Abstract
Pathogenic Yersinia species possess a type III secretion system, which is required for the delivery of effector Yop proteins into target cells during infection. Genes encoding the type III secretion machinery, its substrates, and several regulatory proteins all reside on a 70-Kb virulence plasmid. Genes encoded in the chromosome of yersiniae are thought to play important roles in bacterial perception of host environments and in the coordinated activation of the type III secretion pathway. Here, we investigate the contribution of chromosomal genes to the complex regulatory process controlling type III secretion in Yersinia pestis. Using transposon mutagenesis, we identified five chromosomal genes required for expression or secretion of Yops in laboratory media. Four out of the five chromosomal mutants were defective to various extents at injecting Yops into tissue culture cells. Interestingly, we found one mutant that was not able to secrete in vitro but was fully competent for injecting Yops into host cells, suggesting independent mechanisms for activation of the secretion apparatus. When tested in a mouse model of plague disease, three mutants were avirulent, whereas two strains were severely attenuated. Together these results demonstrate the importance of Y. pestis chromosomal genes in the proper function of type III secretion and in the pathogenesis of plague.
Introduction
Yersinia pestis, the causative agent of plague, is one of three Yersinia species pathogenic to humans. All three species share a tropism for lymphoid tissues and utilize a type III secretion system (TTSS) to undermine the host immune system [1], [2]. The Yersinia TTSS is encoded by a conserved 70-kb virulence plasmid, termed pCD1 in Y. pestis [3]#. In addition to the machinery components and its secreted substrates (YopB, YopD, YopE, YopH, YopM, YopN, YopO, YopP, YopK, YopR, YopT, LcrV, and LcrQ), pCD1 encodes several regulatory proteins that respond to environmental cues. Expression of the TTSS and its substrates are induced upon temperature shift to 37°C by the transcriptional activator LcrF [4], [5], [6], [7]. YopD, LcrH, and YscM1/YscM2 (LcrQ) repress activation of the TTSS in the absence of certain serum amino acids [8]#. YopN, TyeA, SycN, YscB and LcrG prevent transport of Yop effectors until contact with a target cell is made [9], [10], [11], [12], [13], [14], [15], [16], [17], [18]. These conditions can be simulated in vitro, and upon a shift to 37°C in the absence of calcium, yersiniae undergo growth cessation and massively secrete all of the type III substrates into the medium [19], [20], [21], [22], [23]. This phenomenon is referred to as the LCR (low calcium response) phenotype [24]#. The onset of bacteriostasis under TTSS-inducing conditions is particularly severe for Y. pestis compared to the enteric Yersinia and correlates with a loss of aspartase and glucose-6-phosphate dehydrogenase functions [19], [25], [26], [27]. Interestingly, calcium does not prevent growth or Yop expression during cell culture, and indeed bacteriostasis can be uncoupled from induction of the Yersinia TTSS by manipulating levels of glutamate, sodium, and pH to reflect the presumed in vivo conditions of a necrotic lesion [19], [28], [29], [30]. Therefore while the calcium-dependent LCR phenotype may be an artifact of in vitro culture conditions, this Yersinia-specific phenomenon has been utilized as a tool to investigate the details of TTSS regulation and function in Yersinia. It has been suggested that yersiniae sense environmental calcium concentrations in the extracellular fluids of their host and in the cytoplasm of host cells via insertion of the type III secretion needle into the plasma membrane of target host cells [16], [31], [32]; however the mechanism for sensing calcium levels remains a mystery.
Although many of the genes required for activation of the type III pathway in response to environmental signals have been identified, the mechanism by which yersiniae perceive their environment and contact with a target cell and then transmit that information to the cytoplasm and secretion apparatus remains unclear. Also, early work in Y. enterocolitica suggested that the virulence plasmid is necessary, but not sufficient, for expression and/or delivery of effector Yops [33], [34]#. Presumably, additional unidentified genes are involved in the regulation of type III secretion. In 1984, Goguen and colleagues performed a genetic analysis, taking advantage of the LCR growth phenotype that occurs at 37°C in the absence of calcium, and reported that a large number of Y. pestis mutations, which seemed to be chromosomally located, gave rise to an LCR− phenotype [24]. However, these mutants remain uncharacterized.
In an effort to determine the potential role of chromosomal genes in the regulation of the Yersinia TTSS, we undertook a screen similar to that of Goguen et al. Using a Mu transposon derivative [35], we generated insertional mutations in Y. pestis and isolated mutants with an LCR− phenotype, i.e. the variants gained the ability to grow at elevated temperatures in the absence of calcium. These mutants were then evaluated for their ability to secrete Yops when grown in calcium-depleted medium. The location of the transposon insertion was determined for each secretion-defective mutant based on inverse PCR and sequence analysis. After screening 2067 mutants displaying the LCR− growth phenotype, we identified 5 variants (termed CHI strains for chromosomal insertion) harboring a transposon insertion that gave rise to defects in secretion of Yops and/or expression of the TTSS. The ability of the 5 strains to target Yops into host cells during tissue culture infection was also analyzed, revealing a distinction between a secretion defect and a targeting defect in some cases. Further characterization of the mutants in a mouse model of infection confirmed the importance of the identified genes in virulence.
Materials and Methods
Bacterial strains and media
All Yersinia pestis strains are attenuated (non-pigmented) variants of Y. pestis biovar mediaevalis KIM lacking the 102-kb pgm locus [36]. KIM5 (pCD1+, pMT1+, pPCP1+) was obtained from R. Brubaker. KIM8 (pCD1+, pMT1+, pPCP1−) and YP769 (also called KIM8 Δ1234 [37]; pMT1+, pPCP1−, pCD1+ ΔyopM/yopT/sycT/yopK/ylpA/yopE/sycE/ypkA/yopJ/yopH) were obtained from G. Plano. Y. pestis strains were propagated on Heart Infusion or Blood Base agar plates at 26°C for two days. Overnight cultures were grown in Heart Infusion Broth (HIB) at 26°C. Antibiotics were added as appropriate to a final concentration of 20 µg/ml chloramphenicol, 50 µg/ml ampicillin, or 50 µg/ml kanamycin. Escherichia coli strains were propagated in LB broth or agar, supplemented with 20 µg/ml chloramphenicol, 100 µg/ml ampicillin, or 25 µg/ml kanamycin, at 37°C, except CR202, which was grown at 30°C.
Mu Mutagenesis
E. coli CR202 carries the Mucts62pAp5hP1#1 lysogen (also called MuAphP1), a Mu-P1 hybrid [35]#. To obtain a lysate, overnight cultures of CR202 were subcultured 1∶50 into HIB, supplemented with 2.5 mM MgCl2 and 2.5 mM CaCl2, and incubated at 30°C for 1 hour 45 minutes. The culture was then shifted to 42°C for 2 hours, or until lysis occurred. The lysed culture was sterilized by passing through a 0.2 µm filter and then stored on ice until needed. Y. pestis KIM5 overnight cultures were diluted 1∶25 into the same medium and incubated at 26°C until late exponential phase, about 3 hours. Cells were sedimented by centrifugation and suspended in 10 mM MgSO4, followed by incubation at room temperature for 20 minutes. To infect, 5 ml of lysate was added to 2.5 ml of cells and incubated at room temperature for 20 minutes. The cells were again sedimented by centrifugation, suspended in 10 mM MgSO4, and plated on Blood Base agar containing 20 mM sodium oxalate, 20 mM MgCl2 (BB/MgOx agar), and 50 µg/ml ampicillin to select for Mu insertions leading to an LCR− phenotype. During initial testing, the plates were incubated at a range of temperatures (26°C, 30°C, 33°C, 35°C, and 37°C) for 2 days. At temperatures above 33°C, many chromosomal insertion mutants also carried additional Mu insertions on pCD1. We therefore chose 33°C for routine selection of LCR− mutants.
Identification of mutants
Primer sequences are listed in Table S1. To determine the location of the Mu insertion, chromosomal DNA of mutants was digested with either Taqα I or BspHI, followed by large-volume ligation. Ligations were purified and used as the template for inverse-PCR reactions using primers complimentary to the left (BfaI-LT, BfaI-RT) and right (TaqI-LT, TaqI-RT) ends of Mu. PCR products were sequenced using nested primers. The obtained sequences were subjected to the following computational analysis. Sequences were formatted into FASTA format, using only the transposon sequence plus the following 160 nucleotides. If no transposon sequence was found, the first 260 nucleotides were used. These formatted sequences were then used to search the Y. pestis KIM genome using the BLAST algorithm [38]. Positions of significant BLAST hits against the genome were obtained from the BLAST output and the genes surrounding or overlapping these positions were recorded. Significant hits were those with an e-value below 10−7 and percent identity over 30. Functional characterization of disrupted genes was carried out using bioinformatic analysis [39], [40], [41].
