteriocins may provide therapeutic alternatives in the future. The worldwide emergence of pathogens resistant to antibiotics has led to an ever-increasing demand of new antibacterial agents. Enterobacterial microcins could offer exciting new possibilities for prophylaxis and treatment of bacterial infections. Here we present the identification and functional characterization of microcin S, a completely novel plasmid encoded bacteriocin produced by probiotic E. coli G3/10. Microcin S is able to inhibit the adherence of enteropathogenic E. coli (EPEC) strain E2348/69 to intestinal epithelial cells in an in vitro adherence assay and growth of E. coli is hampered by L-arabinose induced recombinant expression of MccS.
Results
Evaluation of the In vitro Adherence Assay
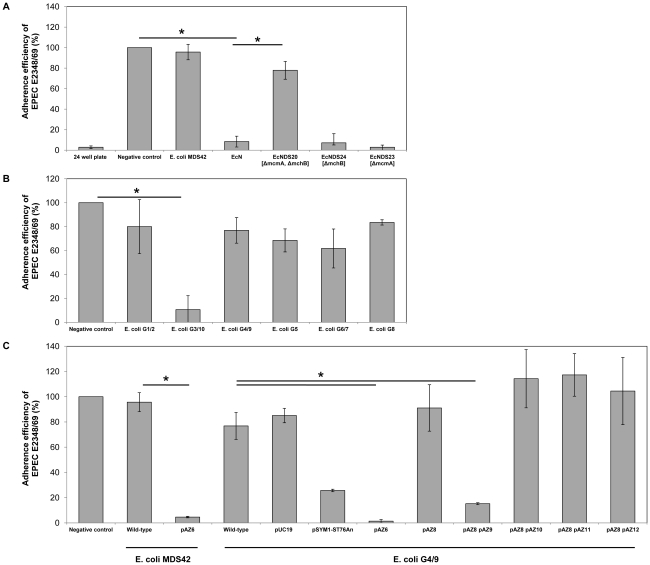
Bacterial adhesion is a crucial first step of many infectious diseases. Therefore, a test system quantifying adherence inhibition of enteropathogens to human intestinal epithelial cells is a suitable model system to evaluate this beneficial effect to the host. Initially, we used enteropathogenic E. coli strain E2348/69 [20] to investigate adherence efficiency to human intestinal epithelial cells (LOVO or CACO-2) in response to a pre-incubation with different probiotic or non-probiotic E. coli isolates. We demonstrated that EcN significantly inhibits EPEC adherence (Fig. 1A), which is consistent with the results of Kleta et al. [21] who used a similar assay with porcine intestinal IPEC-J2 cells. Since EcN produces two different microcins M and H47 [12], we assumed that the observed effect resulted from those antibacterial peptides, which can inhibit bacterial growth and kill target bacteria. Indeed, we were able to show that an EcN deletion strain (EcNDS20), negative for both microcins M and H47, is not able to inhibit EPEC adhesion (Fig. 1A), whereas deletion of only one microcin (EcNDS23 and EcNDS24) resulted in wild-type behavior (Fig. 1A). Therefore, the in vitro adherence assay used here also provides a suitable test environment for detecting bactericidal activity of a given strain specifically directed against the used adherent strain.
Figure 1. Adherence efficiency of EPEC E2348/69 to human intestinal epithelial cells after pre-incubation with E. coli Nissle 1917 (EcN) and EcN deletion strains (EcNDS) (A); E. coli G1/2, E. coli G3/10, E. coli G4/9, E. coli G5, E. coli G6/7 and E. coli G8, the components of Symbioflor 2 (DSM17252) (B); E. coli strains MDS42 and G4/9 wild-type and appropriate mutants (C).
Confluent monolayers of CACO-2 or LOVO cells were pre-incubated with bacterial test strains at an MOI of 100∶1 E. coli to host cells. After two hours of incubation, cells were washed and infected with EPEC E2348/69 using an MOI of 100∶1 EPEC to host cells. Adherence efficiency in % is expressed as adherence of EPEC relative to the adherence without any pre-incubation (negative control), which is set at 100%. Pre-incubation of human intestinal epithelial cells with EcN significantly reduces adherence efficiency of EPEC E2348/69, comparable to the mutant strains EcNDS24 and EcNDS23 which carry single genomic deletions of either microcin H47 precursor (mchB; EcNDS24) or microcin M precursor (mcmA; EcNDS23). However, this phenomenon cannot be observed, when double microcin mutant EcNDS20 is used (A). The adherence efficiency of EPEC E2348/69 is also significantly reduced in the presence of microcin S, either expressed by E. coli G3/10 wild-type (B) or by E. coli strains MDS42 and G4/9, transformed with plasmids pSYM1-ST76An (mcsS, mcsI, mcsA, mcsB), pAZ6 (mcsS, mcsI, mcsA, mcsB) or pAZ9 (mcsS) (C). With all further plasmids, pAZ8 (mcsI, mcsA, mcsB), pAZ10 (ORF1), pAZ11 (ORF2) and pAZ12 (mcsS 193–363), the indicator strains E. coli MDS42 and G4/9 did not inhibit adherence of EPEC E2348/69 to intestinal epithelial cells (C). Data are the mean±SD of at least three separate experiments in duplicate wells. * p≤0.01 compared to negative controls or wild-type strains.
