Abstract
Human history is punctuated by periods of rapid cultural change. Although archeologists have developed a range of models to describe cultural transitions, in most real examples we do not know whether the processes involved the movement of people or the movement of culture only. With a series of relatively well defined cultural transitions, the British Isles present an ideal opportunity to assess the demographic context of cultural change. Important transitions after the first Paleolithic settlements include the Neolithic, the development of Iron Age cultures, and various historical invasions from continental Europe. Here we show that patterns of Y-chromosome variation indicate that the Neolithic and Iron Age transitions in the British Isles occurred without large-scale male movements. The more recent invasions from Scandinavia, on the other hand, appear to have left a significant paternal genetic legacy. In contrast, patterns of mtDNA and X-chromosome variation indicate that one or more of these pre-Anglo-Saxon cultural revolutions had a major effect on the maternal genetic heritage of the British Isles.
Archeologists once assumed that the British Isles were settled by successive waves of continental invaders, from Neolithic times onward (1). Today the pendulum has swung the other way, with archeologists tending to postulate considerable cultural exchange such as the establishment of trading networks, with little or no movement of people (2, 3). It is likely, however, that the extent of genetic continuity in the face of cultural change has varied from case to case.
We have utilized a number of genetic marker systems to determine the genetic legacy of cultural change. Analyses of the nonrecombining part of the Y chromosome are becoming increasingly important in uncovering paternal heritage in human evolutionary studies because of the recent development of a highly informative combination of different genetic markers (4–6). Slowly evolving biallelic markers are used to define distinct genealogical groups [haplogroups (hg)], whereas rapidly evolving microsatellites are used to distinguish more closely related chromosomes within haplogroups. Together, the two sets of markers identify well defined haplotypes, which have proven powerful tools in identifying relationships among populations. Occurrences of particular haplotypes, for example, have been suggested as population-specific signatures, as in the case of a high-frequency haplotype that appears to mark Jewish populations (7). Here we contrast the pattern observed on the Y chromosome with that observed by using multiple genetic systems influenced by female migration (mtDNA and unlinked X-chromosome systems) to evaluate whether cultural changes in the British Isles have involved different demographic roles for males and females.
Identification of genetic changes associated with these transitions requires that the source populations be distinguished with respect to some genetic marker. There are numerous candidate source populations for the British Isles from the pre-Anglo-Saxon British to the Romans, Anglo-Saxons, Scandinavians, and Normans. For tractability, we have focused mainly on two, the pre-Anglo-Saxon British and the Scandinavians. We have achieved this by concentrating on the Celtic-speaking populations and on Orkney, a Northern Scottish archipelago with Viking and pre-Anglo-Saxon British heritage.
Subjects and Methods
Samples and Genotyping.
Buccal swabs were scraped on the inside of the cheek by each subject and replaced in collection tubes to which 0.05 M EDTA/0.5% SDS had been added. In all cases, informed consent was obtained before samples were collected. Standard phenol/chloroform DNA extractions then were performed. Three Y-chromosome multiplex PCR kits were used as described (8). The products of each kit were subjected to electrophoresis on ABI 377 or ABI 310 automated sequencers and analyzed by genescan (Applied Biosystems) software. Conversions to repeat lengths were standardized by using control individuals sequenced by P. de Knijff. To assess the reliability of our data, 876 Y-chromosome microsatellite genotypes were retyped blindly; 6 were found to differ, an error rate of 0.7%. The Irish data do not include DYS388, so this locus was dropped from comparisons involving Ireland. The first hypervariable segment of the mitochondrial control region was amplified and sequenced from nucleotide positions 16090–16365 (9). Thirty-four X-linked microsatellites were genotyped by using multiplex PCR kits (10).
Y-Chromosome Hgs.
The hg and haplotype cluster designations (with unique event polymorphism genotypes in the sY81, SRY4064, YAP, SRY10831, M13, M9, Tat, M20, SRY+465, 92R7, and M17 order, and microsatellite genotypes in the DYS388-DYS393-DYD392-DYS19-DYS390-DYS391 order) are as follows: hg 1—AG-GGGTACT+G, not including the 1.15+ cluster (in the Basque data, the subclade of hg 1 defined by a mutation at SRY-2627 was included in hg 1 as we did not genotype this polymorphism); haplotype cluster 1.15+—hg 1 chromosome with microsatellite genotype 12-13-13-14-24-11 and one-step mutational neighbors (DYS388 was not typed in the Irish but is almost monomorphic at 12 repeats in hg 1. Only ≈3% of the hg 1 chromosomes in this study have different alleles.); hg 2—AG-GGCTACC+G, not including the 2.47+ cluster; haplotype cluster 2.47+—hg 2 chromosome with microsatellite alleles 14–13-11–14-22–10 and one-step network; hg 3—AG-AGGTACT-G, not including the 3.65+ cluster; haplotype cluster 3.65+—hg 3 chromosome with microsatellite alleles 12-13-11-16-25-11 and one-step network; hg 7—AG-ACCTACC+G; hg 8—GA+GGCTACC+G; hg 9—hg 2 chromosome with microsatellite haplotypes found only in 12f2 deleted chromosomes (DYS388*14, DYS393*12, DYS392*11, or 15-12-11, 15-13-11, 17-12-11, or 16-12-11 in the same order); hg 16—AG-GGGCACC+G; hg 21—AA+GGCTACC+G; hg 26—AG-GGGTACC+G; and hg 28—AG-GGGTGCC+G. A tree presenting the genealogical relationships of these hgs (except hg 28, which branches from hg 26) is presented in ref. 11.
mtDNA Hgs.