Complementation of mutants
False positive mutants were eliminated by pCD1 displacement. Y. pestis KIM-3001.P39 carries a tagged pCD1 (referred to here as pCD1::KM) in which the yopE-sycE locus was replaced by a kanamycin resistance cassette [42]. Plasmid DNA isolated from KIM-3001.P39 was electroporated into each chromosomal mutant. Transformants were plated on BB containing kanamycin to select for the tagged pCD1 (pCD1::KM). Displacement of the original pCD1 by pCD1::KM was confirmed by immunoblotting to demonstrate the loss of yopE and the presence of npt. Mutants carrying pCD1::KM were screened by secretion assay and immunoblotting (described below). Mutants retaining the secretion defect were then subjected to gene-specific complementation. Table S1 lists the primers and vectors used to clone and introduce the wild-type gene or operon of interest into each mutant. The PCR products for y0447 and rfaL were cloned into the pDONR-221 vector using the BP Clonase mix as per manufacturer's instructions (Invitrogen) and then transformed into DH5α. The pgsA locus was amplified with the pgsAORFNdeI pgsAORFBamHI primers to clone the open reading frame as an NdeI-BamHI fragment. The native promoter was amplified using the uvrYproEcoRI and uvrYproNdeI primers. The EcoRI-NdeI promoter fragment and NdeI-BamHI ORF fragment were cloned into the EcoRI-BamHI site of pHSG576 [43] by three-way ligation. The regions containing folD and pssA were amplified using the folD.SOE.3, folD.SOE.BamHI.4, pssA.SOE.3, and pssA.SOE.revEcoRI primers. The promoter region encompassing ∼300 bp upstream of nptII was amplified using the npt.pro.SOE.PstI. and npt.pro.folD.SOE.2 or npt.pro.pssA.SOE.2 primer pair. The npt promoter product and folD and pssA products were stitched together using the folD.SOE.3 and folD.SOE.BamHI.4 or the pssA.SOE.3 and pssA.SOE.revEcoRI primer pair. The resulting PCR products were cloned into the SmaI site of pBluescript SK+ and sequenced. The Pnpt-folD and Pnpt-pssA products were then cloned into the PstI and BamHI or PstI and EcoRI sites of pHSG576. The resulting plasmids were named pAH191 and pAH195. All constructs were introduced into their respective CHI strains by electroporation.
Standard Secretion Assays
For routine screening, overnight cultures of Y. pestis strains were diluted 1∶40 into modified M9 (MM9) and secretion assays were performed as previously described [44]. Briefly, cells were grown at 26°C for 2 hours and then at 37°C for 3 hours. The OD600 of each culture was taken following the incubation at 37°C. Cells were sedimented and proteins in the pellet and supernatant fractions were TCA precipitated and visualized by immunoblotting. Samples were loaded onto SDS-PAGE gels based on the OD600 of each culture compared to wild type unless noted otherwise. Additional characterization of secretion phenotypes were conducted by diluting overnight cultures into either HIB, supplemented with 5 mM CaCl2 or 5 mM EGTA, or DMEM tissue culture medium (see below) with or without supplemented 5 mM EGTA.
Flow cytometry to measure Bla reporter injection
Plasmids expressing YopM-Bla (pMM83), Gst-Bla (pMM91), and YopE-Bla (pMM85) from their native promoters have been previously described [44]. pAH83 was created to drive constitutive expression of YopM-Bla. The regions containing yopM and bla were amplified from pCD1 and pMM83, respectively, using the npt.pro.yopM.SOE.3 and yopM.SOE.4.bla.2 or yopM.SOE.4.bla.2 and bla.SOE.4.EcoRI primer pair. The nptII promoter was amplified using the npt.pro.SOE.BamHI.1 and npt.pro.yopM.SOE.2 primers. The resulting yopM, bla, and npt promoter products were stitched together with Phusion using the primers npt.pro.SOE.BamHI.1 and bla.SOE.4.EcoRI. The resulting PCR product was cloned into the SmaI site of pBluescript SK+. The Pnpt-yopM-bla region was then subcloned into the BamHI and EcoRI sites of pHSG576, creating pAH83. All plasmids were introduced into Y. pestis strains by electroporation. The night before the experiment, 3.25×105 CHO (ATCC, CCL-61) cells per well were seeded into a 12 well plate and maintained in F12K supplemented with 10% FBS at 37°C+5% CO2. Y. pestis strains were grown to mid-exponential phase at 26°C in HIB with chloramphenicol and then pre-induced at 37°C for 1.5 hrs. CHO cells were counted and bacteria were added at an MOI of 10, followed by centrifugation at 500×g for 5 min to facilitate cell contact. The infections were incubated at 37°C for 3 hrs after which the cells were washed with PBS and trypsin was added to remove CHO cells from the plate. The trypsin was quenched with the addition of 1 ml of F12K+10% FBS, cells were pelleted at 500×g for 5 min, and the cell pellets resuspended in 300 µL of HBSS-Flow (1× Hank's Buffered Saline Solution, 0.5 mM EDTA, 25 mM HEPES, 2% BSA, pH 7.4). 50 µL of 2× CCF-2AM+probenicid was added to the cells, and the cells were incubated at room temp for 15 min. Following the incubation, 300 µL of HBSS-Flow was added to each tube, and the cells were analyzed on a BD LSRII flow cytometer to quantitate blue and green fluorescence as previously described [44], [45]. Significance was determined using one-way ANOVA with a Tukey post-test in GraphPad Prism 5.
Cytotoxicity Assays
The night before the infection, 2.5×104 HeLa cells were seeded into 12 well plates and maintained in DMEM supplemented with GlutaMAX and 10% FBS at 37°C+5% CO2. Y. pestis strains were grown to mid-exponential phase at 26°C in HIB with ampicillin and then pre-induced at 37°C for one hour. The HeLa cells were washed twice with 1×PBS and counted. The media was replaced with 1 mL of warm DMEM+10% FBS and GlutaMAX containing bacteria at an MOI of 10, followed by centrifugation at 500×g for 5 min to facilitate cell contact. After a 30 minute incubation at 37°C, the medium was removed and replaced with 1 mL of fresh pre-warmed medium and the cells were returned to 37°C for 3.5 hours. Cells were then imaged using an Olympus 1×71 microscope and Olympus MicroSuite Five imaging software. For each well of the triplicate infections, five fields of view were counted (100 cells/field of view) and the average percentage of round cells was determined. The experiment was performed in triplicate and repeated twice. Significance was determined using one-way ANOVA with Tukey post-hoc test in GraphPad.
Trimethoprim Assays
To evaluate the affect of trimethoprim on type III secretion, secretion assays were performed in MM9. Bacteria were grown at 26°C for 2 hours and then at 37°C for 3 hours. 0, 15, or 30 µg/ml of trimethoprim was added when the cultures were shifted to 37°C. Cells were sedimented and proteins in the pellet and supernatant fractions were TCA precipitated and visualized by immunoblotting. Because YP769 (KIM8 Δ1234) lacks all effector Yops, we introduced the YopM expressing plasmid, pMM207, so that we could track secretion of at least one effector. pMM207 was constructed by amplifying a ∼3.8 kb region containing YopM using the primers YopM-upstream and YopM-downstream, followed by cloning the blunt PCR product into the SmaI site of pUC19. pMM207 was introduced into KIM8 and YP769 by electroporation.
To assess the effect of trimethoprim on Yop injection, HeLa cells (ATCC, CCL-2) were infected with Y. pestis strains carrying pMM85 (YopE-Bla). Infections and microscopy were carried out as previously described [44], except that trimethoprim (500 µg/ml) was added during the infection.
Mouse infections
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The work was approved by the Institutional Animal Care and Use Committees at the University of Chicago and Indiana University. All efforts were made to minimize suffering of the mice.Y. pestis overnight cultures were diluted 1∶20 into HIB and grown for 3 hours at 26°C. Cultures were washed and diluted to appropriate densities in PBS. 6–8 week old female BALB/c mice (Charles River Laboratories, Inc., Wilmington MA or Harlan Laboratories, Indianapolis IN) were anesthetized with either avertin (∼400 mg/kg) or a cocktail of ketamine (100–130 mg/kg) and xylazine (3–6 mg/kg) intra-peritoneally. Mice were then injected via the retro-orbital plexus with 0.1 ml suspensions of bacteria. The administered doses consisted of serial dilutions from 100 to 105 cfu for KIM5 and 102 to 107 cfu for each mutant. All mice were monitored for signs of disease for 14 days or until acute disease (hunched posture, severely matted fur, emaciation, immobility) was apparent, at which time mice were euthanized by CO2 inhalation followed by cervical dislocation.
