Abstract
Odorant binding proteins play a crucial role in transporting semiochemicals across the sensillum lymph to olfactory receptors within the insect antennal sensilla. In this study, the general odorant binding protein 2 gene was cloned from the antennae of Loxostege sticticalis, using reverse transcription PCR and rapid amplification of cDNA ends. Recombinant LstiGOBP2 was expressed in Escherichia coli and purified by Ni ion affinity chromatography. Real-time PCR assays indicated that LstiGOBP2 mRNA is expressed mainly in adult antennae, with expression levels differing with developmental age. Ligand-binding experiments using N-phenyl-naphthylamine (1-NPN) as a fluorescent probe demonstrated that the LstiGOBP2 protein has binding affinity to a broad range of odorants. Most importantly, trans-11-tetradecen-1-yl acetate, the pheromone component of Loxostege sticticalis, and trans-2-hexenal and cis-3-hexen-1-ol, the most abundant plant volatiles in essential oils extracted from host plants, had high binding affinities to LstiGOBP2 and elicited strong electrophysiological responses from the antennae of adults.
Introduction
The meadow moth, Loxostege sticticalis (Lepidoptera∶Pyralidae), is one of the worst pests in Asia, Europe, and North America and has caused severe economic damage almost every year [1]. This insect is a polyphagous pest that can feed on 35 plant families and 200 species, but it has an obvious preference for certain host plants [1]–[4]. The meadow moth can find its host using the chemical volatiles emitted by the plant as cues. The plants release volatiles as chemical cues that are diluted and mixed with a myriad of compounds in the environment. With a highly developed olfactory system, the insects can detect low-level signals released from particular plants [5]–[7].
The insects recognize the chemical signals by olfactory receptor neurons in the olfactory sensilla, which are located in the insect antennae and are surrounded by sensillar aqueous lymph that creates a barrier between the external environment and the olfactory receptors, particularly for hydrophobic molecules [8]–[10]. The sensillar lymph contains odorant binding proteins (OBPs), which are believed to carry hydrophobic odorants from the environment to the surface of olfactory receptor neurons [10]–[11]. Odorant binding proteins are the first relay in semiochemical reception in insects, as they enable ligand-receptor interactions. OBPs are water-soluble proteins with a pattern of six conserved cysteine residues. The six conserved cysteines are paired in three disulphide bridges in an interlocking fashion (1–3, 2–5 and 4–6), which are used as a ‘signature’ for identifying insect OBPs [10], [12]–[14]. The first OBP of insects was discovered in the giant moth Antheraea polyphemus [8]. Over the last few years, 400 OBPs have been isolated and cloned from more than 40 insect species belonging to eight different orders [15]–[16]. However, even for species for which multiple OBPs have been identified or cloned, the number of binding proteins in the sensillar lymph is significantly lower than the number of compounds the insects can smell. Based on their amino acid sequences, insect OBPs are divided into Pheromone binding proteins (PBPs), general OBPs (GOBPs), and antennal binding protein X (ABPX) [15], [17]–[18]. In many species, the specific binding of proteins to pheromones has been determined for the PBPs, which are located in the pheromone sensilla, the sensilla trichodea [11], [19]–[26]. GOBPs located in the sensilla basiconica are thought to interact with general odorants (e.g., plant volatiles) and are further classified as GOBP1 and GOBP2 [9], [27]–[31]. GOBP2 plays an important role in the detection of general odorants and has a conserved sequence across different species [27]–[28], [32]–[34]. Surprisingly, GOBP2 has been found to have high binding activity to major pheromones in several insect species, some of which are located in the sensilla trichodea [18], [35]–[38].
More than 100 sex pheromones have been used as potential biological agents to control insects through mating disruption or mass trapping strategies [39]. The relationship between plant volatiles and insect behavior is always a hot focal spot for researchers interested in insect control [40]–[44]. In previous works, we demonstrated that the meadow moth has an obvious preference for Chenopodium glaucum, and the volatiles of Chenopodium glaucum were identified by GC-MS [3] and analyzed the function of a general odorant-binding protein from Loxostege sticticalis [45]. The LstiGOBP1 protein has binding affinity to a broad range of test compounds. In the present study, another gene encoding GOBP2 from Loxostege sticticalis (LstiGOBP2) was identified, and ligand-binding activities were measured by a fluorescence competitive binding assay using the 1-NPN fluorescent probe. Electroantennograms (EAGs) were used to record the reaction of the antennae to different compounds that can bind to GOBP2 of Loxostege sticticalis. The tissue and developmental expression patterns of GOBP2 of Loxostege sticticalis were also detected by real-time quantitative PCR (qPCR). In the last, we also compared the binding affinity of two general odorant-binding proteins to discuss the charactering of LstiGOBP2.
Results
1 Coding and amino acid sequences
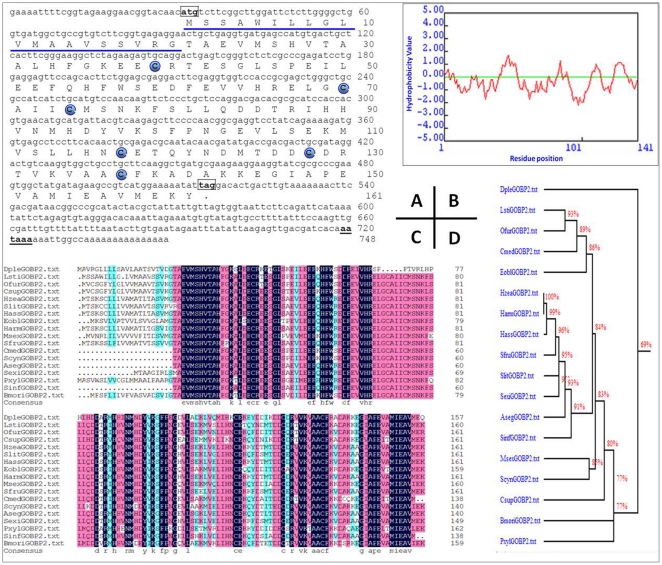
Using rapid amplification of cDNA ends (RACE)-PCR, a full-length cDNA encoding GOBP2 was cloned from L. sticticalis (Fig. 1A) (GenBank EU239360). The open reading frame (ORF) of LstiGOBP2 cDNA consists of 486 nucleotides and encodes a predicted precursor protein containing 161 amino acids (Fig. 1). The ORF is terminated by a TAG stop codon that is followed by a 232 nucleotide 3′ untranslated region, exclusive of the poly A tail. A consensus polyadenylation signal (AATAAA) is found at 202 bp from the stop codon. The deduced protein sequence revealed a 20-amino acid signal peptide as predicted by SignalP software. The calculated molecular weight and the isoelectric point of mature LstiGOBP2 were 16.0 kDa and 4.76, respectively. LstiGOBP2 had the typical six-cysteine signature of OBPs (Fig. 1A). The hydropathic nature of LstiGOBP2, which is very similar to other insect GOBPs and PBPs [46], was calculated and plotted for each residue in the sequence, revealing that four residue regions were hydrophobic (Fig. 1B).