Identification of the Microcin S-Encoding Gene Cluster in E. coli G3/10
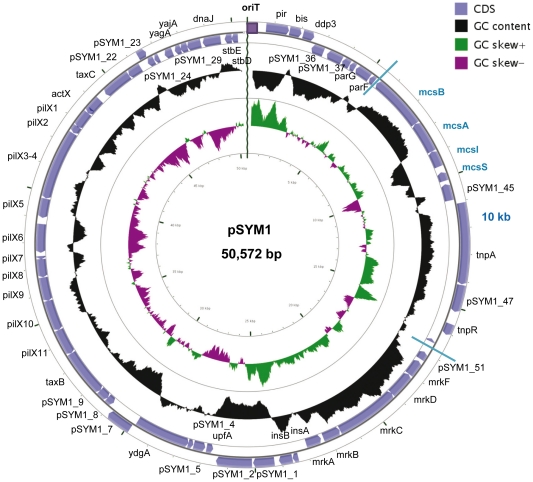
The EPEC in vitro adherence assay was then repeated with all genomotypes of the probiotic drug Symbioflor 2, which are E. coli G1/2, G3/10, G4/9, G5, G6/7 and G8. Surprisingly, we could show that E. coli G3/10 also significantly inhibited EPEC adherence efficiency (Fig. 1B) indicating a bactericidal activity of that strain. The genome of E. coli G3/10 was sequenced in our laboratory (unpublished data). However, neither during manual editing of the automatically annotated sequence nor with a BLAST analysis, could any coding sequences of a known microcin be identified within the genome of E. coli G3/10. Microcins are often encoded by plasmids [5]. E. coli G3/10 contains a large conjugative plasmid pSYM1 (Fig. 2), having a size of 50.6 kb. The plasmid is 99% identical to plasmid pMAS2027 of uropathogenic E. coli MS2027 [22]. However, it additionally contains a 10 kb insertion fragment, which carries only uncharacterized and unnamed genes. To identify the origin of E. coli G3/10s bactericidal action we first tried to cure the strain from its megaplasmid pSYM1. However, while using several common curing procedures such as treatment with mitomycin C or heat, we failed to remove pSYM1 from E. coli G3/10. Therefore, the plasmid was transferred to E. coli G4/9 by conjugation. To allow screening of conjugants, we first integrated an ampicillin resistance cassette into pSYM1 resulting in pSYM1-ST76An (Table 1). E. coli G4/9 transformed with pSYM1-ST76An was able to inhibit EPEC adherence significantly (Fig. 1C). It logically followed that plasmid pSYM1 carries genes responsible for the observed effect. Then, we cloned a 4.7 kb subfragment of plasmid pSYM1 into pBR322, resulting in plasmid pAZ6 (Table 1), which was subsequently transformed into E. coli G4/9. We demonstrated that pAZ6 enables E. coli G4/9 to inhibit EPEC adherence efficiency significantly (Fig. 1C), indicating that the 4.7 kb fragment of pSYM1 (Fig. 3) is responsible for the EPEC adherence inhibition effect of E. coli G3/10. BLAST analysis of automatically annotated open reading frames (ORFs) in this segment of pSYM1 revealed small homologies to characterized proteins or protein families belonging to microcin-encoding operons. Nevertheless, the microcin itself remained undetected. For this reason, we cloned three genes, named mcsI, mcsA and mcsB in the following, into pBR322 leading to pAZ8 (Table 1). Small ORFs upstream of this operon (Fig. 3), which were candidate microcin-encoding genes, were cloned into pACYC184 and subcloned into E. coli G4/9 pAZ8. With this strategy we successfully managed a sequential plasmid-based identification and characterization of potential microcin-coding regions together with the respective microcin-helper proteins. Only E. coli G4/9, containing plasmids pAZ8 and pAZ9 (Table 1) together, significantly inhibited EPEC adherence, whereas E. coli G4/9 pAZ8 affected EPEC adherence similar to that of E. coli G4/9 wild-type (Fig. 1C). We therefore concluded that the small gene cloned into pAZ9, which we termed mcsS, encodes a novel E. coli microcin, named microcin S in the following. Truncated mcsS (pAZ12; Table 1) resulting from an alternative ORF did not show microcin activity in the in vitro adherence assay (Fig. 1C).
Figure 2. Circular diagram of megaplasmid pSYM1.
The outer circle indicates (to scale) the genetic organization of ORFs within the plasmid. The direction of transcription of each ORF is indicated. The middle circle indicates the GC content, and the inner circle indicates the GC skew. The 10 kb region of pSYM1 that is different from pMAS2027 is marked. The image was constructed using cgview [41]. CDS = coding sequences.
Table 1. Plasmids used in this study.
| Plasmid | Resistance | Relevant genotype | Origin |
| pSYM1 | None | mcsS, mcsI, mcsA, mcsB | This work |
| pST76–A | Ampr | oriTS (30°C) | [43] |
| pSYM1–ST76An | Ampr | mcsS, mcsI, mcsA, mcsB | This work |
| pAZ6 | Ampr | mcsS, mcsI, mcsA, mcsB | This work |
| pAZ8 | Ampr | mcsI, mcsA, mcsB | This work |
| pAZ9 | Cmr | mcsS | This work |
| pAZ10 | Cmr | ORF1 | This work |
| pAZ11 | Cmr | ORF2 | This work |
| pAZ12 | Cmr | mcsS 193–363 | This work |
| pAZ13 | Ampr | mcsI | This work |
| pAZ14 | Ampr | mcsI 361–651 | This work |
| pGS1 | Ampr | PBAD | This work |
| pAZ15 | Ampr | PBAD, mcsS | This work |
Ampr = ampicillin resistance; Cmr = chloramphenicol resistance; ORF = open reading frame; PBAD = araC PBAD activator-promoter.