Results
Isolation of LCR− mutants
The design of the screen to identify chromosomal genes involved in type III secretion of Yop proteins is shown in Figure 1. The parent strain (KIM5) used in this study carries a functional pla gene, which encodes a protease known to degrade fibrin clots in vivo and is important for infection [46], [47]. We chose to use this strain, rather than a pla mutant, so that the transposon mutants could be assayed in vivo as well as in vitro. Step 1 of the screen was carried out similar to that of Goguen et al. [24]#. Y. pestis strain KIM5 was mutagenized with Mucts62pAp5hP1#1, a Mu-P1 hybrid also called MuAphP1 [35]. This Mu derivative carries an ampicillin resistance cassette to select for insertions of the transposon into the Y. pestis genome. The pool of mutagenized Y. pestis cells was then plated directly onto Blood Base agar containing oxalate (a calcium chelator), and ampicillin to select for mutants unable to undergo growth restriction (an LCR− phenotype) at elevated temperatures. We found a distribution of insertions on pCD1 within the lcrA (yopN, tyeA, sycN, yscX, yscY, lcrD/yscV, lcrR), lcrB (yscN-U), lcrC (yscA-L, lcrQ), virF, and virG/yscW loci, demonstrating the validity of the screening process. More importantly, mapping of the insertions by inverse PCR showed that for the majority of chromosomal mutants identified under these conditions there was only one insertion site, indicating that the mutants harbored only a single Mu insertion in the genome.
Figure 1. Experimental strategy used to identify Y. pestis chromosomal genes involved in type III secretion of Yops.
In Step 1, transposon mutants were selected based on their ability to grow at 33°C in the absence of calcium. These mutants were subjected to a secondary screen (Step 2) evaluating their ability to secrete Yops. All mutants that displayed a defect in the synthesis or secretion of Yops were then sequenced to identify the location of the Mu insertion (Step 3). To eliminate mutants that carried unmarked mutations on the virulence plasmid, the tagged pCD1::KM was used to displace the original pCD1, and the transformants were then evaluated for their ability to secrete Yops (Step 4). Strains which retained the secretion defect after Step 4 were kept for further characterization.
Screen for secretion defects
Transposon mutants that were able to grow under inducing conditions (called CHI strains for chromosomal insertion) were subjected to a secondary screen to analyze their ability to secrete Yops in vitro (Step 2 in Figure 1). We first evaluated the ability of the KIM5 parent to secrete Yops in various media over different periods of time (data not shown). We chose to use a modified version of M9 (MM9), a minimal medium previously shown to induce secretion of Yops in Y. enterocolitica [48] because it gave the highest level of Yop expression and secretion after three hours at 37°C. Characteristic patterns of degraded Yops were observed, due to the Pla protease [49], [50], and these patterns were consistent during repeat experiments. After analyzing 2067 mutants showing a growth phenotype in step 1 of the screen, we identified 1607 strains with decreased secretion, no secretion, or no expression of YopE.
Locating transposon insertions
Genomic DNA from transposon mutants was used as template for inverse PCR amplification of DNA regions surrounding the Mu insertion. DNA sequences of amplified fragments were subjected to BLAST analysis to locate the position of the insertion (Step 3 in Figure 1). Out of 1607 candidates showing a defect in secretion or expression of YopE, sequences were obtained for 1591 strains. In all but 290 strains, insertions on pCD1 were identified. We were unable to determine the location of insertion for 16 strains. 13 strains harbored insertions in the 100-kb plasmid pMT1 and were not pursued further. The remaining 261 strains carried at least one chromosomal Mu insertion.
To verify that the mutant phenotypes were not due to unmarked spontaneous mutations on pCD1, which would not have been identified by our sequencing analysis, we obtained a pCD1 derivative in which the nptI cassette conferring kanamycin resistance replaces the yopE-sycE locus [42]. This plasmid, which we refer to here as pCD1::KM, was introduced into each of the CHI strains carrying chromosomal insertions (Step 4 in Figure 1). Candidates were screened for their ability to secrete YopM. The replacement of the original pCD1 by the tagged pCD1::KM was verified by immunoblotting to demonstrate the loss of yopE and the presence of nptI (data not shown). Transposon mutants which regained the ability to secrete YopM after acquiring the new pCD1::KM were discarded. This final step of the screen identified 5 strains that carried a chromosomal insertion leading to a defect in expression and/or secretion of Yops. Arrowheads in Figure 2 indicate the locations of the transposon insertions in these 5 CHI strains. The mutants were next evaluated for their ability to secrete Yops in vitro, inject Yops into mammalian cells, and cause disease in a mouse model of infection.
Figure 2. Identification of mutations in CHI strains.
The genetic locus carrying the Mu insertion, indicated by an arrowhead, is depicted for each CHI strain. The genome annotations of the disrupted genes (shaded) are shown. Arrows refer to primer locations used to amplify and clone each locus for complementation analysis. Expression of folD and pssA were driven by the nptII promoter.
Secretion phenotypes of CHI strains
To characterize each of the transposon mutants, secretion assays were conducted using minimal (MM9) and rich laboratory media (HIB with or without EGTA) (Figure 3 and data not shown). RpoA, which is the alpha-subunit of RNA polymerase, was used a fractionation and loading control, since its expression should be independent of the TTSS and it remains in the bacterial cytoplasm. YscD is a TTSS machine component, which is also not secreted and should only be visible in the cell pellets. To evaluate Yop secretion, we used antibodies against the effectors YopM and YopE. A transposon mutant carrying an insertion in the TTSS component yscU, which was isolated from this screen, was included as a negative control. As expected, this mutant did not secrete Yops and displayed lower levels of TTSS proteins. Interestingly, one of the chromosomal mutants displayed a conditional phenotype depending on the medium used in the assay. CHI 1345 (ctgA−) consistently secreted less Yops than the KIM5 parent strain when grown in MM9 (Figure 3). However, in HIB there was a clear absence of Yops in the culture supernatant for the mutant, in addition to decreased expression of TTSS proteins (not shown). The secretion phenotypes of the remaining CHI strains were consistent in both types of media: they did not secrete YopE or YopM in either MM9 or HIB lacking calcium (Figure 3 and data not shown). Similar to the yscU mutant, the chromosomal mutants all had lower expression levels of TTSS related proteins, although to varying degrees. The lack of Yop secretion in these mutants therefore may be due to impaired TTSS expression. When plasmids carrying the respective wild type genes were introduced into each mutant, expression and secretion of Yops was restored, thereby confirming that the mutant phenotypes are the result of the transposon insertions (Figure 3).
Figure 3. Secretion phenotypes of CHI strains.
Complementation plasmids were introduced into each mutant and the KIM5 parent, and the strains were evaluated by performing secretion assays. The TTSS mutant, yscU, was included as a negative control. Y. pestis strains were grown in MM9 at 26°C for 2 hours and then shifted to 37°C for 3 hours to induce secretion. Proteins in the supernatant (S) and pellet (P) fractions were precipitated and visualized by immunoblotting with antibodies to YopM, YopE (TTSS secreted proteins), YscD (TTSS apparatus component) and RpoA (RNA Polymerase alpha subunit, loading and fractionation control). An asterisk indicates truncated Yops due to degradation by Pla protease.
Targeting phenotypes of CHI strains
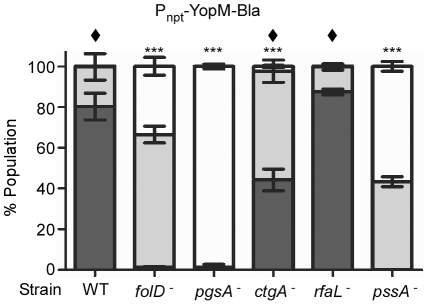
The purpose of the Yersinia TTSS is to inject effector Yops into target host cells during infection. To determine if any of the transposon insertions identified in our screen contribute to this aspect of virulence, we analyzed the fate of two injected effector Yops (YopE and YopM) during infection of tissue culture cells. We introduced plasmids bearing β-lactamase fusions to YopM (pMM83), YopE (pMM85), or glutathione-S-transferase (Gst, pMM91) [44] into the CHI strains and the KIM5 parent (Figure 4). These strains were used to infect CHO cells at an MOI of 10 for three hours. CCF2-AM, a fluorescent substrate for β-lactamase, was added to stain CHO cells prior to flow cytometry. Cells that had not been injected with β-lactamase hybrids fluoresce green as a result of fluorescence resonance energy transfer (FRET) within intact (uncleaved) CCF2-AM substrate. In the presence of β-lactamase, however, FRET is disrupted, resulting in blue fluorescence of cells that are injected with hybrid Yop-Bla fusions via the Yersinia type III pathway. The levels of fluorescence in each cell can be quantified by flow cytometry, and when synchronized, infections by different strains can be quantitatively compared [44], [45]. As previously demonstrated [44], [45], an abundance of blue cells were observed when KIM5 carrying either pMM83 (YopM-Bla) or pMM85 (YopE-Bla), but not pMM91 (Gst-Bla), was used to infect CHO cells (Figure 4 A and B and data not shown). Also as previously demonstrated, the yscU − mutant did not translocate YopM-Bla or YopE-Bla (data not shown). pgsA− had an identical phenotype to yscU − in that it also did not inject either Yop-Bla reporter into host cells. Surprisingly, rfaL− injected both Bla reporters at wild type levels. In addition, the remaining mutants all injected YopM-Bla and YopE-Bla, albeit at significantly lower levels than wild type (*** P<0.001).
