Abstract
The joint consequences of inbreeding, natural selection, and deleterious mutation on mean fitness after population shrinkage are of great importance in evolution and can be critical to the conservation of endangered populations. I present simple analytical equations that predict these consequences, improving and extending a previous heuristic treatment. Purge is defined as the “extra” selection induced by inbreeding, due to the “extra” fitness disadvantage (2d) of homozygotes for (partially) recessive deleterious alleles. Its effect is accounted for by using, instead of the classical inbreeding coefficient f, a purged inbreeding coefficient g that is weighed by the reduction of the frequency of deleterious alleles caused by purging. When the effective size of a large population is reduced to a smaller stable value N (with Nd ≥ 1), the purged inbreeding coefficient after t generations can be predicted as gt ≈ [(1 – 1/2N) gt-1 + 1/2N](1 – 2d ft-1), showing how purging acts upon previously accumulated inbreeding and how its efficiency increases with N. This implies an early fitness decay, followed by some recovery. During this process, the inbreeding depression rate shifts from its ancestral value (δ) to that of the mutation–selection–drift balance corresponding to N (δ*), and standard selection cancels out the inbreeding depression ascribed to δ*. Therefore, purge and inbreeding operate only upon the remaining δ − δ*. The method is applied to the conservation strategy in which family contributions to the breeding pool are equal and is extended to make use of genealogical information. All these predictions are checked using computer simulation.
NATURAL populations of diploid sexual species usually harbor an important genetic load concealed in heterozygosis, which is mainly due to the poor efficiency of natural selection against partially recessive deleterious mutations segregating at low frequencies. Due to the increased expression of recessive effects in homozygosis, this concealed load is expected to be exposed by inbreeding, accounting for most of the inbreeding depression observed for fitness (Crow 2008). However, this increased expression of (partially) recessive deleterious effects also causes an increase of natural selection, known as purge, which limits the actual reduction in mean fitness (see reviews by Crnokrak and Barrett 2002; Keller and Waller 2002; Leberg and Firmin 2007). The fitness reduction resulting from inbreeding and purging is of major importance in population and evolutionary genetics, as well as in their application to conservation and animal breeding.
In a previous article (García-Dorado 2008), I presented a simple heuristic approach to predicting fitness evolution in a panmictic population after a stable reduction in size. This relied upon an inbreeding coefficient gt that was corrected for purging. Here I give a more precise interpretation for gt, which is more meaningful, and I formally deduce a new approximated expression for its evolution that provides more accurate predictions, clearly stating the underlying assumptions of the model. These predictions account for some previously unexplained reversals of the early fitness decline, observed in both simulations (Perez-Figueroa et al. 2009) and experimental studies (Crnokrak and Barrett 2002; Larsen et al. 2011). I then describe an approach that predicts the evolution of mean fitness expected from the joint effect of inbreeding, mutation, and selection, as the population approaches the corresponding mutation–selection–drift (MSD) balance.
This method also provides approximate predictions for the evolution of the concealed load (i.e., of the inbreeding depression rate), and it can be used under varying population sizes as well as under equal family contributions, which extends its practical interest in conservation programs. Furthermore, I adapt these predictive expressions so that they can be applied to genealogical information.
Analytical Predictions
The inbreeding-purge (IP) model
Assume a large ancestral panmictic population with segregating deleterious alleles that are recessive to some extent. The load concealed in the heterozygous condition contributed by these alleles can be quantified by the rate δ at which fitness (w) would be expected to decline, in the absence of selection, for increasing values of Wright’s inbreeding coefficient f. In this section we derive predictions for the evolution of fitness expected from the expression of this load after the effective size of this population shrinks to a new value N, more or less stable but much smaller than the ancestral size.
We first consider a single locus where a deleterious allele m is segregating with small frequency q0 in this ancestral population. The relative fitness for the wild homozygous (++), the heterozygous (+m), and the mutant homozygous (mm) genotypes are 1, 1 − hs, and 1 − s, respectively. Thus, s is the selection coefficient against mm homozygotes, and h is the coefficient of dominance, which represents the proportion of s that is expressed in the heterozygous condition, so that h = 0.5 implies additive gene action. We define d = s(1 − 2h)/2 as half the excess of the selection coefficient against mm homozygotes (s) over its expected value inferred from the heterozygote disadvantage on an additive hypothesis (2hs). Thus, the fitness of mm is 1-2hs - 2d. Note that d equals the heterozygous value for relative fitness in Falconer’s scale (Falconer and Mackay 1996, p. 109). It is not necessary that this ancestral population is at the MSD balance, but we assume that it has been under continued natural selection, so that the larger is hs, the smaller is the expected q0, implying fitness average w0 ≈1.
