Abstract
Here we characterize the relationship between the PRE-2 pheromone receptor and its ligand, CCG-4, and the general requirements for receptors, pheromones, G proteins, and mating type genes during fusion of opposite mating-type cells and sexual sporulation in the multicellular fungus Neurospora crassa. PRE-2 is highly expressed in mat a cells and is localized in male and female reproductive structures. Δpre-2 mat a females do not respond chemotropically to mat A males (conidia) or form mature fruiting bodies (perithecia) or meiotic progeny (ascospores). Strains with swapped identity due to heterologous expression of pre-2 or ccg-4 behave normally in crosses with opposite mating-type strains. Coexpression of pre-2 and ccg-4 in the mat A background leads to self-attraction and development of barren perithecia without ascospores. Further perithecial development is achieved by inactivation of Sad-1, a gene required for meiotic gene silencing. Findings from studies involving forced heterokaryons of opposite mating-type strains show that presence of one receptor and its compatible pheromone is necessary and sufficient for perithecial development and ascospore production. Taken together, the results demonstrate that although receptors and pheromones control sexual identity, the mating-type genes (mat A and mat a) must be in two different nuclei to allow meiosis and sexual sporulation to occur.
DISTINCTION between self and nonself is a fundamental requirement for various biological events in eukaryotes, ranging from somatic growth and mate selection in fungi (Janeway and Medzhitov 2002; Aanen et al. 2010; Casselton and Feldbrugge 2010; Debuchy et al. 2010) to immune defense in vertebrates (Janeway and Medzhitov 2002; Boehm 2006). In heterothallic ascomycete filamentous fungi, non–self-recognition is important during vegetative growth and sexual reproduction (see reviews in Glass and Dementhon 2006; Aanen et al. 2010). During vegetative growth, fusions between cells are constantly formed, generating a network of interconnected hyphae that is important for cell–cell communication and homeostasis in an individual colony. Hyphal fusion between individuals that differ at certain loci results in rejection of heterokaryon formation, leading to programmed death of the fusion cell. This process is referred to as heterokaryon incompatibility and is regulated by genetic differences at het loci (Glass and Dementhon 2006; Aanen et al. 2010).
As opposed to self-fertile homothallic species, which do not require a mating partner to complete sexual reproduction, heterothallic fungi are self-sterile and are only able to mate with a haploid cell of opposite mating type. The genetic barrier and sexual identity of heterothallic strains are established by the mating type (mat) genes. mat genes encode transcriptional regulators that control the expression of many genes required for sexual compatibility and reproduction, including the mating-type–specific pheromone and its G-protein–linked receptor (reviewed by Debuchy et al. 2010). The mating process of heterothallic fungi is best understood in the yeast Saccharomyces cerevisiae (Kurjan 1992; Bardwell 2005). Morphologically identical haploid cells of opposite mating type (MATa and MATα) mutually stimulate each other by secreting small peptide pheromones that are detected by cell-surface receptors. The pheromone–receptor interaction activates a mitogen activated protein kinase (MAPK) cascade, triggering several cellular responses, including cell cycle arrest in G1, elongation toward cells of opposite mating type, and transcription of genes involved in cellular and nuclear fusion. Under appropriate environmental conditions, the resulting diploid cell then undergoes meiosis to form four haploid spores (Cross et al. 1988; Herskowitz 1988).
Neurospora crassa is a heterothallic filamentous fungus with two mating types, mat a and mat A. N. crassa undergoes a more complex process of asexual and sexual reproduction than unicellular yeasts (Raju 1992b; Springer 1993). During vegetative growth, N. crassa produces tubular structures called hyphae and two types of mitotic spores: multicellular macroconidia and uninucleate microconidia. Under nitrogen starvation, the vegetative hyphae undergo complex morphological changes to form a spherical multicellular female reproductive structure (protoperithecium), which extends a female-specific receptive hypha (trichogyne) (Raju 1992b). A trichogyne of one mating type is attracted to and fuses with a male cell of opposite mating type (a macroconidium, microconidium or hyphal fragment). After fusion between the trichogyne and conidium, the male nucleus is transported to the base of the protoperithecium where nuclei of opposite mating type recognize each other and undergo synchronous rounds of mitosis. The male and female nuclei then fuse and the diploid zygote nucleus immediately enters meiosis. During this process, the protoperithecium enlarges greatly to become the flask-shaped perithecium. The haploid meiotic products undergo a round of postmeiotic mitosis and are then packaged into a spore (ascospore) in an eight-spored ascus. When fully mature, the ascospores are ejected through a pore (ostiole) at the tip of the perithecium.
In recent years, our laboratory has demonstrated that the pheromone receptor gene expressed in mat A cells, pre-1, is required for mat A trichogynes to recognize and fuse with mat a male cells (Kim and Borkovich 2004) . The ccg-4 and mfa-1 pheromone genes are necessary for chemotropic attraction and male fertility in mat A and mat a strains, respectively, with the prediction that PRE-1 binds to MFA-1, while PRE-2 recognizes CCG-4 (Bobrowicz et al. 2002; Kim et al. 2002; Kim and Borkovich 2006). Similar to Δpre-1 mat A strains, trichogynes of Δgna-1 (Gα), Δgnb-1 (Gβ), and Δgng-1 (Gγ) mutants are completely unresponsive to male cells of opposite mating type (Kim and Borkovich 2004; Krystofova and Borkovich 2005). In particular, data support coupling of the Gα subunit GNA-1 to pheromone receptors and transduction of the pheromone signal (Kim and Borkovich 2004). Finally, we have shown that although coexpression of a compatible pheromone and receptor in the same strain triggers self-stimulation, only abortive perithecia with no ascospores are produced, suggesting additional requirements for successful sexual reproduction in N. crassa (Kim and Borkovich 2006).
In this study, we generated and characterized mutants lacking pre-2, a gene that encodes the pheromone receptor expressed in mat a cells. Mutants lacking both pheromone receptor genes, pre-1 and pre-2, were also analyzed. We crafted strains with swapped identity through high-level, heterologous expression of pre-2 or ccg-4 (the PRE-2 pheromone) in the absence or presence of both mating-type genes as well as in a strain background that eliminates meiotic silencing. We also determined the minimum requirement for receptors, pheromones, and the Gα gene gna-1 during sexual reproduction in forced heterokaryons between opposite mating type strains. Our data indicate that pheromones and receptors are important for initial recognition of mates. However, additional determinants, such as the presence of mat A and mat a genes in two different nuclei, are required for nuclear fusion, meiosis, and ascospore production.
Materials and Methods
Strains and growth conditions
The N. crassa strains used in this study are listed in Table 1. The strains were grown either on Vogel’s minimal medium (VM) (Vogel 1964) for vegetative growth or on synthetic crossing medium (SCM) (Westergaard and Mitchell 1947) to induce sexual development. Sorbose-containing medium [Fructose-Glucose-Sorbose (FGS)] (Davis and Deserres 1970) was used to facilitate colony formation on plates. Supplements were added for auxotrophic strains as necessary and hygromycin B was used at 200 μg/ml in media where indicated. Conidia from 7-day-old VM agar cultures were used for inoculation of cultures. For submerged cultures, conidia were inoculated in liquid VM at 3 × 106 cells/ml and grown in the dark at 30° for 5, 16, or 24 hr with agitation. For differentiated cultures on solid medium, 1 μl of a conidial suspension was inoculated on the center of VM or SCM plates. VM plate cultures were grown in the dark at 30° for 3 days, while SCM plate cultures were grown for 6 days at 25° under constant light. For growth of cultures for RNA or protein extractions, plates were overlaid with cellophane (Bio-Rad Laboratories, Hercules, CA), and differentiated tissues were collected from the plates and frozen immediately in liquid nitrogen. For perithecial tissues, strains were grown on SCM plates and fertilized with a conidial suspension of opposite mating type. Sexual tissues were scraped from the plates 3 or 6 days after fertilization and frozen immediately. All plasmids were maintained in Escherichia coli strain DH5α (Hanahan 1983).
Table 1. N. crassa strains.
| Strain | Relevant genotype | Comments | Source or reference |
|---|---|---|---|
| 74A; FGSC 987 | 74-OR23-1A | Wild type | FGSC |
| 74a; FGSC 988 | 74-OR-1a | Wild type | FGSC |
| Am44; FGSC 4570 | un-3 ad-3A nic-2 cyh-1 Am44 | mat A sterile mutant | FGSC |
| am1; FGSC 4564 | ad-3B cyh-1 am1 | mat a sterile mutant | FGSC |
| am33; FGSC 5382 | am33 | mat a mutant | FGSC |
| FGSC 4317 | fl mat A | FGSC | |
| FGSC 4347 | fl mat a | FGSC | |
| FGSC 8740 | ΔSad-1::hph mat A | FGSC | |
| FGSC 8741 | ΔSad-1::hph mat a | FGSC | |
| FGSC 6103 | his-3 mat A | FGSC | |
| his3a | his-3 mat a | Krystofova and Borkovich (2005) | |
| pan-2 A | pan-2 mat A | R. L. Weiss (UCLA) | |
| pan-2 a | pan-2 mat a | Kim and Borkovich (2006) | |
| 16A | Δpre-1::hph mat A | Kim and Borkovich (2004) | |
| 16a | Δpre-1::hph mat a | Kim and Borkovich (2004) | |
| 16Ahis3 | Δpre-1::hph his-3 mat A | Kim and Borkovich (2004) | |
| 16ahis3 | Δpre-1::hph his-3 mat a | Kim and Borkovich (2004) | |
| 16Apan2 | Δpre-1::hph pan-2 mat A | Kim and Borkovich (2004) | |
| 16a pan2 | Δpre-1::hph pan-2 mat a | Kim and Borkovich (2004) | |
| c1a | Δccg-4::hph mat a | Kim and Borkovich (2006) | |
| c1ahis3 | Δccg-4::hph his-3 mat a | Kim and Borkovich (2006) | |