Figure 4. Translocation phenotypes of CHI strains.
CHO cells were infected with Y. pestis strains carrying either pMM83 (YopM-Bla, Panel A), pMM85 (YopE-Bla, Panel B) or pMM91 (Gst-Bla, not shown) at 37°C for 3 hours, followed by CCF2-AM staining and flow cytometry to quantify blue and green fluorescence. The level of translocated Bla reporter is indicated by white (no injection, green fluorescence), gray (intermediate level of injection, aqua fluorescence), or black (high level of injection, blue fluorescence) bars. For each experiment, the assays were performed in triplicate, and error bars indicate the standard deviation. The data shown is representative of three independent experiments. One-way ANOVA with Tukey post-hoc test was done to demonstrate significant differences in injection (high-level injection, black bars) for mutants relative to the wild type parent. *** P<0.001. To assess cytotoxicity due to Yop delivery, cell rounding was also evaluated (Panel C). HeLa cells were infected with the indicated Y. pestis strains for 3–4 hours. The percentage of round cells was determined for each infection. The infections were performed in triplicate and error bars represent standard deviation. The data shown is representative of two independent experiments. One-way ANOVA with Tukey post-hoc test was done to determine significance. *** Represents significant difference of mutants compared to WT (P<0.001); n.s. = no significance between indicated strains.
To confirm the translocation phenotypes of the mutants we also evaluated the ability of each mutant to cause cytotoxicity associated with Yop delivery. Toward that end, HeLa cells were infected with Y. pestis strains for 3–4 hours and then visualized by microscopy to determine the percentage of cell rounding. As shown in Figure 4C, the majority of cells infected with KIM5 were round, while cells infected with yscU − appeared similar to uninfected cells. Likewise, cells infected with folD−, pgsA−, and pssA − all showed no significant difference compared to uninfected cells. The ctgA mutant was still able to cause some cytotoxicity, though the percentage of cell rounding was still lower than WT (P<0.001). In contrast, the rfaL mutant behaved identically to WT. These data correlate nicely with the Bla reporter assays and confirm that the rfaL mutant is able to deliver Yops as well as WT, whereas Yop injection by the remaining mutants is impaired.
Heterologous expression cannot bypass the mutant defects
Since several mutants displayed defective expression of TTSS proteins, and since the defect in secretion could be bypassed by cell contact in some cases to allow Yop injection during cell culture infection, we hypothesized that there may be conditional blocks in the TTSS at the level of expression. To determine if heterologous expression of a Yop could bypass the defects observed in the mutants, we introduced a plasmid carrying a yopM-bla fusion located behind the constitutive nptII promoter. As previously demonstrated for other Yop fusions under the control of the this promoter [51]#, expression of YopM-Bla was considerably lower than from the native yopM promoter (Figure 5). Despite the low level of expression, YopM-Bla and its characteristic degradation products were routinely detected in the supernatant of wild type secretion assays (Figure 5). For the CHI strains, all of them consistently expressed large amounts of YopM-Bla; however, the reporter did not appear in culture supernatants (Figure 5). It remains a formal possibility that low levels of Yops or YopM-Bla are secreted in these mutants, which are undetected due to Pla activity; however, after repeated attempts to observe secretion at various times after induction of the TTSS, we never observed YopM-Bla or any apparent degradation products of the reporter in the culture supernatants. This lack of secretion, despite high expression levels of the reporter, supports the conclusion that Yop secretion is impaired in these mutants. Therefore, it is unlikely that the absence of Yops in the supernatants during secretion assays (Figure 3 and 5) is simply due to detection limits of the assay or a result of impaired Yop expression. The defects may however be attributed to decreased expression of the TTSS machine components, at least in the cases of folD−, pgsA−, and pssA− (Figure 5), as these mutants display less YscD and YscF.
Figure 5. Secretion phenotypes of strains carrying a heterologously expressed Yop reporter.
pAH83, which constitutively expresses YopM-Bla, was introduced into each mutant and the KIM5 parent, and the strains were evaluated by performing secretion assays as described in Figure 3. Proteins were visualized by immunoblotting with antibodies to β-lactamase (YopM-Bla), YopM, YopE (TTSS secreted proteins), YscD, YscF (TTSS apparatus components) Ail (outer membrane protein) and RpoA (cytoplasmic protein, loading and fractionation control). An asterisk indicates truncated Yops due to degradation by Pla protease.
It is important to point out that while some Yops could be detected in the supernatant of pgsA− (Figure 5), the cytoplasmic protein RpoA, which is used as a fractionation control, was also found in the supernatant. In contrast, the majority of Ail (an outer membrane protein and additional fractionation control) was found in the cell pellet fraction, as expected. This was a consistent observation, even after filtering supernatants to ensure removal of cells, suggesting that these mutant cells may be leaky (Figure 3 and data not shown). Therefore, we cannot determine from these experiments whether the small amount of Yops occasionally seen in the supernatant for this mutant is due to type III secretion or simple leakage.
Since cell contact was able to bypass the in vitro secretion defect for some mutants, we tested whether heterologous expression of the YopM-Bla reporter could restore injection levels to that of wild type for the transposon mutants. As expected, constitutive expression of YopM-Bla resulted in high levels of its injection into CHO cells by the parent strain (Figure 6). Again, rfaL− displayed a similar ability to inject the YopM-Bla reporter. In fact, there was no significant difference in Bla reporter injection between wild type and rfaL− during any of the translocation assays (Figures 4 and 6). Both strains showed a statistically significant increase (diamonds: P<0.001) in YopM-Bla injection when the reporter was constitutively expressed (compare Figure 4A to Figure 6). Likewise, although injection of YopM-Bla by ctgA− was still impaired (*** P<0.001) compared to wild type (Figure 6), constitutively expressing the reporter did significantly enhance its injection (Figure 4A and 6). In contrast, folD−, pgsA−, and pssA− did not respond to heterologous expression of the reporter. Both folD− and pssA− translocated similar amounts of the YopM-Bla reporter whether expression was driven from the native promoter or the npt promoter, and pgsA− did not translocate any YopM-Bla.
Figure 6. Translocation phenotypes of strains carrying a heterologously expressed Yop reporter.
CHO cells were infected with Y. pestis strains carrying pAH83 at 37°C for 3 hours, followed by CCF2-AM staining and flow cytometry. Cells were analyzed for blue and green fluorescence. The level of translocated Bla reporter is indicated by white (none), gray (intermediate level), or black (high level) bars. For each experiment, the assays were performed in triplicate, and error bars indicate the standard deviation. The data shown is representative of three independent experiments. One-way ANOVA with Tukey post-hoc test was done to demonstrate significant differences in injection (high-level injection, black bars) for mutants relative to the wild type parent. *** P<0.001. Diamonds indicate statistically significant differences (P<0.001) compared to samples in Figure 4, Panel A.