Figure 1. Characterization and Phylogenetic analysis of LstiGOBP2.
(A) Nucleotide sequence and deduced amino acid sequence of the LstiGOBP2 cDNA in L. sticticalis. The six conserved cysteines are indicated in boxes. The predicted signal peptide is underlined. The asterisk marks the translation-termination codon. (B) The predicted hydropathy profiles of the deduced amino acid sequences of the LstiGOBP2. The hydropathy profiles was predicted using DNAman 6.3.0.99. (C) Alignment of GOBP2 from lepidopteran insects. Ostrinia furnacalis (OfurGOBP2, ABG66419)\ Antheraea pernyi (AperGOBP2, Q17075)\ Chilo suppressalis (CsupGOBP2, ACJ07120)\ Helicoverpa zea (HzeaGOBP2, AAG54078)\ Helicoverpa assulta (HassGOBP2, AAQ54909)\ Spodoptera litura (SlitGOBP2, ABM54824)\Mamestra brassicae (MbraGOBP2, AAC05703)\ Manduca sexta (MsexGOBP2, AAG50015)\ Heliothis virescens (HvirGOBP2, Q27288)\ Ectropis oblique (EoblGOBP2, ACN29681)\ Spodoptera frugiperda (SfruGOBP2, AAT74555)\ Cnaphalocrocis medinalis (CmedGOBP2, ACJ07122)\ Epiphyas postvittana (EposGOBP2, Q95VP2)\ Samia cynthia ricini (ScynGOBP2, BAF91328)\ Spodoptera exigua (SexiGOBP2, CAC12831)\ Agrotis segetum (AsegGOBP2, ABI24161)\ Plutella xylostella (PxylGOBP2, ABY71035)\Sesamia inferens (SinfGOBP2, ACJ07121)\ Amyelois transitella (AtraGOBP2, ACX47894)\ (D) Homology tree of GOBP2 amino acid sequences in Lepidoptera. LstiGOBP2 is indicated in boxes.
2 Alignment to orthologous of other species
An alignment of the amino acid sequences of LstiGOBP2 with the corresponding GOBP2s from other species of Lepidoptera is shown in Figure 1C. GOBP2 is highly conserved between species. GOBP2s from all Lepidoptera species, including LstiGOBP2, have the typical six-cysteine signature of OBPs and have a common pattern: X18−Cys−X30−Cys−X3−Cys−X42−Cys−X8–10−Cys−X8−Cys−X24–26, in which X is any amino acid. LstiGOBP2 shares high identity (70–93%) with other Lepidoptera GOBP2s; the highest identities are 93% with Ostrinia furnacalis and 89% with Cnaphalocrocis medinalis, which is in accordance with their phylogenetic relationships based on morphological characters (Fig. 1D).
3 Expression and purification of recombinant LstiGOBP2
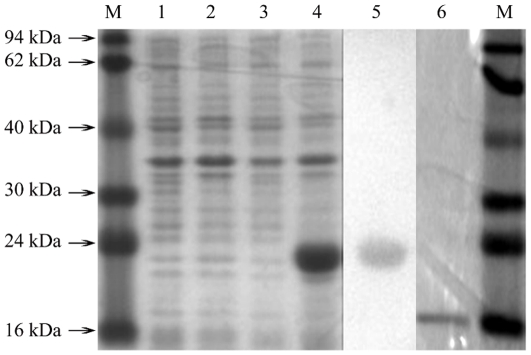
Recombinant LstiGOBP2 was expressed in E. coli as a completely soluble protein with high yields (more than 20 mg/L). The His-tag of the recombinant protein was removed by rEK. The protein was purified by two rounds of Ni ion affinity chromatography: the first round was intended to purify the recombinant protein from total protein and the second round was intended to divide the His-tag and the uncleaved His-tagged proteins (Fig. 2). The purified recombinant proteins were then tested for their binding properties and used for the production of polyclonal antibodies.
Figure 2. SDS-PAGE electrophoretic and western blot analysis of expressed recombine LstiGOBP2.
M. Molecular weight marker; 1. Non-induced E. coli pET30; 2. Induced E. coli pET30; 3. Non-induced E. coli LstiGOBP2; 4. Induced E. coli LstiGOBP2; 5. Western blot; 6. Purified protein cleaved His by rbEK.
4 Expression pattern analysis of LstiGOBP2
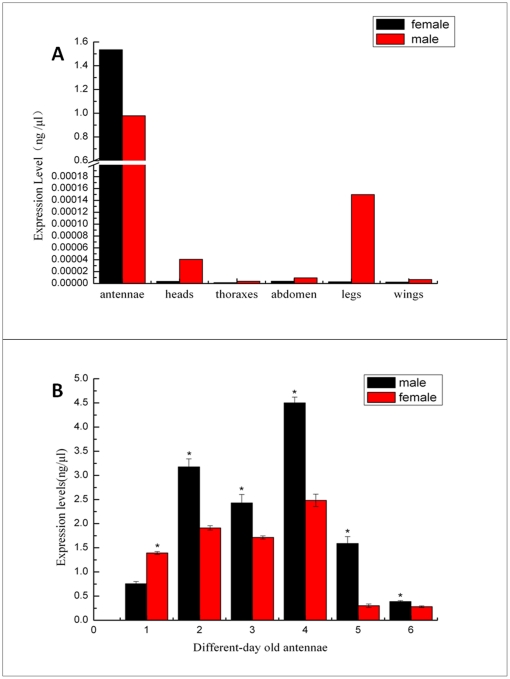
We examined the expression pattern of LstiGOBP2 mRNA in different tissues by qPCR. The desired product was largely amplified from cDNA templates that were reverse-transcribed from total RNA in male and female antennae, with only a few derived from other tissues, suggesting that GOBP2 is mainly expressed in antennae (Fig. 3A). In general, the levels of transcripts were very low in all tissues except the antennae, where LstiGOBP2 was highly expressed. However, the expressed quantity of LstiGOBP2 was different in antennae of different ages and there was a notable difference between males and females, with the quantity in male antennae being significantly higher than in female antennae. The quantity of LstiGOBP2 was the highest for both males and females in four-day-old antennae, which is consistent with the age that adults find host plants (Fig. 3B).
Figure 3. Expression pattern analysis of LstiGOBP2.
(A) qPCR analysis of Loxostege sticticalis general odorant binding protein 2 (LstiGOBP2) expressed in different tissues. cDNAs were amplified with specific primers from antennae, thoraxes, abdomens, wings, legs, tarsite and heads(without antennae). (B) qPCR analysis of Loxostege sticticalis general odorant binding protein 2 (LstiGOBP2) expressed in different-day old antennae.
