Abstract
The association between fitness-related phenotypic traits and an environmental gradient offers one of the best opportunities to study the interplay between natural selection and migration. In cases in which specific genetic variants also show such clinal patterns, it may be possible to uncover the mutations responsible for local adaptation. The malaria vector, Anopheles gambiae, is associated with a latitudinal cline in aridity in Cameroon; a large inversion on chromosome 2L of this mosquito shows large differences in frequency along this cline, with high frequencies of the inverted karyotype present in northern, more arid populations and an almost complete absence of the inverted arrangement in southern populations. Here we use a genome resequencing approach to investigate patterns of population divergence along the cline. By sequencing pools of individuals from both ends of the cline as well as in the center of the cline—where the inversion is present in intermediate frequency—we demonstrate almost complete panmixia across collinear parts of the genome and high levels of differentiation in inverted parts of the genome. Sequencing of separate pools of each inversion arrangement in the center of the cline reveals large amounts of gene flux (i.e., gene conversion and double crossovers) even within inverted regions, especially away from the inversion breakpoints. The interplay between natural selection, migration, and gene flux allows us to identify several candidate genes responsible for the match between inversion frequency and environmental variables. These results, coupled with similar conclusions from studies of clinal variation in Drosophila, point to a number of important biological functions associated with local environmental adaptation.
UNCOVERING the genetic basis of adaptation to heterogeneous environments is a central goal of ecological genomics (Storz 2005). A direct approach to this problem entails quantitative trait locus mapping of experimental crosses to associate genetic variation with fitness-related traits. However, the direct approach relies on measurable phenotypic differences previously implicated in environmental adaptation. When experimental crosses are not feasible, or phenotypic trait differences and their fitness consequences are unknown or uncharacterized, an alternative indirect approach is required. In such cases, genome-wide scanning for regions of elevated sequence divergence between natural populations inhabiting different environments—but connected by gene flow—is a powerful strategy that can be used to search for gene–environment associations and thereby identify candidate loci potentially involved in the adaptive process (e.g., Berry and Kreitman 1993; Bonin et al. 2006; Turner et al. 2008; Hohenlohe et al. 2010; Turner et al. 2010; Ellison et al. 2011; Kolaczkowski et al. 2011). This indirect population genomic strategy (“reverse ecology”; Y. Li et al. 2008) complements phenotype-based or candidate gene-based approaches, as it provides a comprehensive genome-wide view of divergence not otherwise possible.
Environmental gradients, such as those produced by shifts in altitude or latitude, impose spatially varying selective pressures on populations distributed along the gradient. Accompanying clinal variation of fitness-related traits and genotypes in these populations, as observed in a number of organisms, may reflect the action of natural selection (Endler 1977). One of the best-studied examples of clinal variation occurs in Drosophila melanogaster along the east coast of Australia, spanning tropical northern Queensland to temperate Tasmania (reviewed by Hoffmann and Weeks 2007). Fitness-related phenotypic traits such as body size, temperature and desiccation tolerance, and genetic polymorphisms—notably four cosmopolitan chromosomal inversions—vary clinally along the latitudinal gradient. Clinal patterns can arise from demographic processes, independent of selection. However, spatially varying selection is strongly implicated in maintaining the cline in Australia both because of the high rates of gene flow inferred from noncoding genetic markers (Kennington et al. 2003) and because parallel clinal patterns are found on different continents (De Jong and Bochdanovits 2003). This example and similar clines in other organisms provide a powerful context for implicating potentially adaptive genotypes and phenotypes.
As first revealed by Dobzhansky’s pioneering work, the observation that chromosomal inversion frequencies are correlated with latitudinal (and seasonal) climatic transitions suggests that inversion polymorphisms in natural populations are maintained by intense selection pressure imposed by climate-related variables (Dobzhansky 1947; Krimbas and Powell 1992; Powell 1997; Schaeffer 2008). A number of models to explain the spread and maintenance of inversions hinge on the fact that they suppress recombination between the rearranged chromosomal regions and thus preserve linkage disequilibrium between favorable combinations of alleles (Kirkpatrick and Barton 2006; Hoffmann and Rieseberg 2008). However, alternative gene arrangements are not completely isolated. Genetic exchange due to gene conversion and double crossovers, particularly away from the breakpoints toward the middle of an inversion, erodes linkage disequilibrium over time (Navarro et al. 1997; Andolfatto et al. 2001; Laayouni et al. 2003; Schaeffer and Anderson 2005). Accordingly, the ease of detecting footprints left by positive selection on loci inside of inversions depends upon the age of the inversion and the selection–recombination balance at loci conferring adaptation to local conditions.
Few studies have addressed the relative importance of chromosomal inversions vs. collinear genomic regions in facilitating adaptive evolution, and population genomic studies in a variety of organisms have sometimes yielded contradictory results (Hoffmann and Rieseberg 2008; Feder and Nosil 2009). The mosquito Anopheles gambiae, one of the major malaria vectors in Africa, is an excellent model system in which to address this question. The range of this species extends across most of tropical Africa, spanning various ecoclimatic zones from rainforest through savanna and sahel, although always in association with humans in rural or peri-urban settings. Polymorphic chromosomal inversions are abundant in An. gambiae, with seven common inversions segregating on chromosome 2 (Coluzzi et al. 1979; Coluzzi et al. 2002). These inversions are nonrandomly distributed across time and space in a manner that suggests their maintenance by selection. The frequencies of several inversions are positively correlated with aridity, such that frequencies peak and trough between dry and rainy seasons in a repeatable cyclical pattern (Coluzzi et al. 1979; Rishikesh et al. 1985; Petrarca et al. 1990; Toure et al. 1998). In addition, latitudinal clines of inversions in West and Central Africa run from mesic rainforest where the inversions are virtually absent, to xeric sahel where they are fixed, or nearly so (Coluzzi et al. 1979; Simard et al. 2009). Although the precise selective agents have not been established, thermal and desiccation stress are known threats to insects in arid environments (Gibbs 2002), and physiological tests on laboratory colonies of An. gambiae differing only in the arrangement of the 22-Mb 2La inversion (2La/a or 2L+a/+a) demonstrated that the inverted orientation confers greater resistance to thermal and desiccation stress, as predicted by its association with arid environments (Gray et al. 2009; Rocca et al. 2009). The complete genome sequence available for An. gambiae (Holt et al. 2002) facilitates population genomic strategies to identify loci responsible for environmental adaptations, whether inside or outside of chromosomal rearrangements.
In two previous studies, we employed gene-based microarrays in divergence mapping of single sympatric population samples of An. gambiae carrying alternative chromosomal arrangements on chromosome 2 (White et al. 2007a; White et al. 2009), with the goal of identifying candidate genomic targets of natural selection. These studies revealed regions of strikingly elevated divergence between alternative arrangements of inversion 2La and provided compelling evidence that this inversion is maintained by selection, but failed to find significant divergence between most rearrangements on chromosome 2R. Because the microarray-based divergence mapping was geared toward identifying fixed or major frequency differences, and because the mapping platform was not a tiling array, existing differentiation between rearrangements on 2R may have escaped detection. Regions of elevated differentiation in collinear genomic regions was neither expected nor observed within sympatric population samples of the same An. gambiae molecular form (M or S).
Here, we expand the resolution and scope of our population genomics investigation of environmental adaptation in An. gambiae through whole genome resequencing of S form populations sampled near the ends and middle of a latitudinal gradient of aridity in Cameroon. Along this gradient, S form populations show steep clinal variation of inversions 2La and 2Rb, but little genetic differentiation at 12 microsatellite markers on chromosome 3 (average FST = 0.0053) (Slotman et al. 2007), suggesting unrestricted gene flow. By examining spatial patterns of sequence variation, we identify candidate loci within and outside of chromosomal rearrangements that are potentially involved in local adaptation.
Materials and Methods
Population sampling and karyotyping
Indoor resting mosquitoes were sampled in September 2007 by insecticide spray sheet collection inside human dwellings, from seven villages spanning a latitudinal gradient from southern rainforest (3°52’N) to northern arid savanna (9°25’N) in Cameroon, Central Africa (Figure 1; for a more detailed ecogeographic description, see Simard et al. 2009). Specimens were preserved over desiccant in individual numbered microtubes. An. gambiae S form was identified by sequential morphological and molecular taxonomic methods (Gillies and De Meillon 1968; Santolamazza et al. 2004). Both molecular taxonomy and molecular karyotyping of the 2La and 2Rb inversions (White et al. 2007b; Lobo et al. 2010) were performed on DNA extracted from a single leg (Collins et al. 1987); the remaining carcass was held in reserve.