Role of folate synthesis on type III secretion
One of the mutants, CHI 919 (folD−), was unable to secrete Yops in vitro and was only able to inject low levels of Yops into tissue culture cells (Figures 3,4,5,6). To determine whether the effect of the Mu insertion in CHI 919 on type III secretion was specific to the folD gene, or if it was reflective of folate biosysnthesis in general, we tested whether trimethoprim, a dihydrofolate reductase (DHFR) inhibitor, also inhibits injection of Yops by the TTSS (Figure 7). When the wild type strain KIM5 was incubated for three hours with trimethoprim, bacterial growth was inhibited, but the cells were still viable throughout the duration of the experiment (data not shown). We then infected HeLa cells with KIM5 carrying pMM85 (YopE-Bla), in the presence or absence of trimethoprim, for three hours. Because of the growth inhibition that was observed in the presence of drug, we used an MOI of 1 and 10 for the KIM5 infections. As a negative control, HeLa cells were also infected at an MOI of 10 with the TTSS mutant (yscQ −) expressing YopE-Bla. After the infections, cells were stained with CCF2-AM and visualized by microscopy to evaluate injection of the Bla reporter. In the absence of drug at an MOI of 10, large numbers of bacteria could be seen, and most of the HeLa cells displayed blue fluorescence indicating translocation of the YopE-Bla reporter (Figure 7A). When the MOI was decreased to 1, in the absence of drug, there was a mixture of green and light blue cells indicating that the YopE-Bla reporter was injected into host cells to a lesser extent. In contrast, when trimethoprim was added, no blue cells were observed, even at an MOI of 10, which gave a similar bacterial density to infection at an MOI of 1 in the absence of drug (Figure 7A). Likewise, cells that were either uninfected or infected with the yscQ − mutant did not turn blue, confirming that the appearance of blue cells correlates with delivery of YopE-Bla in a TTSS dependent manner. Because no injection of YopE-Bla was observed for cells treated with trimethoprim, we hypothesized that trimethoprim may prevent expression and/or function of the TTSS.
Figure 7. Trimethoprim inhibits the Yersinia TTSS.
A. HeLa cells were infected with KIM5 or CHI 30 (yscQ −) expressing YopE-Bla. Infections were carried out at the indicated MOI for three hours in the presence or absence of 500 µg/ml of trimethoprim. Following infection, cells were stained with CCF2-AM and visualized by microscopy to assess translocation of the YopE-Bla reporter. Blue fluorescence = YopE-Bla injection; green fluorescence = no injection. B. Y. pestis KIM8 and KIM8 Δ1234, carrying pMM207 (yopM expressed from native promoter), were subcultured into MM9 and grown at 26°C for 2 hours. Trimethoprim was then added at the indicated concentrations (0, 15, or 30 µg/ml) and the cultures were shifted to 37°C for 3 hours, followed by TCA precipitation of supernatant (S) and cell pellet (P) fractions and immunoblotting with antibodies against YopM, YopN, YopD, YopR (TTSS secreted proteins), YscD (TTSS machine), RpoA (cytoplasmic protein), Ail (outer membrane protein). Degradation products are not indicated since KIM8 and KIM8 Δ1234 lack Pla protease. C. Secretion assays and western blotting was performed as in Panel B using Y. pestis KIM8 carrying pAH83 (yopM expressed from nptII promoter).
To further assess the effects of trimethoprim on the TTSS, we conducted secretion assays in MM9 in the presence or absence of trimethoprim (0, 15, 30 µg/ml). For these experiments, KIM8 (pla −) was used as the parent strain to ensure that the absence of secreted Yops is not due to degradation by the Pla protease. We also used Y. pestis strain KIM8 Δ1234 [37], which carries a dihydrofolate reductase cassette conferring trimethoprim resistance, thereby allowing us to assess any non-specific effects of the drug on protein expression.
This strain lacks all the effector Yops (ΔyopM/yopT/sycT/yopK/ylpA/yopE/sycE/ypkA/yopJ/yopH), so we introduced a plasmid carrying the yopM locus into both KIM8 and KIM8 Δ1234. This allowed us to track expression and secretion of YopM, as well as YopN, YopR, and YopD, which are still present in KIM8 Δ1234. We also tracked expression of two non-TTSS proteins: Ail, which is an outer membrane protein adhesin that is constitutively expressed [52], [53], and RpoA As shown in Figure 7B, expression of TTSS proteins (YopM, YopN, YopR, YopD) was severely reduced after addition of the drug to the KIM8 parent. Not surprisingly, secretion of those proteins was also severely reduced (at 15 mg/ml) or abolished (at 30 mg/ml) of trimethoprim. However, expression of unrelated, non-TTSS proteins (Ail and RpoA) was only slightly reduced in the presence of the drug. In contrast to KIM8, expression and secretion of all proteins by KIM8 Δ1234 was unaffected even at the highest concentration of trimethoprim (Figure 7B).
Since expression of non-TTSS proteins was less affected by trimethoprim treatment, we asked whether heterologous expression of a Yop could overcome the affects of trimethoprim. We therefore introduced pAH83, which expresses YopM-Bla from the nptII promoter, into KIM8 and performed secretion assays as before. As shown in Figure 7C, expression of YopM-Bla from the nptII promoter was also only slightly reduced, even at the highest trimethoprim concentration. However, despite being expressed, YopM-Bla was not secreted in the presence of trimethoprim, which is probably due to the lack of TTSS proteins. These observations suggest that availability of folate contributes to the regulation of type III secretion in Y. pestis and may be reflective of a link between the TTSS and the metabolic state of yersiniae. Furthermore, since type III proteins were either absent or barely visible within the pellet fractions, it suggests that the effect of a block in the DHFR pathway is at the level of expression of genes in the type III pathway.
Independent mechanisms for activating the TTSS
One of the transposon mutants, CHI 1800 (rfaL−), despite having a secretion defect in vitro (Figures 3 and 5), was able to inject Yops like wild type during infection of CHO cells (Figures 4 and 6). The ability of rfaL− to inject Yops as well as the parent was independently verified using HeLa cells, followed by digitonin fractionation of infected cells and immunoblotting (data not shown). The finding that the rfaL− mutant is able to deliver Yops into host cells is surprising given its inability to secrete Yops in the absence of calcium in vitro. To determine whether the disparity in observed phenotypes could be attributed to the different growth media used in the assays, we conducted in vitro secretion assays in the same medium used to culture HeLa cells (DMEM supplemented with FBS and glutamax). As observed in Figure 8, the mutant was able to both express and secrete the early Yops YopR, YopD, and LcrV as well as wild type when grown in DMEM containing calcium. As expected the late Yops YopE, YopH, and YopM were not secreted in the presence of calcium by either wild type or mutant. However, when EGTA was added to chelate the calcium, YopE, YopH, and YopM were all secreted by wild type KIM5 (Figure 8). In contrast, when calcium was chelated from the medium, rfaL − did not secrete either early or late Yops. The mutant expressed all Yops in the absence of calcium, but several of the Yops appear to be synthesized at lower levels in the mutant compared to wild type (see pellet fractions of YopR, YopD, LcrV, YopE). In addition, the level of the TTSS component YscD is also reduced in the mutant in the absence of calcium (Figure S1). As expected the defects in expression and secretion are restored by complementing the mutant in trans (Figure S1). Since the expression levels of Yops and the TTSS machine are reduced when calcium is depleted, it is possible that the absence of calcium prevents the proper function of assembled needles in rfaL −, leading to the negative feedback on TTSS expression that is typically seen for mutations within TTSS components. The data suggest that although the secretion apparatus in the rfaL mutant is capable of transporting early Yops, it cannot respond normally to the low calcium signal in order to fully activate the TTSS to secrete the effector (late) Yops, and this signaling defect can be overcome during cell contact. Thus, it appears that, while the type III pathway can be induced by either chelating calcium in vitro or by cell contact in vivo, Y. pestis can discern and respond to these two stimuli by different mechanisms.
Figure 8. CHI 1800 cannot induce type III secretion in response to low calcium.
Y. pestis strains KIM5 and CHI 1800 (rfaL −) were subcultured into DMEM with or without EGTA and grown at 26°C for 2 hours and 37°C for 3 hours, followed by TCA precipitation of supernatant and cell pellet fractions and immunoblotting for early Yops (YopR, YopD, LcrV) and late Yops (YopH, YopM, YopE).
Virulence defects of CHI strains
The Yersinia TTSS is an essential virulence component, and mutants lacking a functional TTSS are unable to cause acute disease in mouse models of infection. Therefore, we next investigated the ability of the chromosomal mutants to cause disease in mice. BALB/c mice were infected intravenously with the KIM5 parent or its isogenic mutant CHI strains. Doses were administered in 10-fold increments spanning from 100 to 105 cfu for wild type and from 102 to 107 for the mutants in order to determine the AD50 (dose required to cause acute disease in 50% of the population). The results are summarized in Table 1. As expected, wild type KIM5 produced signs of disease in a dose dependent manner. For mice infected with the highest dose (105 cfu) visible symptoms, such as ruffled fur and squinty eyes, were apparent on day 1. The disease progressed normally and by days 5–6 signs of terminal disease were apparent (severely matted fur and partially or completely closed eyes, severe weight loss, rigid hunched posture, lack of mobility even when touched, labored breathing), at which point the mice were judged to be moribund and were euthanized. The AD50 for the parent was determined to be between 800–3400 cfu in three independent experiments. In contrast, all mutants were reduced in virulence by several orders of magnitude. Infections at the highest dose of 107 cfu with folD−, pgsA−, and pssA− all resulted in the appearance of ruffled fur for 2 to 3 days post-infection, indicating illness relating to the high doses of Gram-negative bacteria. However by days 5 to 6, the symptoms of disease had disappeared and all mice survived. No mice were ever moribund and no deaths occurred at these doses or any of the lower doses for infections with any of these mutants. Identical results were obtained using the yscQ TTSS mutant isolated from this screen. These results demonstrate that these three mutants are indeed avirulent.