5 Fluorescence binding assays
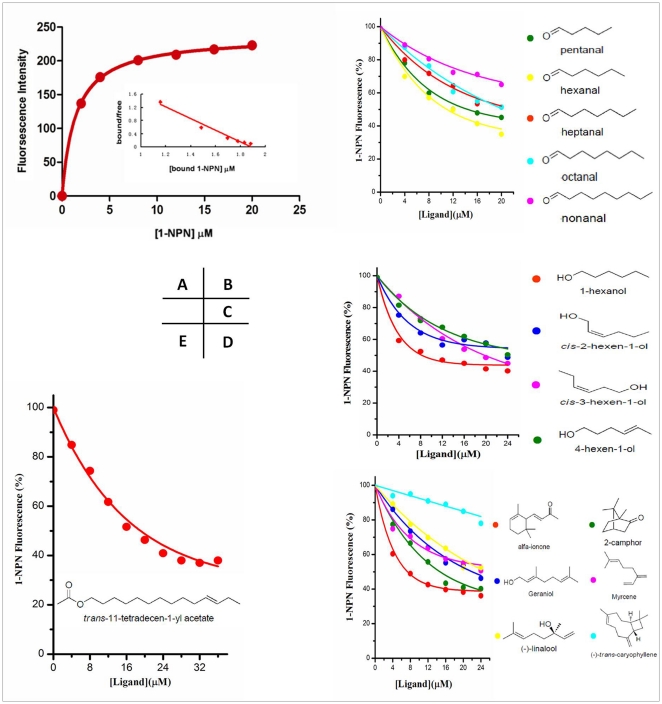
For binding assays with LstiGOBP2, we found that N-phenyl-naphthylamine (1-NPN) was suitable for investigating odorant ligand binding to OBPs. When excited at 337 nm, 1-NPN displayed an emission peak at 380 nm, which shifted to about 420 nm in the presence of LstiGOBP2. The dissociation constant of 1-NPN-bound recombinant LstiGOBP2, approximately 1.44 µM, was calculated according to the changes in fluorescence intensity (Fig. 4A).
Figure 4. Ligand-binding experiments.
(A) Binding curve and relative Scatchard plot. (B) Competitive binding curves of different-carbon-aldehyde ligands to the LstiGOBP2. The chemical structures of the ligands are shown on the right. Mixtures of proteins and 1-NPN, at a 2 µM concentration, were titrated with a 1 mM ligand solution in methanol. (C, D) Competitive binding curves of different structural arrangement ligands to the LstiGOBP2. (E) Competitive binding curves of the sex pheromone component to the LstiGOBP2.
Using competitive binding assays, we tested 51 synthetic potential ligands as competitors, including compounds from the leaves of green plants, plant volatiles, high binding affinity ligands of LstiGOBP1 and a sex pheromone component [6], [45], [47]–[48]. Curves for a few representative ligands tested are shown in Figure 4B–4E. Table 1 lists the IC50 values (the concentration of ligand halving the initial fluorescence value) and the calculated inhibition constants (K i) where possible for each OBP/ligand combination. When IC50 values of low-affinity ligands could not be calculated, we report the fluorescence intensity (Int) measured at the ligand concentration (20 µM) as a percent of the initial fluorescence in the absence of competitor.
Table 1. Binding of pure organic compounds to selected recombinant LstiGOBP2.
| Ligands | LstiGOBP2 | Ligands | LstiGOBP2 | ||||
| IC50 | Int | Ki | IC50 | Int | Ki | ||
| Aliphatic alcohols | Aromatic compounds | ||||||
| 1-Hexanol | 11 | 42 | 6.5 | Benzaldehyde | 19 | 48 | 11.2 |
| Cis-3- hexen-1-ol | 19 | 49 | 11.2 | Cinnamaldehyde | 17 | 45 | 10 |
| Cis-2-hexen-1-ol | 24 | 59 | 14.1 | Phenyl acetaldchyde | 20 | 50 | 11.8 |
| 4-Hexen-1-ol | 26 | 58 | 15.3 | 2,4-Di-tert-butylphenol | 20 | 50 | 11.8 |
| 1-Heptanol | 10.5 | 40 | 6.2 | Dimethyl phthalate | 36 | 64 | 21.2 |
| 6-Methyl-5-hepten-2-ol | 28 | 63 | 16.5 | Methyl salicylate | - | 83 | - |
| Iso-octanol | 28 | 58 | 16.5 | Terpenoids | |||
| Aliphatic aldehydes | α-ionone | 8 | 39 | 4.7 | |||
| 1-Pentanal | 14 | 46 | 8.2 | β-ionone | 11 | 37 | 6.5 |
| 1-Hexanal | 12.5 | 36 | 7.3 | Myrcene | 25 | 56 | 14.7 |
| Trans-2-hexenal | 10.5 | 50 | 6.2 | Camphene | 23 | 54 | 13..5 |
| 1-Heptanal | 24 | 52 | 14.1 | Camphor | 14 | 41 | 8.2 |
| 1-Octanal | 24 | 52 | 14.1 | (-)-Linalool | 28 | 53 | 16.5 |
| 1-Nonanal | 39 | 66 | 22.9 | Geraniol | 22 | 54 | 12.9 |
| Aliphatic ketones | (-)-Trans-caryophyllene | - | 86 | - | |||
| 6-Methyl-5-hepten-2-one | 28 | 64 | 16.5 | Heterocyclic compound | |||
| Aliphatic ester derivatives | Benzothiazole | 22 | 52 | 12.9 | |||
| Cis-3-hexenyl acetate | 29 | 57 | 17.1 | Sex pheromone component | |||
| Trans-2-hexenyl acetate | 28 | 59 | 16.5 | Trans-11-tetradecen-1-yl acetate | 18 | 48 | 10.6 |
| Dimethyl phthalate | 20 | 50 | 11.8 | ||||
Solution of protein was at 2 µM, and the added concentration of 1-NPN was in line with the dissociation constants of LstiGOBP2/1-NPN complex calculated. Then the mixed solution was titrated with 1 mM solution of each ligand in methanol to final concentrations of 2–50 µM. For the protein, we report the fluorescence intensity (Int) measured at the ligand concentration (20 µM) as percent of the initial fluorescence, the concentration of ligand halving the initial fluorescence intensity (IC50), where applicable, and the relative dissociation constant (Ki) calculated as described in “Materials and methods”. Dissociation constants of ligands whose IC50 exceeded 50 mM are represented as “-”. Other potential ligands were tested, but the remaining 17 potential ligands did not bind LstiGOBP2. These compounds included1-Octen-3-ol, Trans-2-hexen-1-ol, R-(+)-Limonene, α-Phellandrene, α-Terpineol, Nerolidol, α-Pinene, Octadecene, Methyl anthranilate, Methyl palmitate, Undecane, Dodecane, Tridecane, Tetradecane, Pentadecane, Hexadecan and Heptadecane.