Haplotypes were assigned to hgs according to the West Eurasian mtDNA genealogy (12). hg assignment proceeded by using the following algorithm (all numbering is according to ref. 13 minus 16,000 in the control region for brevity): 069T 126C 223C assigned to hg J (note in all but four cases 069 information was available); 126C 223C 294T assigned to T; 129A 223T 391A assigned to I (391 information was available); 223T 292T assigned to W; 189C 223T 278T assigned to X; 223C 224C 311C assigned to K; 223C 249C and either 189C or 327T assigned to U1; 129C 223C assigned to U2 (051G, if information available); 223C 343G assigned to U3; 223C 356C assigned to U4; 223C 270T assigned to U5; 172C 219G 223C assigned to U6; 223C 318T assigned to U7; 223C 298C assigned to V; 067T 223C assigned to HV1 (067 information usually available); 126C 223C 362C assigned to preHV; 145A 176G 223T assigned to N1b; 223T 278T 390A assigned to L2; and 187T 189C 223T 278T 311C assigned to L1. For sequences not matching any of those above, the algorithm used was the following: if 223T, test for +10397 AluI (where + indicates restriction site presence and − indicates absence) for M; −10871 MnlI and −10397 AluI for L1, L2, or L3; if 223C, test for −7025 AluI for H; −14766 MseI, +7025 AluI, −4577 NlaIII for HV*; +12308 HinfI for U*, otherwise assign to R*. The first hypervariable section (HVS-1) sequences were checked also for matches with common East Asian hg motifs. Recurrent mutations may cause ambiguities by eliminating part of a diagnostic motif or recreating it in another part of the tree. In many cases, the presence of substitutions defining subclades within the major hgs allowed sequences to be assigned even when reversion had occurred at an hg motif site. In the case of hybrid motifs, PCR-restriction fragment length polymorphism (RFLP) testing was used to assign hg (14). Particularly in the Welsh and Irish data, HVS-1 sequences matching a unique haplotype in an RFLP-defined hg were assigned to that hg.
Analysis.
Exact tests and analyses of molecular variance were calculated by using arlequin (15). Principal components analyses were performed on hg and allele frequencies by using popstr (H. Harpending, personal communication). Population structure was assessed by using the model-based clustering method implemented in structure (16). The admixture model was used with a burn-in of 50,000 steps and a run length of 106 steps. All loci within 2 centimorgans of another locus were excluded from the structure analysis, leaving 23 loci.
Results and Discussion
Genetic History of Orkney.
When the Norsemen invaded (about A.D. 800), Orkney was populated by the Picts, little-understood pre-Anglo-Saxon inhabitants. Orkney remained a Norse colony while an increasing number of Scottish settlers arrived in the islands, which were pledged to Scotland in 1468 (17). As the place-names of Orkney are almost entirely Old Norse in origin (18) and a Nordic language replaced the earlier tongue, linguists have assumed that the Viking invaders completely replaced the native population (19). Modern archeological interpretations, however, suggest continuities in both artifacts and lifestyle, which are more compatible with considerable integration between native Picts and incoming Norsemen (20, 21). To investigate whether Orkney's Viking heritage is genetic as well as cultural, we sampled 71 adult males claiming at least three unrelated paternal generations in Orkney, and all with surnames found on the islands before 1700 (22). For comparison, we used analogous criteria to sample 78, 88, and 94 individuals from Norway, Anglesey (North Wales), and West Friesland (The Netherlands), respectively. Data on 146 Irish males with Irish Gaelic surnames also were included (23).