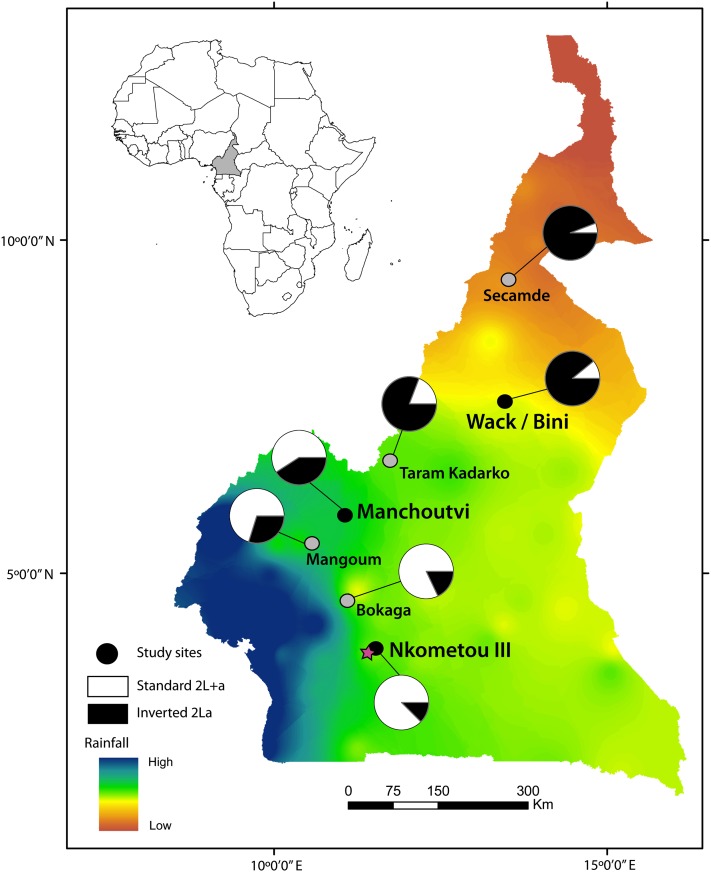
Figure 1 .
Sampling sites along a latitudinal transect in Cameroon. Mean annual precipitation, from high (blue) to low (orange), is based on data from FAO SDdimensions (http://www.fao.org/sd/2002/EN1203a_en.htm). Pies show the frequency of the standard (2L+a, white) and inverted (2La, black) arrangement of inversion 2La in An. gambiae S populations sampled at each locality (circles). Black circles indicate the three localities chosen for population resequencing. The capital Yaoundé is represented by a star.
Population pools and genome resequencing
Four population pools were constructed for genomic resequencing, each consisting of DNA from 34 An. gambiae S form females (to avoid undersampling of the X chromosome in heterogametic males). Pooling was based on collection locality and karyotype of the 2La inversion and was blind to karyotype of other inversions on 2R, including 2Rb. Two equal pools—comprising homokaryotypes of the 2La or 2L+a arrangement, respectively—originated from the same central locality (Manchoutvi: 5°52’48” N, 11°06’49” E) with a high level of 2La/+a polymorphism (2L+a frequency, 59%). The other two pools originated from localities at opposite ends of the cline, where 2La/+a polymorphism is low. At the southern end where 2L+a predominates, a pool of 2L+a/+a homokaryotypes was constructed from Nkometou III (3°51’56” N, 11°30’56” E; 2L+a frequency, 88%). At the northern end where 2La predominates, a single pool of 2La/a homokaryotypes was constructed from two nearby villages (Wack and Bini: respectively, 7°41’02” N, 13°32’56” E and 7°23’55” N, 13°33’02” E; average 2L+a frequency, 6%) (Figure 1).
DNA was isolated from individual carcasses using the Wizard SV 96 genomic DNA purification system (Promega, Madison, WI). To prepare pools, 50 ng of DNA from each of 34 female mosquitoes (68 chromosomes) was combined (1.7 μg total). Sequencing libraries were constructed from each of the four pools by the Center of Genomics and Bioinformatics (CGB) at Indiana University, Bloomington, according to standard Illumina protocols. Each library was sequenced on an Illumina Genome Analyzer IIx at the CGB to generate 72-bp single-end reads, with the goal of achieving ∼10× average genome coverage (over 260 Mb) while avoiding oversampling of the same chromosome sequence in the pools of 68 chromosomes (Futschik and Schlotterer 2010). Across all four libraries, this produced 245,586,259 reads (approximately 18.7 Gb of sequence data). These reads are available from the Short Read Archive (SRA) of the European Nucleotide Archive (ENA) under accession no. ERP000955.
Read alignment
Illumina sequencing reads were aligned to the AgamP3 (PEST) assembly of the An. gambiae genome (http://www.vectorbase.org; Lawson et al. 2009). With the goal of maximizing the number of mapped reads, we conducted two separate alignments to AgamP3 using different programs, and compared metrics between the two. The first program, Mapping and Assembly with Qualities (MAQ, v. 0.7.1; H. Li et al. 2008), is computationally efficient, but was developed for genomes with relatively low levels of polymorphism (e.g., human) and it does not allow for mapping of reads with insertions or deletions (indels). Among the 120,297,539 reads uniquely mapped by MAQ (reads with two or more equally likely positions were excluded), the average mismatch rate to the AgamP3 reference was 2.8%. Average genome coverage was 39.7-fold summed across all four pools.
The second program, SHort Read Mapping Package (SHRiMP, v. 1.3.0; Rumble et al. 2009), can capture higher levels of polymorphism including indels, but is computationally intensive. To reduce runtime, we parallelized SHRiMP jobs through Condor, a distributed batch computing system at the University of Notre Dame. Reads mapped with SHRiMP were processed with the PROBCALC utility. After excluding reads with multiple equally good matches to the reference (alignments with pchance <0.05, or normodds <0.8), 137,185,394 uniquely mapped reads were piled up to the reference genome using custom Perl scripts. The average mismatch and indel rates between reads and reference were 2.6 and 0.4%, respectively. The 44.9-fold average genome coverage achieved by SHRiMP across the four pools was ∼10% higher than MAQ. Supporting information, Figure S1 (left), shows read coverage for MAQ and SHRiMP across the four pools by chromosome arm. In Figure S1, right, a detailed plot of read coverage along the length of chromosome 2L based on the northern (2La/a) population pool indicates the substantial increase in coverage achieved by SHRiMP in the rearranged region of 2L, where a much higher level of read mismatch (to the uninverted 2L+a AgamP3 reference) is expected. Average read coverage inside the rearranged region is 12× with SHRiMP compared to 8.5× with MAQ for the northern pool. Because SHRiMP mapping provided higher read coverage both inside and outside inversions, all downstream analysis of genetic diversity and differentiation was based on SHRiMP alignments.
Data filtering
Only uniquely aligned reads were retained for analysis. To be considered, a site was required to have at least 10× read coverage in each population being compared, but no more than 30× coverage (to mitigate against repetitive sequences). For window-based analyses, only windows containing a minimum number of sites meeting coverage thresholds were retained. The minimum number of sites required per 1-kb window (317) was arbitrarily set on the basis of empirical distribution, to exclude windows containing the smallest 25% of usable sites. Both site-based and window-based analyses excluded low recombination regions of the genome designated as heterochromatic (Sharakhova et al. 2010), including pericentromeric heterochromatin (X—20,009,764-24,393,108; 2L—1-2,431,617; 2R—58,984,778-61,545,105; 3L—1-1,815,119; 3R—52,161,877-53,200,684) and intercalary heterochromatin (2L—5,078,962-5,788,875; 3L—4,264,713-5,031,692; 3R—38,988,757-41,860,198).
SNP identification and frequency inference
To reduce the effect of random sequencing error, singleton polymorphisms (those found only in a single read in a single population) were discarded. Although this treatment should also remove true low-frequency polymorphisms, it should have little consequence on the ability to detect the most extreme frequency differences between populations—the focus of this study. For polymorphisms supported by at least two reads, SNP frequency estimates were weighted according to their quality scores (given by the Illumina GA pipeline software), using Holt et al.’s (2009) Equation 6.
Population genomic and outlier analysis
Following Akey et al. (2010) and Kolaczkowski et al. (2011), we adopted an empirical outlier approach to identify exceptionally diverged regions of the genome that are potentially affected by selection. In comparing populations sampled at opposite ends of the cline, we found that average levels of divergence between rearranged (2La, 2Rb) genomic regions were drastically higher than those between collinear genomic regions (see Figure 2). Given our interest in identifying outliers outside as well as within the 2La and 2Rb rearrangements, outlier analysis was performed separately for each rearrangement and for the remaining (collinear) regions exclusive of heterochromatin. For each of these three data partitions, measures of diversity and divergence were estimated from nonoverlapping 1-kb windows. Specifically, bias-corrected estimates of Tajima’s π for each 1-kb window were calculated following Futschik and Schlötterer (2010). These estimates were used to derive FST values, calculated as FST = (πBetween − πWithin)/πBetween (Hudson et al. 1992) and averaged over all segregating SNPs in each 1-kb window. For each data partition, outlier windows were defined as those with mean FST values falling in the top 1% of the empirical distribution. For some analyses, individual outlier SNPs were defined as those with FST values falling in the top 0.1% of the empirical distribution (collinear region and 2Rb rearrangement) or FST = 1 (2La).
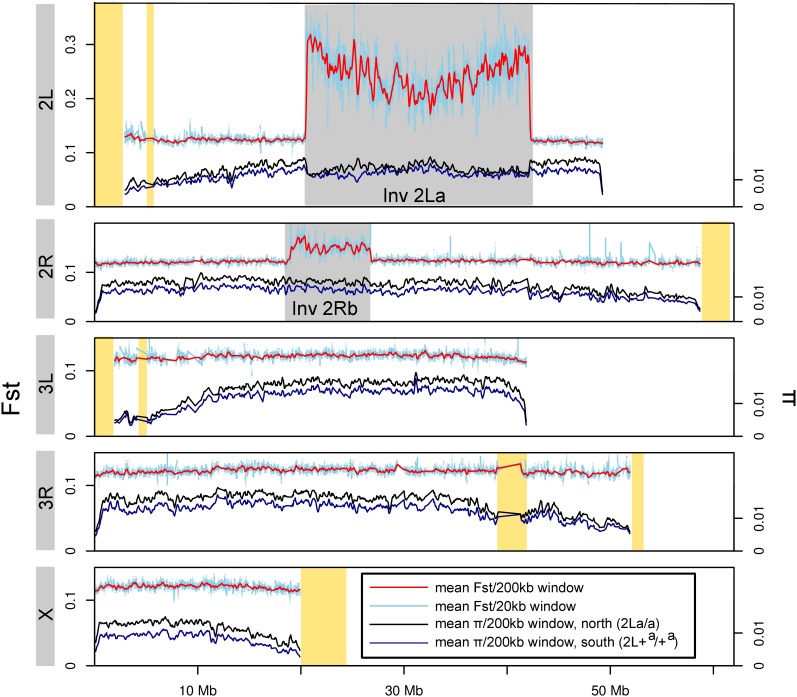
Figure 2 .