For this one-locus model, under relaxed selection, fitness would be expected to decline linearly on f with a small slope δ = 2dp0q0 (Falconer and Mackay 1996, p. 249, Equation 14.2), so that w = w0 − δ f ≈ w0 exp[−δ f]. Considering that fitness is an essentially multiplicative trait, the exponential expression holds for a multilocus model, where the inbreeding depression rate δ is the sum of the δ values of the individual loci, and can be roughly interpreted as the percentage reduction in fitness that would be expected, in the absence of selection, per 1% increases of f.
Now assume that, at generation t = 0, this population undergoes a reduction in size and, thereafter, remains small and panmictic. To account for the evolution expected for the properties of this shrunk population, we consider it as one of the infinite subpopulations in the classical Wright’s “subdivided population” model, where we assume that the proportion of homozygotes by descent (ft) increases at the same rate as in the absence of selection (Crow and Kimura, 1970, p. 101, Equation 3.11.1). Thus, two copies of a gene, sampled at generation t from the same ith subpopulation, have a probability fit of being identical by descent and, then, a probability qit of producing an mm homozygote (where qit is the frequency of m in subpopulation i at generation t). Note that, due to natural selection, the average frequency of m over subpopulations after t generations of reduced size will be smaller than in the ancestral population (E(qt) < q0, where E stands for expected value over subpopulations). Those two gene copies also have a small probability ζt of not being identical by descent and still produce an mm homozygote. Therefore, the overall proportion of mm homozygotes at generation t is E(qt2) = E(qt ft) + ζt . Then, since q0 is small, it can be shown (see Appendix A) that the expected reduction in average fitness (w) after t generations is
| (1) |
The purged inbreeding coefficient is defined as
| (2) |
so that, for q0 so small that δ ≈ 2dq0, the inbreeding depression contributed by the locus can be predicted by using gt instead of ft into the standard neutral prediction, giving
| (3) |
Therefore, assuming multiplicative fitness and equivalent loci contributing an overall inbreeding depression rate δ, the expected overall fitness is
| (4) |
If deleterious effects vary among mutations, this prediction should be averaged over the corresponding joint distribution for s and h.
The evolution of gt is due to the increase of ft caused by inbreeding and to the reduction of qt induced by selection. As shown in Appendix A, if, in a given subpopulation i, the frequency of a deleterious allele at generation t before selection is qit, then the frequency expected after natural selection can be expressed as
| (5) |
In Equation 5, the first term within brackets (sh pitqit) accounts for the reduction in frequency that natural selection would cause for an additive allele with deleterious effect hs in the heterozygote and 2hs in the homozygote, and the second term (2dqit2) represents the “extra” reduction of frequency expected from the “extra” selection coefficient against mm homozygotes (2d), which is due to nonadditive gene action.
I use the term “purge” to denote the increase of natural selection caused by inbreeding. Purge is due to the increase of the frequency of mm homozygotes for (partially) recessive deleterious alleles, and it occurs because these homozygotes have a 2d extra selection coefficient.
In this section I consider that the reduction of the effective size is so drastic that it causes a relatively rapid increase in the frequency of mm homozygotes, due to inbreeding. Therefore, after size reduction, purge is the main selective factor against the deleterious alleles segregating in the ancestral population. Thus, neglecting ζt (see Appendix A, Equation A3), we obtain the approximated expression
| (6) |
Then, a new generation is produced through panmictic mating, for which gi t+1 = E(ft+1 qt+1)/q0, so that, as shown in Appendix A, gt can be predicted as
| (7a) |
This prediction also holds if the effective population size varies through generations (t), as long as Nt is always large enough that the fixation of the deleterious alleles contributing to the ancestral concealed load can be disregarded. Then,
| (7b) |
Thus, the evolution of fitness can be approximately predicted using gt in Equations 3–4. Equations 7 show that purge cannot occur until some inbreeding has accumulated and formally describes how it increases with the probability of homozygosis by descent (ft-1).