| c1apan2 | Δccg-4::hph pan-2 mat a | Kim and Borkovich (2006) | |
| m1a70 | Δmfa-1::hph Pccg-1::ccg-4+::his-3+ mat a | Kim and Borkovich (2006) | |
| 1B4 | Δgna-1::hph mat A | Ivey et al. (1999) | |
| B3 | Δgna-1::hph Δgna-2::pyrG+ mat a | Kays and Borkovich (2004) | |
| Δ1his3 | Δgna-1::hph his-3 mat A | Progeny of FGSC 6103 × B3 | This study |
| Δ1pan2 | Δgna-1::hph pan-2 mat a | Progeny of pan-2 A × B3 | This study |
| 23 | Δpre-2::hph mat A heterokaryon | 74A pHK15 transformant | This study |
| 32 | Δpre-2::hph mat A heterokaryon | 74A pHK15 transformant | This study |
| 23A | Δpre-2::hph mat A | Progeny of 23 × 74a | This study |
| 23a | Δpre-2::hph mat a | Progeny of 23 × 74a | This study |
| 32A | Δpre-2::hph mat A | Progeny of 32 × 74a | This study |
| 32a | Δpre-2::hph mat a | Progeny of 32 × 74a | This study |
| 23Ahis3 | Δpre-2::hph his-3 mat A | Progeny of 23a × FGSC 6103 | This study |
| 23ahis3 | Δpre-2::hph his-3 mat a | Progeny of 23a × FGSC 6103 | This study |
| 32ahis3 | Δpre-2::hph his-3 mat a | Progeny of 32a x FGSC 6103 | This study |
| 23Apan2 | Δpre-2::hph pan-2, mat A | Progeny of 23a × pan-2 mat A | This study |
| 23apan2 | Δpre-2::hph pan-2, mat a | Progeny of 23a × pan-2 mat A | This study |
| 23-1 | Δpre-2::hph his-3+::pre-2+ mat a | 23ahis3 pHK12 transformant | This study |
| 32-4 | Δpre-2::hph his-3+::pre-2+ mat a | 32ahis3 pHK12 transformant | This study |
| P1P2A | Δpre-1::hph Δpre-2::hph mat A | Progeny of 16a × 23A | This study |
| P1P2a | Δpre-1::hph Δpre-2::hph mat a | Progeny of 16a × 23A | This study |
| P1P2h3A | Δpre-1::hph Δpre-2::hph his-3 mat A | Progeny of 16a × 23Ahis3 | This study |
| P1P2h3a | Δpre-1::hph, Δpre-2::hph his-3 mat a | Progeny of 16a × 23Ahis3 | This study |
| P1P2pan2A | Δpre-1::hph, Δpre-2::hph pan-2 mat A | Progeny of 16a × 23Apan2 | This study |
| P1P2pan2a | Δpre-1::hph Δpre-2::hph pan-2, mat a | Progeny of 16a × 23Apan2 | This study |
| P1A17 | Δpre-1::hph Pccg-1::pre-2-GFP::his-3+ mat A | 16Ahis3 pHK17 transformant | This study |
| P2a17 | Δpre-2::hph Pccg-1::pre-2-GFP::his-3+ mat a | 23ahis3 pHK17 transformant | This study |
| P1A35 | Δpre-1::hph Ppre-1::pre-2-GFP::his-3+ mat A | 16Ahis3 pHK35 transformant | This study |
| P1A46 | Δpre-1::hph Pccg-1::pre-2-FLAG::his-3+ mat A | 16Ahis3 pHK46 transformant | This study |
| P2a46 | Δpre-2::hph Pccg-1::pre-2-FLAG::his-3+ mat a | 23ahis3 pHK46 transformant | This study |
| P1Gα1h3A | Δpre-1::hph Δgna-1::hph his-3 mat A | 16ahis3 × 1B4 progeny | This study |
| P1Gα1A46 | Δpre-1::hph Δgna-1::hph Pccg-1::pre-2-FLAG::his-3+ mat A | P1Gα1AH3 pHK46 transformant | This study |
| P1C4a46 | Δpre-1::hph Δccg-4::hph Pccg-1::pre-2-FLAG::his-3+ mat a | Progeny of c1a × P1A46 | This study |
| P1C4A46 | Δpre-1::hph Δccg-4::hph Pccg-1::pre-2-FLAG::his-3+ mat A | Progeny of P1C4a46 × P1A46 | This study |
| P1A46-68 | Δpre-1::hph, Pccg-1::pre-2-FLAG::his-3+ mat a-1+::bar mat A | P1A46 pHK68 transformant | This study |
| P1A68 | Δpre-1::hph mat a-1+::bar+ mat A | Progeny of FGSC 8741 × P1A46-68 | This study |
| P1P2C4A116 | Δpre-1::hph Δpre-2::hph Δccg-4::hph mat A | Progeny of c1apan2 × P1P2pan2A | This study |
| P1P2C4a48 | Δpre-1::hph Δpre-2::hph Δccg-4::hph mat a | Progeny of c1apan2 × P1P2pan2A | This study |
| P1A46-Sad1 | Δpre-1::hph Pccg-1::pre-2-FLAG::his-3+ ΔSad-1::hph mat A | Progeny of FGSC 8741 × P1A46-68 | This study |
| P1A68-Sad1 | Δpre-1::hph mat a-1+::bar ΔSad-1::hph mat A | Progeny of FGSC 8741 × P1A46-68 | This study |
| P1A46-68-Sad1 | Δpre-1::hph Pccg-1::pre-2-FLAG::his-3+ mat a-1+::bar ΔSad-1::hph mat A | Progeny of FGSC 8741 × P1A46-68 | This study |
FGSC, Fungal Genetics Stock Center, Kansas City, MO.
Isolation of Δpre-2 and Δpre-2::hph, pre-2+ complemented strains
A construct (pHK15) was generated to replace the 1.7-kb pre-2 open reading frame (ORF) with the 1.4-kb hph cassette encoding E. coli hygromycin B phosphotransferase under the control of the Aspergillus nidulans trpC promoter (pCSN44) (Staben et al. 1989). The 1.2-kb 5′ and 3.2-kb 3′ flanking DNAs for the pre-2 ORF were amplified from a genomic cosmid clone (G18A5) (Orbach 1994) in polymerase chain reactions (PCRs). Primers 1 and 2 were used for the 5′ flanking DNA and primers 3 and 4 were for the 3′ flanking DNA (Table 2). To facilitate the cloning process, BamHI, HindIII, and SpeI restriction enzyme sites were introduced into primers 2, 3, and 4, respectively. The PCR products were cloned into the pGEM-T vector (Promega, Madison, WI), yielding pHK13 (5′) and pHK14 (3′). The 1.4-kb hph cassette was released from pCSN44 using BamHI and HindIII, and pHK14 was digested with HindIII and SpeI to excise the 3′ flanking DNA. Finally, pHK13 (containing the 5′ flank) was linearized using BamHI and SpeI and the hph cassette and the 3′ flank were inserted, yielding pHK15.
Table 2. Oligonucleotides used in this study.
| Name | Sequence (5′–3′) |
|---|---|
| 1 | TACATCCGCATACCTGCCTTCATA |
| 2 | GTGTTGATGGGATCCCCAATGAAT |
| 3 | GTGCTCATGTCCATGAAGCTTG |
| 4 | GTGCTGGTGGTAGCACTAGTTA |
| 5 | TCACTGGCTGGGCTACTATTACAG |
| 6 | GATGTGTGGAAGGGATCTAGAGCT |
| 7 | GAGGATTCTAGACAGTGTGGGAGA |
| 8 | CATCGAATCTAGACCGTACGCACC |
| 9 | ATCTTCAGCGGCATAACCCA |
| 10 | TCAAAAGACCTCGGCTTCGT |
| 11 | GGCCCGGGTAGAAGGAGCAGTCCATCTGCGTG |
| 12 | GCCCGGGCTACTTATCGTCGTCATCCTTGTAATCCTCAAA AGACCTCGGCTTCGTGAC |
| 13 | TTTCTAGATCACCCGTGCATC |
| 14 | CCAGATGGATCCATGTGATGG |
| 15 | ATCGTGCGGCCGCTGACTATACAC |
| 16 | GGTGTACTAGTAACTCGCGAAAGTCCG |
| 17 | CCACCACCATGCTGAGGATCTCGGG |
| 18 | GTTACCTAACCATCCACCCTTGTCTTGTCC |
| 19 | CTCCACTAGCTCCAGCCAAGCCC |
| 20 | CACCATCTCCCACGCTTACTGCATATTAGGG |
pHK15 (2 μg) was linearized with SpeI and used to electroporate wild-type N. crassa strain 74A as described (Ivey et al. 1996). Transformants were isolated by plating on FGS plates containing hygromycin. Genomic DNA was isolated using the Puregene DNA kit (Gentra Systems, Minneapolis, MN) and subjected to Southern analysis after digestion with BamHI. Transformants with the Δpre-2 mutation were identified by using the 1.9-kb NcoI fragment from pHK7 as a probe. pHK7 was constructed by PCR-amplifying (using primers 5 and 6; Table 2) and cloning a 3.5-kb 5′ flanking DNA for the pre-2 ORF into the pGEM-T vector. All probes were labeled using the Prime-a-Gene labeling system (Promega).
Two heterokaryotic Δpre-2 strains (strains 23 and 32; Table 1) were selected and crossed to wild-type strain 74a, and progeny were selected on FGS hygromycin plates. Southern analysis was used to verify homokaryotic Δpre-2::hph strains (data not shown). The homokaryons were subsequently crossed to his-3 or pan-2 strains of opposite mating type (see Table 1) to obtain auxotrophic Δpre-2::hph strains. To isolate prototrophic and auxotrophic Δpre-1 Δpre-2 double mutants, the Δpre-2 his-3 and Δpre-2 pan-2 strains were individually crossed to Δpre-1::hph strains of opposite mating type (Kim and Borkovich 2004; Table 1).
The pre-2 complementation construct pHK12 was generated by cloning a 4.4-kb wild-type pre-2+ fragment into the XbaI site of the his-3 targeting vector pRAUW122 (Aramayo and Metzenberg 1996). The pre-2+ genomic fragment extends 1.9 kb upstream to 0.8 kb downstream of the pre-2 ORF and was amplified from the G18A5 cosmid in PCRs using primers 7 and 8, both of which contained an introduced XbaI site to facilitate the cloning process (Table 2). pHK12 was electroporated into Δpre-2 mutant strains 23ahis3 and 32ahis3, and transformants were selected on FGS plates with no added histidine. Heterokaryotic transformants that contained the desired integration event at the his-3 locus were identified by Southern analysis of genomic DNA, using the 8.8-kb HindIII fragment from pRAUW122 as a probe. All genomic DNAs were digested with HindIII and BamHI. Homokaryons were isolated using the microconidiation procedure as described (Ebbole and Sachs 1990) and verified by Southern analysis.
Northern analysis
For Northern analysis, total RNA was isolated using the Purescript RNA isolation kit (Gentra Systems) following the manufacturer’s instructions. Samples containing 10 or 30 µg of total RNA were subjected to Northern analysis (Tsui et al. 1994). All probe templates were labeled using the random primer method as described above. For the pre-2 probe, the entire 1.7-kb ORF was amplified from the G18A5 cosmid using primers 9 and 10 (Table 2). For the mat a-1 probe, the gene was excised from pCSN4 as a 1.9-kb XhoI–EcoRI fragment (Staben et al. 1989). The pre-1, ccg-4, and mfa-1 probes were prepared as described previously (Kim and Borkovich 2004, 2006).
Expression and localization of PRE-2–FLAG and PRE-2–GFP fusion proteins
Expression of a C-terminal FLAG-epitope–tagged version of PRE-2 was driven by the highly expressed and mating-type–independent ccg-1 promoter (Pccg-1) (Loros et al. 1989). The construct was cloned into pHK40, a his-3 targeting vector (Kim and Borkovich 2006). The Pccg-1::pre-2 fragment was amplified from pHK17 (see below) by PCR using primers 11 and 12. Primers 11 and 12 each contained a SmaI site to facilitate the cloning process (Table 2) and primer 12 was also designed with a 24-bp sequence encoding the FLAG epitope (DYKDDDDK) (Brizzard et al. 1994) followed by a stop codon (TAG; Table 2). The resulting Pccg-1::pre-2-FLAG fragment was then digested with SmaI and inserted into pHK40 cut with XbaI and then made blunt using Klenow. A Δpre-2 his-3 mat a strain (23ahis3; Table 1) was electroporated with pHK46, and transformants with the desired integration of pHK46 at the his-3 locus were selected for histidine prototrophy and verified by Southern analysis. All genomic DNAs were digested with HindIII and the 8.8-kb HindIII fragment from pRAUW122 was used as a probe. Genomic DNA of heterokaryotic transformants contained 9.5-kb, 8.2-kb, and 3.9-kb hybridizing fragments while that of the Δpre-2 recipient strain contained only a 9.5-kb hybridizing fragment (data not shown). Homokaryons were isolated using the microconidiation method (Ebbole and Sachs 1990) and verified by Southern analysis. Strain P2a46 (Table 1) was chosen for further analysis. The P1A46 strain (Table 1) was isolated by transformation of Δpre-1 mat A strain 16A using the procedures described for P2a46, above.