Divergence (FST) between and diversity (π) within An. gambiae S populations from opposite ends of the latitudinal cline in Cameroon. Across each chromosome arm, mean divergence is plotted over 200-kb windows slid every 50 kb (red), and 20-kb windows slid every 5 kb (light blue). Mean diversity is plotted for 200-kb windows slid every 50 kb for Nkoumetu in the south (2La/a; dark blue) and Wack/Bini in the north (2L+a/+a; black). Gray and yellow shaded boxes indicate chromosomal rearrangements or heterochromatic regions, respectively. Only windows containing ≥317 sites meeting coverage thresholds are plotted, causing reduced representation of heterochromatic regions.
Copy-number variation
To identify possible copy-number differences between the two populations at opposite ends of the cline, average read depth coverage was estimated in 1-kb windows across nonheterochromatic regions of the genome. Window-based ratios of read depth between the two populations were normalized by the median genome-wide ratio and plotted following arctan transformation. We considered windows in the top 0.5% or bottom 0.5% of the empirical distribution of normalized depth ratios to represent candidate copy-number differences (Kolaczkowski et al. 2011).
Functional clustering
Lists of candidate genes were assembled from the set that overlapped the outlier FST or read depth ratio 1-kb windows. For this purpose, a gene was defined as the predicted transcribed region plus 1 kb upstream and downstream. The resulting gene lists were explored for possible functional relationships based on annotation profiles built from terms derived from multiple sources [e.g., Gene Ontology (GO), SMART and InterPro Domains, SwissProt/Uniprot, and PIR keywords], using the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) (Huang et al. 2009). We employed DAVID’s Functional Annotation Clustering utility with default settings to identify groups of genes (“annotation clusters”) whose annotation profiles suggest common function. The enrichment score assigned to each annotation cluster represents the relative importance of that functional gene group on the basis of the fraction of its members associated with highly enriched annotation terms. It is measured by the geometric mean of the EASE Scores (a modified Fisher exact P-value) associated with each enriched annotation term in the gene group (Hosack et al. 2003; Huang et al. 2007) and is intended to rank the importance of the groups, rather than to provide a rejection/acceptance threshold customary of a traditional statistical analysis. For this reason, enrichment scores are presented in the form of minus log-transformed geometric means instead of an absolute P-value (Huang et al. 2007).
Results
We resequenced whole genomic DNA from four population pools each composed of 34 An. gambiae S form mosquitoes of known 2L homokaryotype (2La/a or 2L+a/+a), derived from three localities along an arid-to-mesic latitudinal gradient in Cameroon: two from the extremes of the gradient and one in the center (Figure 1). Because the climatic gradient is associated with a steep cline in 2La inversion frequency, from near fixation in the arid north to near absence in the mesic south, our study design included samples of each homokaryotype from either endpoint and in the center. Specifically, we sampled a homokaryotypic 2La population in the north and a homokaryotypic 2L+a population in the south and separately sequenced pools of homokaryotypic 2La and 2L+a individuals from the central population (where heterokaryotypes are common in nature).
Two flow cells of Illumina GA IIx resequencing produced ∼45-fold coverage of the genome across all four population pools (12.2× and 9.6× from the north 2La/a and south 2L+a/+a populations; 10.3× and 12.8× from the central 2La/a and 2L+a/+a populations). After retaining only uniquely aligned reads, filtering based on read coverage, and eliminating 1-kb windows with insufficient data meeting coverage thresholds (Materials and Methods), the average 1-kb window contained 779 sites. In the combined populations, 17,077,264 nonsingleton SNPs were detected outside of regions designated as centromeric or intercalated heterochromatin. Importantly, population-pairwise estimates of FST based on 1-kb windows indicate only moderate genome-wide differentiation between any population pair (Table S1), and differentiation between the northern and southern samples (FST = 0.123 excluding rearranged regions) is only marginally greater than that between subsamples of a single central population (FST = 0.114). Taken with the absence or very low level of significant microsatellite differentiation between comparable population samples of An. gambiae S from Cameroon (Slotman et al. 2007), these data suggest sufficient genetic connectivity to merit a reverse ecology approach for mapping candidate genes underlying local adaptation.
Genomic patterns of divergence between endpoint populations
Nucleotide divergence between populations sampled from the northern and southern ends of the climatic gradient and nucleotide diversity within these populations were estimated from nonoverlapping 1-kb windows across the euchromatic chromosome arms (Figure 2; Table S2). Excluding the two rearranged regions on chromosome 2, mean FST is relatively low (0.123) and similar among all five chromosome arms. In contrast, mean FST is strikingly elevated—although to different extents—inside the regions spanned by the chromosome 2 rearrangements (FST inside 2La = 0.247; FST inside 2Rb = 0.149). Elevated FST does not persist outside of the rearrangements beyond 20 kb from the breakpoints, in contrast to the larger extent of divergence (1–2 Mb) observed outside the breakpoints of inversions distinguishing Drosophila pseudoobscura and D. persimilis (Machado et al. 2007).
More pronounced differentiation between alternative arrangements of 2La relative to alternative arrangements of 2Rb has been noted previously in natural populations of An. gambiae (White et al. 2009). However, in the present data set the distinction between levels of divergence observed in the two rearrangements is also partly due to the fact that population pooling took only the 2La karyotype into account and was blind to 2Rb karyotype. Molecular karyotyping of 2Rb (Lobo et al. 2010) performed on individual mosquito DNA aliquots after Illumina libraries had been prepared from pooled DNA revealed a frequency of 2Rb ranging from ∼7% in the southern to ∼90% in the northern population pools, respectively (and ∼67% in the central locality). Thus, as expected (Coluzzi et al. 1979; Lee et al. 2009), 2Rb is clinally distributed along the arid-mesic latitudinal gradient in a pattern that parallels the 2La inversion. These data also indicate that although alternative 2Rb arrangements predominate at opposite ends of the cline, both arrangements are present in the two population pools, presumably resulting in some dampening of observed levels of divergence in the region spanned by 2Rb between northern and southern population samples.
Across the genome, nucleotide diversity is consistently lower in the southern than in the northern population (Figure 2 and Table S2). However, the general trends are very similar between the two. Most notable are markedly lower diversity toward telomeric and especially centromeric ends of the chromosomes, and lower diversity inside the 2La, but not the 2Rb, rearrangement relative to flanking chromosomal regions.
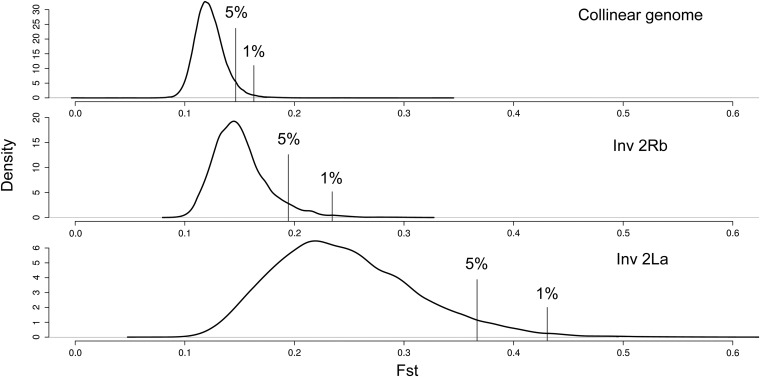
Given the heterogeneous levels of divergence in collinear and rearranged regions of 2L and 2R, and our interest in identifying elevated divergence within each of these three regions, we treated all three as separate data partitions when assessing outlier 1-kb windows of FST. Histograms of FST values for windows in each data partition are provided in Figure 3. The threshold FST values for windows in the top 1% of the three distributions differed greatly, and the rank order was: collinear genome (0.163) < 2Rb (0.234) < 2La (0.431).
Figure 3 .
Distribution of FST values for 1-kb windows spanning the collinear euchromatic genome, the 2Rb and the 2La rearrangements (between north and south populations). Positions in the distributions marking the top 5% and top 1% FST values are indicated by vertical lines.