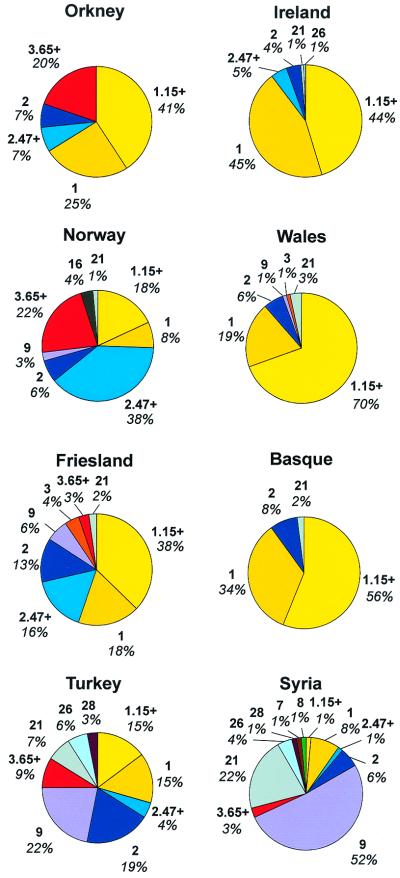
The Irish and Welsh are not significantly differentiated from each other at the hg level [P = 0.16 (24)] and will hereafter be called “Celtic.” However, Celtic, Frisian, Norwegian, and Orcadian Y chromosomes are all highly differentiated at the hg level (P < 0.0001) (Fig. 1 and Table 1). The Orkney sample seems intermediate between the Celtic and Norwegian samples, consistent with an origin by admixture between two such populations—hg 1 decreases from the Celtic populations through Orkney to Norway, whereas hgs 2 and 3 show the opposite trend. With respect to microsatellite variation within hgs, the Celtic samples are very homogeneous—the modal haplotype [microsatellite haplotype 15 within hg 1 (haplotype 1.15)] has a frequency of 26% in Wales and 18% in Ireland, and along with its one-mutational-step neighbors (7) constitutes 70% of the Welsh and 44% of the Irish chromosomes [as well as 56% of a Scottish sample (25)]. Frequencies of haplotype 1.15 (and its neighbors) in Orkney and Norway are 11% (41%) and 6% (18%), respectively.
Figure 1.
Charts of Y chromosome hg and haplotype cluster frequencies in each population. Haplotype clusters 1.15+, 2.47+, and 3.65+ are within hgs 1, 2, and 3, respectively.
Table 1.
Pairwise comparisons of Y-chromosome hg distributions
| Basque | Wales | Ireland | Scot Ork | Friesland | Orkney | Norway | Indig Ork | Turkey | |
|---|---|---|---|---|---|---|---|---|---|
| Wales | 0.96 | ||||||||
| Ireland | 0.75 | 0.16 | |||||||
| Scot Orkney | 0.50 | 0.45 | 0.21 | ||||||
| Friesland | 0.00 | 0.00 | 0.00 | 0.19 | |||||
| Orkney | 0.00 | 0.00 | 0.00 | N/A | 0.00 | ||||
| Norway | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||
| Indig Orkney | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | N/A | 0.00 | ||
| Turkey | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Syria | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
P values are from a test (24) on R × C contingency tables analogous to Fisher's exact test for a 2 × 2 table.
Scot Ork, Scottish Orkney surnames; Indig Ork, Indigenous Orkney surnames; P > 0.05 in bold; N/A, not applicable; 0.00, P < 0.001.
Other types are common in Norway and rare in the Celtic populations. Although hg 2 is much more diverse than hg 1, there is a subcluster found at high frequency only in Norway (network not shown)—haplotype 2.47 and one-step network constitute 38% of the sample. This mini-network, however, also occurs at a frequency of 16% in the Frisians, who may be similar to an Anglo-Saxon source population. The appearance of this cluster in mainland Britain, therefore, could be explained by either Scandinavian or Anglo-Saxon influence. Another one-step network, within hg 3 (centered on haplotype 3.65), however, is also present at high frequency in Norway (22%) and in Orkney (20%) but is rare in Friesland [and in The Netherlands (26)]. These frequency distributions suggest that both haplotypes 2.47 and 3.65 are diagnostic of Viking invaders in parts of Britain in which the only candidate parental populations are Celtic and Scandinavian, such as Orkney. In mainland Britain, however, it seems that only haplotype 3.65 would distinguish Norse and Anglo-Saxon contributions.
Correlation Between Y Chromosomes and Surnames in Orkney.
Orcadian surnames present a second method for identifying markers of Scandinavian contributions in the British Isles. Orcadian names can be divided into two classes: indigenous names endemic to the islands and those brought to the islands with Scottish settlers (22). As Y chromosomes cosegregate with surnames, haplotypes might be expected to reflect this partition to the extent that Norwegian and Scottish Y-chromosome types can be distinguished. In fact, the distribution of chromosome types between the surname classes is significantly different at the hg (P = 0.029, Table 1) and the haplotype (P = 0.035; ref. 24) levels. Moreover, the putative Norse (2.47, 3.65) and pre-Anglo-Saxon British (1.15) types clearly are concentrated in the expected classes (indigenous and Scottish, respectively, data not shown). This distribution confirms the heavier Viking component in the indigenous surname class and the increased pre-Anglo-Saxon British contribution to the Scottish surname class.
The frequency distribution in the indigenous Orkney chromosomes is consistent with a substantial Scandinavian contribution to the Orcadian Y-chromosome pool. As the Scottish Orkney surname class is statistically indistinguishable from the Welsh and Irish (P > 0.2), it cannot have a significant Norse component. Considering, therefore, only the indigenous surnames, 38% of the Y chromosomes can be identified as Scandinavian in origin (hg 3 and the 2.47 cluster), whereas those in hg 1 are not of obvious provenance. Thus, the legacy of the Viking age in Orkney was both cultural and genetic.
Genetic Continuity in the British Isles.