Most of the volatiles tested succeeded in displacing 1-NPN from the LstiGOBP2/1-NPN complex at concentrations up to 40 mM. The compounds α-ionone, β-ionone, trans-2-hexenal, 1-hexanol and 1-heptanol had high binding affinities to LstiGOBP2 with Ki values of 4.7, 6.5, 6.2, 6.5 and 6.2 µM, respectively. Interestingly, C11–C17 alkanes did not bind to LstiGOBP2, even though pentadecane has been reported to bind LstiGOBP1 [45]. Most of the aromatic compounds tested in these experiments had a medium binding affinity.
The binding activity of linear aliphatic aldehydes and alcohols to LstiGOBP2 were also tested in our study. We observed that C5–C9 aliphatic aldehydes had different binding activities, and the binding capacities decreased as of the number of carbon atoms increased (Fig. 4B). The binding activities were also different when the compounds differed in structural arrangement, even when the number of carbon atoms remained the same (Fig. 4C, 4D). Surprisingly, trans-11-tetradecen-1-yl acetate, a sex pheromone component, could bind strongly to LstiGOBP2 (Fig. 4E).
6 Electroantennogram (EAG) recordings
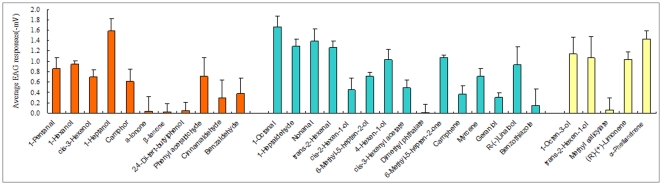
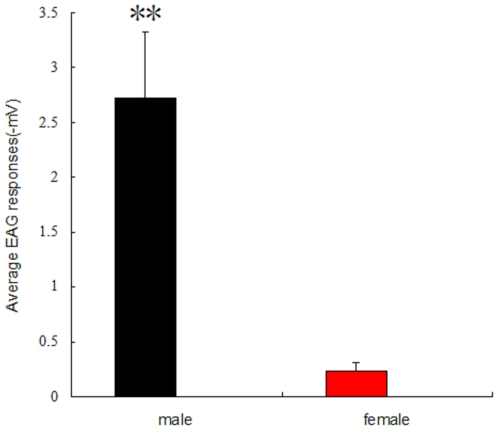
Using the results of the competitive binding assays, we selected 31 volatiles to perform EAGs based on their different affinities to LstiGOBP2. The EAG recordings demonstrated that most plant volatiles elicited strong electrophysiological responses from the antennae of female L. sticticalis adults at 10 mg/ml (Fig. 5). 1-Heptanol and 1-octanal gave the highest EAG responses and induced a depolarization of −1.6 mV. However, α-ionone, β-ionone and 2,4-di-tert-butylphenol, which were high-affinity competitive binding assay ligands, had only weak effects on both male and female antennae. Conversely, compounds 1-octen-3-ol, E-2-hexeno1, (R)-(+)-limonene and α-phellandrene elicited strong responses from the antennae in spite of their weak affinities to LstiGOBP2. The EAG responses were not statistically different between males and females (data not shown), except in the case of trans-11-tetradecen-1-yl acetate. The EAG responses excited by the sex pheromone component were significantly different between males and females (Fig. 6).
Figure 5. The EAG activity of L. sticticalis antennae to different plant volatiles (10 mg/ml).
The volatiles in green were the higher competitive binding assay ligand; These in blue showed the moderate affinity and in red could not demonstrate affinity to LstiGOBP2.
Figure 6. The EAG response difference of L. sticticalis antennae to the sex pheromone component between male and female.
Discussion
Full-length GOBP2 cDNA was cloned from antennae of L. sticticalis using RT-PCR and RACE-PCR techniques. The deduced amino acid sequence suggested that the protein should be classified under the insect GOBP2 subfamily, according to the nomenclature proposed by Vogt et al. [9]. LstiGOBP2 shares a high sequence similarity to other insect GOBP2s, especially those of the same family, Pyralidae, Ostrinia furnacalis and Cnaphalocrocis medinalis, while LstiGOBP1, the other GOBP found in the antennae of L. sticticalis, is much less conserved [49]. Based on the complete genome annotation, 61, 72 and 44 OBPs were identified in Drosophila melanogaster, the malaria mosquito Anopheles gambiae and the silkworm Bombyx mori, respectively [46], [50]–[52]. Fourteen OBPs have been identified independently from antennal cDNA libraries of the cotton bollworm, Helicoverpa armigera (Hübner) and the lucerne plant bug Adelphocoris lineolatus (Goeze) [53], [54]. Therefore, we infer that more than two GOBPs might exist in L. sticticalis and diversified functions of these OBPs could be discovered later.
Quantitative examination of transcript levels has revealed that LstiGOBP2 is expressed at a rather high level in the antennae, which implies that LstiGOBP2 is likely to be involved in chemoreception [18], [38]. The transcript levels of LstiGOBP2 are different among male and female moths and also differ between developmental ages. It has been reported that the preoviposition period of female adults lasts for only 4–5 days at 25°C, so having the highest quantity of LstiGOBP2 in 4-day old antennae is beneficial to specific behaviors like oviposition host selection and mating [2]–[3].
Previous research has demonstrated that proteins in the GOBP2 class share high sequence similarity and can bind to a wide range of odors with a broad specificity [18], [27]–[28], [34], [38], [46]. In this study, LstiGOBP2 not only can bind green leaf volatiles, including aliphatic alcohols and aldehydes, but also can bind aromatic compounds and terpenoids. Comparing with the previous study of LstiGOBP1 [45], we found that LstiGOBP2 had a different binding activity compared to LstiGOBP1. In the 50 compounds that we tested, 14 volatiles could displace half 1-NPN from the LstiGOBP2/1-NPN complex at a ligand concentration of 20 µM, but only 4 volatiles could do that from the LstiGOBP1/1-NPN. The binding activity of most of the volatiles tested to LstiGOBP2 was higher than to LstiGOBP1.
Many other researchers have also demonstrated that the length of the carbon chain is critical to the affinities between proteins and ligands [32], [34], [55]–[57]. In our fluorescence competition assays, we found that the binding affinity of these ligands decreased when the number of carbon atoms increased. Both hexanal and pentanal exhibited high affinities to LstiGOBP2, while nonanal had a very weak binding affinity, even at a concentration of 20 µM (Fig. 4B).
Other compounds tested include a collection of different chemicals with the same number of carbon atoms but different functional groups (double bonds, hydroxyl group, aldehyde group, acetyl group or carbonyl group) (Fig. 4C, 4D). Ligands exhibited different binding activities when the position of double bonds was changed in these compounds (Fig. 4C). At the same time, we found that α-ionone and camphor were more competitive than myrcene, geraniol and (-)-linalool (Fig. 4D). There is no obvious difference between these ligands except for the formation of a ring in α-ionone and camphor. Therefore, the spatial structure of ligands can also affect the binding affinity to LstiGOBP2. A ring in the compound perhaps better modulates its interactions with the protein by reducing the number of possible conformations [55]. Thus, the high affinity of 2, 4-di-tert-butylphenol can easily be explained. However, (-)-trans-caryophyllene had only a very weak binding affinity to LstiGOBP2, which could be attributed to the size of the ring in (-)-trans-caryophyllene, which is too big to enter the binding cavity of LstiGOBP2. The binding activities of α-ionone and β-ionone were significantly different, which indicates that isomeric differences can influence affinity in fluorescence binding experiments. Therefore, LstiGOBP2 might discriminate among odorants on the basis of chain length, functional group, and alkene geometry [58].