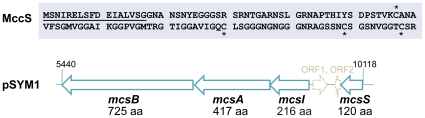
Figure 3. The MccS gene cluster.
The upper panel shows the amino acid sequence of the microcin S precursor. A probable leader peptide is underlined. Asterisks indicate cysteines possibly involved in the formation of a disulfide bond typical for class II microcins. The microcin S gene cluster on megaplasmid pSYM1 (lower panel) consists of the four clustered genes mcsS, mcsI, mcsA and mcsB. ORF1 and ORF2 revealed no bactericidal activity.
Microcin S Immunity
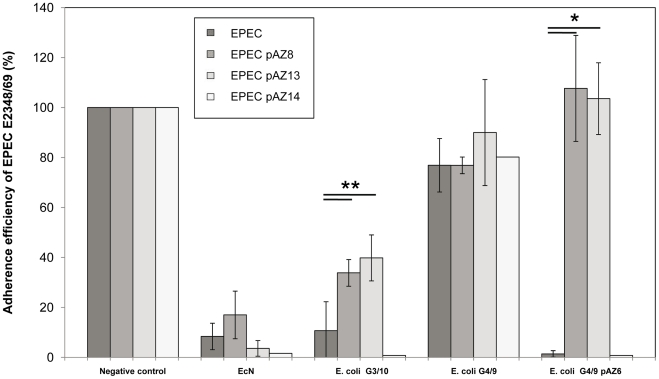
Class II microcins are characterized by a dedicated self-immunity protein. Gene mcsI adjacent to mcsS shows homology to the CAAX amino terminal protease family potentially involved in microcin self-immunity. No signal sequence or transmembrane helices are predicted. We transformed mcsI into EPEC strain E2348/69 using either pAZ8 (mcsI, mcsA, mcsB), pAZ13 (mcsI ) or pAZ14 (mcsI, truncated). We could show that the EPEC strain carrying the complete mcsI gene (pAZ8 or pAZ13) is resistant to microcin S-producing E. coli G3/10 or E. coli G4/9 pAZ6 in the in vitro adherence assay. Truncated mcsI (pAZ14) was not able to confer microcin S resistance. Therefore we identified gene mscI, which encodes a 216 amino acid protein, as responsible for microcin S self-immunity. Additionally, we demonstrated that mcsI is not effective against EcN microcins M and H47 (Fig. 4).
Figure 4. Adherence efficiency of wild-type and mutant strains of EPEC E2348/69 to human intestinal epithelial cells after pre-incubation with different microcin positive and negative E. coli strains.
Confluent monolayers of CACO-2 or LOVO cells were pre-incubated with bacterial test strains EcN, E. coli G3/10, microcin-negative E. coli G4/9 wild-type or microcin S expressing E. coli G4/9 pAZ6 at an MOI of 100∶1 E. coli to host cells. After two hours of incubation, cells were washed and infected with EPEC E2348/69 using an MOI of 100∶1 EPEC to host cells. Adherence efficiency in % is expressed as adherence of EPEC relative to the adherence without any pre-incubation (negative control), which is set at 100%. In EPEC E2348/69 plasmids pAZ8 (mcsI, mcsA, mcsB) and pAZ13 (mcsI) containing full-length gene mcsI confer immunity to mcsS expressing strains E. coli G3/10 and E. coli G4/9 pAZ6. However, EPEC indicator strain transformed with pAZ14 carrying truncated mcsI361-651 is not resistant to microcin S. None of the plasmids used procures immunity against EcN microcins H47 and M. Data are the mean±SD of at least three separate experiments in duplicate wells. * p ≤ 0.01, ** p ≤ 0.05.
Investigation of Microcin S Activity in E. coli MDS42
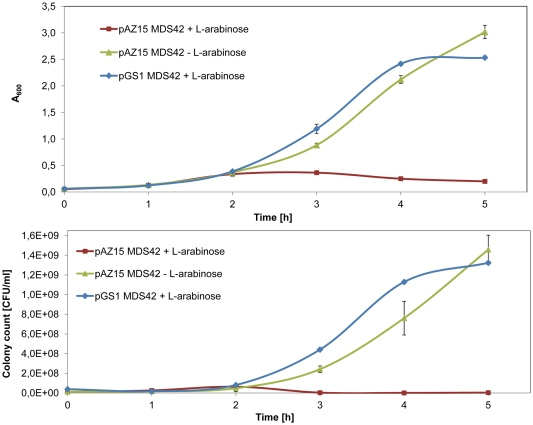
In order to directly demonstrate microcin S activity against a susceptible E. coli strain, mcsS was cloned into pGS1 resulting in the plasmid pAZ15 (Table 1). In this construct, mcsS expression is controlled by an araC PBAD activator-promoter, rendering microcin S expression inducible by L-arabinose. When E coli MDS42 [23] growing in liquid culture after being transformed with pAZ15 was treated with 0.2% v/v L-arabinose at the beginning of the logarithmic phase after 1.5 h, A600 remained almost stable around 0.3 while absorbance of control cultures increased constantly (Fig. 5A). Counts of colony forming units (CFU) taken at various time points during this experiment revealed a sharp drop in the number of viable bacteria in the L-arabinose induced culture of E. coli MDS42+pAZ15 while CFU of the other cultures rose to a maximum of 1.4 × 109/ ml (Fig. 5B).