Given the similarity of the Irish and Welsh samples (Table 1), the Y-chromosome distributions shown in Fig. 1 seem to represent the pre-Anglo-Saxon population of the British Isles and Ireland. If extensive genetic drift had occurred, there is no reason why these communities would remain so similar, especially as Wales and Ireland represent two different branches of the Celtic languages, P-Celtic and Q-Celtic, respectively. Two extreme possibilities regarding the demographic nature of early cultural transitions in the British Isles can be contrasted: (i) demic diffusion models such as the wave-of-advance model (27) proposed for the arrival of farming in Europe (2), which predicts considerable genetic discontinuity; and (ii) cultural diffusion models, which predict genetic continuity, as they involve little or no movement of people, only the diffusion of technology. For example, the arrival of a Celtic material culture including Hallstatt and La Tène elite goods and skills in the late Bronze Age and early Iron Age once was interpreted as reflecting waves of immigrants but is now usually explained without invoking folk migrations (3). As with the Neolithic, however, no solid evidence is available.
Basque Population History.
To investigate the degree of paternal genetic continuity in the British Isles through the Neolithic and the development of Iron Age cultures, we compared the Welsh and Irish samples with 50 Basques (28, 29). The Basques are widely believed to be descended from the Paleolithic inhabitants of Europe for reasons including the following: (i) Basque is a non-Indo-European language with some features suggesting a distant relationship with the North Caucasian language family (30, 31). (ii) Analyses of classical markers consistently place the Basques as genetic outliers in Europe. For example, the Basques have the highest frequency in Europe of the blood group O and of rhesus cde, which is thought to represent the contribution of Paleolithic Europeans (32). (iii) An analysis of European mtDNA estimates the Neolithic component in the Basques to be the lowest for any region in Europe. Although the criteria used to identify Near Eastern founder types are somewhat heuristic and involve many assumptions, the relative number of types in different European populations should still be informative, and the Basque component, estimated at 7%, clearly lies outside the distribution for the rest of Europe, estimated to range between 9% and 21% (33). We also sampled 68 and 72 unrelated, adult male Anatolian Turks and Syrians, respectively. The former were representative of the source population for the European Neolithic and the latter were representative of the Near East more generally. If the pre-Anglo-Saxon British, therefore, trace genetically to the European Paleolithic, we might expect a similarity between the Irish and Welsh Y chromosomes and those of the Basques.
Basque and Celtic Y Chromosomes.
The Y chromosome complements of Basque- and Celtic-speaking populations are strikingly similar (Fig. 1). Haplotype 1.15 is also modal in the Basques and constitutes 41% of the sample, rising to 56% for the cluster of one-step neighbors. We call this the Atlantic modal haplotype (AMH). In each of the Basque, Welsh, and Irish populations, a total of 89–90% of the chromosomes are in hg 1, which contains the M173-defined Eu18 hg in Semino et al. (34), with the majority of the remainder in hg 2. The Turkish sample, however, is much more diverse at the hg level (Fig. 1). The AMH and one-step neighbors are present (15%) but only one chromosome from this group is found in the Syrian sample (Fig. 1), and it is absent in India (unpublished data) and Central Asia (35). There is no evidence, therefore, that incoming Neolithics or later immigrants originating in the Near East carried the AMH at frequencies as high as those characterizing the Atlantic populations.
Other studies have suggested the possibility of a Basque–Celtic connection, most notably the synthetic maps of Cavalli-Sforza et al. (32) that show Irish and Basque populations falling very near one another on the first principal component axis, which is thought to reflect the spread of Neolithic farmers from the Near East. The relative proximity of the Basque and Irish on this axis may therefore reflect the relatively small Neolithic component in these populations. More recently, Hill et al. (23) have used a northwest to southeast cline through Europe of p49a, f haplotype XV (36) [which forms a subclade of hg 1 (37)] to argue that hg 1 in Ireland must be old. We know of no other study, however, that provides direct evidence of a close relationship in the paternal heritage of the Basque- and the Celtic-speaking populations of Britain. In fact, treating Orkney as a single population, all pairwise comparisons of hg distributions between the populations included here are significantly different (Table 1) except for those within the Atlantic group—Welsh, Irish, and Basques—none of which are distinguishable, showing that they form a Y-chromosome community with members more closely related to one another than they are to the other European populations. Within Orkney, the Scottish surnames are not distinguishable from the Atlantic group but neither are they from the Frisians (Table 1), which may reflect an Anglo-Saxon component in the Scottish incomers. It should be noted that Basque-Celtic similarity not only implies that Basque- and Celtic-speaking populations derive from common paternal ancestors, but that genetic drift in these communities has not been sufficiently great to differentiate them.
Analysis of molecular variance was used to apportion Y-chromosome genetic diversity among individuals within populations, among populations within groups, and between groups in a hierarchical manner. When the Atlantic community form one group and the Frisians and Norwegians form the other, the between-populations within-groups variance component is lowest (4.3%) and the between-groups component is highest (12.1%), consistent with the pattern of differentiation seen in Table 1. Moving the Basques to the Frisian-Norwegian group almost doubles the between-populations within-groups variance component (to 8.0%) at the expense of the between-groups component. Swapping the Irish or Welsh across groups increases the within-groups component even more (to 10.6% and 12.4%, respectively).