Interestingly, we found that the long chain chemical trans-11-tetradecen-1-yl acetate, a sex pheromone component of L. sticticalis [48], had a high binding affinity to LstiGOBP2. This is consistent with previous studies showing that recombinant MsexGOBP2 could be labeled by pheromone analogue (6E, 11Z)-hexadecadienyl diazoacetate [35], that CsupGOBP2 has a high specificity for a major pheromone component 11Z-hexadecenal [46] and that BmorGOBP2 can bind to the B. mori sex pheromone component (10E,12Z)-hexadecadien-1-ol (bombykol) [18], [38]. In addition, the native form of GOBP2 from Mamestra brassicae (purified from male antennae) did not show affinity to the pheromone components, but displayed a highly specific affinity for cis-11-hexadecenol, an antagonist of pheromone-mediated male attraction [59]. Besides, the expression level of LstiGOBP2 in the antennae of male moths was significantly higher than that in antennae of female moths (Fig. 3B) and the EAG response of the male antennae to trans-11-tetradecen-1-yl acetate was the highest among all the volatiles tested. Therefore, LstiGOBP2 may also involve in the detection of sex pheromone.
In our study, LstiGOBP2 has been shown to have high binding affinities to trans-2-hexenal and cis-3- hexen-1-ol, the most abundant plant volatiles in essential oils extracted from the host plant of L. sticticalis [3]. During EAG recordings, these chemicals have been found to elicit a high EAG response on their antennae. However, although α-ionone and β-ionone had the strongest affinities for LstiGOBP2, they failed to elicit a strong electrophysiological response on the antennae. Similar results were also reported for 2,4-di-tert-butylphenol, cinnamaldehyde and benzaldehyde, which only exhibited a weak response in spite of their high affinities. It is well known that EAG only represents an overall activity of all the sensilla on the antenna and, therefore, even highly sensitive specialist olfactory receptor neurons may not show up if their total number is low [34]. Conversely, 1-octen-3-ol and trans-2-hexen-1-ol induce intensive responses on the antenna, but only present an affinity to LstiGOBP1 [45]. Moreover, antennae have strong responses to volatiles such as (R)-(+)-limonene and α-phellandrene, which do not show affinities to LstiGOBP1 or LstiGOBP2 [45]; similar results have been observed in Microplitis mediator OBPs, Holotrichia oblita Fald OBPs and the Drosophila melanogaster OBP LUSH [34], [46], [60]–[63].
In fact, many moths have shown a plasticity of olfactory-guided behavior, dependent not only on the nature of the chemical but also on the physiological status (e.g., age/hormone or mating status) of the individual [64]. Laughlin et al. also found that a mutation of OBP causing it to “lock” into a very specific conformation can cause receptor and sensillum activation without the presence of a ligand [26]. The relationship between the transcription of GOBP genes and behavioral plasticity in L. sticticalis is very interesting. LstiGOBP2 has been shown to have specialized characteristics, therefore understanding its function is a very important direction in the future. These studies may assist in developing artificial biosensors to monitor odors and devising strategies to disrupt the aggregation behavior of this species.
Materials and Methods
1 Insects and reagents
The meadow moth, Loxostege sticticalis, was fed Chenopodium glaucum in a laboratory at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. The antennae, heads (without antennae), thoraxes, abdomens, legs, and wings of the moths were dissected in an insect saline solution containing 0.75% NaCl and stored at −70°C until use.
2 RNA extraction, cloning and sequencing
Total RNA was extracted from 20 mg of antennae from L. sticticalis females using an RNeasy Mini kit (TIANGEN, China). First-strand cDNA was synthesized using a Prime Script first-strand cDNA synthesis kit (TaKaRa Co., Dalian, China), according to the manufacturer's instructions. Primers were designed by aligning OBP gene sequences from other moths in Lepidoptera.
Based on the sequences of the OBP gene from other moths in Lepidoptera, two degenerate primers, gps4 and gpa4 (gps4: 5′-GTC(G/T)ATGAAA(G)GAC(T)GTCACC(G/T)CTA(C/G/T)GG-3′; gpa4: 5′-AGGTTA(G)AAGTGC(G/T)CG(T)GCTCAT-3′), were synthesized by TaKaRa Company (Dalian, China) for amplification of the partial fragment of LstiGOBP2 cDNA. PCR was performed under the following conditions: three cycles of 40 sec at 94°C, 40 sec at 45°C, and 45 sec at 72°C, and then 30 cycles of 40 sec at 94°C, 40 sec at 47°C, and 45 sec at 72°C. The PCR product was gel purified (TaKaRa Co., Dalian, China), ligated into T-vector (TaKaRa Co., Dalian, China), and the recombinant plasmid DNA was transformed into XL-1 blue competent bacteria. Positive clones were sequenced using the dideoxynucleotide chain termination method (TaKaRa Co., Dalian, China).
Based on the partial sequences of the LstiGOBP2 cDNA, one specific primer, 3R-go1 (5′-GACGGAGGAGTTCTTCCACTTCTG-3′), was synthesized and used with 3 sites adaptor primer (5′-CTGATCTAGAGGTACCGGATCC-3′) for 3′-RACE. Two pairs of nest primers (OP1: 5′-GCTCCAGAAGTGGAAGAACTCCTC-3′; IP1: 5′-CTGTCCGAAGCCGAGGGTGACATCTTT-3′; OP2: 5′-CATGGCTACATGCTGACAGCCTA-3′; IP2: 5′-CGCGGATCCACAGCCTACTGATGATCAGTCGATG-3′) were synthesized for 5′-RACE. 5′- and 3′-RACE were performed following the manufacturer's protocol (SMART™ kit, Clontech). PCR products were sequenced after being inserted into T-vector (TaKaRa Co., Dalian, China).
3 Recombinant expression of LstiGOBP2
pGEM plasmid DNAs containing the ORF of LstiGOBP2 were digested with Noc I and Sac I enzymes for 3 h at 37°C, and the target products were separated on agarose gels. The purified targets were ligated into the expression vector pET30a (Novagen, Germany) and transformed into E. coli BL21 (DE3) pLysS cells. A positive clone of LstiGOBP2 in pET30a (pET30a-LstiGOBP2) was identified by PCR and sequencing.