For increasing t, ft approaches 1, and gt approaches an asymptotic value that implies gt+1 = gt, i.e.,
| (8) |
These results do not coincide with the approximate predictions in García-Dorado (2008), where gt was easily solved for t; see Equation 1 in García-Dorado (2008), or the expression given a few lines below it. The reason is that those predictions were based on the intuitive argument that the frequency of mm homozygotes undergoes each generation a reduction by a (1 − d) factor due to purge and an increase at a rate 1/2N due to the finite population size. However, although the direct target of purging selection at generation t is the frequency of mm homozygotes, the purged inbreeding coefficient at generation t + 1 does not depend upon that frequency, but upon the average coancestry between individuals of generation t. Thus, purging is better understood by considering the reduction it causes on the deleterious frequency. Notwithstanding, as pointed out below, equations in García-Dorado (2008) often give reasonably approximated predictions.
Finally, as inbreeding progresses, the ancestral concealed load is exhausted. I denote by δ0t the inbreeding depression rate at generation t that is expected from deleterious alleles segregating in the ancestral population. As derived in Appendix A, this is approximately given by
| (9) |
instead of by the neutral prediction
| (10) |
If the new effective population size is very small, no substantial concealed load accumulates from new mutations arisen through the process, so that Equation 9 approximately describes the evolution of the inbreeding depression rate.
In practice, the deleterious effect (s, h, and, therefore, d) is different for different mutations. Therefore, the above predictions should be averaged over the joint distribution of s and h. However, I explore the reliability of predictions obtained by using a single empirical d value, which I denote effective purging coefficient de. Here, de will be the d value that, when used in Equations 7, produces the same long-term IP prediction that is obtained by averaging Equations 7 predictions over the joint distribution of s and h. The reliability of these predictions is of great relevance, because it is more feasible to obtain estimates for de (see below) than for the joint distribution of s and h.
Computing purged inbreeding from genealogical information
The above model can also be used to predict the consequences of purging from genealogical information. Consider an individual X that is sampled from the population at a given generation before natural selection acts. Let A and B be the parents of X, C and D the parents of A, and E and H the parents of B. The inbreeding and the purged inbreeding coefficients expected for X are f(X) and g(X), and the coancestry and the purged coancestry coefficients between individuals A and B, which are sampled at their corresponding generation after natural selection, are c(AB) and γ(AB), respectively.
Equation 7 implies that the purged inbreeding coefficient increases each generation at the same rate as Wright’s inbreeding coefficient, but is reduced by purging at the same rate 2dft as qt (see Equation 6). In genealogical terms, this yields
Since purging acts upon the accumulated inbreeding, we have
and, since purge acts upon the inbreeding coefficient of A and B arising from the coancestry between their parents, for A ≠ B
The recurrent use of these expressions gives the purged inbreeding coefficient for the individuals in the genealogy, which in practice requires an appropriate estimate of de.
The full model (FM) with inbreeding, purge, mutation, and standard selection
We now consider an ancestral panmictic population at the MSD balance, with inbreeding depression rate δ, whose effective size is reduced at generation 0 to a new stable arbitrary value N, after which it shifts toward a new MSD balance with inbreeding depression rate δ*. Here, for any population size, I define standard selection as the amount of selection that would cancel out the per-generation fitness decline from inbreeding depression (δ*/2N) and from new deleterious mutations at the corresponding MSD balance. If the new effective size N is relatively large, the new MSD balance can conceal a relevant load in the heterozygous condition (i.e., δ* can be relevant), and standard selection can be important. In fact, under nonadditive gene action, a reduction in population size can lead to a new MSD balance with a larger additive variance for fitness (García-Dorado 2003; Perez-Figueroa et al. 2009), where standard selection is more intense than in the ancestral larger population.
During the shift toward the new balance, the ancestral inbreeding depression rate that is exhausted by inbreeding and purge (Equation 9) is partly replaced with concealed load arising from new deleterious mutations, so that the inbreeding depression rate expected in the subpopulation at generation t is
As illustrated below using simulation, this expression improves Equation 19 in García-Dorado (2007), which assumed that δ was replaced with δ* at a neutral rate ft. Rearranging the above expression, we can write
| (11) |
The δ* value at the new MSD balance (as well as in the ancestral one) can be computed to a good approximation as
| (12) |
where
is the proportion of deleterious copies that undergo selection in the homozygous conditions at the MSD balance (i.e., K = E(q2)/E(q)), and where λ is the rate of deleterious mutation (assumed non recurrent) per gamete and generation (García-Dorado 2007). In Equation 12, the three terms in the denominator account, respectively, for the rate at which heterozygosity is lost at the MSD balance due to: (i) drift (1/2N); (ii) standard selection, as would operate against an additive deleterious allele that determines disadvantage hs in heterozygosis and 2hs in homozygosis; and (iii) standard selection ascribed to dominance (2dK).