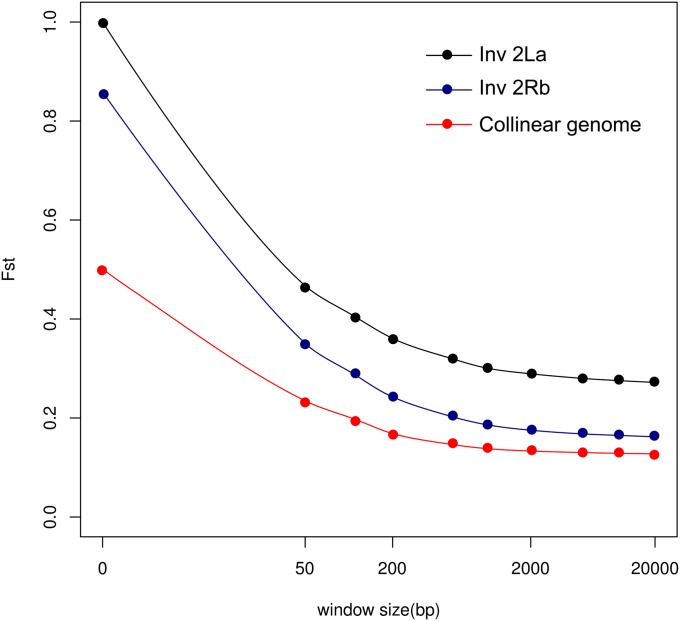
Our ability to identify candidate genes for local adaptation depends upon the degree of linkage disequilibrium between the polymorphisms most strongly associated with populations at the endpoints of the climatic gradient (i.e., SNPs with the highest FST values between our northern and southern population pools). As haplotype information is lost in pooled sequencing, we approached this question by assessing the rate of decay of FST with increasing distance from a focal SNP (Turner et al. 2010), treating each of the three data partitions separately. Focal SNPs for this analysis were defined as those carrying the highest 0.1% of FST values for the collinear genome and the 2Rb rearrangement (respectively, 9458 SNPs with FST > 0.43 and 560 SNPs with FST > 0.77), and those with FST = 1 for the 2La rearrangement (25,915 SNPs). Beginning with a window size of 1 bp (the focal SNP itself), windows centered on each focal SNP were incrementally increased in size up to 20,000 bp. Mean FST was recalculated per window and averaged across windows of the same size. As seen in Figure 4, FST declines precipitously within 500 bp, not only in the collinear genome but also inside both rearrangements, suggesting that the most differentiated SNPs are not colocalized.
Figure 4 .
The decay of average FST with increasing window size for windows centered on the most differentiated SNPs between northern and southern Cameroon populations in the collinear genome (red), the 2Rb rearrangement (blue), and the 2La rearrangement (black). Note that the x-axis is on a logarithmic scale.
Clinal patterns of divergence
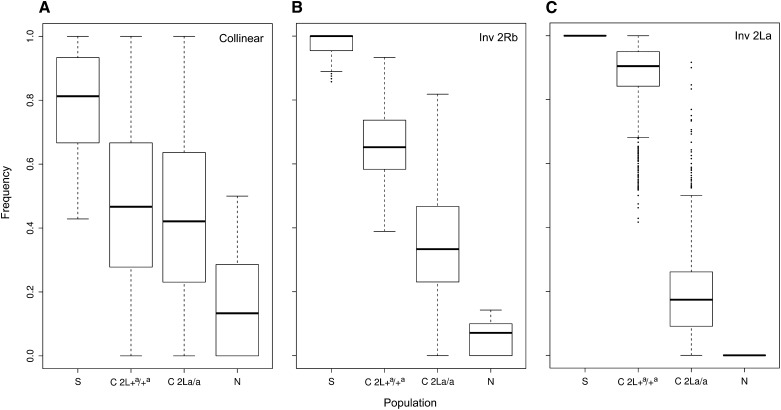
We focused on the three sets of SNPs whose north–south FST values were in the top 0.1% for the 2Rb and collinear regions, and maximal (FST = 1) for 2La (the same SNPs used to explore the decay of FST with increasing distance, described above). Allele frequency at each SNP, measured with respect to the major allele in the southern population, was estimated for northern, southern, and central populations. In the case of the central population, allele frequencies in each genomic region were estimated for both (2La/a and 2L+a/+a) subsamples, as these were sequenced separately. As shown in Figure 5, the SNP frequencies in all three genomic regions—2La, 2Rb and collinear—are clinal; mean frequencies in the central population are intermediate to the end populations.
Figure 5 .
Individual SNP frequencies along the latitudinal gradient in Cameroon, based on loci that most strongly differentiated populations at opposite ends of the gradient. Frequencies are plotted with respect to the allele predominating in the southern population in (A) the collinear genome, (B) the 2Rb rearrangement, and (C) the 2La –75th percentile); the horizontal line marks the median. Upper (lower) whiskers include the maximum (minimum) values unless the distance from the first (third) quartile exceeds 1.5× that of the IQR. Outlier values smaller or larger than the whiskers are indicated by dots. N, northern population; S, southern population; C, central population, partitioned by karyotype into 2La/a and 2L+a/+a homozygotes.
A cross-comparison of patterns among rearranged and collinear regions demonstrates the power of following the separate fates of the 2La/a and 2L+a/+a samples in the center of the cline. Although differences in allele frequencies between the two samples are significantly different by Wilcoxon signed rank test in each case (P-value < 2.2 × 10−16), the magnitude of difference is substantially less for collinear as compared to rearranged regions. SNPs in the collinear genome that differentiate these mosquitoes at opposite ends of the cline are effectively homogenized between alternative karyotype subsamples in the central population (Figure 5A). These data indicate that the two groups of mosquitoes carrying alternative homozygous arrangements of 2La are not genetically isolated from each other, and in fact exchange alleles across the majority of the genome. By contrast, and in keeping with the association between 2Rb and 2La karyotypes along the cline in Cameroon, 2Rb-associated SNPs appear more likely to be found in 2La/a samples in the central population (while 2R+b-associated SNPs are more likely in 2L+a/+a samples), despite independent segregation of the two inversions (Figure 5B).
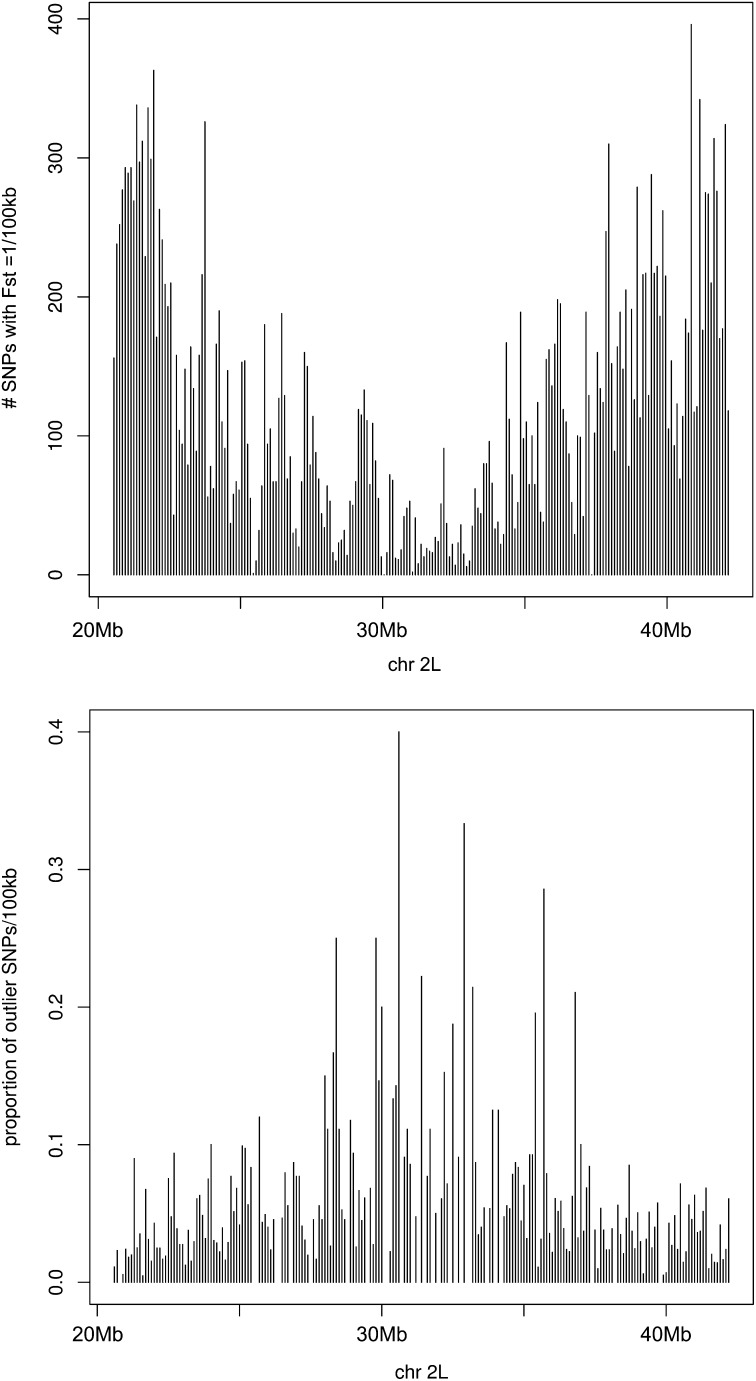
The most compelling result concerns the ∼26,000 SNPs inside the 2La rearrangement that are completely fixed for alternative states at opposite ends of the cline. In the center of the cline, considering 2La and 2L+a chromosomes separately, there is a clear shift in allele frequencies on both arrangements such that many SNPs are no longer fixed between alternative karyotypes and some show radical changes in frequency (Figure 5C). Gene flux (the transfer of alleles between alternative arrangements by crossing over and gene conversion) is the most reasonable explanation for this pattern, given that it occurs orders of magnitude more frequently than new mutations (Navarro et al. 1997; Andolfatto et al. 2001; Laayouni et al. 2003). Of particular note is the relatively large number of 2La and 2L+a SNPs whose frequencies are extreme (indicated by dots in 5C) relative to the distribution of frequency values for the central population; these represent outlier SNPs that have been homogenized between inversion arrangements. In Figure 6, the location and density of these outlier SNPs in the center of the cline can be compared to the distribution and density of all fixed SNP differences between alternative 2La arrangements at opposite ends of the cline. The outlier SNPs are distributed across the length of the inversion, but not uniformly. They are more rare toward the breakpoints and more frequent in the center of the inversion. This pattern is consistent with gene flux, given that gene conversion is expected to predominate closer to the breakpoints while double crossovers and gene conversion both operate in the center of an inversion (Navarro et al. 1997; Andolfatto et al. 2001; Laayouni et al. 2003). If gene flux explains the admixture of 2La- and 2L+a-associated alleles in the central population, as seems likely, the implication is that selection is maintaining the clinal pattern of differentiation at many SNPs against the potent homogenizing forces of gene flow and gene flux.