For the expression of recombinant proteins, pET30a-LstiGOBP2 plasmid DNAs were transformed into E. coli BL21 (DE3) pLysS cells. After a 3 hr preincubation, recombinant LstiGOBP2 was induced by adding isopropyl-beta-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM for 4 hr. The cells (1L) were harvested by centrifugation, and the pellets were homogenized in phosphate-buffered saline (PBS, 0.04 M, pH 7.0). After centrifugation at 12,000 g for 20 min at 4°C, the supernatants were purified by Ni ion affinity chromatography (GE-Healthcare). To prevent the His-tag from affecting LstiGOBP2 functional studies, the His-tag was removed by recombinant enterokinase (rEK) (Bio Basic Inc.). Uncleaved His-tagged proteins were removed by a second round of Ni ion affinity chromatography. Recombinant LstiGOBP2 was identified with antibodies to 6×His-tag (Abcam, USA) by western blot analysis [65].
4 Expression pattern of LstiGOBP2
Total RNA isolated from different tissues was prepared in triplicate. The quality and concentration of the RNA was estimated by determining A260/A280 ratios and then modified to the same (0.1 µg/µl) using DEPC. For RT-PCR, cDNA was synthesized by using a first strand cDNA synthesis kit (TaKaRa Co., Dalian, China). Two primers (GOBP2-FP: 5′-TCCAACAAGTTCTCCCTGCTCC-3′ and GOBP2-RP: 5′-TCCTATCGCAGTCGTCGGTCAT-3′) were used to amplify the cDNA templates of LstiGOBP2. To quantify each mRNA absolute expression level of LstiGOBP2, a credible Standard Curve was constructed using a series 10× diluted standard samples. The PCR reaction conditions were as follows: 95°C for 2 min followed by 42 cycles of 95°C for 15 sec, 61.9°C for 15 sec and 72°C for 30 sec. The expected length of the LstiGOBP2 PCR product was 166 bp. Ct values are presented as mean ± SD for three independent experimental repeats. Real-time qPCR was performed using an iCycleriQ fast real-time PCR system (Bio-Rad, USA), which measures increased fluorescence of the fluorescent dye SYBR (TaKaRa Co., Dalian, China). The Ct value for each tissue and the standard curve were used to calculate the difference of each tissue.
5 Fluorescence binding assays
There were 51 compounds used in binding assays, which were purchased from Sigma-Aldrich (Chemie Gmbh, Steinheim, Germany) and had chemical purities >97% (determined by gas chromatography). Fluorescence spectra were recorded in a right angle configuration on a Lengguang 970 CRT spectrofluorimeter (Shanghai Jingmi, China) at room temperature using a 1-cm light path fluorimeter quartz cuvette. Slit widths of 10 nm were selected for both excitation and emission. The spectra data were processed using 970 CRT 2.0l software.
N-phenyl-1-naphthylamine (1-NPN) was dissolved in methanol to yield a 1 mM stock solution. The binding affinity for 1-NPN was determined by adding aliquots of 1-NPN into a 2 µM protein sample to final concentrations of 1 to 20 µM. 1-NPN was excited at 337 nm and emission spectra were recorded between 350 and 600 nm. Spectra were recorded with high-speed scanning. All ligands used in competition experiments were dissolved in spectrophotometric-grade methanol. Binding data were collected in three independent measurements.
Bound ligand was evaluated from the values of fluorescence intensity assuming that the protein was 100% active, with a stoichiometry of 1∶1 (protein∶ligand) at saturation. The K1-NPN values were estimated using Prism 5 (GraphPad Software, Inc.) by nonlinear regression for a unique site of binding. The curves were linearized using Scatchard plots. Dissociation constants of the competitors were calculated according to Campanacci et al. from the corresponding IC50 values in the equation: Ki = [IC50]/1+[1-NPN]/K1-NPN, where [1-NPN] is the free concentration of 1-NPN and K1-NPN is the dissociation constant of the complex protein/1-NPN [66].
6 Electroantennogram (EAG) recording
Antennae of adult moths were excised at the base and immediately placed on an EAG Micromanipulator MP-15 (Syntech) platform for EAG. Antennae were attached to two electrode holders with non-drying clay (Spectra 360 Electrode Gel). The binding affinity of the compounds to LstiGOBP2 proteins was tested. Pure chemicals were diluted with hexane to a final concentration of 10 mg/ml. EAG signals were amplified, monitored, and analyzed with EAG-Pro software (Syntech). The preparation was held in a humidified air stream delivered by a Syntech stimulus controller (CS-55 model; Syntech) at 500 ml/min, to which a stimulus pulse of 40 ml/min was added for 0.5 min. Signals were recorded for 10 sec, beginning at 2 sec before the onset of the stimulus pulse. An aliquot (10 µl) of each stimulus was loaded onto a filter paper strip. After each sample was tested, hexane was tested as a control. EAG responses of at least three antennae were recorded.
Acknowledgments
We very appreciate that the EAG supporting by Prof. Chen Luo, gratefully acknowledge Prof. Zhaojun Wei for valuable proposals in improving our manuscript and Prof. Yongjun Zhang for useful discussion. We also want to thank anonymous editors of Elsevier Language Editing Services for providing language editing help.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Special Fund for Agro-Scientific Research in the Public Interest (No. 201003079) from the Chinese government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Qu XF, Shao ZR, Wang JQ. Analysis of periodic outbreak of meadow moth in agricultural and pastoral area of North China. Entomological Knowledge. 1999;36:11–14. [Google Scholar]
- 2.Yin J, Cao YZ, Luo LZ, Hu Y. Effects of host plants on population increase of meadow moth, Loxostege sticticalis L. Journal of Plant Protection Research. 2004;31:173–178. [Google Scholar]
- 3.Yin J, Cao YZ, Luo LZ, Hu Y. Oviposition preference of the meadow moth, Loxostege sticticalis L., on different host plants and its chemical mechanism. Acta Ecologica Sinica. 2005;25:1844–1852. [Google Scholar]
- 4.Zhang LX, Fan JS, Wang GQ. Research Advances on Loxostege sticticalis L. (Lepidoptera: Pyralidae) in China. Chinese Agricultural Science Bulletin. 2010;26:215–218. [Google Scholar]
- 5.Visser JH. Differential sensory perceptions of plant compounds by insects, In: Hedin PA, editor. Plant resistance to insects, American Chemical Society Symposium Series. Washington: 1983. pp. 215–230. [Google Scholar]
- 6.Du YJ, Yan FS. The role of plant volatiles in tritrophic interactions among phytophagous insects, their host plants and natural enemies. Acta Entomology Sinica. 1994;31:233–249. [Google Scholar]
- 7.Yan SC, Zhang DD, Chi DF. Advances of studies on the effects of plant volatiles on insect behavior. Chinese Journal of Applied Ecology. 2003;14:310–313. [PubMed] [Google Scholar]