The signal of Basque-Celtic similarity depends to a large degree on the AMH, which has much higher frequency in these populations than in other European populations. With one-step neighbors, the AMH composes only 38% of the Frisian sample (significantly different, P < 0.05), consistent with the view that the Basques are genetically distinguishable from continental populations generally. As three alleles within this six-locus haplotype are known to follow a southeast to northwest cline in Europe (38), it is likely that most other European populations will have even lower frequencies than the Frisians. Both the Basque and the Celtic populations show high frequencies of the AMH. Because the former are generally considered to have received a very limited input of Near Eastern genes in the Neolithic, that similarity also suggests that in the British Isles the Neolithic transition did not entail a major demographic shift. Accordingly, farming may have spread in Britain more through cultural transmission than through migration.
Coalescent Times.
Genealogical depths in hg 1 were estimated to investigate whether coalescent times are consistent with its presence in Britain since the Paleolithic. We used the average squared distance (ASD), which is the average across loci of the squared difference in the microsatellite repeat numbers between two haplotypes (39, 40). Under the single stepwise mutation model, the expectation of the ASD calculated between the inferred ancestral type and all observed haplotypes is equal to the product of the mutation rate and the average coalescence time in generations (39). In hg 1, we designated haplotype 1.15 as ancestral, because it is modal and has modal alleles at all of its constituent loci as well as being the haplotype connected to the most other haplotypes in a network. By using a mutation rate of 1.2 × 10−3 per locus per generation (41) and a generation time of 27 years, the estimated coalescent times in the British Isles and the continent are 6,800 and 7,100 years, respectively. The similarity of these values indicates that the populations in the Isles have not undergone extensive drift during colonization or afterward. However, the confidence intervals on the mutation rate alone (41) widen both estimates to between ≈2,900 and 18,400 years. This uncertainty associated with the estimated mutation rate is compounded by a likely systematic bias caused by the misspecification of the mutation model (42). Finally, it should be noted that the real uncertainty is influenced not only by these factors, but also by the variation associated with the stochastic distribution of mutations through the hg 1 genealogy. Because we do not know the shape of the hg 1 genealogy, it is difficult to assess this source of error (43). For these reasons, the coalescent calculations are consistent with almost any historical scenario. Unfortunately, it is not possible to calculate coalescence times for hg 2, because it is made up of many divergent genealogical clades.
Beyond their similarity, the lack of variation within the Atlantic populations is also remarkable. The Basque, Welsh, and Irish samples have mean microsatellite repeat count variances of 0.39–0.42, less than half that of Turkey (0.92) and much lower than Friesland, Norway, Syria, and Orkney (0.62–0.72). The similarity and homogeneity suggest one of two explanations: (i) preagricultural European Y chromosomes were homogeneous or (ii) there was a specific connection between the Basques, the pre-Anglo-Saxon British, and the Irish. With regard to the latter hypothesis, it is interesting that a northward expansion from a glacial refugium in Iberia has been postulated from the diffusion of Magdalenian industries (44) and patterns of Y-chromosome (34) and mtDNA variation (ref. 45; but see ref. 46). More detailed investigation of the diversity present in and around Europe may allow these hypotheses to be distinguished.
Maternal and Biparental Genetic Systems.
Given the extraordinary similarity of the Atlantic Y chromosomes compared with those in other European populations, it is important to assess whether a similar pattern is observed in other genomic regions. In particular, we shall use a comparison of Y chromosome and mtDNA patterns of variation to evaluate whether cultural change in the British Isles has affected differentially male and female patterns of movement. To assess whether any differences are caused by demographic factors as opposed to other differences between the two uniparental systems, we also include X-chromosome markers influenced by both male and female patterns of movement.
Mitochondrial DNA.
To investigate whether mtDNA variation showed the same patterns as the Y-chromosome data, we sequenced the first hypervariable section of the control region (HVS-1) and genotyped coding region variants as necessary to assign hg (see Subjects and Methods) in 90 Frisians and 59 Orcadians and compared these with 231 Norwegians (33, 47), 92 Welsh (9), 156 Basques (33, 48), 101 Irish (33), 218 Turks (33), and 69 Syrians (33). Slowly evolving coding-region variants and control-region sites are used to assign mtDNAs into genealogical clades or hgs, whereas more quickly evolving control-region sites define haplotypes within hgs. The hg distributions in all of the European populations are very similar (9, 49). Turkey and Syria, however, are distinct with much lower frequencies of the most common European hg (H) and large proportions of hgs not present or extremely rare in the European samples. The lack of structure is also evident at the haplotype level of resolution; analysis of molecular variance apportions 99% of the variance in our European populations between individuals within populations, regardless of the grouping scheme.
Principal Components (PC) Analysis.