Figure 6 .
The position and density of SNPs inside the 2La rearrangement with fixed differences between 2La and 2L+a at opposite ends of the cline. Counts are based on nonoverlapping 100-kb windows slid between the breakpoints of 2La. Top: The total set of SNPs with FST = 1 between northern and southern populations. Bottom: The outlier SNPs in the central 2La/a and 2L+a/+a populations (see Figure 5C).
Genic patterns of divergence
To obtain a genic view of elevated differentiation between northern and southern populations, we compiled three sets of genes that overlapped 1-kb windows falling within the top 1% of FST values in the rearranged (2La, 2Rb) and collinear regions of the genome (listed in Table S3, Table S4, and Table S5 with their putative Drosophila orthologs obtained from the Ensembl Metazoa Genes 10 database). Considering the gene sets separately, we subjected each to a functional annotation clustering analysis using the DAVID software package (Huang et al. 2009), with the goal of condensing the gene lists into groups potentially associated with common biological processes or functions.
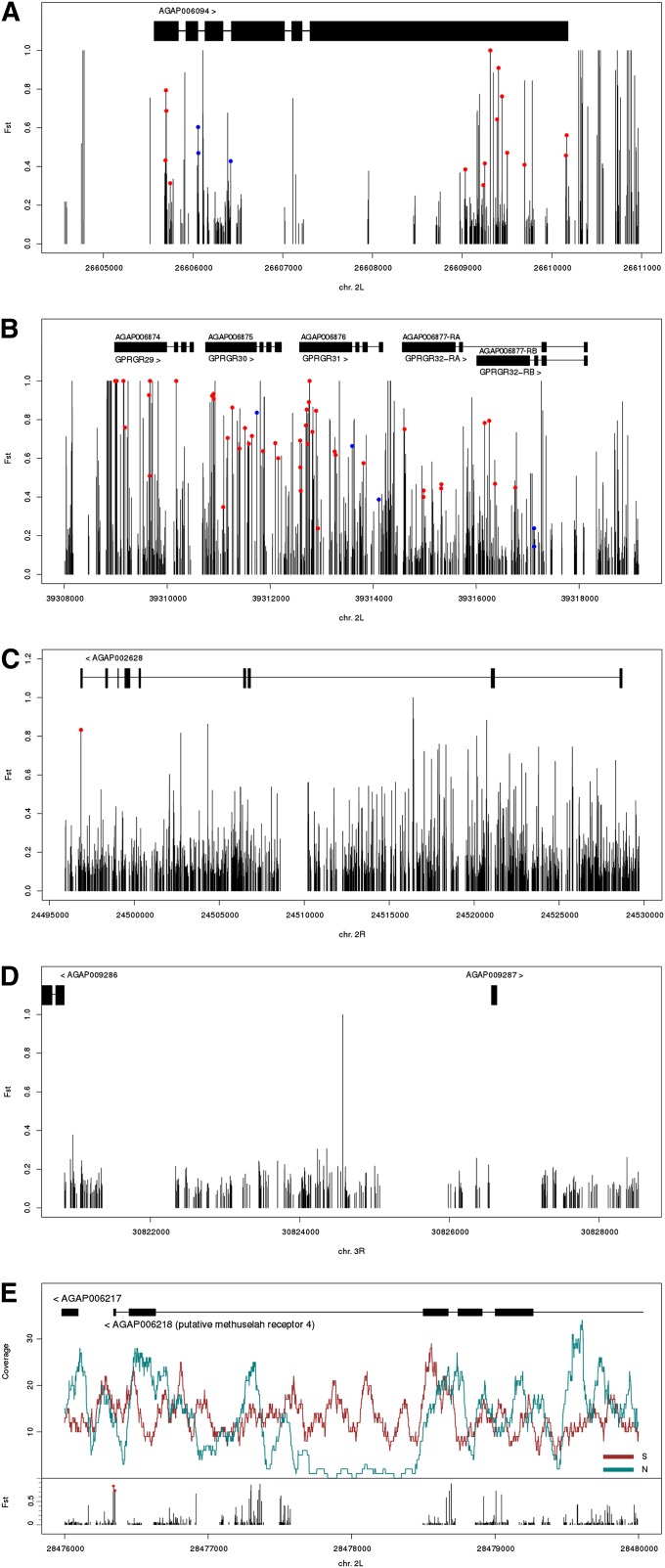
The 2La rearrangement spans 1281 genes, of which 52 overlapped the most divergent 1-kb windows. Functional annotation clustering attempted on the 52 genes revealed four small groups whose enriched annotation terms classified them as cuticle proteins (annotation cluster 1), serine/threonine protein kinases (2), and ion channels/GPCRs (clusters 3 and 4 contained the same 8 genes) (Table 1). Annotation cluster 1 contains mainly cuticle protein (CPR) genes of the RR-2 family, which are often found in close proximity in different regions of the An. gambiae genome. Indeed, the physical cluster of CPR genes inside 2La is the largest, containing ∼40 CPR genes, which overlap a ∼1-Mb region previously identified through microarray-based divergence mapping as one of the most diverged between alternative arrangements of 2La (White et al. 2007a). Among the serine/threoine protein kinase genes in cluster 2 with identifiable homologs are several putative Drosophila orthologs that regulate aspects of growth, development, and innate behavioral responses. These include cyclin-dependent kinase 4 and nimA-like kinase (growth regulation and mitotic control), nemo (specification of polarity and development of wing size/shape), frayed (axon ensheathment), and alan shepard (geotaxis). Another is JIL-1, a gene whose protein kinase product is proposed to reinforce the status of active chromatin through phosphorylation of histone H3 at serine 10 (Regnard et al. 2011). Divergence between northern and southern populations mapped onto the An. gambiae gene model of JIL-1 (AGAP006094) is illustrated in Figure 7A. Within clusters 3 and 4 are ion channel-related genes of the potassium voltage-gated channel family (KCNQ and shal2 orthologs) implicated in nervous system development and function. In addition, there are chemosensory receptors: three originally classified as G protein-coupled receptors (GPCRs) in the gustatory receptor family (GPRGR29, GPRGR30, GPRGR31; Figure 7B) and another ionotropic glutamate receptor (GPRMGL1). Unsurprisingly, the DAVID tool did not cluster all 52 genes overlapping the top 1% FST windows. Manual appraisal of the complete 2La gene list in conjunction with putative Drosophila ortholog assignments uncovered several unclustered candidates with potential roles in the regulation of nervous system development and function, roles that cut across some of the DAVID annotation clusters (diacylglycerol kinase 1, follistatin, multiplexin, pebble, peptidylamidoglycolate lyase 2, still life, syntrophin-like 2, and unc-13-4A). Also unclustered, but functionally related to JIL-1 in the context of chromatin regulation of transcription, was asf1 (a histone chaperone with a role in SWI/SNF-mediated chromatin assembly and remodeling).
Table 1. Functional annotation clusters of genes overlapping the top 1% windows of FST between northern and southern populations in rearranged (2La, 2Rb) and collinear genome regions.
| Genome partition | Annotation cluster | Representative annotation term(s) | Gene count | Enrichment score |
|---|---|---|---|---|
| 2La | 1 | Insect cuticle protein | 5 | 1.56 |
| 2 | Serine/threonine protein kinase | 9 | 1.52 | |
| 3 | Ion channel/transport | 8 | 1.51 | |
| 4 | GPCR | 8 | 1.17 | |
| 2Rb | 5 | Immunoglobulin-like fold | 4 | 2.50 |
| Collinear | 6 | Cell-surface receptor-linked signal transduction | 63 | 2.97 |
| 7 | Calcium-dependent membrane targeting | 6 | 1.84 | |
| 8 | Transcription regulator/homeodomain | 32 | 1.45 | |
| 9 | GPCR/neurotransmitter receptor activity | 7 | 1.30 | |
| 10 | ETS domain/winged helix repressor DNA binding | 3 | 1.12 | |
| 11 | Postembryonic development/morphogenesis | 14 | 1.03 |
Figure 7 .
Elevated FST or copy-number variation between northern and southern Cameroon populations in protein coding genes. FST is plotted for individual SNPs along the gene models (solid rectangles, exons; horizontal lines, introns). Nonsynonymous and splice site mutations are indicated by red and blue circles, respectively. Divergence in (A) the JIL-1 ortholog, AGAP006094 in the 2La rearrangement; (B) the cluster of gustatory receptors GPRGR29-32 in the 2La rearrangement; (C) AGAP002628 in the 2Rb rearrangement, a putative member of the immunoglobulin superfamily; (D) an intergenic region upstream and nearby two genes in a collinear region on 3R; (E) the Methuselah receptor 4 ortholog AGAP006218 in the 2La rearrangement. In addition to FST at individual SNPs, read coverage is shown for each population (northern, green line; southern, red line) in relation to the gene model.