Figure 5. Growth curve of E. coli MDS42 with pAZ15 or pGS1 showing A600(A) and colony count of viable cells (B).
pAZ15 is a vector containing mcsS under control of the L-arabinose-induced araC PBAD activator-promoter. Induction with L-arabinose of the respective cultures, as indicated in the legend to panel A and B, was carried out after 90 minutes. pGS1 is the empty vector with the araC PBAD activator-promoter. E. coli MDS42 with pAZ15 shows a significant reduction of its A600 as well as of its colony counts after induction with L-arabinose (red squares) compared to growth without induction (green triangles) and also with the vector devoid of mcsS (pGS1, blue rhombi). Data are the mean±SD of three independent experiments.
Presence of Microcin S Gene in Enterobacteriaceae
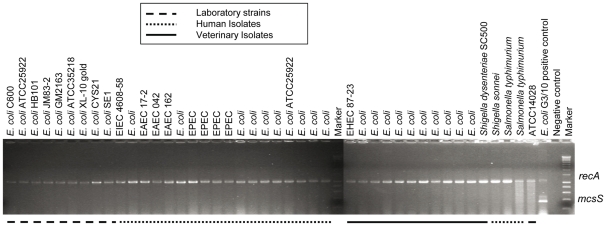
The sequences of mcsS and mcsI were listed very recently (04.09.2011) as new entries in the NCBI database, being part of the 54 kb megaplasmid pMO17_54 (accession # HE578057) of Shigella sp. MO17. The proteins derived from these coding sequences, termed pMO17_54_21 and pMO17_54_23, were described as hypothetical proteins and no function was assigned to them. Nevertheless, a complete MccS operon is not listed in NCBI (as of 21.10.2011). A further 38 E. coli, two Shigella and two Salmonella strains were screened for mcsS using a multiplex PCR protocol. We were unable to detect the mcsS gene in any of the 42 human and veterinary isolates or the laboratory strains tested (Fig. 6).
Figure 6. Multiplex PCR to screen for presence of mcsS using recA as inhibition control.
The tested strains are common laboratory strains, human clinical isolates and veterinary isolates of different origin. mcsS could not be detected in any of the tested strains while the amplified inhibition control indicated that all PCR reactions were truly negative.
Discussion
Microcins are small ribosomally synthesized peptides produced by enterobacteria. We could show that E. coli G3/10, an E. coli genomotype of Symbioflor 2 (DSM17252), encodes and produces a completely novel class II microcin, named microcin S. The term microcin was introduced to distinguish this class of antibacterial peptides with a size <10 kDa from colicins with a higher molecular mass [24]. Genome sequencing of E. coli G3/10 enabled us to indentify the gene mcsS, located on plasmid pSYM1, encoding the 120 amino acid (aa) microcin S precursor peptide. It has a calculated molecular weight of 11.67 kDa and is therefore the largest of all currently known microcins. Using the Pfam database [25] microcin S can be classified as a class II bacteriocin with a double-glycine leader peptide. Indeed, its amino acid sequence reveals a glycine-rich peptide with a double-glycine cleavage site and cysteines probably involved in formation of a disulfide bond (Fig. 3). These characteristics, together with the organization of the microcin gene cluster (Fig. 3), indicate MccS to be a member of the class IIa microcins. The current classification follows a discussion of Duquesne et al. [26] and Rebuffat [27]. MccV (formerly colicin V), MccL and Mcc24 are three members assigned to the microcin class IIa. All are composed of four plasmid-borne genes, mostly harbored by large conjugative low-copy-number plasmids [7], [10]. The genes for MccV and MccL, cvaC and mclC, encode 103 and 105 aa precursors, which also constitute higher-molecular mass microcins. Both peptides form disulfide bonds [28], [29]. Class IIa microcins have leader peptides whose cleavage results in the mature biologically active peptide. Furthermore, they are not subject to post-translational modification. Gene clusters of all class IIa microcins consist of four genes organized in one or two transcription units: the gene encoding the microcin precursor, a self-immunity protein, an accessory protein involved in the secretion of the microcin, and an ATP-binding cassette (ABC)-transporter (for reviews see [26], [30]). The self-immunity genes code for small peptides that protect the producing strain from its own microcin. Here we can show that gene mcsI cloned into EPEC E2348/69 confers immunity to MccS. Gene mcsI encodes a 216 aa protein of the CAAX amino terminal protease protein family. The two genes comprising the MccS export