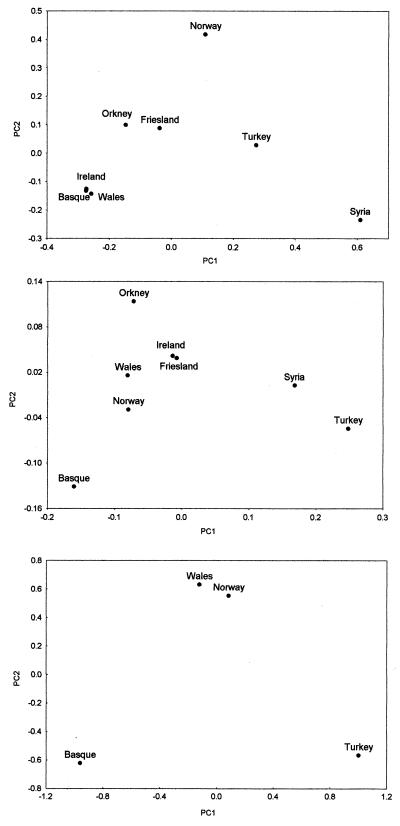
PC analyses were performed on both Y chromosome and mtDNA hg frequencies (Fig. 2). In each case, the first PC (explaining 65% and 54% of the variation, respectively) depicts a general East–West population gradient; a pattern usually interpreted as indicating the Neolithic component (32, 50). In line with this interpretation, the poles of the first PC of both systems are defined on the one hand by the Basques, and on the other by Turkey and Syria. As may be expected, in the Y-chromosome plot, the Celtic-speaking populations fall extremely close to the Basques, and Orkney falls midway between the Atlantic cluster and Norway. This pattern is in sharp contrast to that for mtDNA, in which the Celtic-speaking populations are closer to the center of the plot, indicating that they have undergone more female-mediated gene flow from other European populations than the Basques have. Thus, at least one of the cultural transitions in the British Isles since the Upper Paleolithic must have involved a demic component on the female side. The similarity of the non-Basque European populations means that there is no power to apportion the Orcadian maternal heritage into Scandinavian and pre-Anglo-Saxon British components by using the available mtDNA data.
Figure 2.
Plots of the first and second PC of Y chromosome hg (Top), mtDNA hg (Middle), and X microsatellite (Bottom) allele frequency distributions. All Y hgs were included while the following common European mtDNA hgs were included: H, V, J, T, I, W, X, U3, U4, U5, and K. In the Y-chromosome data, the proportion of the variation explained by PC1 is 65% and that explained by PC2 is 28%, whereas in the mtDNA and X microsatellite data, these figures are 54% and 15%, and 46% and 33%, respectively.
X-Chromosome Microsatellites.
To assess which of the two uniparentally inherited genetic systems more closely reflects the history of the genome more widely and to check that the lack of differentiation among the British and non-Basque European populations is not caused by a lack of resolution in the mtDNA data, we analyzed microsatellites on the X chromosome. Although having far less genealogical information at each genetic locus than is available for completely linked systems such as mtDNA and the Y chromosome, multilocus genotypes are known to provide a sensitive test of population structure (16, 51). Thirty-four dinucleotide markers located across the length of the X chromosome were genotyped in the Basques, Norwegians, Welsh, and Turks. Population structure was assessed by using a model-based clustering approach implemented in the structure program (16). Briefly, the model assumes K populations, each characterized by a set of allele frequencies at each locus, and individuals are assigned to these populations on the basis of their genotypes. We estimated Pr(X|K), where X is the data, for K = {1, 2, 3, 4}. By using Bayes' theorem and assuming a uniform prior on K between 1 and 4, we can then approximate the posterior distribution, Pr(K|X). For the Basque, Welsh, Norwegian, and Turkish data, all of the posterior probability is on K = 1, i.e., there is no detectable genetic structure.
However, when we performed a PC analysis on the allele frequencies at these 34 X-linked microsatellites, we observed a pattern essentially identical to that seen for mtDNA (Fig. 2). Once more, the Basques and Turks occupy opposite poles of PC1 and the Welsh and Norwegians fall in the center of the plot. Despite there being no statistical support for genetic structuring in the X-microsatellite data considered on their own, the similarity of the patterns observed across different genetic systems provides robust evidence that the Basques are differentiated from the other European populations, specifically in having a lower input from the Near East.
Female-mediated gene flow between the Celtic-speaking populations and other North European populations has thus homogenized the variation, not only for mtDNA but also for other parts of the genome affected by female migration. There are two extreme scenarios that could account for the sharp differences observed between the genetic systems that are and that are not (Y chromosome) affected by female movement (mtDNA, X chromosome, and the Y chromosome, respectively). First, the pre-Anglo-Saxon British source populations may have been different from the current European population for the Y chromosome but less so for other regions of the genome. This explanation is inconsistent with the position of the Basques, however, which is distinctive for both the Y chromosome and the systems affected by female migration. The second explanation is that the European Paleolithic populations were originally distinct from the current European population for both the Y chromosome and other parts of the genome, but this distinctiveness was eroded subsequently by female movements between the Celtic-speaking and non-Basque European populations. In other words, at least one of the Neolithic or Iron Age cultural transitions in the British Isles involved some female immigration.