Immunoprecipitation of the PRE-2–FLAG protein was performed as described with minor modifications (Krystofova and Borkovich 2005). Whole cell extracts were prepared from P2a46 and wild-type strains and immunoprecipitated protein samples were resolved on a 10% SDS–PAGE gel. The gel was blotted onto a PVDF membrane (Millipore) and subjected to Western analysis. A mouse anti-FLAG antibody (Sigma; 1:2000) was used as the primary antibody and a HRP-conjugated goat antimouse IgG (Bio-Rad; 1:10,000) was used as the secondary antibody. Immunoreactive proteins were detected by chemiluminescence (ECL; Pierce, Rockford, IL) with a Biochemi system (UVP BioImaging Systems, Upland, CA). A duplicate gel was Coomassie stained to verify equal protein loading (Sambrook and Russell 2001).
The his-3 targeting vector pMF272 has been used for overexpression of green fluorescent protein (GFP) fusion proteins under the control of the N. crassa ccg-1 promoter (Folco et al. 2003). For overexpression of PRE-2–GFP, the entire pre-2 ORF was amplified from the G18A5 cosmid in PCRs using primers 13 and 14. To facilitate cloning, XbaI and BamHI sites were introduced into primers 13 and 14, respectively. pMF272 was digested with XbaI and BamHI, creating an opening between Pccg-1 and the GFP gene. The pre-2 PCR product was digested using XbaI and BamHI and ligated with XbaI–BamHI-digested pMF272 to yield pHK17. The stop codon (TAG) in the pre-2 ORF was replaced with a Trp codon (TAT) to create the carboxyterminal translational fusion of pre-2 and GFP (Table 2).
Plasmid pHK35 was constructed to express the PRE-2–GFP fusion protein under the control of the mating-type–specific pre-1 promoter in mat A strains. The pre-1 promoter region was amplified as a 2-kb fragment from pHK6, the pre-1 rescue construct (Kim and Borkovich 2004), by PCR using primers 15 and 16. Primers 15 and 16 contain NotI and SpeI sites, respectively (Table 2). The 2-kb PCR product was digested with NotI and SpeI and cloned into NotI–XbaI-digested pHK17 as a replacement of the ccg-1 promoter, yielding pHK35.
pHK17 and pHK35 were separately electroporated into Δpre-2 his-3 mat a strain 23ahis3 and Δpre-1 his-3 mat A strain 16Ahis3, respectively. Transformants with the desired integration of pHK17 or pHK35 at the his-3 locus were isolated on the basis of histidine prototrophy and confirmed by Southern analysis. All genomic DNAs were digested with HindIII and blots probed with the 5.2 kb HindIII–BamHI fragment from pRAUW122. When subjected to Southern analysis, genomic DNA from all heterokaryotic pHK17 or pHK35 transformants contained a 9.5-kb hybridizing fragment corresponding to the wild-type gene (data not shown). In addition, pHK17 transformants contained a 5.2-kb fragment, while those from pHK35 possessed 4.5- and 3.9-kb fragments. Homokaryotic pHK17 (strain P2a17) or pHK35 (strain P1A35) strains obtained using the microconidiation method (see above) did not contain the 9.5-kb wild-type fragment.
Western analysis was used to verify expression of the PRE-2–GFP fusion protein. Whole cell extracts (Turner and Borkovich 1993) were prepared from liquid VM submerged or SCM plate cultures of the P2a17 and wild-type strains and protein concentrations were determined as described above. Samples containing 15 µg of total protein were resolved on 7.5% SDS–PAGE gels and transferred onto PVDF membranes (Millipore). Blots were probed with a rabbit anti-GFP antibody (Abcam, Cambridge, MA; 1:2000 dilution) followed by a horseradish peroxidase (HRP)-conjugated goat antirabbit secondary antibody (Bio-Rad; 1:10,000 dilution), as recommended by the manufacturer (Abcam). Detection of immunoreactive proteins was as described above for PRE-2–FLAG.
To determine the tissue localization of the PRE-2–GFP protein, strains were grown on 2% water agar plates and observed microscopically as described (Kim and Borkovich 2004). Protoperithecia were visualized using a Leica TCS SP2 laser-scanning confocal microscope (Leica, Wetzlar, Germany) at the University of California Riverside Core Instrumentation Facility, while trichogynes and conidia were imaged using a BX41 fluorescent microscope with UM Plan Fluorite objective lenses and a PM-C35B camera (Olympus America, Lake Success, NY).
Generation of self-stimulating strains
For coexpression of ccg-4 and its receptor, pre-2, in the same strain, pHK46 was transformed into Δpre-1 his-3 mat A strain 16Ahis3. Isolation and purification of Δpre-1 Pccg-1::pre-2::his-3 mat A transformants using the microconidiation procedure (representative strain P1A46; Table 1) was as described above. The involvement of GNA-1 (Gα) and CCG-4 in self-stimulation was analyzed by introducing the Δgna-1 and Δccg-4 mutations into the P1A46 strain background. For the Δgna-1 strain, a Δpre-1 his-3 mat a female was crossed with a Δgna-1 mat A male, and histidine-requiring hygromycin-resistant progeny were selected. The presence of the Δpre-1 and Δgna-1 mutations was verified by Southern analysis as described previously (Ivey et al. 1996; Kim and Borkovich 2004). A Δpre-1 Δgna-1 his-3 mat A strain (P1Gα1h3A; Table 1) was then electroporated with pHK46. Transformants with pHK46 integrated at the his-3 locus were isolated, verified, and purified using the microconidiation procedure as described above. Representative strain P1Gα1A46 was chosen for further analysis.
Strains lacking ccg-4 were isolated in both mating types in the P1A46 background. To generate a mat a strain lacking ccg-4, P1A46 was crossed as a male to a Δccg-4 mat a strain c1a as a female (Table 1). Hygromycin-resistant progeny were selected, tested for mating type, and genomic DNAs were subjected to Southern analysis to determine Δpre-1, ectopic pre-2 (integrated at his-3 locus), and Δccg-4 genotypes (see above; Kim and Borkovich 2006). Δpre-1 Δccg-4 Pccg-1::pre-2::his-3 mat a strain P1C4a46 was chosen for further analysis. To isolate a mat A strain, P1C4a46 was crossed as the female parent to strain P1A46 as the male. Progeny were isolated, tested for mating type, and the presence of the Δccg-4 mutation was verified by Southern analysis (Kim and Borkovich 2006). Representative strain P1C4A46 was subjected to phenotypic analysis.
Effects of the mat a-1 and Sad-1 genes in self-stimulating strains
To express the mat a-1 gene in the mat A background, pHK68 was constructed as follows. The mat a-1+ gene was obtained from pCSN4 as a 1.9-kb XhoI–EcoRI fragment (Staben et al. 1989) and ligated into XhoI–EcoRI digested pTJK1. pTJK1 is a pBluescript-based plasmid carrying the bar+ phosphinothricin-resistance gene (Pall and Brunelli 1994; Jones et al. 2007). The resulting construct pHK68 was then transformed into P1A46, a strain that inappropriately expresses pre-2 in the mat A background (Table 1). Transformants were isolated on FGS plates containing 200 μg/ml phosphinothricin and 0.5% proline as the sole nitrogen source (Pall and Brunelli 1994). The presence of the full-length ectopic mat a-1 gene in these transformants was verified by Southern analysis. Genomic DNAs were digested with XhoI and EcoRI and blots were probed with the 1.9-kb mat a-1 fragment. Representative strain P1A46-68 was chosen for phenotypic analysis (Table 1).
A possible influence of the Sad-1 gene (Shiu et al. 2001) on expression of the ectopic pre-2 and mat a-1 genes in the P1A46 background was investigated. ΔSad-1 mat a strain FGSC 8741 was crossed as a female to strain P1A46-68 (Δpre-1::hph Pccg-1::pre-2-FLAG::his-3+ mat a-1+::bar+ mat A) as a male (Table 1). Progeny were screened by plating on FGS medium ± hygromycin to isolate all possible combinations of genotypes. All isolates were then spot tested to check for resistance to phosphinothricin. The hygromycin- and/or phospinothricin-resistant progeny were examined for the presence of the Δpre-1, ΔSad-1, pre-2ec, and mat a-1ec alleles using Southern analysis (see above; Shiu et al. 2001). Expression of pre-2 and mat-a1 was determined using Northern analysis, as described above.
Phenotypic analysis
Growth rate, hyperosmotic sensitivity, conidiation, heterokaryon formation, and CAT fusion assays:
Apical extension rates, hyperosmotic sensitivity, morphology of submerged cultures, and growth in standing liquid cultures were examined as previously described (Ivey et al. 1996; Kays et al. 2000; Kim and Borkovich 2004). Formation and fusion of conidial anastomosis tubes were assayed (Fleissner et al. 2005; Kim and Borkovich 2006). The ability of two strains to undergo hyphal fusion and form a heterokaryotic colony was tested (Davis and Deserres 1970; Kim and Borkovich 2004). For these experiments, the wild-type, am1, Δpre-2, Δpre-1 Δpre-2, and Δgna-1 strains contained different auxotrophic markers (his-3, pan-2, or ad-3; see Table 1).
Fertility tests:
Crosses between N. crassa strains were conducted using standard techniques and mating types of progeny from such crosses were determined using fl mat a and fl mat A females (Davis and Deserres 1970). To examine the involvement of the pheromone receptor genes during sexual development, Δpre-2 and Δpre-1 Δpre-2 strains were crossed to wild type as either the female (protoperithecial) or male (fertilizing) parent to detect dominant mating-specific defects. The Δpre-2 and Δpre-1 Δpre-2 strains were crossed to sibling mutant strains to detect recessive traits for pre-2 (alone or with pre-1) affecting sexual development.
Trichogyne assay:
Chemotropic responses of trichogynes to conidia of opposite mating type were examined as described previously (Bistis 1981; Kim and Borkovich 2004). Orientation and growth of trichogynes were monitored and photographed using a BX41 fluorescent microscope with UM Plan Fluorite objective lenses and a PM-C35B camera (Olympus America).
Sexual development of forced heterokaryons between opposite mating-type strains:
Forced heterokaryons formed between opposite mating type strains of various genotypes (see Figure 6) were examined for sexual development by cultivation on SCM plates. Each plate was prepared using 50 ml of medium for prolonged growth of heterokaryons. Formation of protoperithecia and development of perithecia were observed microscopically using a SZX9 stereomicroscope with an ACH 1X objective lens, and images were recorded using a PM-C35B camera (Olympus America).
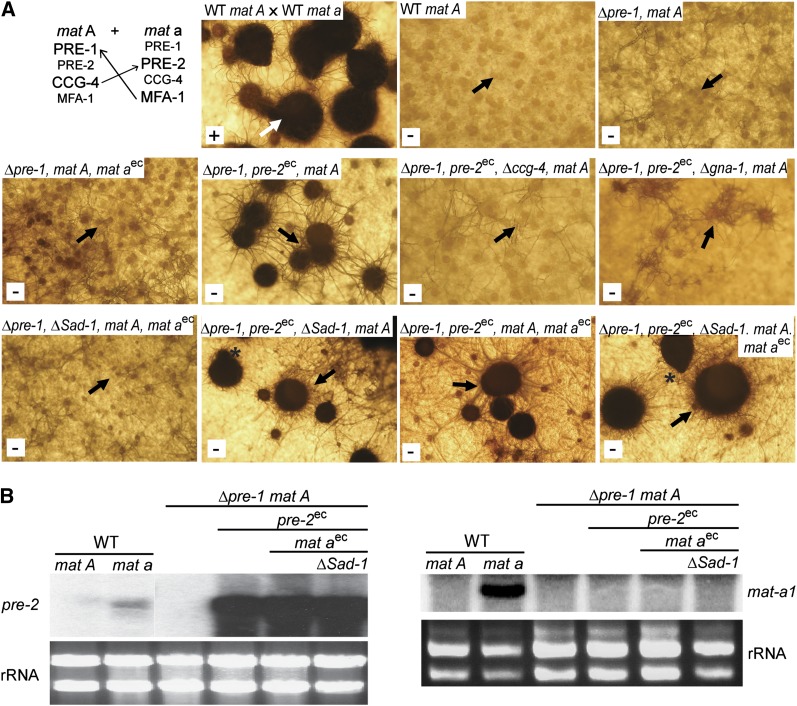
Figure 6 .