Table 1. Mutant phenotypes.
| Strain | Mutation | Doubling time (hours)a | AD50 | Median survivalb |
| KIM5 | WT | 1.34 (0.03) | 800–3400 cfu | 5.5 days |
| CHI 919 | folD | 1.95 (0.01) | >107 cfu | ND |
| CHI 1065 | pgsA | 1.60 (0.03) | >107 cfu | ND |
| CHI 1345 | y0447 (ctgA) | 1.31 (0.02) | 4×106 cfu | 7 days |
| CHI 1800 | y3762 (rfaL) | 1.28 (0.09) | = 107 cfu | 7 days |
| CHI 1965 | pssA | 1.62 (0.03) | >107 cfu | ND |
In vitro growth curves were done using Heart Infusion Broth.
Median survival was calculated based on infections with WT at 105 cfu, the ctgA mutant at 107 cfu, or the rfaL mutant at 107 cfu. No deaths occurred during infections with other mutants, so survival times are not indicated (ND).
In contrast, ctgA− and rfaL− were extremely attenuated but did retain some ability to cause disease at the highest doses tested. Five out of five mice infected with 107 cfu of ctgA− became moribund and were euthanized by days 4 to 7. All mice infected with lower doses of ctgA− survived. From this, we estimate an AD50 of ∼4×106 cfu. Similarly, all mice infected with 107 cfu of rfaL− became ill, but only 3 of 5 mice progressed to moribundity and were euthanized 5–7 days post-infection. Mice infected with lower doses of rfaL− did not become ill. From this, we estimate that the AD50 is equal to or greater than 107 cfu. Because it is not possible to test higher doses, we can only estimate an AD50 for these two mutants and therefore cannot determine whether there is statistically significant difference in virulence between them. However when the data is evaluated to determine time to death, there does not appear to be any significant difference. Figure 9 shows the survival curves for mice infected with 105 cfu of WT, 105 or 107 cfu of ctgA−, and 105 or 107 cfu of rfaL−. The median survival time was 5.5 days for mice infected with WT, compared to 7 days for mice infected with either mutant at the 107 cfu dose. The Log-rank test was used to compare the survival curves between WT and the two mutants. There was no significant difference between the two mutants, indicating that they are similarly attenuated (P = 0.2725). When each mutant (107 cfu) was compared to WT (105 cfu), there was also no significant difference (P = 0.0629 for ctgA− and P = 0.0361 for rfaL−), which indicates that at the highest doses tested the mutants resemble WT in their ability to cause terminal disease. Thus, while ctgA− and rfaL− display severely attenuated phenotypes in this mouse model of infection, these strains are not completely avirulent and retain some ability to cause lethal disease in mice.
Figure 9. In vivo attenuation of CHI strains.
BALB/c mice were infected intravenously with serial dilutions Y. pestis KIM5 or transposon mutants. Moribund mice were euthanized and time to death was recorded. Survival curves are shown for KIM5 (WT) and two mutants (ctgA − and rfaL −).
Discussion
The Yersinia TTSS is an essential component of virulence, and loss of the TTSS, either by mutational inactivation or loss of the pCD1 virulence plasmid, corresponds with a reduction in virulence by six orders of magnitude or more [54]#. Therefore, the TTSS represents an important target for vaccine and therapeutic approaches. Previous work alluded to the presence of chromosomal loci, which contribute to the regulation of type III secretion of Yops by Yersinia pestis [24]. However these genes have remained unidentified. We sought to identify these chromosomal loci as they represent potential targets for vaccine development.
CHI 1065 (pgsA) and CHI 1965 (pssA)
After screening over 2000 strains carrying transposon insertions and displaying the LCR− growth phenotype, we identified five mutants that displayed a strong defect in type III secretion. Three of those mutants (CHI 1065, 1800, 1965) carried insertions in genes involved in the biosynthesis or integrity of the cell envelope. CHI 1065 carries an insertion in pgsA, which encodes a phosphatidylglycerophosphate synthase. Phosphatidylglycerol (PG) is an essential component of membranes, and E. coli mutants lacking pgsA are not viable [55]. While it appears that pgsA is not an essential gene in Y. pestis, CHI 1065 does have a growth defect as shown by a ∼20% longer doubling time compared to wild type (Table 1). PG is essential for lipid modification and the proper localization and/or function of proteins such as DnaA and SecA. It is also a precursor for membrane-derived-oligosaccharides (MDO's), which serve as an osmoprotectant. In Y. enterocolitica, the lipoprotein VirG [56] is required for the proper localization of the YscC secretin in the outer membrane [57]. The lipid modification of VirG is essential for its interaction with YscC and is therefore necessary for type III secretion of Yops [58]. Because of the involvement of PG in localization of membrane proteins, and its requirement for the Sec-dependent translocation pathway, it is likely that the effect of the pgsA mutation in Y. pestis is pleiotropic. Further work is necessary to determine if components of the type III secretion machinery, as well as other membrane proteins which may indirectly contribute to Yop secretion, are mislocalized in CHI 1065.
Likewise, the pssA mutation in CHI 1965 may affect membrane structure or localization of membrane proteins. pssA is required for the synthesis of phosphatidylserine (PS) and subsequently phosphatidylethanolamine (PE), which comprises 70–80% of membrane phospholipids [55]. PE constituents also decorate MDO's and LPS, thereby potentially contributing to membrane structure and integrity. As in the case of pgsA−, the loss of pssA function is not a lethal event, but it does result in approximately 20% longer doubling time in vitro (Table 1).
CHI 1800 (y3762 or rfaL)
Surprisingly, in CHI 1800 we identified an insertion in a gene predicted to be involved in LPS biosynthesis (y3762). Although Y. pestis harbors several mutations in the O Antigen biosynthetic cluster [59], [60], it does produce a lipo-oligosaccharide (LOS) with pyrogenic properties [61]#. The core component of Y. pestis LOS is based on a pentasaccharide containing 4 heptose residues attached to lipid A via Kdo [62]. CHI 1800 carries an insertion in y3762. Although there were no BLAST hits with any significant homology, y3762 is annotated as a putative lipid A-O Antigen ligase. Further comparison of Y3762 and RfaL (the corresponding protein in E. coli [63]#) reveals that both of these proteins are largely transmembrane and the largest contiguous periplasmic stretch of the proteins contain the O-antigen polymerase domain. Moreover, y3762 is located in a predicted operon with rfaQ, which is involved in completion of the inner core of LPS in E. coli. Therefore, we propose to refer to y3762 as rfaL.
The lack of O Antigen in Y. pestis makes the physiological function of RfaL unclear. Interestingly, in E. coli RfaL is also involved in the transfer of alternative saccharides, including enterobacterial common antigen (ECA), to the LPS core [64], [65]. The Y. pestis genome carries the genes for ECA biosynthesis, and several mutations in that pathway were isolated in our screen that lead to only a slight decrease in Yop secretion (unpublished observations). Therefore RfaL may be required for the attachment of an unidentified saccharide moiety to the LOS core.
Low calcium response versus cell contact
Interestingly, rfaL− exhibited the ability to distinguish between activation of the type III pathway in response to low calcium versus cell contact. A similar phenotype was observed in Pseudomonas aeruginosa, where it was found that there are two independent pathways for inducing expression of TTSS genes [66]#. These observations suggest that although a low calcium environment can trigger secretion, it is not a mimic of cell contact. Additionally, our results indicate that there are different mechanisms for activation of the secretion machinery in Y. pestis. It is possible that chelating environmental calcium induces a conformational change in the secretion apparatus, thereby allowing secretion substrates to pass. In the context of a cell culture infection, where there is calcium present in the medium, protein-protein interactions during cell contact could also lead to a conformational change leading to secretion of TTSS effectors. Further work is required to determine how the conformational change in the secretion apparatus is sensed and which proteins are involved in the hypothetical protein-protein interaction during target cell contact.