system are mcsA and mcsB (Fig. 3). Gene mcsA is a member of the E. coli HlyD family and shows little homology to the colicin secretion protein CvaA. Gene mcsB encodes an ABC-transporter consisting of a transmembrane domain, an ATP-binding domain and a peptidase domain [25]. The nucleotide lengths of mcsA and mcsB from the MccS operon, 1254 bp and 2178 bp respectively, are comparable to export genes of the other microcins MccV (cvaA, 1242 bp; cvaB, 2097 bp), MccL (mclA, 1242 bp; mclB, 2097 bp) and Mcc24 (mtfA, 1245 bp; mtfB, 2124 bp) [26]. The sensitivity of microorganisms to microcins can generally be shown by a standard agar diffusion test, for example with the microcin-producing strain spotted onto an agar plate and an indicator strain inoculated into a soft agar overlay [12]. However, E. coli G3/10 does not produce real inhibition zones in agar diffusion tests (data not shown). This phenomenon can possibly be explained by the widely unknown conditions for effective MccS expression. Motivated by the fact that synthesis of MccV is repressed by excess iron [31], we added the iron chelator Desferal to the solid media growing E. coli G3/10 in the diffusion test. However, microcin production on agar plates could still not be shown. We therefore assume that other factors, such as pH or nutrients, may be involved in the regulation of microcin S expression. Because the adherence assay provides only indirect proof of the antibacterial activity of MccS-producing E. coli strains and the above mentioned agar diffusion test did not work properly with microcin S, we decided to subclone mcsS under the control of an araC PBAD activator-promoter. When microcin S production was induced with L-arabinose in E. coli MDS42 growing in liquid culture, viable counts dropped by several logs, clearly indicating a direct toxic activity of MccS against a susceptible E. coli strain. MccS from E. coli G3/10 has the great advantage that the microcin does not necessarily have to be purified from the strain. Since E. coli G3/10 is set out in a probiotic formulation that can be used for treatment of gastrointestinal disorders, MccS is very much likely to also be expressed effectively in vivo, rendering this beneficial microorganism a promising biological drug with prophylactic capacities against enteric pathogens such as Salmonella, Shigella or diarrheagenic E. coli.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains used in this work are listed in table 2. Bacteria were grown in Luria Bertani (LB) medium or on LB agar plates (Becton Dickinson, BD, Heidelberg, Germany). Culture media were supplemented with ampicillin (100 µg ml–1), chloramphenicol (25 µg ml–1), kanamycin (50 µg ml–1), tetracyclin (12.5 µg ml–1) or streptomycin (30 µg ml–1) as required. Antibiotics were purchased from AppliChem, Darmstadt, Germany.
Table 2. Bacterial strains used in this study.
| Strain | Relevant genotype | Origin |
| E. coli Nissle 1917 DSM6601 [42] | Wild-type | Mutaflor, Ardeypharm |
| EcNDS20 | ?mcmA, ?mchB | This work |
| EcNDS23 | ?mcmA | This work |
| EcNDS24 | ?mchB | This work |
| E. coli G1/2 | Wild-type | Symbioflor 2a, SymbioPharm |
| E. coli G3/10 | Wild-type | Symbioflor 2a, SymbioPharm |
| E. coli G4/9 | Wild-type | Symbioflor 2a, SymbioPharm |
| E. coli G5 | Wild-type | Symbioflor 2a, SymbioPharm |
| E. coli G6/7 | -type Wild | Symbioflor 2a, SymbioPharm |
| E. coli G8 | Wild-type | Symbioflor 2a, SymbioPharm |
| E. coli MDS42 [23] | E. coli K-12 multiple deletion strain | F. Blattner, University of Wisconsin - Madison, USA |
| EPEC E2348/69 [20] | Ampr (pUC19), Kanar (pUC4k) or Cmr (pACYC184) | Human isolate |
DSM17252; EcNDS = E. coli Nissle 1917 deletion strain; Ampr = ampicillin resistance; Kanar = kanamycin resistance; Cmr = chloramphenicol resistance.
Construction of EcN Microcin Deletion Mutants
EcN mutants negative for chromosomally encoded microcins M and H47 (mcmA, mchB) were constructed with a PCR-based one-step inactivation method [32], [33]. Deletions were controlled by PCR using primer pairs outlined in table 3 as well as automated DNA sequencing based on dideoxy chain termination method [34], using an ABI 3100 sequencing instrument (Life Technologies, Darmstadt, Germany).
Table 3. Oligonucleotide primers used in this study.