Perithecial development in the absence of opposite mating-type males. (A, top left) Cross of wild-type strains 74A and 74a (positive control for normal perithecial development). Protoperithecia from 6-day-old SCM cultures of strain 74A were fertilized with 74a and resulting perithecia were photographed 4 days later. The white arrow indicates an example of a normal perithecium. For all other panels, SCM plate cultures were grown for 10 days without fertilization. Strains are wild-type 74A (WT mat A), 16A (Δpre-1, mat A), P1A68 (Δpre-1, mat A, mat aec), P1A46 (Δpre-1, pre-2ec, mat A), P1C4A46 (Δpre-1, Δccg-4, pre-2ec, mat A), P1Gα1A46 (Δpre-1, pre-2ec, Δgna-1, mat A), P1A46-68-Sad1 (Δpre-1, ΔSad-1, mat aec, mat A), P1A46-Sad1 (Δpre-1, pre-2ec, ΔSad-1, mat A), P1A46-68 (Δpre-1, pre-2ec, mat aec, mat A), and P1A46-68-Sad1 (Δpre-1, pre-2ec, ΔSad-1, mat aec, mat A). The white box in the lower left corner of each panel indicates whether the strain produced ascospores (+ or −). Black arrows point to examples of protoperithecia or barren perithecia (enlarged bodies) developing in unfertilized cultures. The asterisks indicate beak formation in self-stimulated perithecia from the P1A46-68–Sad1 strain. Note that pre-2ec is Pccg-1::pre-2-FLAG::his-3+, while mat aec is mat a::bar. (B) Expression of pre-2 and mat a in self-stimulating strains. Total RNA was isolated from 6-day-old SCM plate cultures of wild-type mat A (74A), wild-type mat a (74a), Δpre-1, mat A (16A), Δpre-1, pre-2ec, mat A (P1A46), Δpre-1, pre-2ec, mat A, mat aec (P1A46-68), and Δpre-1, pre-2ec, mat A, mat aec, ΔSad-1 (P1A46-68-Sad1). Note that pre-2ec is Pccg-1::pre-2-FLAG::his-3+, while mat aec is mat a::bar. Samples containing 30 μg of total RNA were used to prepare Northern blots that were probed with pre-2 (left) and mat a (right). rRNA is used as a loading control.
Results
The pre-2 gene is preferentially expressed in mat a strains
To characterize the functions of the pheromone receptor pre-2 in N. crassa, we generated Δpre-2 and Δpre-1 Δpre-2 strains (see Materials and Methods). Replacement of the pre-2 gene in all nuclei was confirmed by the correct hybridizing bands during Southern analysis and by the lack of pre-2 transcript as assessed using Northern analysis (Figure 1A). Δpre-1 Δpre-2 double mutants were generated through sexual crosses between single mutants. The wild-type pre-2+ genomic fragment was used to complement the Δpre-2 mutation. Confirmation that the pre-2+ gene was successfully integrated at the his-3 locus was achieved using Southern analysis and by observation of pre-2 mRNA (Figure 1A; strains 23-1 and 32-4). Complemented strains exhibited wild-type phenotypes for all functions tested.
Figure 1 .
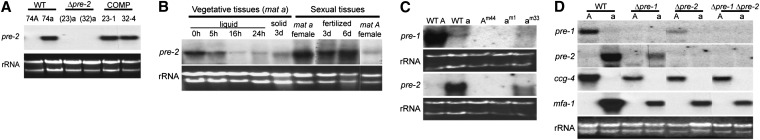
Northern analysis. (A) Verification of Δpre-2 and rescued strains. Total RNA was isolated from 6-day-old SCM plate cultures of mat A and mat a wild-type (WT) strains 74A and 74a; Δpre-2::hph knockout mutants 23a and 32a (Δpre-2), and Δpre-2::hph+, pre-2+ complemented strains 23-1 and 32-4 (COMP). Approximately 20 μg of total RNA was subjected to Northern analysis using the pre-2 ORF as a probe (see Materials and Methods). rRNA is the loading control. (B) pre-2 mRNA levels in various tissues. Strains were wild-type mat a (74a) or mat A (74A), as indicated. RNA samples representing vegetative tissues were isolated from 74a grown under the following conditions: freshly harvested conidia (0 hr), conidia germinated in liquid VM for 5, 14, and 24 hr, and on solid VM plates for 3 days. RNA representing unfertilized sexual tissues was prepared from 74A or 74a grown on solid SCM plates for 6 days. RNA for fertilized sexual tissues was from perithecia harvested 3 and 6 days after fertilization, respectively, from a cross of 74a with 74A. Northern blots were probed as in A. rRNA is used as a loading control. (C) Expression of pheromone receptor genes in mating-type mutants. Total RNA was prepared from 6-day-old SCM plate cultures of wild-type mat A strain 74A (WT A), wild-type mat a strain 74a (WT a), the mat Am44 and mat am1 sterile mutants, and the vegetative incompatibility mutant mat am33. Northern blots were probed using the pre-2 (A) or pre-1 ORF fragments (see Materials and Methods). rRNA is a loading control. (D) Pheromone receptor and pheromone precursor mRNA levels in strains lacking pre-1 and/or pre-2. Strains are wild-type (WT) 74A (WT mat A), 74a (WT mat a), 16A (Δpre-1, mat A), 16a (Δpre-1, mat a), 23A (Δpre-2, mat A), 23a (Δpre-2, mat a), P1P2A (Δpre-1 Δpre-2, mat A), and P1P2a (Δpre-, Δpre-2, mat a). Total RNA was isolated from 6-day-old SCM plate cultures. Northern blots were probed using pre-1, pre-2, ccg-4, and mfa-1 specific probes (see Materials and Methods). rRNA is the loading control.
Our previous studies have shown that expression of the mat A pheromone receptor gene pre-1 and the ccg-4 and mfa-1 pheromone precursor genes is mating-type specific and also developmentally regulated (Kim and Borkovich 2004, 2006). For example, pre-1 is abundantly expressed in protoperithecia, perithecia, and conidia from mat A strains (Kim and Borkovich 2004). Therefore, we examined the mating type and developmental expression pattern of pre-2 in N. crassa. Limiting nitrogen (SCM medium) stimulates development of female reproductive structures, protoperithecia. Fertilization of protoperithecia with male cells (usually conidia) of opposite mating type leads to perithecial development and sexual spore (ascospore) production. Northern analysis using RNA isolated from a mat A wild-type strain (74A) showed that pre-2 mRNA is not expressed in vegetative cultures (data not shown) and that very low levels are present in SCM cultures producing protoperithecia (Figure 1B). In contrast, pre-2 was expressed to relatively high levels in a mat a wild-type strain (74a). Amounts were highest in sexual tissue (protoperithecia), followed by conidia (0 hr) and germling (5 hr) cultures (Figure 1B). Lower levels were detected in older vegetative tissues (16 or 24 hr of submerged growth) and 3-day vegetative plate cultures (Figure 1B). High amounts of pre-2 transcript were also detected in developing perithecia after fertilization with wild type. These results indicate that pre-1 and pre-2 share a similar pattern of developmental expression, but are regulated by different mating-type genes in N. crassa (Kim and Borkovich 2004, Figure 1B).
The mating-type–dependent expression of pre-1 and pre-2 was further analyzed using three previously characterized N. crassa mating-type mutants: mat A mutant Am44 and mat a mutants am1 and am33 (Griffiths and Delange 1978; Griffiths 1982). While Am44 and am1 are both male and female sterile and do not produce pheromone (Bistis 1981; Bobrowicz et al. 2002; Kim et al. 2002), the am33 mutant is fertile as a male or female and expresses both pheromones (Griffiths and Delange 1978; Bobrowicz et al. 2002). The major defect of am33 is loss of mating-type incompatibility functions during vegetative growth. Consistent with pheromone receptor genes being regulated by mating type to control sexual fertility, pre-1 and pre-2 messages could not be detected in the Am44 and am1 strains, respectively (Figure 1C; Kim and Borkovich 2004). In contrast, low but detectable levels of pre-1 and pre-2 are present in the am33strain that has retained sexual fertility functions (Figure 1C). These results support the mating-type specificity of pheromone receptor gene expression observed in wild-type strains and the requirement for pheromone receptors during mating in N. crassa.
In a previous study, we demonstrated that deletion of pre-1 greatly affected expression of each pheromone precursor gene in the mating type that normally supports high message levels (Kim and Borkovich 2004). Therefore, we investigated the effect of pre-2 on expression of ccg-4 and mfa-1. We also determined whether loss of one pheromone receptor gene influences expression of the remaining receptor. As observed for Δpre-1 mutants (Kim and Borkovich 2004), deletion of pre-2 alone or together with pre-1 reduced expression of ccg-4 to a great extent in the mat A background (Figure 1D). Similarly, mfa-1 expression was significantly decreased in Δpre-1 and Δpre-2 single mutants and Δpre-1 Δpre-2 double mutants in the mat a mating type (Figure 1D). Surprisingly, pre-1 message levels were greatly reduced in sexually differentiated tissues of the Δpre-2 mat A strain and pre-2 expression was drastically downregulated by the deletion of pre-1 in the mat a background (Figure 1D). Taken together, these and our previous results demonstrate that loss of either pheromone receptor leads to reduced expression of both pheromones and the other pheromone receptor.
The PRE-2 protein is localized in protoperithecia, trichogynes, and conidia
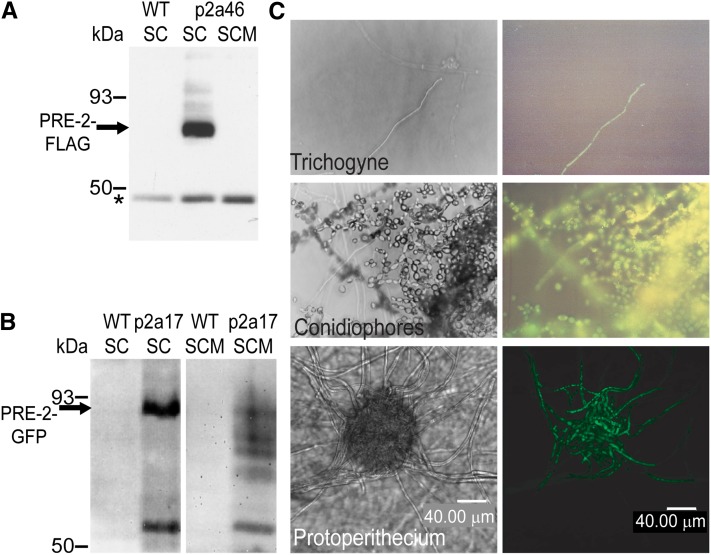
Strains carrying a pre-2 allele with a C-terminal FLAG epitope tag were generated in the Δpre-2 background (strain P2a46; Table 1). Expression of the pre-2–FLAG allele was driven by the highly expressed and mating-type–independent ccg-1 promoter (Arpaia et al. 1995) to accumulate high levels of PRE-2–FLAG protein in either mating type. The FLAG epitope did not interfere with the function of the PRE-2 receptor, as the pre-2–FLAG allele complemented the defects of the Δpre-2 mutation. Immunoprecipitation of whole cell extracts from vegetative submerged (liquid VM) cultures with a FLAG antibody demonstrated that a reactive species of ∼60 kDa could be detected in the P2a46 strain, but not in the untransformed wild-type control (Figure 2A). The size of the immunoreactive protein is in agreement with the molecular mass of PRE-2 (61.6 kDa) predicted using ExPASy Proteomics tools (http://web.expasy.org/compute_pi/). No PRE-2–FLAG could be immunoprecipitated from extracts obtained from P2a46 SCM plate cultures (Figure 2A). This result is consistent with rapid degradation of PRE-2 in sexually differentiated tissues, the site where pheromone receptors are assumed to be most active. Such a scenario is observed in S. cerevisiae, with ubiquitination leading to rapid turnover of pheromone receptors (Yin et al. 2005).