Population parameters such as estimates of divergence times inferred from one-locus systems always have a high variance, because information is only incorporated from one realization of the evolutionary process. Certain evolutionary questions, however, are less subject to this source of variation and can be addressed profitably with only a single genetic locus. For example, identification of related lineages in different populations could be taken as secure evidence of some kind of connection between the populations such as gene flow or common ancestry, even though genetic drift at a single locus would make it impossible to estimate accurately parameters reflecting the quantitative relationship (e.g., migration rate or population-separation time). Despite these problems, in cases where female migrations have homogenized the variation in other parts of the genome, the Y chromosome may be the only signal of certain historical relationships.
In summary, we have identified markers of paternal Scandinavian influence in the British Isles that suggest the Viking settlement of Orkney involved substantial genetic as well as cultural replacement. Accepting the widely held view that the Basques are representative of pre-Neolithic European Y chromosomes (32), we have also shown that Neolithic, Iron Age, and subsequent cultural revolutions had little effect on the paternal genetic landscape of the Celtic-speaking populations (there has been continuity from the Upper Paleolithic to the present). However, comparison with mtDNA and X-linked microsatellites reveals that at least one of these cultural revolutions had a major effect on the maternal genetic heritage of the Celtic-speaking populations.
Acknowledgments
We thank the subjects who provided samples; M. A. Strøksnes, S. Jones, and R. Jager for collecting samples; J. Betranpetit for the Basque DNA; and E. Hill, D. Bradley, C. Tyler-Smith, L. L. Cavalli-Sforza, M. Jobling, T. Zerjal, and F. Calafell for access to unpublished results and useful discussions.
Abbreviations
- hg
haplogroup
- PC
principal component(s)
- AMH
Atlantic modal haplotype
Note Added in Proof.
Basque, Welsh, Norwegian, and Orcadian hg 1 chromosomes also were genotyped at DYS194469 and 25/25, 72/75, 18/20, and 45/46, respectively, carried the derived A allele [i.e., were hg 1L in the nomenclature of Hammer et al. (52)].
Footnotes
See commentary on page 4830.
References
- 1.Hawkes C. Antiquity. 1931;5:60–97. [Google Scholar]
- 2.Renfrew C. Archaeology and Language. London: Penguin; 1987. [Google Scholar]
- 3.Cunliffe B. The Ancient Celts. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 4.Jobling M A, Tyler-Smith C. Trends Genet. 1995;11:449–456. doi: 10.1016/s0168-9525(00)89144-1. [DOI] [PubMed] [Google Scholar]
- 5.Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos F R, Schiefenhovel W, Fretwell N, Jobling M A, Harihara S, et al. Am J Hum Genet. 1997;60:1174–1183. [PMC free article] [PubMed] [Google Scholar]
- 6.Karafet T M, Zegura S L, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, et al. Am J Hum Genet. 1999;64:817–831. doi: 10.1086/302282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M G, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein D B. Nature (London) 1998;394:138–140. doi: 10.1038/28083. [DOI] [PubMed] [Google Scholar]
- 8.Thomas M G, Bradman N, Flinn H M. Hum Genet. 1999;105:577–581. doi: 10.1007/s004399900181. [DOI] [PubMed] [Google Scholar]
- 9.Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt H J, Sykes B. Am J Hum Genet. 1996;59:185–203. [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson J F, Goldstein D B. Am J Hum Genet. 2000;67:926–935. doi: 10.1086/303083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerjal T, Pandya A, Santos F R, Adhikari R, Tarazona E, Kayser M, Evgrafov O, Singh L, Thangaraj K, Destro-Bisol G, et al. In: Genomic Diversity: Applications in Human Population Genetics. Papiha S S, Deka R, editors. New York: Plenum; 1999. pp. 91–102. [Google Scholar]
- 12.Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 14.Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus M L, Wallace D C. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider S, Kueffer J-M, Roessli D, Excoffier L. arlequin, A Population Genetic Data Analysis Program. Univ. of Geneva, Switzerland: Genetics and Biometry Laboratory; 1997. [Google Scholar]
- 16.Pritchard J K, Stephens M, Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson W P L. In: The People of Orkney. Berry R J, Firth H N, editors. Kirkwall: Orkney Press; 1986. pp. 209–224. [Google Scholar]
- 18.Lamb G. Testimony of the Orkneyingar. Kirkwall: Byrgisey; 1993. [Google Scholar]
- 19.Barnes M P. The Norn Language of Orkney and Shetland. Lerwick: Shetland Times; 1998. [Google Scholar]