CHI 1345 (y0447 or ctgA)
One of the mutants, CHI 1345, isolated in this screen carried a mutation in a hypothetical gene. Y0447 is predicted to have 4 transmembrane helical domains and localize to the cytoplasmic face of the membrane. It is annotated as belonging to the DedA family of membrane proteins. Proteins from this family can be found in members of the α-, β-, and γ- proteobacteria. Little is known about the function of the proteins, and these proteins often share little similarity in their amino acid sequences. However, screens performed in E. coli, Salmonella, and Neisseria have shown that proteins in the DedA family are associated with cell division, response to divalent cations, and resistance to cationic antimicrobial peptides, all of which are linked to membrane biology [67], [68], [69]. The phenotypes of the mutants isolated in these screens suggest that they may have structural defects in their membranes. Mutations in genes encoding YghB and YqjA of the DedA family in E. coli result in an altered phospholipid ratio in the membranes of the mutants [67]. Interestingly, CHI 1965 (pssA −) and CHI 1065 (pgsA −) isolated from our screen would also be predicted to have altered phospholipid ratios. We suspect that y0447, pgsA and pssA may play a role in maintaining the structure and stability of the bacterial membranes. Defects in membrane biology in these mutants could consequently affect the structure or stability of the TTSS machinery. It is not clear at this point if any such membrane defect would prevent the proper assembly of the TTSS machinery or if these alterations would activate cellular stress response systems that could lead to a regulatory block on expression of the TTSS genes. Because this is the first characterization of y0447, we will refer to it as ctgA (chromosomal type III secretion gene), for its role in type III secretion.
CHI 919 (folD)
In contrast to several other screen isolates with mutations affecting membrane structure, the transposon insertion in folD in CHI 919 likely causes an effect on metabolic functions. folD is annotated as a bifunctional enzyme (5, 10-methylene-tetrahydrofolate dehydrogenase and cyclohydrolase) similar to that of E. coli [70]# which is involved in tetrahydrofolate (THF) production. The Y. pestis genome harbors two additional genes, glyA and gcvT, whose products are predicted to bypass the FolD-catalyzed reactions, thus explaining the viability of CHI 919 in media lacking folate. Despite this, CHI 919 had the most severe growth defect of the mutants with a ∼65% longer doubling time (Table 1). THF is an important coenzyme and provides one-carbon donors to several metabolic pathways including purine, methionine and thymidylate synthesis [71]. The observation that trimethoprim inhibits type III secretion (Figure 7), likely at the level of expression, confirms the importance of folD and/or THF in type III secretion. Therefore, the lack of secretion in a folD mutant (CHI 919) may reflect a connection between type III secretion and the metabolic state of the cell. This hypothesis is supported by studies in Pseudomonas aeruginosa, which showed that histidine levels and the two-component system, cbrAB, contribute to the regulation of type III secretion [72]. Similarly, the aceAB operon, which encode subunits of pyruvate dehyrogenase, are also necessary for type III secretion in P. aeruginosa [73].
Perspectives
In conclusion, Y. pestis genes identified in a Mu insertional mutagenesis screen for LCR phenotypes point to hitherto unappreciated roles for LPS biosynthesis, envelope structure and integrity, and intermediary metabolism in the Yersinia TTSS. Further, unknown genes that have never been linked to type III secretion may reveal novel regulatory or assembly mechanisms for the type III pathway. Several other genetic screens using STM or IVET to identify virulence genes have been conducted in Yersinia species [74], [75], [76], [77], [78], yet there is no overlap between the genes identified previously and those identified in our study. One possible explanation is that we did not saturate the screen, and presumably there are additional chromosomal genes that may contribute to type III secretion. Additionally, the STM and IVET screens are used to identify genes that are important in vivo, and the approaches are fundamentally different than ours. It is interesting to note that all of the mutants tested in our screen demonstrated a clear virulence defect. Perhaps a more thorough STM screen in Y. pestis would also identify these or similar genes. In P. aeruginosa, several screens have identified environmental sensors and two-component systems, such as soxR, copRS, and retS, as important regulatory factors [79], [80], [81], [82]. Although we did not identify any of those genes in this study, it is not unlikely that similar signal transduction cascades exist in Yersinia species, and this is a prospect that warrants further investigation.
Finally, as one goal of our screen was to identify candidates for vaccines or new antimicrobials, we must point out the affect of trimethoprim on the TTSS (Figure 7). Trimethoprim, which is among the treatment options for plague infections, targets FolD. And while the antimicrobial properties of trimethoprim are independent of its affect on the Yersinia TTSS, the isolation of a folD mutant (CHI 919) in our screen along with the observation that trimethoprim prevents proper expression of TTSS components, may indicate an additional indirect method for the drug to inhibit disease progression. It is possible that other genes identified by this study would also make good targets for antimicrobials. A particularly interesting possibility for a broad-spectrum target is CtgA, which appears to be widely conserved. Since loss of CtgA impairs TTSS without affecting growth, targeting it may be useful in reducing virulence without putting a direct selective pressure on growth and correspondingly on the selection of drug-resistant mutants.
Supporting Information
Secretion phenotype of CHI 1800. Y. pestis strains KIM5 and CHI 1800 (rfaL −), in the presence or absence of complementing plasmid, were subcultured into DMEM with or without EGTA and grown at 26°C for 2 hours and 37°C for 3 hours, followed by TCA precipitation of supernatant and cell pellet fractions and immunoblotting for YopE and YscD
(TIF)
Primers used in this study.
(DOC)
Acknowledgments
We thank Greg Plano for providing strains, and Bob Brubaker, Deb Anderson, and members of the Schneewind and Marketon laboratories for helpful discussions. We would also like to thank Christopher Cole for help with the DMEM secretion assays.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors acknowledge membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE, National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153). MM acknowledges funding for this work through the Career Development Program at the Great Lakes RCE (CDP6). In addition, this work was partially funded by the Office of the Vide Provost of Research at Indiana University-Bloomington through the Faculty Research Support Program (MM). This work was also partially funded by grant awards to OS, AI055545 and AI42797, from the National Institute of Allergy and Infectious Diseases, Infectious Diseases Branch. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, et al. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yother J, Chamness TW, Goguen JD. Temperature controlled plasmid regulon associated with low calcium response in Yersinia pestis. J Bacteriol. 1986;165:443–447. doi: 10.1128/jb.165.2.443-447.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoe NP, Minion FC, Goguen JD. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol. 1992;174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoe NP, Goguen JD. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wattiau P, Cornelis GR. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee VT, Mazmanian SK, Schneewind O. A program of Yersinia enterocolitica type III secretion reactions is triggered by specific host signals. J Bacteriol. 2001;183:4970–4978. doi: 10.1128/JB.183.17.4970-4978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skrzypek E, Straley SC. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J Bacteriol. 1993;175:3520–3528. doi: 10.1128/jb.175.11.3520-3528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, et al. Status of YopM and YopN in the Yersinia yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 11.Nilles ML, Williams AW, Skrzypek E, Straley SC. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iriarte M, Cornelis GR. YopT, a new Yersinia effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;29:915–929. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 13.Jackson MW, Day JB, Plano GV. YscB of Yersinia pestis functions as a specific chaperone for YopN. J Bacteriol. 1998;180:4912–4921. doi: 10.1128/jb.180.18.4912-4921.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilles ML, Fields KA, Straley SC. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J Bacteriol. 1998;180:3410–3420. doi: 10.1128/jb.180.13.3410-3420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng LW, Schneewind O. Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for the specific targeting of YopE, YopH, YopM and YopN into the cytosol of eukaryotic cells. J Bacteriol. 2000;182:3183–3190. doi: 10.1128/jb.182.11.3183-3190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng LW, Kay O, Schneewind O. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J Bacteriol. 2001;183:5293–5301. doi: 10.1128/JB.183.18.5293-5301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBord K, Lee VT, Schneewind O. On the Role of LcrG and LcrV during the type III targeting of effector Yops by Yersinia enterocolitica. J Bacteriol. 2001;183:4588–4598. doi: 10.1128/JB.183.15.4588-4598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferracci F, Schubot FD, Waugh DS, Plano GV. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol. 2005;57:970–987. doi: 10.1111/j.1365-2958.2005.04738.x. [DOI] [PubMed] [Google Scholar]
- 19.Brubaker RR. Influence of Na(+), dicarboxylic amino acids, and pH in modulating the low-calcium response of Yersinia pestis. Infection and immunity. 2005;73:4743–4752. doi: 10.1128/IAI.73.8.4743-4752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brubaker RR, Surgalla MJ. The Effect of Ca++ and Mg++ on Lysis, Growth, and Production of Virulence Antigens by Pasteurella Pestis. J Infect Dis. 1964;114:13–25. doi: 10.1093/infdis/114.1.13. [DOI] [PubMed] [Google Scholar]