Figure 2 .
Expression and localization of the PRE-2 protein. (A) Immunoprecipitation and Western analysis of a PRE-2–FLAG fusion protein. Whole cell extracts were prepared from 16-hr submerged cultures (SC) of untransformed wild-type mat a strain 74a (WT) and a Δpre-2::hph, Pccg-1::pre-2-FLAG::his-3+ mat a strain (P2a46), and from sexually differentiated plate cultures (SCM) of strain P2a46. Samples containing 2 mg of total protein were immunoprecipitated using anti-FLAG M2-agarose (see Materials and Methods). Precipitated proteins were resolved on 10% SDS–PAGE gels and subjected to Western analysis using a FLAG-specific antiserum. The PRE-2–FLAG protein (∼62 kDa) is indicated by an arrow and a nonspecific band is shown by an asterisk. (B) Expression of a PRE-2–GFP fusion protein. Whole cell extracts were prepared from 16-hr liquid SCs and sexually differentiated plate cultures (SCM) of untransformed wild-type (WT) strain 74a and a Δpre-2::hph Pccg-1::pre-2-GFP::his-3+ mat a strain (P2a17). Samples containing 15 μg of total protein were resolved on 7.5% SDS–PAGE gels and subjected to Western analysis using GFP-specific antiserum as described in Materials and Methods. The arrow indicates the full-length PRE-2–GFP fusion protein in submerged cultures of strain P2a17 (∼90 kDa). (C) In vivo localization of the PRE-2–GFP protein. The P2a17 strain (Δpre-2::hph, Pccg-1::pre-2-GFP::his-3+, mat a) was cultivated on 2% water agar plates and observed under a fluorescent compound microscope (top two rows) or a laser-scanning confocal microscope (bottom). The distribution of the PRE-2–GFP protein was analyzed in trichogynes (top right) and conidia (middle right). A protoperithecium expressing the fusion protein was scanned at 5-µm intervals and the 3-dimensional image was reconstructed (bottom right). The left panels are control images viewed in phase contrast for panels on the right side of the figure. Bar, 40 µm.
PRE-2–FLAG could be immunoprecipitated from solubilized plasma membrane but not cytosolic fractions, consistent with a plasma membrane localization for PRE-2 in N. crassa (data not shown). To further investigate the localization of PRE-2, we created a strain that expresses a PRE-2–GFP fusion protein under the control of the ccg-1 promoter (strain P2a17; Table 1) in the Δpre-2 background. This construct complemented the defects of the Δpre-2 mutant (data not shown). Western analysis using a GFP-specific polyclonal antibody revealed a band of approximately 90 kDa was detected in submerged and SCM plate cultures of strain P2a17, but was absent from controls (Figure 2B). This size corresponds to GFP (29 kDa) plus the PRE-2 protein (61 kDa). In addition to the full-length PRE-2–GFP protein, lower molecular weight species were also detected (particularly in SCM plate cultures) of strain P2a17 (Figure 2B). These smaller proteins may result from rapid turnover of the PRE-2 protein in sexually differentiated tissue (see above).
We next analyzed the localization pattern of the PRE-2–GFP fusion protein in N. crassa tissues. Using a fluorescent compound microscope, the PRE-2–GFP protein was shown to be present in protoperithecia, trichogynes, and conidia (Figure 2C; data not shown). When protoperithecia were examined at higher resolution using a laser-scanning confocal microscope, PRE-2–GFP could only be detected in hyphae within the protoperithecium or in individual trichogynes extending from the structure (Figure 2C).
pre-2 has no apparent roles in vegetative functions or male fertility, but is essential for female fertility in mat a strains
Deletion of pre-2 singly or in combination with pre-1 did not cause any apparent defects during vegetative growth or development in either mating type. Δpre-2 and Δpre-1 Δpre-2 strains displayed hyphal extension rates similar to those of wild-type strains on solid VM or SCM medium. Differentiation of aerial hyphae and conidia; sensitivity to elevated levels of NaCl, KCl, or sorbitol; and morphology in submerged cultures were also normal in Δpre-2 and Δpre-1 Δpre-2 strains. Δpre-2 and Δpre-1 Δpre-2 strains were able to undergo hyphal fusion and establish stable heterokaryons between same or opposite mating-type strains. Conidia of pre-1 and pre-2 single or double mutants also formed conidial anastomosis tubes (CATs) normally during germination and were able to fuse with neighboring conidia. Δpre-2 and Δpre-1 Δpre-2 strains were fertile as males in both mating types.
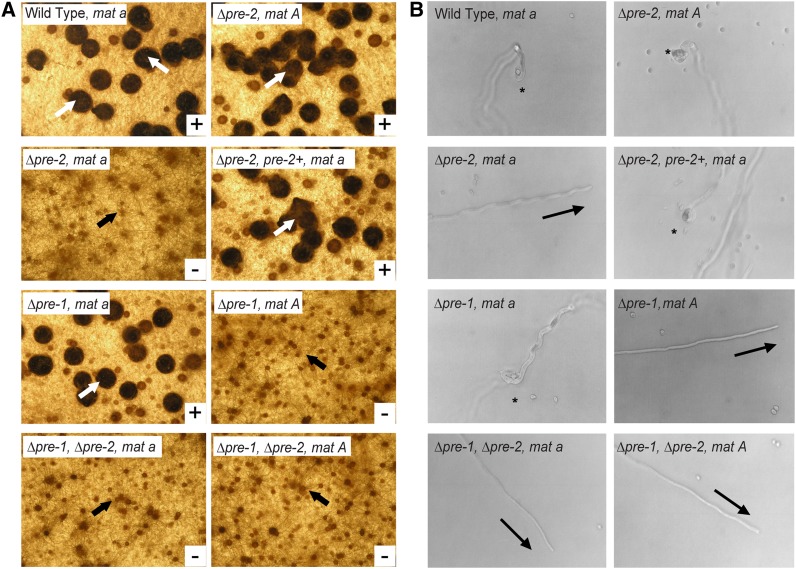
As mentioned above, mating in the heterothallic species N. crassa involves chemotropic interaction and fusion between female-specific hyphae (trichogynes) and male cells (typically conidia) of opposite mating type. This process is mediated by pheromones and pheromone receptors (Bistis 1981; Kim and Borkovich 2004, 2006). One way in which female fertility can be tested is by direct application of conidia of opposite mating type onto fully developed protoperithecia and subsequent assessment of perithecial development and ascospore production (Figure 3A; Davis and Deserres 1970). A second method investigates actual chemotropic interactions by assessing the ability of the trichogyne to track, coil around, and fuse with a conidium of opposite mating type (Figure 3B; Bistis 1981; Kim and Borkovich 2004, 2006). We used both of these assays to examine Δpre-2 and Δpre-1 Δpre-2 mutants for defects in trichogyne attraction and perithecial development. In crosses to wild-type mat A males, Δpre-2 mat a strains were completely unable to develop perithecia, although they formed fully differentiated protoperithecia with trichogynes (Figure 3A). When examined microscopically, the trichogynes of Δpre-2 mat a strains did not display chemotropic growth toward, coiling, or fusion with mat A male cells (Figure 3B). In contrast, Δpre-2 mat A strains underwent normal sexual development (Figure 3A) and exhibited directional growth, coiling, and fusion with mat a conidia (Figure 3B). Reintroduction of pre-2+ in trans into a Δpre-2 mat a strain restored female fertility (Figure 3A) and chemotropic growth of trichogynes toward mat A males (Figure 3B). These observations are consistent with the proposed function of pre-2 as a pheromone receptor specific to mat a strains of N. crassa. In accordance with previous results (Kim and Borkovich 2004), Δpre-1 strains displayed mat A-specific defects in female fertility and chemotropic interaction with male cells (Figure 3, A and B). Δpre-1 Δpre-2 double mutants were female sterile in both mating types (Figure 3, A and B). The double mutants did not produce perithecia (Figure 3A) and their trichogynes failed to grow toward, coil around, or fuse with male cells of opposite mating type (Figure 3B).
Figure 3 .
Female fertility assays. (A) Formation of perithecia. Protoperithecia from the indicated strains were fertilized with conidia from wild-type strains of opposite mating type and perithecial development was evaluated after 6 days. White arrows show examples of mature perithecia, while black arrows indicate unfertilized protoperithecia. Strains are wild-type 74a (WT mat a), wild type 74A (WT mat A), 23a (Δpre-2, mat a), 23A (Δpre-2, mat A), 16a (Δpre-1, mat a), 16A (Δpre-1, mat A), P1P2a (Δpre-1, Δpre-2, mat a), and P1P2A (Δpre-1, Δpre-2, mat A). (B) Trichogyne attraction. Trichogynes from the indicated strains were tested for their ability to grow toward and fuse with microconidia from a wild-type strain of opposite mating type. Asterisks indicate positive chemotropic response and coiling events, while arrows point to the direction of trichogyne growth in cases of nonattraction. Strains are the same as in A.
Pheromone and pheromone receptor determine the identity of a gamete in N. crassa
In nature, successful mating and sexual development in N. crassa require two strains of opposite mating type (e.g. wild-type crosses in Figures 3, 4, and 6). In a previous study, we used a genetically engineered mat a strain that inappropriately expresses high levels of ccg-4 (m1a70) (Kim and Borkovich 2006) to demonstrate that pheromones and receptors are initial determinants for sexual identity. Conidia from the m1a70 strain (Δmfa-1 ccg-4ec mat a) attracted trichogynes from mat a protoperithecia naturally expressing pre-2 and induced early signs of fertilization, but they ultimately failed to produce ascospores.