| Oligonucleotide | Sequence (5′→ 3′) | Function |
| mcm-H1 | cttaaagcgttacataggcaccattatcatataatgaagcaccgattgtgtaggctggagctgcttc | EcN deletion primer |
| mcm-H2 | gaatttttacttcttcacaaatcttatagcgaaggtgttgaaatggtccatatgaatatcctcctta | EcN deletion primer |
| mch-H1 | atcaacgactgtaaatcatatcttcatcagtaaagtgttgaacgattgtgtaggctggagctgcttc | EcN deletion primer |
| mch-H2 | ggtcaggctggaaaaacggaagttaaatatgatggagtttatatggtccatatgaatatcctcctta | EcN deletion primer |
| mcm_for | cgttcggaggagcctaac | EcN deletion primer/sequencing |
| mcm_rev | gattcatgggattcgaagg | EcN deletion primer/sequencing |
| Contig49_for | cagctggatatcctgcgcg | pSYM1 sequencing primer |
| Contig49_rev | ggttgcccggcatccaacg | pSYM1 sequencing primer |
| pSYM1-SalIHF | tcaattgtgtcgactcaattactcttgtgag | pAZ6/pAZ8 cloning primer |
| pSYM1-NheI | catgtaatagtgctagcatgttaaaatttataag | pAZ6 cloning primer |
| pSYM1-NheIac | caaaaataatagctagcaagtgatgttttgtaatg | pAZ8, pAZ12 cloning primer |
| ac-EcoRI | ctcgaattcatccattacaaaacatcac | pAZ9 cloning primer |
| ac-PstI | ctggctgcagtaattgttcaggaagtaacg | pAZ9 cloning primer |
| pSYM1_44–EcoRI | taggaattcagaggaactattggtggg | pAZ10 cloning primer |
| pSYM1_44–PstI | ctccgctgcagacttacttatcgactacaggtaccac | pAZ10 cloning primer |
| ab-EcoRI | gttagaattcataagagggatttttatgtcaaatatc | pAZ11 cloning primer |
| ab-PstI | gttgatactgcagcttatcgactacaggtaccacc | pAZ11 cloning primer |
| pAZ9-HindIII | cccaagcttagttaaatgtgctaatgctgtc | pAZ12 cloning primer |
| pAZ9-SalI | ggcatcggtcgacgcaac | pAZ12 cloning primer |
| pSYM1_43–NheI | cattgctagccatcacagataaactggataac | pAZ14 cloning primer |
| pSYM1_43–SalI | ccctgagtcgactcatggttataaaatattttg | pAZ13, pAZ14 cloning primer |
| recA–ff | atggctatcgacgaaaacaaac | Multiplex PCR inhibition control |
| recA–rev | ttaaaaatcttcgttagtttctgc | Multiplex PCR inhibition control |
| mcsS–ff | atgtcaaatatcagagaattgag | mcsS PCR screening primer |
| mcsS–rev | ttatcgactacaggtaccacc | mcsS PCR screening primer |
Oligonucleotides were synthesized by Biomers, Ulm, Germany.
Construction of Plasmids Used for Functional Characterization of the Microcin S Operon
The targeted sequence was amplified by PCR using oligonucleotides with 5′ restriction sites (Table 3). Due to their proofreading capacity, PCR was performed with Phusion High-Fidelity DNA-Polymerase (Thermo Fisher Scientific/Finnzymes, Vantaa, Finland) according to the recommended protocol. Following purification of PCR products with columns containing silica membranes (Qiagen, Hilden, Germany), fragments were digested with the respective restriction enzymes (New England Biolabs, NEB, Frankfurt (Main), Germany) and ligated into an appropriate vector using T4 DNA-Ligase (NEB). Ligated DNA constructs were then transformed to competent bacteria by electroporation (2.5 kV; 400 Ω; 25 µFD) using a Bio-Rad Gene Pulser II and Pulse Controller Plus (Munich, Germany). Clones were selected on LB agar plates containing the appropriate antibiotic and were confirmed by PCR, DNA sequencing and by restriction digest using the respective enzymes.
Conjugation Experiments
The conjugative plasmid pSYM1 allows the transfer to a recipient strain. pSYM1 was mobilized from E. coli G3/10 into E. coli G4/9 following a protocol published by Miller et al. [35] with slight modifications. E. coli G4/9 was chosen as the recipient strain because it is closely related to E. coli G3/10 and harbors only one natural plasmid (unpublished data). Since donor and recipient require dissimilar antibiotic resistance, suicide vector pST76-A, containing an ampicillin resistance cassette and a temperature-sensitive origin of replication, was integrated into pSYM1 by homologous recombination, resulting in pSYM1-ST76An. Then, recipient strain E. coli G4/9 was transformed with pACYC184 conferring it with chloramphenicol resistance. Donor E. coli G3/10 and recipient E. coli G4/9 were grown in LB medium to an absorbance of A600 ∼ 0.5 to 1, each containing the appropriate antibiotic. Cells were pelleted in a bench top centrifuge at 5000 g for 10 min, washed twice with phosphate buffered saline (PBS) and resuspended in LB medium. Donor and recipient were mixed 2∶1, pelleted and resuspended in a small volume of LB medium (∼100 µl). The cell suspension was plated on blood agar (BD) and incubated overnight at 37°C. Bacteria were resuspended in 1 ml LB medium, diluted in PBS and plated on MacConkey agar (BD) containing 25 µg ml-1 chloramphenicol and 100 µg ml-1 ampicillin (AppliChem). The resulting clones were confirmed by PCR.
Sequencing and Annotation of pSYM1
Plasmid pSYM1 was discovered as one of six wild-type plasmids in E. coli strain G3/10. The genome of the strain was obtained by pyrosequencing with a 454 GS-FLX System (Roche, Basel, Switzerland) (unpublished data). About ninety-five percent of the plasmid sequence was amassed during genome sequencing and assembly. Then, primers for an outwardly directed primer walking strategy were designed (Table 3) until a completely closed plasmid sequence was obtained. Annotation was performed automatically at CeBiTec followed by manual revision using GenDB 2.4 software [36]. BLASTn and BLASTp searches were done using the National Center for Biotechnology Information (NCBI) website [37].