- 21.Straley SC, Plano GV, Skrzypek E, Haddix PL, Fields KA. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 22.Michiels T, Wattiau P, Brasseur R, Ruysschaert J-M, Cornelis G. Secretion of Yop proteins by yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higuchi K, Kupferberg LL, Smith JL. Studies on the nutrition and physiology of Pasteurella pestis. J Bacteriol. 1959;77:317–321. doi: 10.1128/jb.77.3.317-321.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goguen JD, Yother J, Straley SC. Genetic analysis of the low calcium response in Yersinia pestis Mud1(Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter PB, Zahorchak RJ, Brubaker RR. Plague virulence antigens from Yersinia enterocolitica. Infection and immunity. 1980;28:638–640. doi: 10.1128/iai.28.2.638-640.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreyfus LA, Brubaker RR. Consequences of aspartase deficiency in Yersinia pestis. Journal of bacteriology. 1978;136:757–764. doi: 10.1128/jb.136.2.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortlock RP, Brubaker RR. Glucose-6-Phosphate Dehydrogenase and 6-Phosphogluconate Dehydrogenase Activities of Pasteurella Pestis and Pasteurella Pseudotuberculosis. Journal of bacteriology. 1962;84:1122–1123. doi: 10.1128/jb.84.5.1122-1123.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields KA, Nilles ML, Cowan C, Straley SC. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infection and immunity. 1999;67:5395–5408. doi: 10.1128/iai.67.10.5395-5408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosdent N, Maridonneau-Parini I, Sory MP, Cornelis GR. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infection and immunity. 2002;70:4165–4176. doi: 10.1128/IAI.70.8.4165-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portnoy DA, Moseley SL, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infection and immunity. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoiczyk E, Blobel G. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc Natl Acad Sci USA. 2001;98:4669–4674. doi: 10.1073/pnas.071065798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torruellas J, Jackson MW, Pennock JW, Plano GV. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 33.Heesemann J, Laufs R. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J Bacteriol. 1983;155:761–767. doi: 10.1128/jb.155.2.761-767.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heesemann J, Algermissen B, Laufs R. Genetically manipulated virulence of Yersinia enterocolitica. Infect Immun. 1984;46:105–110. doi: 10.1128/iai.46.1.105-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roncero C, Darzins A, Casadaban MJ. Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. J Bacteriol. 1990;172:1899–1904. doi: 10.1128/jb.172.4.1899-1904.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brubaker RR. Mutation rate to non-pigmentation in Pasteurella pestis. J Bacteriol. 1969;98:1404–1406. doi: 10.1128/jb.98.3.1404-1406.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartra SS, Jackson MW, Ross JA, Plano GV. Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infection and immunity. 2006;74:1381–1386. doi: 10.1128/IAI.74.2.1381-1386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucl Acid Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acid Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day JB, Ferracci F, Plano GV. Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol Microbiol. 2003;47:807–823. doi: 10.1046/j.1365-2958.2003.03343.x. [DOI] [PubMed] [Google Scholar]
- 43.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for LacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 44.Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. Plague bacteria target immune cells during infection. Science. 2005;309:1739–1741. doi: 10.1126/science.1114580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dewoody R, Merritt PM, Houppert AS, Marketon MM. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol. 2011;79:1445–1461. doi: 10.1111/j.1365-2958.2011.07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathem WW, Price PA, Miller VL, Goldman WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–513. doi: 10.1126/science.1137195. [DOI] [PubMed] [Google Scholar]
- 47.Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci U S A. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson DM, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 49.Sodeinde OA, Goguen JD. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect Immun. 1988;56:2743–2748. doi: 10.1128/iai.56.10.2743-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sodeinde OA, Sample AK, Brubaker RR, Goguen JD. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramamurthi KS, Schneewind O. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J Bacteriol. 2002;184:3321–3328. doi: 10.1128/JB.184.12.3321-3328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolodziejek AM, Sinclair DJ, Seo KS, Schnider DR, Deobald CF, et al. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology. 2007;153:2941–2951. doi: 10.1099/mic.0.2006/005694-0. [DOI] [PubMed] [Google Scholar]
- 53.Bartra SS, Styer KL, O'Bryant DM, Nilles ML, Hinnebusch BJ, et al. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infection and immunity. 2008;76:612–622. doi: 10.1128/IAI.01125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry RD, Fetherston JD. Yersinia pestis - etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 56.Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis GR. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J Bacteriol. 1995;177:4230–4237. doi: 10.1128/jb.177.15.4230-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis GR, et al. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 58.Burghout P, Beckers F, de Wit E, van Boxtel R, Cornelis GR, et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol. 2004;186:5366–5375. doi: 10.1128/JB.186.16.5366-5375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng W, Burland V, Plunkett Gr, Boutin A, Mayhew GF, et al. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhill J, Wren BW, Thompson NR, Titball RW, Holden MT, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 61.Davies DA. A specific polysaccharide of Pasteurella pestis. Biochem J. 1956;63:105–116. doi: 10.1042/bj0630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hitchen PG, Prior JL, Oyston PC, Panico M, Wren BW, et al. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: regulation of LOS structure by the PhoPQ system. Mol Microbiol. 2002;44:1637–1650. doi: 10.1046/j.1365-2958.2002.02990.x. [DOI] [PubMed] [Google Scholar]
- 63.Schnaitman CA, Klena JD. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klena JD, Schnaitman CA. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 65.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick PD. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 66.Dasgupta N, Ashare A, Hunninghake GW, Yahr TL. Transcriptional induction of the Pseudomonas aeruginosa type III secretion system by low Ca2+ and host cell contact proceeds through two distinct signaling pathways. Infect Immun. 2006;74:3334–3341. doi: 10.1128/IAI.00090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompkins K, Chattopadhyay B, Xiao Y, Henk MC, Doerrler WT. Temperature sensitivity and cell division defects in an Escherichia coli strain with mutations in yghB and yqjA, encoding related and conserved inner membrane proteins. Journal of bacteriology. 2008;190:4489–4500. doi: 10.1128/JB.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol Microbiol. 2004;53:229–241. doi: 10.1111/j.1365-2958.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- 69.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, et al. Cationic antimicrobial peptide resistance in Neisseria meningitidis. Journal of bacteriology. 2005;187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dev IK, Harvey RJ. A complex of N5,N10-methylenetetrahydrofolate dehydrogenase and N5,N10-methenyltetrahydrofolate cyclohydrolase in Escherichia coli. Purification, subunit structure, and allosteric inhibition by N10-formyltetrahydrofolate. J Biol Chem. 1978;253:4245–4253. [PubMed] [Google Scholar]
- 71.Maden BE. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem J. 2000;350 Pt 3:609–629. [PMC free article] [PubMed] [Google Scholar]
- 72.Rietsch A, Wolfgang MC, Mekalanos JJ. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun. 2004;72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dacheux D, Epaulard O, de Groot A, Guery B, Leberre R, et al. Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect Immun. 2002;70:3973–3977. doi: 10.1128/IAI.70.7.3973-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darwin AJ, Miller VL. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;51–62:32. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 75.Karlyshev AV, Oyston PC, Williams K, Clark GC, Titball RW, et al. Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associated genes. Infect Immun. 2001;69:7810–7819. doi: 10.1128/IAI.69.12.7810-7819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flashner Y, Mamroud E, Tidhar A, Ber R, Aftalion M, et al. Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect Immun. 2004;72:908–915. doi: 10.1128/IAI.72.2.908-915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mecsas J, Bilis I, Falkow S. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect Immun. 2001;69:2779–2787. doi: 10.1128/IAI.67.5.2779-2787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young GM, Miller VL. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 79.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci U S A. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ha U, Jin S. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect Immun. 1999;67:5324–5331. doi: 10.1128/iai.67.10.5324-5331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ha UH, Kim J, Badrane H, Jia J, Baker HV, et al. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol Microbiol. 2004;54:307–320. doi: 10.1111/j.1365-2958.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- 82.Zolfaghar I, Angus AA, Kang PJ, To A, Evans DJ, et al. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 2005 doi: 10.1016/j.micinf.2005.04.017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secretion phenotype of CHI 1800. Y. pestis strains KIM5 and CHI 1800 (rfaL −), in the presence or absence of complementing plasmid, were subcultured into DMEM with or without EGTA and grown at 26°C for 2 hours and 37°C for 3 hours, followed by TCA precipitation of supernatant and cell pellet fractions and immunoblotting for YopE and YscD
(TIF)
Primers used in this study.
(DOC)