The smaller 2Rb rearrangement spans only 548 genes. Among the 24 genes overlapping the most diverged 1-kb windows, a single annotation cluster (5) of 4 genes shared annotation terms suggesting that they could be members of the immunoglobulin superfamily. Members of this superfamily play important roles in development, and cell–cell adhesion and communication (Vogel et al. 2003). Only two genes in cluster 5 had named homologs in Drosophila (klingon, defective proboscis response 18), and only klingon has a known functional role in neurogenesis, participating in the development of the R7 receptor neuron. Two other candidate genes that were not clustered also potentially participate in neural development on the basis of their Drosophila orthologs: the homeobox gene rough (required in photoreceptors R2 and R5 for inductive interactions in the developing eye) and twisted. Figure 7C shows divergence in the vicinity of AGAP002628, a gene of unknown function in cluster 5.
Of the considerably larger set of genes (11,262) in the collinear (euchromatic) genome, 485 overlapped the top 1% FST windows and were placed into a number of different clusters. Six of the most important annotation clusters are given in Table 1. Interestingly, these clusters and their component genes reflect many of the same functional themes evoked by genes inside the rearrangements: development (including neural development and differentiation in the eye and elsewhere), receptor-mediated signaling, and transcriptional regulation. Clusters 6, 7, and 9 contain genes whose products putatively function as ion channels and receptors, analogous to clusters 3–4 for the 2La rearrangement: genes encoding odorant and gustatory receptors, an insulin-like receptor, a muscarinic acetyl choline receptor, and two transient receptor potential cation channels (trpl and TrpA1 orthologs). Scattered across annotation clusters, or in some cases not clustered, are genes whose Drosophila orthologs are involved in the signaling pathways Wnt (e.g., Wnt 2; frizzled 4), EGF (star), and TGF-B (Smad on X), or genes whose products control response to ecdysone (dopamine/ecdysteroid receptor DopEcR, ecdysone receptor-interacting factor smrter, ecdysone-induced protein 74E eip74E). Of particular note is another chromatin-modifying gene encoding a histone deacetylase (HDAC4).
One intergenic region in the collinear genome captured our attention, because it contains the sole instance of a SNP with FST = 1 between northern and southern populations outside of the two rearranged regions (Figure 7D). Located on chromosome 3R, this SNP is surrounded by SNPs with far lower FST values (not exceeding FST = 0.4) and is in the neighborhood of two genes. It is ∼2 kb upstream of a tRNA(Arg) and ∼4 kb upstream of a zinc finger protein whose predicted Drosophila ortholog earmuff (erm) is involved in the regulation of neurogenesis—a functional theme enriched among the set of genes that overlap the most diverged windows genome-wide. However, it is currently unknown whether this SNP is located in a regulatory region.
Aside from inferences based on functional annotation clustering, we mined a newly available gene expression atlas of sex and tissue specificity in An. gambiae (Baker et al. 2011) for additional clues about the nature of candidate genes. Overall, the 620 candidates were interrogated by 761 probes in the MozAtlas microarray experiments (http://www.tissue-atlas.org). However, gene expression was not detected for almost half of these probes (>57% of those inside rearrangements were not detected as expressed). In Table S3, Table S4, and Table S5, we have indicated which candidate genes had at least one probe whose expression was detected, and of those, which had gender-biased expression or expression limited to one sex and one tissue. Although it is difficult to make robust quantitative statements on the basis of these data, we note a similar level of gender-biased expression as that reported by MozAtlas for the genome as a whole (∼43% of expressed probes), and an intriguing enrichment of sex- and tissue-specific expression (29% of expressed probes), particularly testis-specific expression.
Copy number variation
In addition to nucleotide divergence, we also scanned the genome for potential genic regions of copy-number variation (CNV). Two caveats should be noted about detecting CNVs in our data set. Our data-filtering excluded reads with multiple equally good alignments, a precaution intended to minimize problems in distinguishing orthology from paralogy in repetitive DNA, but also a step that may have removed loci with CNVs. Additionally, if sequence divergence from the reference genome is more extreme for one of the endpoint populations, the overall effect on read mapping might mimic the pattern expected for CNVs (i.e., lower coverage in one population relative to the other), leading to false positives. Bearing in mind these caveats, we identified outlier 1-kb windows in the distribution of normalized read depth coverage (0.5% upper and lower tails) (Figure S2). The set of 415 genes overlapping windows of unusually skewed read coverage were compiled (Table S6) and submitted for functional annotation clustering using DAVID (Table S7). Although the resulting candidate CNV genes do not generally overlap the genes associated with high FST 1-kb windows, similar functional themes emerged from the most important clusters, including transmembrane receptor activity/tyrosine protein kinase signaling, development/morphogenesis, and EGF-responsive genes. In Figure 7E is a representative example from the putative Methuselah receptor 4 gene inside the 2La rearrangement, showing SNP-based divergence and normalized fold coverage differences in both populations. Surprisingly, there is some suggestion of copy-number variation in the penultimate exon of this gene, a result that will require follow-up validation by PCR and sequencing. Considering the CNV gene list as a whole, we noted a significant excess of candidate genes on the X chromosome (139) relative to the autosomes (χ2 = 346.7, d.f. = 1, P-value < 2.2 × 10−16). If this result is largely driven by greater sequence divergence on the X chromosome between the two populations, we might have expected to see an excess of X-linked candidate genes overlapping the top 1% FST windows in the collinear genome, but we did not (χ2 = 1.2, d.f. = 1, P-value = 0.27). Also intriguing is the apparent enrichment of CNV inside the rearranged genome (114 candidate genes) relative to the collinear genome (χ2 = 31.2, d.f. = 1, P-value = 2.35 × 10−8), but in this case we cannot rule out sequence divergence as the driving factor.
Array genotyping vs. population resequencing
In a previous study (White et al. 2007a), we used a gene-based Affymetrix microarray with 25-bp probes to perform single-feature polymorphism (SFP) mapping (e.g., Borevitz et al. 2003; Turner et al. 2005; Turner et al. 2008) of divergence between alternative arrangements of 2La in a single population sample from Cameroon, near the center of the inversion cline. Significantly elevated divergence inside the rearrangement was mapped to two ∼1.5-Mb regions near the inversion breakpoints, a finding broadly consistent with the present resequencing approach (see Figure 6, top). Nevertheless, much detail is missed with microarray-based mapping. Population resequencing revealed 25,915 total SNPs with FST = 1 inside 2La, only 92 (0.36%) of which were interrogated by probes on the microarray. Moreover, of the 92 interrogated SNPs, only 8 (8.7%) were detected as differentiated SFPs on the microarray platform. Of the 84 SNPs that went undetected by the microarray, many may be true negatives in the previous population sampled due to gene flux in the center of the cline. However, 38 of the 84 SNPs fall outside the central (i.e., 6–21 bp) region of the 25-bp probes on the array, and if they escaped detection for this reason, may have instead been false negatives in the earlier study.
Discussion
Population resequencing of the An. gambiae S form along a climatic cline in Cameroon revealed low overall genomic differentiation between populations near its endpoints, a distance of ∼500 km. This is consistent with evidence from microsatellite and mtDNA markers, which indicate little or no population structure in Cameroon (Slotman et al. 2007) and, more generally, only very shallow population structure across the entire African continent (Lehmann et al. 2003) except for that imposed by the Great Rift Valley (Lehmann et al. 1999). Nevertheless, polymorphic chromosomal inversions 2La and 2Rb are nearly fixed at the northern end and virtually absent at the southern end of the Cameroon cline. At the sequence level, alternative arrangements of both chromosomal inversions are strongly differentiated in contrast to most of the collinear genome. Compelling circumstantial evidence from polytene chromosome analysis has long suggested that these chromosomal inversions are targets of selection in An. gambiae, based on their frequency in relation to seasonal, latitudinal, and even microspatial gradients of aridity (Coluzzi et al. 1979; Rishikesh et al. 1985; Petrarca et al. 1990; Powell et al. 1999). By sequencing separately the alternative 2La/a and 2L+a/+a homokaryotypes sampled from a single population near the center of the Cameroon cline, we were able to show that they are not genetically isolated. In collinear genomic regions, SNPs whose frequencies were distinctive at opposite ends of the cline are homogenized in the center, and the same occurs—although to a lesser degree—in rearranged regions of the genome through gene flux in inversion heterozygotes. Without selection acting to maintain the cline, the pattern of genetic differentiation would quickly erode. Taken together, this evidence is the strongest indication to date that spatially varying selection—not demographic history—is responsible for the clinal pattern of genetic differences in An. gambiae from Cameroon.
Judging from the drastically higher levels of divergence in rearranged vs. collinear genomic regions, our findings suggest that inversions play a disproportionate role in ecological adaptation in An. gambiae. This notion is not altogether surprising, when considered in the context of the eponymous An. gambiae complex, a group that includes An. gambiae and at least six other closely related and morphologically indistinguishable African sibling species (Coluzzi et al. 1979; White et al. 2011 for review). Many of these species are thought to have radiated through a process of ecological speciation, driven by larval habitat competition (Costantini et al. 2009; Simard et al. 2009). In this group, more than 120 polymorphic chromosomal inversions and 10 fixed inversions are nonrandomly distributed physically (among the five chromosome arms) and taxonomically (among the member species) (Coluzzi et al. 2002). Chromosome 2R contains 58% (18/31) of the common polymorphic inversions, although it represents <30% of the polytene (euchromatic) complement, while the X chromosome harbors 50% of the fixed inversions (5/10) despite its relatively small (11%) share of the polytene complement. Most species in the complex have relatively limited distributions and little or no inversion polymorphism. The two species that can be considered the most “successful” on the basis of their dominance across much of sub-Saharan Africa—An. gambiae and An. arabiensis—carry an abundance of polymorphic inversions on chromosome 2, although they are distinguished by five fixed inversion differences on the X chromosome (Coluzzi et al. 2002). The polymorphic inversions, some of which are shared through hybridization between An. gambiae and An. arabiensis (see below), are presumed to be responsible for the ecological flexibility of the two species.