LOVO and CACO-2 Cell Culture Conditions
The human intestinal epithelial cell lines LOVO [38] and CACO-2 [39] , purchased from DSMZ [40], were grown in RPMI-1640 cell culture medium (Biochrom, Berlin, Germany) supplemented with 10% FCS (Biochrom) and maintained in an atmosphere of 5% CO2 at 37°C. Cells reached confluency after 3–4 days and were used consistently within 3–4 days from seeding. Cell cultures were tested routinely and found to be free from mycoplasma contamination.
In vitro Adherence Assay
An in vitro adherence assay was performed according to a modified protocol from Kleta et al. [21]. Briefly, human intestinal epithelial LOVO or CACO-2 cells were seeded in 24-well plates and grown to confluency. Bacterial strains to be tested were grown in LB broth to an absorbance of A600 ∼ 0.6, whereas the adherent enteropathogenic E. coli strain E2348/69 was grown to A600 ∼ 1, each supplemented with the appropriate antibiotic. One ml of each bacterial strain was pelleted, washed with PBS and resuspended in cell culture medium. LOVO and CACO-2 cells were infected with the bacteria to be tested at a multiplicity of infection (MOI) of 100∶1 E. coli to host cells. After an incubation period of two hours at 37°C and 5% CO2 bacteria were washed away with PBS for three times and host cells were infected with EPEC. Bacteria were allowed to adhere for six hours in total (37°C, 5% CO2), while the cell culture medium was replaced after three hours of incubation. Cells were washed with PBS and lysed with 0.01% Triton-X-100 in PBS (Sigma-Aldrich, Munich, Germany). The number of adherent EPEC was determined by plating serial dilutions on LB agar plates containing the appropriate antibiotic. Adherence efficiency in percentage is expressed as adherence of EPEC relative to the adherence without any pre-incubation (negative control), which is set at 100%. All strains were tested in at least three independent experiments.
Screening of mcsS Presence in Enterobacteria
Gene mcsS encoding microcin S was amplified by a multiplex PCR protocol from culture material, using recA as an inhibition control. Oligonucleotides were used as indicated in table 3. E. coli G3/10 served as positive control. Thirty-eight different E. coli, two Shigella and two Salmonella strains were screened. To our knowledge, none of the isolates tested has been sequenced. Strains are common lab strains as well as isolates of human or animal origin as indicated in figure 6.
Statistical Analysis
Data are expressed as mean±SD. Student’s t-test was used to determine the statistical significance. p≤0.05 was considered statistically significant.
Nucleotide Sequence Accession Number of pSYM1
The pSYM1 plasmid sequence has been deposited in the GenBank sequence database with the accession number JN887338.
Acknowledgments
We would like to thank F. Ch. Bange for very helpful discussions and Michael J. Schubert for critically reading the manuscript.
Footnotes
Competing Interests: The authors have read the journal’s policy and they have the following conflicts. KZ is an employee of SymbioPharm. FG, AZ and KZ have submitted a patent application disclosing the discovery of "Bacterially formed microcin S, a new antimicrobial peptide, effective against pathogenic microorganisms" (EP 11177451). This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work has been funded by restricted research grants of the Federal Ministry of Education and Research (BMBF GenomikTransfer, PROTumor consortium, # 0315590A) and of SymbioPharm GmbH(# 100_2847) to FG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chiuchiolo MJ, Delgado MA, Farias RN, Salomon RA. Growth-phase-dependent expression of the cyclopeptide antibiotic microcin J25. J Bacteriol. 2001;183:1755–1764. doi: 10.1128/JB.183.5.1755-1764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connell N, Han Z, Moreno F, Kolter R. An E. coli promoter induced by the cessation of growth. Mol Microbiol. 1987;1:195–201. doi: 10.1111/j.1365-2958.1987.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 3.Fomenko D, Veselovskii A, Khmel I. Regulation of microcin C51 operon expression: the role of global regulators of transcription. Res Microbiol. 2001;152:469–479. doi: 10.1016/s0923-2508(01)01220-7. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Chico C, San Millan JL, Kolter R, Moreno F. Growth phase and ompR regulation of transcription of microcin B17 genes. J Bacteriol. 1986;167:1058–1065. doi: 10.1128/jb.167.3.1058-1065.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baquero F, Bouanchaud D, Martinez-Perez MC, Fernandez C. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J Bacteriol. 1978;135:342–347. doi: 10.1128/jb.135.2.342-347.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novoa MA, Diaz-Guerra L, San Millan JL, Moreno F. Cloning and mapping of the genetic determinants for microcin C7 production and immunity. J Bacteriol. 1986;168:1384–1391. doi: 10.1128/jb.168.3.1384-1391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien GJ, Mahanty HK. Colicin 24, a new plasmid-borne colicin from a uropathogenic strain of Escherichia coli. Plasmid. 1994;31:288–296. doi: 10.1006/plas.1994.1030. [DOI] [PubMed] [Google Scholar]