- 20.Ritchie A. Viking Scotland. London: Historic Scotland; 1993. [Google Scholar]
- 21.Morris C D. In: The Prehistory of Orkney. Renfrew C, editor. Edinburgh: Edinburgh Univ. Press; 1990. pp. 210–242. [Google Scholar]
- 22.Lamb G. Orkney Surnames. Edinburgh: Paul Harris; 1981. [Google Scholar]
- 23.Hill E W, Jobling M A, Bradley D G. Nature (London) 2000;404:351–352. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]
- 24.Raymond M, Rousset F. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 25.Helgason A, Sigureth ardottir S, Nicholson J, Sykes B, Hill E W, Bradley D G, Bosnes V, Gulcher J R, Ward R, Stefansson K. Am J Hum Genet. 2000;67:697–717. doi: 10.1086/303046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Knijff P. Am J Hum Genet. 2000;67:1055–1061. doi: 10.1016/s0002-9297(07)62935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ammerman A, Cavalli-Sforza L. The Neolithic Transition and the Genetics of Populations in Europe. Princeton: Princeton Univ. Press; 1984. [Google Scholar]
- 28.Perez-Lezaun A, Calafell F, Seielstad M, Mateu E, Comas D, Bosch E, Bertranpetit J. J Mol Evol. 1997;45:265–270. doi: 10.1007/pl00006229. [DOI] [PubMed] [Google Scholar]
- 29.Bosch E, Calafell F, Santos F, Perez-Lezaun A, Comas D, Benchemsi N, Tyler-Smith C, Bertranpetit J. Am J Hum Gen. 1999;65:1623–1638. doi: 10.1086/302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamkrelidze T, Ivanov V. Sci Am. 1990;262 (March):110–116. [Google Scholar]
- 31.Bengtson J D. In: Sino-Caucasian Languages. Shevoroshkin V, editor. Bochum, Germany: Brockmeyer; 1991. pp. 67–172. [Google Scholar]
- 32.Cavalli-Sforza L L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 33.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, et al. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 34.Semino O, Passarino G, Oefner P J, Lin A A, Arbuzova S, Beckman L E, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, et al. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Lezaun A, Calafell F, Comas D, Mateu E, Bosch E, Martinez-Arias R, Clarimon J, Fiori G, Luiselli D, Facchini F, et al. Am J Hum Genet. 1999;65:208–219. doi: 10.1086/302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti A S. Am J Hum Genet. 1996;59:964–968. [PMC free article] [PubMed] [Google Scholar]
- 37.Jobling M A. Hum Mol Genet. 1994;3:107–114. doi: 10.1093/hmg/3.1.107. [DOI] [PubMed] [Google Scholar]
- 38.Quintana-Murci L, Semino O, Minch E, Passarimo G, Brega A, Santachiara-Benerecetti A S. Eur J Hum Genet. 1999;7:603–608. doi: 10.1038/sj.ejhg.5200347. [DOI] [PubMed] [Google Scholar]
- 39.Slatkin M. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein D B, Pollock D D. J Hered. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- 41.Bianchi N O, Catanesi C I, Bailliet G, Martinez-Marignac V L, Bravi C M, Vidal-Rioja L B, Herrera R J, Lopez-Camelo J S. Am J Hum Genet. 1998;63:1862–1871. doi: 10.1086/302141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldstein, D. B., Zerjal, T., Wilson, J. F., Pandya, A., Santos, F. R., Thomas, M. G., Bradman, N. & Tyler-Smith, C., Genetics, in press.
- 43.Goldstein D B, Reich D E, Bradman N, Usher S, Seligsohn U, Peretz H. Am J Hum Genet. 1999;64:1071–1075. doi: 10.1086/302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otte M. In: The World at 18,000 BP. Soffer O, Gamble C, editors. Vol. 1. London: Unwin Hyman; 1990. pp. 54–68. [Google Scholar]
- 45.Torroni A, Bandelt H J, D'Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus M L, Bonne-Tamir B, Scozzari R. Am J Hum Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G. Am J Hum Genet. 2000;66:262–278. doi: 10.1086/302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opdal S H, Rognum T O, Vege A, Stave A K, Dupuy B M, Egeland T. Acta Paediatr Scand. 1998;87:1039–1044. doi: 10.1080/080352598750031347. [DOI] [PubMed] [Google Scholar]
- 48.Bertranpetit J, Sala J, Calafell F, Underhill P A, Moral P, Comas D. Ann Hum Genet. 1995;59:63–81. doi: 10.1111/j.1469-1809.1995.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 49.Pult I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Pääbo S. Biol Chem Hoppe-Seyler. 1994;375:837–840. [PubMed] [Google Scholar]
- 50.Cavalli-Sforza L L, Minch E. Am J Hum Genet. 1997;61:247–254. doi: 10.1016/S0002-9297(07)64303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldstein D B, Roemer G W, Smith D A, Reich D E, Bergman A, Wayne R K. Genetics. 1999;151:797–801. doi: 10.1093/genetics/151.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammer M F, Redd A J, Wood E T, Bonner M R, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling M A, Jenkins T, et al. Proc Natl Acad Sci USA. 2000;97:6769–6774. doi: 10.1073/pnas.100115997. (First Published May 9, 2000; 10.1073/pnas.100115997) [DOI] [PMC free article] [PubMed] [Google Scholar]