Not only in the An. gambiae complex, but also in other major malaria vectors in the same Anopheles subgenus (Cellia), similar nonuniform distributions of fixed and polymorphic inversions are observed (Kitzmiller 1977; Xia et al. 2010; Sharakhova et al. 2011). The enrichment of polymorphic inversions on chromosome 2 in An. gambiae is observed on the homologous chromosome arms in Anopheles stephensi (2R, 3L) and An. funestus (2R, 3R) (Xia et al. 2010; Sharakhova et al. 2011). Even more remarkable, at least some of the independently derived inversions on 2R nonrandomly share common genes between species (this is not true of 2L and its homologs). The colocalization of similar sets of genes inside 2R inversions is not simply due to retention of ancestral gene order, because many of the genes have been extensively reshuffled into new gene combinations in the three lineages (Sharakhov et al. 2002; Sharakhova et al. 2011). Thus, the possibility exists that shared gene content may reflect similar ecological adaptations to common environmental pressures. An. stephensi is an Asian vector that does not occur in Africa. However, An. funestus is second only to An. gambiae as a major vector of malaria in Africa and is sympatric with An. gambiae over much of its continent-wide distribution (Coetzee and Fontenille 2004), including over the wide range of eco-climatic settings in Cameroon (Ayala et al. 2009). Chromosomal inversions in An. funestus are correlated with degree of aridity, just as they are in An. gambiae. Along the same latitudinal gradient in Cameroon featured in the present study of An. gambiae, An. funestus shows analogous clinal patterns of inversion frequency (Cohuet et al. 2005; Ayala et al. 2011). Two recent studies present strong evidence supporting the role of environmental selection in shaping the distribution of An. funestus inversions in Cameroon (Ayala et al. 2011; D. Ayala, R. F. Guerrero, and M. Kirkpatrick, unpublished data). The sequencing and assembly of an An. funestus reference genome in the framework of a larger anopheline sequencing project (Besansky 2008) will provide an unprecedented opportunity for comparative genomics of adaptation along the same environmental gradient in Cameroon.
Kolaczkowski et al. (2011) recently compared D. melanogaster populations sampled from opposite ends of the Australian climatic cline by population resequencing. Like this study, they observed lower heterozygosity at the ends of chromosome arms near telomeres and centromeres, a pattern that presumably reflects perennially reduced recombination in these regions coupled with linked selection (Begun and Aquadro 1992) (note that recombination inside inversions is reduced only in heterokaryotypes, but is normal within arrangement classes). In addition, they observed comparable levels of average genome-wide sequence divergence between populations at the ends of the cline (mean FST based on 1-kb windows = 0.112 in D. melanogaster and 0.123 in An. gambiae excluding rearranged regions). However, a striking contrast between studies is the considerably lower FST in the region spanned by the clinal chromosomal rearrangements In3RP and In2Lt (0.129, 0.116), compared to levels estimated for 2La and 2Rb in An. gambiae (0.247, 0.149). The correspondingly smaller difference in divergence between rearranged and collinear genomic regions in D. melanogaster led the authors to emphasize the genome-wide distribution of candidates for environmental adaptation, whereas in An. gambiae, outliers of divergence are clearly concentrated in the rearranged regions. One logistical difference between studies is that the D. melanogaster populations were not sorted by karyotype prior to sequencing. The pooling of the predominant arrangement together with the less frequent arrangement in both population samples may factor into the lower divergence estimates for rearrangements in Drosophila. However, it seems unlikely that this technical consideration alone can account for the large differences, as can be seen for 2Rb, which was not selected for but showed the expected clinal pattern.
One possible population genetic explanation for differences in the degree of genetic differentiation between collinear and rearranged regions in D. melanogaster and An. gambiae may have to do with the balance between gene flux and the strength of selection maintaining inversions in these species, although little information is available in this regard from either species. All other things being equal, the apparently younger age estimations for inversions in Drosophila (on the order of Ne generations) (Andolfatto et al. 2001) compared to the estimated ages for the 2La and 2Rb inversion polymorphisms in An. gambiae (∼2.6–2.7 Ne) (White et al. 2007a, 2009) should have led to higher rates of divergence in rearranged regions of Drosophila (Feder and Nosil 2009), the opposite of what was observed. Aside from differences in the selection–migration balance, a second explanation seems at least as likely, and has its basis in hybridization and introgression between An. gambiae and An. arabiensis (Besansky et al. 2003). An. arabiensis, hypothesized to be basal in the An. gambiae complex phylogeny (Ayala and Coluzzi 2005), is an arid-adapted species fixed for the 2La arrangement (and polymorphic for 2Rb/+b). An. gambiae is proposed to have arisen more recently in the humid rainforests of Central Africa, with a karyotype similar to present-day rainforest populations (2L+a; 2R+b) (Ayala and Coluzzi 2005). Acquisition of the 2La and 2Rb arrangements by An. gambiae through secondary contact and hybridization with An. arabiensis would have allowed it to considerably expand its range into the arid savannas (Besansky et al. 2003; Ayala and Coluzzi 2005). The codistribution of inversions 2La and 2Rb in contemporary populations of An. gambiae and An. arabiensis across Africa is considered to be the product of at least one interspecific introgression event (White et al. 2007a, 2009). If this interpretation is correct, the relatively high levels of divergence associated with these rearrangements in An. gambiae, in contrast to rearrangements in D. melanogaster, may be due to the fact that 2La and 2Rb were “captured” in their entirety from a different species. The higher divergence observed between alternative arrangements of 2La compared to 2Rb in An. gambiae may also have something to do with An. arabiensis, in that 2La is fixed in that species whereas 2Rb is polymorphic and subject to gene flux. How often ecological adaptation is aided by interspecific transfer of inversions remains to be seen, but a conceptually related process of introgression between Mexican and North American populations of Rhagoletis pomonella may have facilitated the host shift from hawthorn to apple (Feder et al. 2003).
The Australian populations of Drosophila studied by Kolaczkowski et al. (2011) were tropical in the north and temperate in the south, whereas the Cameroon populations of An. gambiae were all tropical, although spanning a steep gradient of aridity. Although the precise combination of selective agents could differ between these two examples, temperature, humidity, and rainfall appear to be important common factors that vary along both clines (Umina et al. 2005). As such, there is an intriguing correspondence of functional themes among candidate genes identified in the two studies, and in some cases orthologous genes are implicated. For example, genes that function in signaling pathways were enriched in both studies. Candidate genes functioning in EGFR, TGF-β, and EcR pathways could influence clinal variation in body size, metabolism, developmental processes, and other life-history traits (Kolaczkowski et al. 2011). Anopheles orthologs of several Drosophila candidate genes in this category were identified. Another striking commonality was the large number of gustatory receptors, ionotropic receptors, and ion-channel-related genes, including cyclic nucleotide-gated channels, implicated in chemo- and thermo-sensation. Finally, both studies identified a number of the same genes implicated in regulation of chromatin and transcription. It will be of great interest to learn if any of these shared candidate Drosophila and Anopheles genes represent parallel adaptive responses to similar environmental selective pressures. Reverse population genomic analyses such as these are powerful for generating hypotheses about the adaptive process. Yet the daunting challenge remains to directly connect these specific candidate genes to traits that vary clinally. Our finding of substantial amounts of gene flux at the center of the cline in Cameroon suggests that one particularly promising approach to infer adaptation at specific loci would be to perform genome-wide association studies at middle latitudes, to test whether SNP variants are associated with traits already implicated in climatic adaptation, such as thermal and desiccation resistance (Gray et al. 2009; Rocca et al. 2009).
Climate strongly influences the genetic constitution of several Drosophila species, such that adaptive polymorphisms (chromosomal inversions) associated with climate have been proposed as tools for monitoring climate change (Umina et al. 2005; Balanya et al. 2009). Similar associations of adaptive polymorphisms and climate exist in anopheline vectors of malaria. However, in the case of these human disease vectors, the adaptive polymorphisms that facilitate survival in otherwise inhospitable territory have public health as well as evolutionary importance, because expansion of the species range or seasonal activity is accompanied by increased malaria transmission. An understanding of the mechanisms underlying adaptation to climate can potentially provide a much-needed new arsenal of tools targeted against vectors.
Supplementary Material
Acknowledgments
We thank D. Ayala for producing Figure 1; I. Lanc and S. Emrich for programming to execute SHRiMP by batch processing via Condor; D. Thain for assistance with storage of large data files; J. Ford for sample pooling and sequencing; the entomology staff at OCEAC for expert technical assistance; and D. Schrider for early contributions to the analysis. C.C. was supported by a Fellowship from the Eck Institute for Global Health and the University of Notre Dame. Funding was provided by the National Institutes of Health grant R01AI076584 (to N.J.B.).