- 8.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 9.Vogt RG, Prestwich GD, Lerner MR. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. Journal of Neurobiology. 1991a;22:74–84. doi: 10.1002/neu.480220108. [DOI] [PubMed] [Google Scholar]
- 10.Leal WS. Pheromone reception. Topics in Current Chemistry. 2005;240:1–36. [Google Scholar]
- 11.Kim MS, Smith DP. The invertebrate Odorant-binding protein Lush is required for Normal Olfactory Behavior in Drosophlia. Chemical Senses. 2001;26:195–199. doi: 10.1093/chemse/26.2.195. [DOI] [PubMed] [Google Scholar]
- 12.Steinbrecht RA. Odorant-binding proteing: expression and function. Annals of the New York Academy of Sciences. 1998;855:323–332. doi: 10.1111/j.1749-6632.1998.tb10591.x. [DOI] [PubMed] [Google Scholar]
- 13.Vogt RG, Callahan FE, Rogers ME, Dickens JC. Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris(Hemiptera, Heteroptera). Chemical Senses. 1999;24:481–495. doi: 10.1093/chemse/24.5.481. [DOI] [PubMed] [Google Scholar]
- 14.Tegoni M, Pelosi P, Vincent F. Mammalian odorant binding proteins. Biochimica et Biophysica Acta. 2000;1482:229–240. doi: 10.1016/s0167-4838(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 15.Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cellular Molecular Life Sciences. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou JJ. Odorant-binding proteins in insect. Vitamins and Hormones. 2010;83:241–271. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- 17.Krieger J, Nickisch-Rosenegk EV, Mameli M, Pelosi P, Breer H. Binding proteins from the antennae of Bombyx mori. Insect Biochemistry and Molecular Biology. 1996;26:297–307. doi: 10.1016/0965-1748(95)00096-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhou JJ, Robertson G, He X, Dufour S, Hooper AM, et al. Characterisation of Bombyx mori Odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. Journal of Molecular Biology. 2009;389:529–545. doi: 10.1016/j.jmb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Feixas J, Prestwich GD, Guerrero A. Ligand specificity of pheromone-binding proteins of the processionary moth. European Journal of Biochemistry. 1995;234:521–526. doi: 10.1111/j.1432-1033.1995.521_b.x. [DOI] [PubMed] [Google Scholar]
- 20.Prestwich GD, Du G, Laforest S. How is pheromone specificity encoded in proteins? Chemical Senses. 1995;20:461–469. doi: 10.1093/chemse/20.4.461. [DOI] [PubMed] [Google Scholar]
- 21.Clyne P, Grant A, O'Connell R, Carlson JR. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- 22.Maida R, Krieger J, Gebauer T, Lange U, Ziegelberger G. Three pheromone-binding proteins in olfactory sensilla of the two silkmoth species Antheraea polyphemus and Antheraea pernyi. . European Journal of Biochemistry. 2000;267:2899–2908. doi: 10.1046/j.1432-1327.2000.01303.x. [DOI] [PubMed] [Google Scholar]
- 23.Benton R. Sensitivity and specificity in Drosophila pheromone perception. Trends Neurosci. 2007;30:512–519. doi: 10.1016/j.tins.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Smith DP. Odor and pheromone detection in Drosophila melanogaster. Pflugers Arch. 2007;454:749–758. doi: 10.1007/s00424-006-0190-2. [DOI] [PubMed] [Google Scholar]
- 25.Ebbs ML, Amrein H. Taste and pheromone perception in the fruit fly Drosophila melanogaster. Pflugers Arch. 2007;454:735–747. doi: 10.1007/s00424-007-0246-y. [DOI] [PubMed] [Google Scholar]
- 26.Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromonesensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breer H, Krieger J, Raming K. A novel class of binding proteins in the antennae of the silkmoth Antheraea pernyi. Insect Biochemistry. 1990;20:735–740. [Google Scholar]
- 28.Vogt RG, Robert R, Lerner MR. Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: Comparisons with other insect OBPs and their signal peptides. The Journal of Neuroscience. 1991b;17:2972–2984. doi: 10.1523/JNEUROSCI.11-10-02972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laue M, Steinbrecht RA, Ziegelberger G. Immunocytolochemical localization of general odorant-binding proteins in olfactory sensilla of the silk moth Antheraea polyphemus. Naturwissenschafen. 1994;81:178–181. [Google Scholar]
- 30.Steinbrecht RA, Laue M, Ziegelberger G. Immunolocalization of pheromone- binding protein and general odorant-binding protein in olfactory sensilla of the silkmoths Antheraea and Bombyx. Cell and Tissue Research. 1995;282:203–217. [Google Scholar]
- 31.Steinbrecht RA, Laue M, Maida R, Ziegelberger G. Odorant-binding proteins and their role in the detection of plant odours. Entomologia Experimentalis et Applicata. 1996;80:15–18. [Google Scholar]
- 32.Yu F, Zhang SA, Zhang L, Pelosi P. Intriguing similarities between two novel odorant-binding proteins of locusts. Biochemical and Biophysical Research Communications. 2009;385:369–374. doi: 10.1016/j.bbrc.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 33.Zhang ZC, Wang MQ, Lu YB, Zhang G. Molecular Characterization and Expression Pattern of Two General Odorant Binding Proteins from the Diamondback Moth, Plutella xylostella. Journal of Chemical Ecology. 2009;35:1188–1196. doi: 10.1007/s10886-009-9697-2. [DOI] [PubMed] [Google Scholar]
- 34.Deng SS, Yin J, Zhong T, Cao YZ, Li KB. Function and Immunocytochemical Localisation of Two Novel Odorant-Binding Proteins in Olfactory Sensilla of the Scarab Beetle Holotrichia oblita Fald (Coleoptera: Scarabaeidae). Chemical Senses. 2012;37:141–150. doi: 10.1093/chemse/bjr084. [DOI] [PubMed] [Google Scholar]
- 35.Feng L, Prestwich GD. Expression and characterization of a Lepidoptera general odorant binding protein. Insect Biochemistry and Molecular Biology. 1997;27:405–412. doi: 10.1016/s0965-1748(97)00012-x. [DOI] [PubMed] [Google Scholar]
- 36.Maíbèche-Coisné M, Longhi S, Jacquin-Joly E, Brunel C, Egloff MP, et al. Molecular cloning and bacterial expression of a general odorant-binding protein from the cabbage armyworm, Mamestra brassicae. European Journal of Biochemistry. 1998;258:768–774. doi: 10.1046/j.1432-1327.1998.2580768.x. [DOI] [PubMed] [Google Scholar]
- 37.Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of Silkworm, Bomyx mori. BMC Genomics. 2009;10:332–332. doi: 10.1186/1471-2164-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He XL, Tzotzos G, Woodcock C, Pickett JA, Hooper T, et al. Binding of the general odorant binding protein of Bombyx mori BmorGOBP2 to the moth sex pheromone components. Journal of Chemical Ecology. 2010;36:1293–1305. doi: 10.1007/s10886-010-9870-7. [DOI] [PubMed] [Google Scholar]