- 8.Salomon RA, Farias RN. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J Bacteriol. 1992;174:7428–7435. doi: 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Millan JL, Hernandez-Chico C, Pereda P, Moreno F. Cloning and mapping of the genetic determinants for microcin B17 production and immunity. J Bacteriol. 1985;163:275–281. doi: 10.1128/jb.163.1.275-281.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waters VL, Crosa JH. Colicin V virulence plasmids. Microbiol Rev. 1991;55:437–450. doi: 10.1128/mr.55.3.437-450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poey ME, Azpiroz MF, Lavina M. Comparative analysis of chromosome-encoded microcins. Antimicrob Agents Chemother. 2006;50:1411–1418. doi: 10.1128/AAC.50.4.1411-1418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149:2557–2570. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 13.Enck P, Zimmermann K, Menke G, Klosterhalfen S. Randomized controlled treatment trial of irritable bowel syndrome with a probiotic E.-coli preparation (DSM17252) compared to placebo. Z Gastroenterol. 2009;47:209–214. doi: 10.1055/s-2008-1027702. [DOI] [PubMed] [Google Scholar]
- 14.Martens U, Enck P, Zieseniss E. Probiotic treatment of irritable bowel syndrome in children. Ger Med Sci. 2010;8:Doc07. doi: 10.3205/000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarner F, Schaafsma GJ. Probiotics. Int J Food Microbiol. 1998;39:237–238. doi: 10.1016/s0168-1605(97)00136-0. [DOI] [PubMed] [Google Scholar]
- 16.Kuipers A, Rink R, Moll GN. Genetics, Biosynthesis, Structure, and Mode of Action of Lantibiotics. In: Drider D, Rebuffat S, editors. Prokaryotic Antimicrobial Peptides: From Genes to Applications. Berlin: Springer; 2011. pp. 147–169. [Google Scholar]
- 17.Hetz C, Bono MR, Barros LF, Lagos R. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc Natl Acad Sci U S A. 2002;99:2696–2701. doi: 10.1073/pnas.052709699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dicks LMT, Heunis TDJ, van Staden DA, Brand A, Sutyak Noll K, et al. Medical and Personal Care Applications of Bacteriocins Produced by Lactic Acid Bacteria. In: Drider D, Rebuffat S, editors. Prokaryotic Antimicrobial Peptides: From Genes to Applications. Berlin: Springer; 2011. pp. 392–421. [Google Scholar]
- 19.Gillor O, Kirkup BC, Riley MA. Colicins and microcins: the next generation antimicrobials. Adv Appl Microbiol. 2004;54:129–146. doi: 10.1016/S0065-2164(04)54005-4. [DOI] [PubMed] [Google Scholar]
- 20.Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleta S, Steinrück H, Breves G, Duncker S, Laturnus C, et al. Detection and distribution of probiotic Escherichia coli Nissle 1917 clones in swine herds in Germany. J Appl Microbiol. 2006;101:1357–1366. doi: 10.1111/j.1365-2672.2006.03019.x. [DOI] [PubMed] [Google Scholar]
- 22.Ong CL, Beatson SA, McEwan AG, Schembri MA. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 2009;75:6783–6791. doi: 10.1128/AEM.00974-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pósfai G, Plunkett G, III, Fehér T, Frisch D, Keil GM, et al. Emergent Properties of Reduced-Genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 24.Asensio C, Perez-Diaz JC. A new family of low molecular weight antibiotics from enterobacteria. Biochem Biophys Res Commun. 1976;69:7–14. doi: 10.1016/s0006-291x(76)80264-1. [DOI] [PubMed] [Google Scholar]
- 25.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 27.Rebuffat S. Bacteriocins from Gram-Negative Bacteria: A Classification? In: Drider D, Rebuffat S, editors. Prokaryotic Antimicrobial Peptides: From Genes to Applications. Berlin: Springer; 2011. pp. 55–80. [Google Scholar]
- 28.Havarstein LS, Holo H, Nes IF. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology 140 (Pt. 1994;9):2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 29.Pons AM, Delalande F, Duarte M, Benoit S, Lanneluc I, et al. Genetic analysis and complete primary structure of microcin L. Antimicrob Agents Chemother. 2004;48:505–513. doi: 10.1128/AAC.48.2.505-513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway. Mol Membr Biol. 2005;22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- 31.Chehade H, Braun V. Iron-regulated synthesis and uptake of colicin V. FEMS Microbiology Letters. 1988;52:177–181. [Google Scholar]
- 32.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. 1977. Biotechnology. 1992;24:104–108. [PubMed] [Google Scholar]
- 35.Miller HJ. Harbor ColdSpring., editor. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. N.Y.: Cold Spring Harbor Laboratory Press. 1992. 876
- 36.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, et al. GenDB–an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Drewinko B, Yang LY, Barlogie B, Romsdahl M, Meistrich M, et al. Further biologic characteristics of a human carcinoembryonic antigen-producing colon carcinoma cell line. J Natl Cancer Inst. 1978;61:75–83. doi: 10.1093/jnci/61.1.75. [DOI] [PubMed] [Google Scholar]
- 39.Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–1040. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 40.Drexler HG, Dirks W, MacLeod RAF, Quentmeier H, Steube KG, et al. 8th ed; 2001. DSMZ Catalogue of human and animal cell lines. [Google Scholar]
- 41.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nissle A. Über die Grundlagen einer neuen ursächlichen Bekämpfung der pathologischen Darmflora. Dt Med Wschr. 1916;42:1181–1184. [Google Scholar]
- 43.Pósfai G, Koob MD, Kirkpatrick HA, Blattner FR. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]