To understand the role of standard selection after a stable reduction of population size occurring at t = 0, it is useful to consider the particular case in which the new size N equals the ancestral one, so that no inbreeding depression is expected despite how large gt might become. The reason is that, from generation t = 0 onward, standard selection cancels out the inbreeding depression expected from δ*, as it will in the new MSD balance. Since, in this particular case, δ* = δ, this implies that standard selection cancels out the whole inbreeding depression. Therefore, when using gt to predict the evolution of fitness, we should discount the inbreeding depression ascribed to δ* to compensate for having ignored nonpurging selection in the prediction of gt (see Appendix A, where selection upon 2d for the ζ noninbred fraction of mm homozygotes, as well as that upon sh, are neglected to derive Equation A3).
Therefore, to account for standard selection during the shift process, we consider that the net fitness reduction caused by inbreeding occurs at the expense of (δ − δ*), which, according to Equation 11, represents the net inbreeding depression rate that is being exhausted by inbreeding and purge. Thus, letting aside for the moment the fitness loss ascribed to fixation of new deleterious mutation, the net consequences of inbreeding, purge, standard selection, and new mutation during the approach to the new MSD balance can be predicted as wt ≈ w0 exp[– (δ − δ*) gt] (or as wt ≈ w0 – (δ − δ*) gt for a single locus). Below I use computer simulation to check the validity of this heuristic expression.
In addition, at the MSD balance, standard selection does not cancel out the whole fitness decline from inbreeding depression and mutational deleterious input, as there is some rate Dm of fitness decline due to deleterious fixation. While the population shifts from the ancestral balance to the new one, the rate of fitness decline from deleterious fixation also increases from its ancestral value to the new one. Here we assume that the ancestral population was so large that its rate of fitness decline was negligible. Thus, the expected fitness decline accumulated at generation t from fixation of new deleterious mutation can be approximated as (t − 2Nft)Dm (García-Dorado 2008). Thus, taking into account inbreeding and purge, standard selection, new mutation, and drift, the mean fitness expected at generation t is, approximately,
| (13) |
for a single locus (using a per-locus Dm), or
| (14) |
for overall fitness. This expression replaces Equation 5 in García-Dorado (2008). At the new MSD balance, assuming constant deleterious effects across loci and ignoring reverted or beneficial mutation, the overall Dm can be predicted as 2N λUs, where U is the probability of final fixation for a new deleterious mutation, which can be computed from standard diffusion theory (Kimura 1969). This equation is valid no matter how small the reduction in population size, predicting no inbreeding depression for the particular case when N equals the ancestral size, even though gt could reach appreciable values if the ancestral population was relatively small.
The above approach also applies when Nt shows some variation between generations, under the condition settled above for Equation 7b. Then, the prediction of wt should be iterated each generation. For one locus,
| (15) |
where δt* and Dmt are the δ * and Dm values at the MSD balance corresponding to Nt. Similarly, considering all the loci over the whole genome, we have
| (16) |
However, if Nt is always small enough that (δ − δt*) ≈ δ, Equations 3 (one locus) or 4 (overall fitness) provide good approximations while fixation of new deleterious mutation can be ignored (either because we are interested in short-term predictions or because the strength of selection is large compared to drift, i.e., Ns >>1).
Predictions under equal family contributions (EFC)
The evolution of fecundity and viability when, after size reduction, the population is maintained with equal family contributions (EFC), was also considered by García-Dorado (2008). In conservation management, EFC is often recommended (Frankham et al. 2002) because, compared to no management (NM) it roughly doubles the effective population size (Wright 1938).
For fecundity, EFC prevents natural selection, as far as it is perfectly accomplished. The evolution of mean fecundity (ϕ) was predicted by Equation 28 in García-Dorado 2008 (which contains a misprint) and 2007, and can be rewritten as
| (17) |
where the mutation rates and effects refer to fecundity, subscript E stands for EFC (i.e., and δN is the expected rate of inbreeding depression for fecundity at the neutral mutation–drift balance that will be attained under EFC (δN = 4Nλs(1 − 2h)). Since purging does not occur, those predictions remain appropriate.