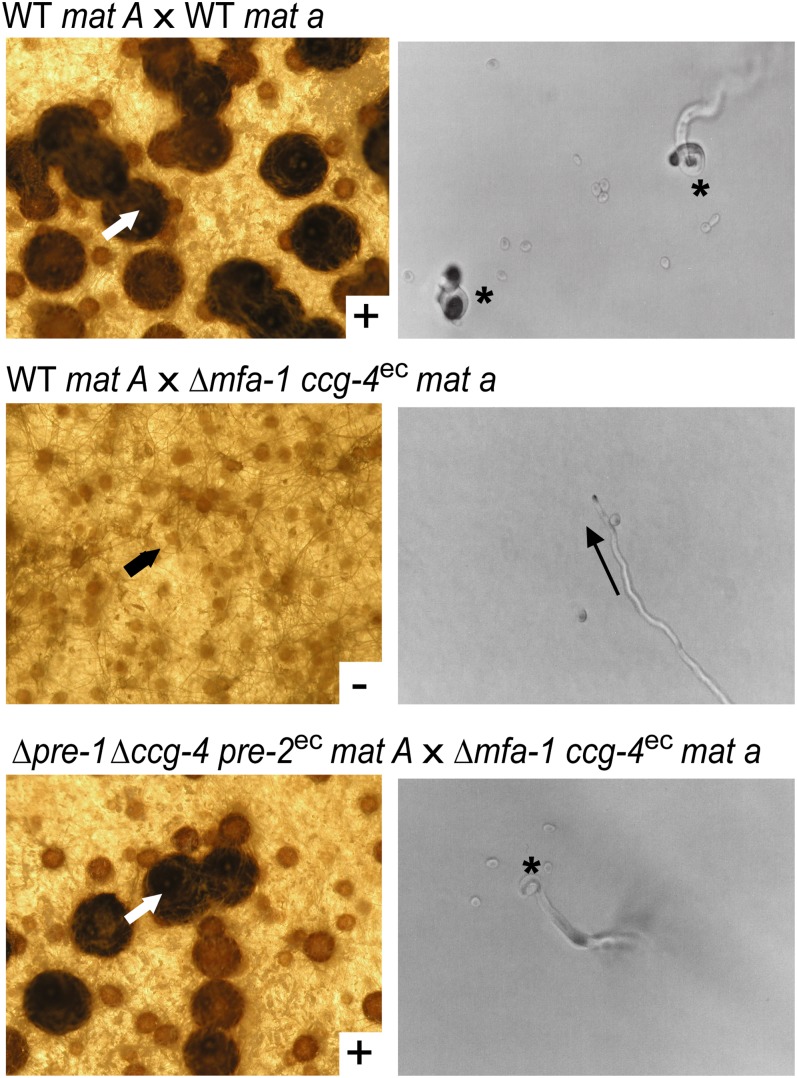
Figure 4 .
Perithecial development of strains with switched identity due to heterologous expression of ccg-4 or pre-2. Strains used are wild-type (WT) 74A (WT mat A), wild-type 74a (WT mat a), m1a70 (Δmfa-1, ccg-4ec, mat a), and P1C4A46 (Δpre-1, Δccg-4, pre-2ec, mat A). Note that ccg-4ec is Pccg-1::ccg-4::his-3+ and pre-2ec is Pccg-1::pre-2-FLAG::his-3+. (Left) Perithecial development in crosses between the indicated strains. The first strain listed was cultured as the female, while the second was used as the male. White arrows indicate ascospore-producing normal perithecia (top and bottom), while the black arrow indicates an unfertilized protoperithecium (middle). (Right) Strains used as females in the crosses shown on the left were cultured on 2% water agar plates and trichogynes analyzed for attraction to microconidia from the second strain listed for each cross. Chemotropic responses were monitored and photographed. Asterisks (top and bottom) indicate directional growth, coiling, and fusion events involving trichogynes, while the arrow (middle) indicates the direction of trichogyne growth in cases of nonattraction.
In this set of experiments, we explore the requirements for PRE-2 and CCG-4 in strains with swapped identity. We implemented two genetically engineered strains of opposite mating type in these studies: P1C4A46 (Δpre-1 Δccg-4 pre-2ec mat A), which inappropriately expresses pre-2 in the mat A background, and m1a70, which produces ccg-4 transcript in the mat a background (see above; Table 1; Kim and Borkovich 2006).
The control cross involving two wild-type strains of opposite mating type resulted in production of perithecia and ascospores (Figure 4, top). In a cross between a wild-type mat A female and a m1a70 male, conidia from the m1a70 strain failed to induce perithecial development or chemotropic growth of trichogynes from wild-type mat A protoperithecia, despite the presence of different mating types (Figure 4, middle). On the other hand, in a cross of P1C4A46 as a female to m1a70 as a male, trichogynes produced by P1C4A46 displayed normal directional growth toward, coiling around, and fusion with m1a70 conidia; underwent normal perithecial development; and produced viable ascospores (Figure 4, bottom). These results indicate that an attraction signal between male and female cells can be transmitted if a pheromone and a pheromone receptor are compatibly paired, but successful perithecial development and ascospore production requires the presence of both mating type loci.
Coexpression of PRE-2 and CCG-4 in the mat A background induces self-stimulation but does not lead to meiosis and ascospore production
We have shown that the pre-2 transcript is expressed to high levels in the mat a background (Figure 1), while ccg-4 mRNA is abundant in mat A strains (Bobrowicz et al. 2002; Kim and Borkovich 2006). The expression profile and deletion phenotypes for pre-2 and ccg-4 are consistent with a model in which the PRE-2 protein interacts with the CCG-4 peptide to facilitate mate recognition and sexual development in N. crassa (Poggeler and Kuck 2001; Kim and Borkovich 2006). One way to test this hypothesis is to determine whether spontaneous sexual development occurs when pre-2 and ccg-4 are coexpressed in the same strain. Therefore, we constructed mat A strains (expressing ccg-4) that also produce pre-2 mRNA under the control of the mating-type–regulated pre-1 promoter or the constitutive ccg-1 promoter. The Δpre-1 mat A background was used to ensure that pre-2 was the only pheromone receptor expressed in these strains.
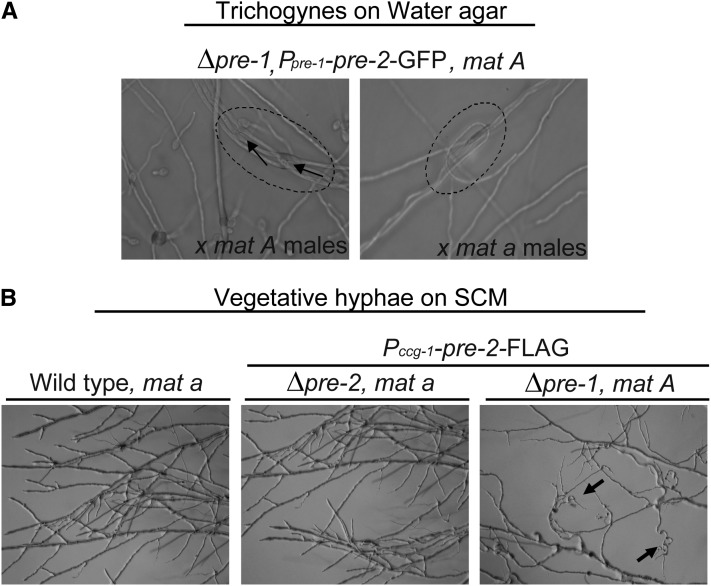
When pre-2 was expressed under the control of the pre-1 promoter in a Δpre-1 mat A background, trichogynes exhibited self-attraction by adhering to one another and forming loops and bundles (strain P1A35; Figure 5A). Production of higher levels of pre-2 mRNA using the ccg-1 promoter in the same background (P1A46; see Figure 6B) resulted in more intense self-stimulation (Figures 5B and 6A). Even vegetative hyphae of strain P1A46 showed signs of self-recognition, including formation of looping hyphae on VM or SCM solid medium (Figure 5B; only SCM culture is shown). When starved for nitrogen, the P1A46 strain initiated protoperithecial development. With continued incubation (and in the absence of male cells of opposite mating type), the protoperithecia darkened and enlarged to the size of 2- to 3-day-old perithecia observed in wild-type crosses (Figure 6A). However, the structures in the P1A46 strains lacked defining features of mature perithecia, such as beaks and ostioles (Figure 6A), and did not develop ascogenous hyphae or ascospores (Figure 6A). Deletion of ccg-4 or gna-1 in the P1A46 background (strains P1A46-ccg-4 and P1A46-gna-1; Table 1) blocked enlargement and melanization of protoperithecia (Figure 6A), resulting in a phenotype similar to that of wild type or Δpre-1 controls (Figure 6A). These results are consistent with a requirement for the appropriate pheromone and the Gα subunit GNA-1 to initiate perithecial development.
Figure 5 .
Self-stimulation of trichogynes or vegetative hyphae in Δpre-1, mat A strains with coexpression of pre-2 and ccg-4. (A) Self-attraction and adhesion of trichogynes from a strain expressing pre-2–GFP under control of the pre-1 promoter. The P1A35 strain (Δpre-1::hph, Ppre-1::pre-2-GFP::his-3+, mat A; abbreviated Δpre-1, pre-2–GFP in figure) was cultured on 2% water agar plates and subjected to a trichogyne assay (see Materials and Methods) using microconidia from wild-type strain 74A (mat A, left) or 74a (mat a, right) as the male. While displaying no response to either mat A or mat a male cells, the trichogynes from P1A35 expressing pre-2 under the control of the pre-1 promoter in the mat A background (producing normal levels of ccg-4 pheromone) were attracted to and fused with one other, forming bundles or loops (circled area). The arrows indicate fusion events between trichogynes. (B) Altered growth of vegetative hyphae from a strain expressing pre-2–FLAG under control of the ccg-1 promoter when cultivated under nitrogen starvation conditions. Strain 74A (wild-type mat A), P2a46 (Δpre-2::hph, Pccg-1::pre-2-FLAG::his-3+, mat a), and P1A46 (Δpre-1::hph, Pccg-1::pre-2-FLAG::his-3+, mat A) strains were grown on SCM plates for 12–20 hr and observed for apical extension of vegetative hyphae. The arrows indicate the curled vegetative hyphae in P1A46 that result from normal expression of the ccg-4 pheromone coupled with high-level production of the pre-2 pheromone receptor (under control of the ccg-1 promoter; see also Figure 6B).
Influence of the opposite mating type gene and/or elimination of meiotic silencing on perithecial development in self-stimulating strains
We next explored whether introduction of the opposite mating-type gene would impact perithecial development in strains expressing a compatible pheromone–receptor pair. Interestingly, ectopic expression of the mat a (mat-a-1) gene in the P1A46 background (strain P1A46-68; Δpre-1 pre-2ec mat aec mat A; Table 1) led to production of slightly larger perithecia than those observed in P1A46 alone, although ascospore development was not observed (Figure 6A). This result suggests that the presence of both mating-type genes may allow further progression of perithecial development, as long as the genetic background contains a compatible pheromone–receptor pair. However, the observation that the mat a-1 transcript could not be detected in strain P1A46-68, containing an ectopic copy of mat a-1 (Figure 6B), indicates that this effect is either independent of, or results from, very low levels of mat-a1 expression.
It has been shown that the Sad-1 gene, encoding a RNA-dependent RNA polymerase, is required for meiotic silencing in N. crassa (Shiu et al. 2001). Sad-1 silences genes that are in different positions in the genomes of the two parents involved in a sexual cross. Silencing is also observed when an extra copy of a gene is present at an ectopic site in one parent, but not the other. Our self-stimulating strains are not involved in a sexual cross with another partner. However, these strains do contain extra copies of pre-2 (at the endogenous and his-3 loci) and/or an ectopic copy of mat a in the mat A background. We reasoned that the presence of an extra copy of pre-2 or the ectopic copy of mat a might be subject to silencing. Therefore, we examined a possible effect of the Sad-1 gene on perithecial development and expression of pre-2.