- 39.Fan XJ, Li Y, Li Y, Li LS, Zhang CF. Research Progress of Insect Sex Pheromone. Journal of Anhui Agriculture Sciences. 2010;38:4636–4638. [Google Scholar]
- 40.Krasnoff SB, Dussourd DE. Dihydropyrrolizine attractants for arctiid moths that visit plants containing pyrrolizidine alkaloids. Journal of Chemical Ecology. 1989;15:47–60. doi: 10.1007/BF02027773. [DOI] [PubMed] [Google Scholar]
- 41.Teulon DAJ, Penman DR, Ramakers PMJ. Volatiles chemicals for thrips (Thysanoptera: Thripidae) host-finding and applications for thrips pest management. Journal of Economic Entomology. 1993;86:1405–1415. [Google Scholar]
- 42.Verkerk RHJ, Wright DJ. Interactions between the diamondback moth, Plutella xylostella L. and glasshouse and outdoor-grown cabbage cultivars. Annals of Applied Biology. 1994;125:477–488. [Google Scholar]
- 43.Hammack L, Hesler LS. Phenylpropanoids as attractants for adult Stomoxys calcitrans (Diptera: Muscidae). Journal of Medical Entomology. 1996;33:859–862. doi: 10.1093/jmedent/33.5.859. [DOI] [PubMed] [Google Scholar]
- 44.Mo S, Zhao DX, Chen Q. Advances on relationships between plant volatiles and insect behavior. Chinese Journal of Tropical Agricultrue. 2006;26:84–89. [Google Scholar]
- 45.Sun HY, Yin J, Feng HL, Li KB, Xi JH, et al. Expression, Purification and Binding characteristic analysis of general odorant binding protein I (GOBP1) from the meadow moth, Loxostege sticticalis (Linnaeus). Acta Entomologica Sinica. 2011;54:381–389. [Google Scholar]
- 46.Gong ZJ, Zhou WW, Yu HZ, Mao CG, Zhang CX, et al. Cloning, expression and functional analysis of a general odorant-binding protein 2 gene of the rice striped stem borer, Chilo suppressalis(Walker) (Lepidoptera: Pyralidae). Insect Molecular Biology. 2009;18:405–417. doi: 10.1111/j.1365-2583.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 47.Du YJ, Poppy GM, Powell W, Pickett JA, Wadhams LJ, et al. Identification of semiochemicals released during aphid feeding that attracts parasitoid Aphidius ervi. Journal of Chemical Ecology. 1998;24:1355–1368. [Google Scholar]
- 48.Struble DL, Lilly CE. An attractant for the meadow moth, Loxostege sticticalis (Lepidoptera: Pyralidae). The Canadian Entomologist. 1977;109:261–266. [Google Scholar]
- 49.Zhong T, Yin J, Liu H, Wang JJ, Li KB, et al. Cloning and sequence analysis of one general odorant binding protein gene from the meadow moth, Loxostege sticticalis (Linnaeus). Plant Protection. 2008;34:31–36. [Google Scholar]
- 50.Hekmat-Scafe DS, Scafe CR, Mckinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Research. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biessmann H, Walter MF, Dimitratos S, Woods D. Isolation of cDNA clones encoding putative odourant binding proteins from the antennae of the malariatransmitting mosquito, Anopheles gambiae. Insect Molecular Biology. 2002;11:123–132. doi: 10.1046/j.1365-2583.2002.00316.x. [DOI] [PubMed] [Google Scholar]
- 52.Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Molecular Biology. 2003;12:549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- 53.Gu SH, Wang SP, Zhang XY, Wu KM, Guo YY, et al. Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze). Insect Biochemistry and Molecular Biology. 2011;41:254–263. doi: 10.1016/j.ibmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Zhang TT, Gu SH, Wu KM, Zhang YJ, Guo YY. Construction and analysis of cDNA libraries from the antennae of male and female cotton bollworms Helicoverpa armigera (Hübner) and expression analysis of putative odorant-binding protein genes. Biochemical and Biophysical Research Communications. 2011;407:393–399. doi: 10.1016/j.bbrc.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 55.Li S, Picimbon JF, Ji S, Kan YC, Qiao CL, et al. Multiple functions of an odorantbinding protein in the mosquito Aedes aegypti. Biochemical and Biophysical Research Communications. 2008;372:464–468. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- 56.Gu SH, Wang WX, Wang GR, Zhang XY, Guo YY, et al. Functional characterization and immunolocalization of odorant binding protein 1 in the lucerne plant bug, Adelphocoris lineolatus (Goeze). Archives of Insect Biochemistry and Physiology. 2011;77:81–98. doi: 10.1002/arch.20427. [DOI] [PubMed] [Google Scholar]
- 57.Jiang QY, Wang WX, Zhang ZD, Zhang L. Binding specificity of locust odorant binding protein and its key binding site for initial recognition of alcohols. Insect Biochemistry and Molecular Biology. 2009;39:440–447. doi: 10.1016/j.ibmb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Prestwich GD. Bacterial expression and photoaffinity labeling of a pheromone binding protein. Protein Science. 1993;2:420–428. doi: 10.1002/pro.5560020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacquin-Joly E, Bohbot J, Francois MC, Cain AH, et al. Characterization of the general odorant-binding protein 2 in the molecular coding of odorants in Mamestra brassicae. European Journal of Biochemistry. 2000;267:6708–6714. doi: 10.1046/j.1432-1327.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- 60.Honson N, Johnson MA, Oliver JE, Prestwich GD, Plettner E. Structure-activity studies with pheromonebinding proteins of the gypsy moth, Lymantria dispar. Chemical Senses. 2003;28:479–489. doi: 10.1093/chemse/28.6.479. [DOI] [PubMed] [Google Scholar]
- 61.Zhou JJ, Zhang GA, Huang W, Birkett MA, Field LM. Revisiting odorant-binding protein LUSH of Drosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Letters. 2004a;558:23–26. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhou JJ, Huang W, Zhang GA, Pickett JA, Field LM. “Plus-C”odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004b;327:117–129. doi: 10.1016/j.gene.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Zhang S, Chen LZ, Gu SH, Cui JJ, Gao XW, et al. Binding characterization of recombinant odorant-binding proteins from the Parasitic Wasp, Microplitis mediator (Hymenoptera: Braconidae). Journal of Chemical Ecology. 2011;37:189–194. doi: 10.1007/s10886-010-9902-3. [DOI] [PubMed] [Google Scholar]
- 64.Anton S, Dufour MC, Gadenne C. Plasticity of olfactory-guided behaviour and its neurobiological basis: lessons from moths and locusts. Entomologia Experimentalis et Applicata. 2007;123:1–11. [Google Scholar]
- 65.Yin J, Wei ZJ, Li KB, Cao YZ, Guo W. Identification and Molecular Characterization of a New Member of the Peritrophic Membrane Proteins from the Meadow Moth, Loxostege Sticticalis. International Journal of Biological Sciences. 2010;6:491–498. doi: 10.7150/ijbs.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A, et al. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. Journal of Biological Chemistry. 2001;276:20078–20084. doi: 10.1074/jbc.M100713200. [DOI] [PubMed] [Google Scholar]