Footnotes
Communicating editor: M. Long
Literature Cited
- Akey J. M., Ruhe A. L., Akey D. T., Wong A. K., Connelly C. F., et al. , 2010. Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. USA 107: 1160–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Depaulis F., Navarro A., 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8 [DOI] [PubMed] [Google Scholar]
- Ayala D., Costantini C., Ose K., Kamdem G. C., Antonio-Nkondjio C., et al. , 2009. Habitat suitability and ecological niche profile of major malaria vectors in Cameroon. Malar. J. 8: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Fontaine M. C., Cohuet A., Fontenille D., Vitalis R., et al. , 2011. Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol. Biol. Evol. 28: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala F. J., Coluzzi M., 2005. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 102(Suppl 1): 6535–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. A., Nolan T., Fischer B., Pinder A., Crisanti A., et al. , 2011. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 12: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanya J., Huey R. B., Gilchrist G. W., Serra L., 2009. The chromosomal polymorphism of Drosophila subobscura: a microevolutionary weapon to monitor global change. Heredity 103: 364–367 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520 [DOI] [PubMed] [Google Scholar]
- Berry A., Kreitman M., 1993. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134: 869–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky N. J., 2008. Genome analysis of vectorial capacity in major Anopheles vectors of malaria parasites, http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/AnophelesGenomesProposal_Aug3.pdf
- Besansky N. J., Krzywinski J., Lehmann T., Simard F., Kern M., et al. , 2003. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100: 10818–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A., Taberlet P., Miaud C., Pompanon F., 2006. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria). Mol. Biol. Evol. 23: 773–783 [DOI] [PubMed] [Google Scholar]
- Borevitz J. O., Liang D., Plouffe D., Chang H. S., Zhu T., et al. , 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M., Fontenille D., 2004. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem. Mol. Biol. 34: 599–605 [DOI] [PubMed] [Google Scholar]
- Cohuet A., Dia I., Simard F., Raymond M., Rousset F., et al. , 2005. Gene flow between chromosomal forms of the malaria vector Anopheles funestus in Cameroon, Central Africa, and its relevance in malaria fighting. Genetics 169: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. H., Mendez M. A., Rasmussen M. O., Mehaffey P. C., Besansky N. J., et al. , 1987. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 37: 37–41 [DOI] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., Petrarca V., Di Deco M. A., 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans. R. Soc. Trop. Med. Hyg. 73: 483–497 [DOI] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., Della Torre A., Di Deco M. A., Petrarca V., 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418 [DOI] [PubMed] [Google Scholar]
- Costantini C., Ayala D., Guelbeogo W. M., Pombi M., Some C. Y., et al. , 2009. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 9: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong G., Bochdanovits Z., 2003. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J. Genet. 82: 207–223 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1947. Adaptive changes induced by natural selection in wild populations of Drosophila. Evolution 1: 1–16 [Google Scholar]
- Ellison C. E., Hall C., Kowbel D., Welch J., Brem R. B., et al. , 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. USA 108: 2831–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler J. A., 1977. Geographic variation, speciation, and clines. Monogr. Popul. Biol. 10: 1–246 [PubMed] [Google Scholar]
- Feder J. L., Nosil P., 2009. Chromosomal inversions and species differences: When are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution 63: 3061–3075 [DOI] [PubMed] [Google Scholar]
- Feder J. L., Berlocher S. H., Roethele J. B., Dambroski H., Smith J. J., et al. , 2003. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA 100: 10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik A., Schlotterer C., 2010. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics 186: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. G., 2002. Water balance in desert Drosophila: lessons from non-charismatic microfauna. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133: 781–789 [DOI] [PubMed] [Google Scholar]
- Gillies M. T., De Meillon B., 1968. The Anophelinae of Africa South of the Sahara. South African Institute for Medical Research, Johannesburg, South Africa [Google Scholar]
- Gray E. M., Rocca K. A., Costantini C., Besansky N. J., 2009. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar. J. 8: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Weeks A. R., 2007. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129: 133–147 [DOI] [PubMed] [Google Scholar]
- Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., et al. , 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Teo Y. Y., Li H., Nair S., Dougan G., et al. , 2009. Detecting SNPs and estimating allele frequencies in clonal bacterial populations by sequencing pooled DNA. Bioinformatics 25: 2074–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R. A., Subramanian G. M., Halpern A., Sutton G. G., Charlab R., et al. , 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149 [DOI] [PubMed] [Google Scholar]
- Hosack D. A., Dennis G., Jr, Sherman B. T., Lane H. C., Lempicki R. A., 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Collins J. R., Alvord W. G., et al. , 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P., 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington W. J., Gockel J., Partridge L., 2003. Testing for asymmetrical gene flow in a Drosophila melanogaster body-size cline. Genetics 165: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmiller J. B., 1977. Chromosomal differences among species of Anopheles mosquitoes. Mosquito Systematics 9: 112–122 [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas C. B., Powell J. R., 1992. Introduction, pp. 1–52 Drosophila Inversion Polymorphism, edited by Krimbas C. B., Powell J. R. CRC Press, Boca Raton, FL [Google Scholar]
- Laayouni H., Hasson E., Santos M., Fontdevila A., 2003. The evolutionary history of Drosophila buzzatii, XXXV: inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol. Biol. Evol. 20: 931–944 [DOI] [PubMed] [Google Scholar]
- Lawson D., Arensburger P., Atkinson P., Besansky N. J., Bruggner R. V., et al. , 2009. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 37: D583–D587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Cornel A. J., Meneses C. R., Fofana A., Andrianarivo A. G., et al. , 2009. Ecological and genetic relationships of the Forest-M form among chromosomal and molecular forms of the malaria vector Anopheles gambiae sensu stricto. Malar. J. 8: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T., Hawley W. A., Grebert H., Danga M., Atieli F., et al. , 1999. The Rift Valley complex as a barrier to gene flow for Anopheles gambiae in Kenya. J. Hered. 90: 613–621 [DOI] [PubMed] [Google Scholar]
- Lehmann T., Licht M., Elissa N., Maega B. T., Chimumbwa J. M., et al. , 2003. Population structure of Anopheles gambiae in Africa. J. Hered. 94: 133–147 [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Costello J. C., Holloway A. K., Hahn M. W., 2008. “Reverse ecology” and the power of population genomics. Evolution 62: 2984–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo N. F., Sangare D. M., Regier A. A., Reidenbach K. R., Bretz D. A., et al. , 2010. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar. J. 9: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C. A., Haselkorn T. S., Noor M. A., 2007. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics 175: 1289–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Betran E., Barbadilla A., Ruiz A., 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146: 695–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrarca V., Sabatinelli G., Di Deco M. A., Papakay M., 1990. The Anopheles gambiae complex in the Federal Islamic Republic of Comoros (Indian Ocean): some cytogenetic and biometric data. Parassitologia 32: 371–380 [PubMed] [Google Scholar]
- Powell J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, Oxford [Google Scholar]
- Powell J. R., Petrarca V., della Torre A., Caccone A., Coluzzi M., 1999. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia 41: 101–113 [PubMed] [Google Scholar]
- Regnard C., Straub T., Mitterweger A., Dahlsveen I. K., Fabian V., et al. , 2011. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishikesh N., Di Deco M. A., Petrarca V., Coluzzi M., 1985. Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Trop. 42: 165–170 [PubMed] [Google Scholar]
- Rocca K. A., Gray E. M., Costantini C., Besansky N. J., 2009. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar. J. 8: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble S. M., Lacroute P., Dalca A. V., Fiume M., Sidow A., et al. , 2009. SHRiMP: accurate mapping of short color-space reads. PLOS Comput. Biol. 5: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F., Della Torre A., Caccone A., 2004. Short report: a new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am. J. Trop. Med. Hyg. 70: 604–606 [PubMed] [Google Scholar]
- Schaeffer S. W., 2008. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution 62: 3082–3099 [DOI] [PubMed] [Google Scholar]
- Schaeffer S. W., Anderson W. W., 2005. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics 171: 1729–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhov I. V., Serazin A. C., Grushko O. G., Dana A., Lobo N., et al. , 2002. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science 298: 182–185 [DOI] [PubMed] [Google Scholar]
- Sharakhova M. V., George P., Brusentsova I. V., Leman S. C., Bailey J. A., et al. , 2010. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics 11: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova M. V., Xia A., Leman S. C., Sharakhov I. V., 2011. Arm-specific dynamics of chromosome evolution in malaria mosquitoes. BMC Evol. Biol. 11: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F., Ayala D., Kamdem G. C., Etouna J., Ose K., et al. , 2009. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M. A., Tripet F., Cornel A. J., Meneses C. R., Lee Y., et al. , 2007. Evidence for subdivision within the M molecular form of Anopheles gambiae. Mol. Ecol. 16: 639–649 [DOI] [PubMed] [Google Scholar]
- Storz J. F., 2005. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 14: 671–688 [DOI] [PubMed] [Google Scholar]
- Toure Y. T., Petrarca V., Traore S. F., Coulibaly A., Maiga H. M., et al. , 1998. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia 40: 477–511 [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42: 260–263 [DOI] [PubMed] [Google Scholar]
- Umina P. A., Weeks A. R., Kearney M. R., McKechnie S. W., Hoffmann A. A., 2005. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308: 691–693 [DOI] [PubMed] [Google Scholar]
- Vogel C., Teichmann S. A., Chothia C., 2003. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development 130: 6317–6328 [DOI] [PubMed] [Google Scholar]
- White B. J., Hahn M. W., Pombi M., Cassone B. J., Lobo N. F., et al. , 2007a Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PLoS Genet. 3: e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Santolamazza F., Kamau L., Pombi M., Grushko O., et al. , 2007b Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am. J. Trop. Med. Hyg. 76: 334–339 [PubMed] [Google Scholar]
- White B. J., Cheng C., Sangare D., Lobo N. F., Collins F. H., et al. , 2009. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics 183: 275–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Collins F. H., Besansky N. J., 2011. Evolution of Anopheles gambiae in relation to humans and malaria. Annu. Rev. Ecol. Evol. Syst. 42: (in press). [Google Scholar]
- Xia A., Sharakhova M. V., Leman S. C., Tu Z., Bailey J. A., et al. , 2010. Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS ONE 5: e10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.