Elimination of Sad-1 in the Δpre-1 pre-2ec mat A and Δpre-1 pre-2ec mat aec mat A strain backgrounds enhanced perithecial development, with enlargement to the size observed in wild-type heterozygous crosses and formation of beaks and ostioles (Figure 6A). However, ascospores were never formed. To determine whether pre-2 or mat a-1 were being silenced at the transcriptional level, we analyzed mRNA amounts using Northern blots (Figure 6B). Compared to the levels observed in the wild-type mat a background, the amount of pre-2 mRNA was greatly elevated in strain Δpre-1 pre-2ec mat A, expressing pre-2 under control of the ccg-1 promoter. Introduction of mat a-1 and/or deletion of Sad-1 did not influence the expression of pre-2 in the Δpre-1 pre-2ec mat A background (Figure 6B). mat a-1 mRNA could not be detected in any of the mat A strains containing ectopic copies of mat a-1, indicating that the low level of expression of mat a-1 is not due to meiotic silencing. These results suggest that while perithecial development is enhanced in the Δpre-1 pre-2ec mat aec ΔSad-1 mat A strain when meiotic silencing is eliminated, this is not due to an effect on the expression of pre-2 or mat-a-1.
Sexual development of heterokaryons requires one compatible receptor–pheromone pair and GNA-1
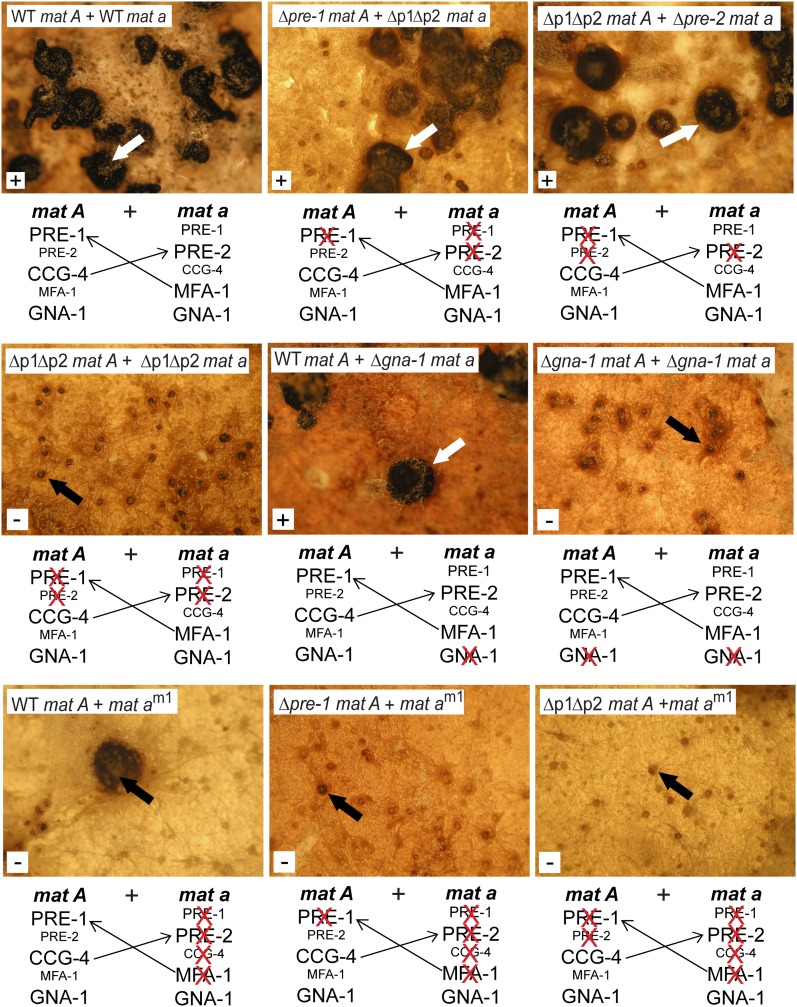
In the experiments described above, we were unable to achieve high levels of expression of both mating-type genes in the same cell. We reasoned that expression of mat A and mat a from different nuclei may be necessary for a single cell to complete sexual development in N. crassa. In addition, this study and previous work indicate that the PRE-1 receptor signals through the Gα protein GNA-1 (Kim and Borkovich 2004). Therefore, we explored the requirement for receptors, pheromones, and the Gα protein GNA-1 in cells containing nuclei of both mating types. In N. crassa, hyphae from two strains can fuse to form a single mycelium (heterokaryon) containing nuclei from the two strains. Heterokaryons can be isolated by mixing strains containing two different auxotrophic markers on minimal medium (forcing conditions; reviewed in Davis and Deserres 1970). The mating-type genes are involved in this process, as efficient fusion (near–wild-type growth rates) is only observed between same mating-type strains (Beadle and Coonradt 1944). However, heterokaryons can be forced between two auxotrophic strains of opposite mating type, with growth rates ∼23% of same–mating-type heterokaryons (Beadle and Coonradt 1944). Our analysis included the Δpre-1, Δpre-2, Δpre-1 Δpre-2, Δgna-1, and/or am1 genotypes and the forcing markers were his-3 (histidine auxotroph) and pan-2 (requires pantothenate). All pairs of opposite mating-type strains formed heterokaryons that grew on minimal medium, while control auxotrophic homokaryons did not exhibit any visible growth or colony formation under these conditions.
Heterokaryons between his-3 and pan-2 strains (used as controls) formed protoperithecia ∼7 days after inoculation onto SCM plates. Many of these protoperithecia grew darker (melanized) and enlarged during days 8–10 and continued development to normal-sized perithecia (Figure 7, top). The perithecia produced and ejected viable ascospores within 5 weeks of inoculation. Heterokaryons formed between Δpre-1 mat A and Δpre-1 Δpre-2 mat a or between Δpre-2 mat a and Δpre-1 Δpre-2 mat A strains also developed ascospore-producing perithecia, albeit more slowly than controls (Figure 7, top), and the perithecia were fewer in number. The observation of fertility in these heterokaryons is significant, as crosses performed between two homokaryons of the indicated genotypes are sterile. Therefore, the results cannot be explained by heterokaryon breakdown, as the remaining receptor in the Δpre-1 or Δpre-2 nucleus does not recognize the predicted highly expressed pheromone produced by the other nucleus. These observations are more consistent with a bypass of the pheromone-mediated chemotropic response that leads to cell fusion in these heterokaryons. In heterokaryons produced by two Δpre-1 Δpre-2 double mutants, protoperithecia darkened without enlargement and perithecial development and ascospore production were not observed (Figure 7, top). This finding indicates that at least one nucleus in the heterokaryon needs to express a pheromone receptor for meiosis and ascospore production to occur.
Figure 7 .
Sexual development of heterokaryons formed between opposite mating-type strains. Conidial suspensions of two opposite mating-type strains with different auxotrophic markers (pan-2 and his-3) were mixed and used to inoculate the center of SCM plates lacking pantothenate and histidine. The plates were incubated to facilitate fusion of hyphae from the two strains and formation of a heterokaryon that can grow in the absence of pantothenate or histidine. All mat A strains are in the his-3 background, while all of the mat a strains (with the exception of mat am1) are pan-2 auxotrophs. The mat am1 strain carries the ad-3B allele and is an adenine auxotroph. Perithecial development was monitored and photographs were taken 45 days after inoculation. White arrows indicate mature perithecia that produced viable ascospores, while black arrows point to melanized protoperithecia or enlarged but barren perithecia. The white box in the lower left-hand corner of each photo indicates whether the heterokaryon produced ascospores (+ or −). WT indicates wild-type alleles of the pre-1, pre-2, gna-1, and mat a genes. Δp1Δp2 = Δpre-1, Δpre-2. Strains (and relevant genotypes) are FGSC 6103 (WT mat A), pan-2 a (WT mat a), 16Ahis3 (Δpre-1, mat A), 23apan2 (Δpre-2, mat a), P1P2h3A (Δpre-1, Δpre-2, mat A), P1P2h3a (Δpre-1, Δpre-2, mat a), Δ1his-3 (Δgna-1, mat A), Δ1pan-2 (Δgna-1, mat a), and am1.
We next examined the involvement of GNA-1 (Gα) in perithecial development of heterokaryons. As observed for heterokaryons of Δpre-1 and Δpre-2 single and double mutants, heterokaryons formed between a wild-type and a Δgna-1 strain underwent slow and less efficient, but ultimately successful, perithecial development and produced viable ascospores (Figure 7, middle). On the other hand, protoperithecia from heterokaryons of two Δgna-1 strains became melanized, but were unable to develop into perithecia or produce ascospores (Figure 7, middle), similar to heterokaryons formed between two Δpre-1 Δpre-2 strains. These findings indicate that at least one nucleus in the heterokaryon must express gna-1 to allow completion of sexual development.
The mat am1 strain does not express any pheromone or pheromone receptor genes and is both male and female sterile (Figure 1C; Bobrowicz et al. 2002; Kim et al. 2002; Kim and Borkovich 2004). Heterokaryons formed between mat am1 and a wild-type strain occasionally formed barren perithecia (Figure 7, bottom). However, mat am1 + Δpre-1 or Δpre-2 single or double mutant heterokaryons completely lacked perithecia and did not produce ascospores (Figure 7, bottom). These results suggest that each nucleus must contribute a compatible receptor or pheromone for meiosis and ascospore development to occur.
Taken together, our results demonstrate that meiosis and perithecial development can proceed in a heterokaryon-containing nuclei of opposite mating type as long as GNA-1 and one compatible pheromone–receptor pair are expressed, even if the pheromone is produced at low levels. Furthermore, forced fusion of normally sterile strains bypasses the requirement for cell fusion mediated by chemotropism. The data support roles for pheromones, receptors, and GNA-1 in postfertilization events in N. crassa.
Discussion
In this study, we have characterized the relationship between the pheromone receptor, PRE-2, and its pheromone CCG-4. These results, along with previous work, also delineate the basic requirements for receptors, pheromones, mating-type loci, and the Gα protein GNA-1 during attraction and fusion of male and female cells and postfertilization events in N. crassa. Our prior work showed that strains lacking the Gα protein GNA-1 are female sterile, with a block in attraction of trichogynes by males and that loss of pheromone causes male sterility. In an earlier study, we also showed that coexpression of a pheromone–receptor pair leads to perithecial development in homokaryons (Kim and Borkovich 2006). Here, we show that while a strain inappropriately expressing the opposite mating-type pheromone receptor is self-attracting and produces perithecia, deletion of either the pheromone gene or gna-1 blocks this attraction. We demonstrate a possible role for meiotic silencing in self-stimulation, as introduction of the ΔSad-1 mutation (Shiu et al. 2001) leads to production of beaks and ostioles in these perithecia. However, in no case are ascospores produced, suggesting that the presence of opposite mating-type nuclei is required for meiosis.
Using forced heterokaryons containing nuclei of both mating types, we demonstrate that meiosis and ascospore production depend on the presence of one compatible receptor–pheromone pair and the gna-1 gene. These requirements do not involve a pheromone response between two cells, as some of the compatible combinations contain strains that are sterile in crosses as homokaryons. Instead, they point to roles for receptors, pheromones, and GNA-1 in postfertilization events. Our results are relevant to a related Neurospora species with a pseudohomothallic mode of sexual reproduction, N. tetrasperma. During sexual development, N. tetrasperma predominantly produces ascospores containing two nuclei, each of a different mating type (Raju 1992a). Heterokaryons containing these two nuclei are self-fertile and do not require an outside mating partner for sexual reproduction. This situation is very similar to the fertility we observed in forced heterokaryons between opposite mating type strains of N. crassa.
A complete replacement of the mat A gene region with mat a has been accomplished in N. crassa, with production of a fertile strain that behaves as a mat a parent in sexual crosses (Chang and Staben 1994). Likewise, gene replacement of mat a with mat A leads to fertility and ascospore production (Glass et al. 1988). In contrast, ectopic integration of the mat A gene in a genome containing a defective mat a allele (am1 mutant) led to production of perithecia, but no ascospores (Glass et al. 1988). Similarly, ectopic introduction into N. crassa of opposite mating-type genes from Podospora anserina that are homologs of N. crassa mat A and mat a leads to self-mating and perithecial enlargement, but ascospores are not produced (Arnaise et al. 1993). These last two findings are consistent with our results testing the effect of an ectopic copy of mat a in the self-stimulated strains. We observed that sexual development is blocked at a stage prior to meiosis, suggesting a requirement for the mating-type genes to be expressed by two different nuclei for completion of sexual development in N. crassa. Our ability to force perithecial development by heterologous expression of a compatible receptor–pheromone pair suggests that the perithecial development observed in the earlier studies in strains containing two mating-type genes may have resulted from elevated expression of the cognate receptor or pheromone. This hypothesis is consistent with our observation that introduction of the mat a gene did not result in a large increase in the size of the barren perithecia produced in the self-stimulated strains in our study.
The results of our attempts to engineer N. crassa to homothallism can be compared to the situation in the true homothallic Neurospora species N. africana and N. terricola. Hybridization experiments indicate that N. terricola contains both a mat A and mat a gene in the haploid genome (Glass et al. 1988). In contrast, N. africana does not contain a mat a gene, but possesses mat A-1, encoding a protein that is 93% identical to, and can substitute functionally for, the N. crassa mat A-1 gene (Glass and Smith 1994). In contrast to our results with N. crassa, these findings indicate that sexual fertility can be observed in Neurospora species containing one or both mating-type genes in the same nucleus. In the future, it would be interesting to investigate expression of pheromones, receptors, and other sexual development genes in N. terricola and N. africana, to identify regulatory proteins that are expressed in N. africana and/or N. terricola, but not in N. crassa. The results of such experiments may shed light on the evolution of homo- and heterothallism in Neurospora.
Requirements for pheromone receptors and mating-type genes have also been explored in the homothallic self-fertile fungus Aspergillus nidulans. A. nidulans contains two different mating-type genes in its genome, on two different chromosomes, and both are essential for self-fertility and sexual spore production (Paoletti et al. 2007). Self-fertility also requires the presence of the GprA and GprB (PRE-2 and PRE-1 homologs, respectively) pheromone receptors; deletion of either receptor gene significantly reduces fruiting body production and viable sexual spores, and loss of both receptors abolishes homothallic reproduction (Seo et al. 2004). Thus, the requirement for receptors for sexual fertility and meiosis is shared between N. crassa and A. nidulans. However, in contrast to our results with N. crassa, the two mating-type genes can function at different positions in the genome to regulate self-fertility and meiosis in A. nidulans.
In this and an earlier study, we observed that mutation of a pheromone receptor gene in the mating type with low expression level leads to reduced production of the other normally highly expressed receptor. Here, we further show that the effect of the receptor mutations also influences expression of both pheromone precursor genes. These results suggest that the low amount of receptor mRNA produced is physiologically relevant and may influence expression of the other normally abundant receptor and the pheromones. However, the low expression of the non–mating-type favored receptor may result from cross-talk and is not sufficient to allow mating as a female. Furthermore, the reduction in the highly expressed receptor does not interfere with sexual fertility. In fact, we only observe a function for the low-expressed pheromone in forced heterokaryons where cell–cell communication has been bypassed, suggesting that low levels of receptor may be adequate for postfertilization events such as nuclear fusion.
Taken together, our data indicate that receptors and pheromones function in postfertilization events in N. crassa. Such a scenario has been observed in basidiomycete mushroom fungi, with these components needed subsequent to fertilization, for nuclear migration in the dikaryon and clamp cell formation (Casselton and Feldbrugge 2010). In yeast, experiments exploring fusion of spheroplasts from same mating-type cells showed that nuclear fusion requires prior exposure to mating pheromone (Rose et al. 1986). Elucidation of the mechanism by which receptors and pheromones regulate postfertilization events is an area ripe for future investigation and one that will yield much information about the extent to which self- and non–self-discrimination modulates sexual reproduction in ascomycete fungi.
Acknowledgments
We acknowledge members of the Borkovich laboratory and Gloria Turner for many helpful discussions. We are indebted to the late Robert Metzenberg for his insights regarding a possible role for Sad-1 in the sexual fertility of homokaryons. This work was supported by National Institutes of Health grants GM048626 and GM086565 to K.A.B.
Footnotes
Communicating editor: E. U. Selker
Literature Cited
- Aanen D. K., Debets A. J. M., Glass N. L., Saupe S. J., 2010. Biology and genetics of vegetative incompatibility in fungi, pp. 274–288 Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Aramayo R., Metzenberg R. L., 1996. Gene replacements at the his-3 locus of Neurospora crassa. Fungal Genet. Newsl. 43: 9–13 [Google Scholar]
- Arnaise S., Zickler D., Glass N. L., 1993. Heterologous expression of mating-type genes in filamentous fungi. Proc. Natl. Acad. Sci. USA 90: 6616–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia G., Loros J. J., Dunlap J. C., Morelli G., Macino G., 1995. Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol. Gen. Genet. 247: 157–163 [DOI] [PubMed] [Google Scholar]
- Bardwell L., 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., Coonradt V. L., 1944. Heterocaryosis in Neurospora crassa. Genetics 29: 291–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistis G. N., 1981. Chemotropic interactions between trichogynes and conidia of opposite mating-type in Neurospora crassa. Mycologia 73: 959–975 [Google Scholar]
- Bobrowicz P., Pawlak R., Correa A., Bell-Pedersen D., Ebbole D. J., 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45: 795–804 [DOI] [PubMed] [Google Scholar]
- Boehm T., 2006. Quality control in self/nonself discrimination. Cell 125: 845–858 [DOI] [PubMed] [Google Scholar]
- Brizzard B. L., Chubet R. G., Vizard D. L., 1994. Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques 16: 730–735 [PubMed] [Google Scholar]
- Casselton L., Feldbrugge M., 2010. Mating and sexual morphogenesis in Basidiomycete fungi, pp. 536–555 Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Chang S., Staben C., 1994. Directed replacement of mt A by mt a-1 effects a mating type switch in Neurospora crassa. Genetics 138: 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F., Hartwell L. H., Jackson C., Konopka J. B., 1988. Conjugation in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 4: 429–457 [DOI] [PubMed] [Google Scholar]
- Davis R. H., deSerres F. J., 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 71A: 79–143 [Google Scholar]
- Debuchy R., Berteaux-Lecellier V., Silar P., 2010. Mating systems and sexual morphogenesis in Ascomycetes, pp. 501–535 Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Ebbole D. J., Sachs M. S., 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37: 17–18 [Google Scholar]
- Fleissner A., Sarkar S., Jacobson D. J., Roca M. G., Read N. D., et al. , 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4: 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H. D., Freitag M., Ramon A., Temporini E. D., Alvarez M. E., et al. , 2003. Histone H1 Is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot. Cell 2: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Dementhon K., 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9: 553–558 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Smith M. L., 1994. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol. Gen. Genet. 244: 401–409 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Vollmer S. J., Staben C., Grotelueschen J., Metzenberg R. L., et al. , 1988. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241: 570–573 [DOI] [PubMed] [Google Scholar]
- Griffiths A. J. F., 1982. Null mutants of the A and a mating-type alleles of Neurospora crassa. Can. J. Genet. Cytol. 24: 167–176 [Google Scholar]
- Griffiths A. J. F., DeLange A. M., 1978. Mutations of the a mating-type gene in Neurospora crassa. Genetics 88: 239–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Herskowitz I., 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52: 536–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey F. D., Hodge P. N., Turner G. E., Borkovich K. A., 1996. The Galpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7: 1283–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey F. D., Yang Q., Borkovich K. A., 1999. Positive regulation of adenylyl cyclase activity by a galphai homolog in Neurospora crassa. Fungal Genet. Biol. 26: 48–61 [DOI] [PubMed] [Google Scholar]
- Janeway C. A., Jr, Medzhitov R., 2002. Innate immune recognition. Annu. Rev. Immunol. 20: 197–216 [DOI] [PubMed] [Google Scholar]
- Jones C. A., Greer-Phillips S. E., Borkovich K. A., 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell 18: 2123–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays A. M., Borkovich K. A., 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G alpha proteins. Genetics 166: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays A. M., Rowley P. S., Baasiri R. A., Borkovich K. A., 2000. Regulation of conidiation and adenylyl cyclase levels by the Galpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20: 7693–7705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Borkovich K. A., 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52: 1781–1798 [DOI] [PubMed] [Google Scholar]
- Kim H., Borkovich K. A., 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Metzenberg R. L., Nelson M. A., 2002. Multiple functions of mfa-1, a putative pheromone precursor gene of Neurospora crassa. Eukaryot. Cell 1: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystofova S., Borkovich K. A., 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gbetagamma dimer required for normal female fertility, asexual development, and galpha protein levels in Neurospora crassa. Eukaryot. Cell 4: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J., 1992. Pheromone response in yeast. Annu. Rev. Biochem. 61: 1097–1129 [DOI] [PubMed] [Google Scholar]
- Loros J. J., Denome S. A., Dunlap J. C., 1989. Molecular cloning of genes under control of the circadian clock in Neurospora. Science 243: 385–388 [DOI] [PubMed] [Google Scholar]
- Orbach M. J., 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150: 159–162 [DOI] [PubMed] [Google Scholar]
- Pall M. L., Brunelli J. P., 1994. New plasmid and lambda/plasmid hybrid vectors and a Neurospora crassa genomic library containing the bar selectable marker and the Cre/lox site-specific recombination system for use in filamentous fungi. Fungal Genet. Newsl. 41: 63–65 [Google Scholar]
- Paoletti M., Seymour F. A., Alcocer M. J., Kaur N., Calvo A. M., et al. , 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17: 1384–1389 [DOI] [PubMed] [Google Scholar]
- Poggeler S., Kuck U., 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280: 9–17 [DOI] [PubMed] [Google Scholar]
- Raju N. B., 1992a. Functional heterothallism resulting from homokaryotic conidia and ascospores in Neurospora tetrasperma. Mycol. Res. 96: 103–116 [Google Scholar]
- Raju N. B., 1992b. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96: 241–262 [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rose M. D., Price B. R., Fink G. R., 1986. Saccharomyces cerevisiae nuclear fusion requires prior activation by alpha factor. Mol. Cell. Biol. 6: 3490–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. A., Han K. H., Yu J. H., 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53: 1611–1623 [DOI] [PubMed] [Google Scholar]
- Shiu P. K., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916 [DOI] [PubMed] [Google Scholar]
- Springer M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays 15: 365–374 [DOI] [PubMed] [Google Scholar]
- Staben C., Jensen B., Singer M., Pollock J., Schechtman M., et al. , 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36: 79–81 [Google Scholar]
- Tsui H.-C. T., Pease A. J., Koehler T. M., Winkler M. E., 1994. Detection and quantitation of RNA transcribed from bacterial chromosomes and plasmids, pp. 197–200 Methods in Molecular Genetics, edited by Adolph K. W. Academic Press, San Diego [Google Scholar]
- Turner G. E., Borkovich K. A., 1993. Identification of a G protein alpha subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268: 14805–14811 [PubMed] [Google Scholar]
- Vogel H. J., 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98: 435–446 [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577 [Google Scholar]
- Yin D., Gavi S., Shumay E., Duell K., Konopka J. B., et al. , 2005. Successful expression of a functional yeast G-protein-coupled receptor (Ste2) in mammalian cells. Biochem. Biophys. Res. Commun. 329: 281–287 [DOI] [PubMed] [Google Scholar]