For viability, EFC reduces selection to that occurring within full-sib families, which uses half the viability additive variance (Falconer and Mackay 1996, p. 232). Therefore, the “effective” selection coefficient s determining the evolution of gene frequency (and, therefore, the d value determining purge) is reduced by a factor. Thus, the evolution of mean viability (V) under inbreeding depends on the purged inbreeding coefficient corresponding to EFC,
| (18) |
where d refers to viability.
As in the NM case, the population shifts toward a new MSD balance, and inbreeding depression occurs at the expense of the net inbreeding depression rate that is being exhausted by inbreeding and purge, which can be computed as under NM (i.e., as δ − δ*; see Appendix B).
In addition, due to the cumulative fixation of deleterious mutations newly arisen through the shift process, the mean viability at generation t is expected to be reduced by a factor Exp[− (t − 4NfEt)DmE] (García-Dorado 2007, 2008), where DmE is the rate of mean viability decline due to deleterious fixation at the new MSD balance . Note that, since EFC doubles the effective population size and reduces the effective deleterious effects for viability by a factor, the effective value of the Ns parameter (which is critical to determine the deleterious fixation rate; Kimura 1969) is increased by a factor. This implies that, for any given population size, the rate of viability decline from deleterious fixation at the MSD balance is smaller under EFC than under NM (DmE < Dm).On the contrary, at this new MSD balance, the expressed segregating load (Ls) is larger than under NM, due to partially relaxed selection (see Appendix B).
Putting all this together, Appendix B shows that, after a reduction in size such that NhsfEt is small, the expected viability under EFC is
| (19) |
where mutational rates and effects refer to viability and where the second term in the exponent accounts for the increase of segregating load for h > 0. Thus, Equation 19 replaces Equation 8 in García-Dorado (2008). However, as larger NhsfEt values are considered, this prediction is bounded by the condition
| (20) |
which takes into account that, for increasing N values, the segregating load in the new MSD balance is bounded by the value corresponding to the mutation–selection (MS) balance (see Appendix B).
During the initial period, while the term on Dm can be neglected in Equations 13 or 14, DmE can also be neglected in Equations 19 and 20. Therefore, if the reduction in population size was so drastic that (δ − δE*) ≈ δ, and if the accumulated segregating load can be neglected (small values for λ or for NλhsfEt), the evolution of mean viability under EFC can be roughly predicted by
| (21) |
or by its corresponding linear one-locus version when appropriate.
Simulation Methods
One-locus case
To explore the conditions for the validity of the above predictions, I first obtain simulation results for a single biallelic locus under recurrent deleterious mutation. I simulate populations of size N sampled from an ancestral population at the MS balance (initial frequency q0 computed from Crow and Kimura 1970, Equation 6.2.6, p. 260). At each generation t, individuals are formed with alleles randomly sampled from individuals that survived to selection in the previous generation t − 1. Each individual obtained this way survives with probability equal to its genotypic fitness, and the process is repeated until N surviving individuals are obtained. Then, each wild allele mutates to the deleterious allele with probability μ. The fitness average and the inbreeding depression rate are computed each generation before selection. Genealogies are recorded. A large number of replicates r are simulated (r × N > 100000).
Simulation results are compared to one-locus predictions from the IP (Equation 3) model and full model (FM, Equation 13). Predictions obtained using gt values computed both from Equation 7a and from genealogies were always so similar that they cannot be visually distinguished in the graphics. The ancestral inbreeding depression rate (δ) is computed as 2d(1 − q0) q0, and δ* is computed from Equation 12 using μ instead of λ. Although Equation 12 was derived under a nonrecurrent mutation model with unlimited potentially mutating sites (García-Dorado 2007), it also gives good predictions for the MSD balance under the recurrent mutation model, as far as the expected per-locus heterozygosity is small, as in the cases simulated here. Dm is numerically computed from standard diffusion theory (Kimura 1969). For comparison, we also give predictions obtained using, into Equation 3, gt values computed from Equations 1 and 2 in García-Dorado (2008). Neutral predictions are obtained substituting ft instead of gt into Equation 3.
The fraction of the ancestral inbreeding depression rate that is left at generation t (δ0t), is predicted taking purging into account (Equation 9), and also under the neutral hypothesis (Equation 10). The overall inbreeding depression rate expected at generation t (δt), taking into account overall selection and new mutation, is computed from Equation 11.
Simulation results are also obtained for populations that, after a number of generations with reduced size N1, undergo a single additional change in size to a new stable value N2. Results are compared to predictions from Equations 3 (IP predictions) and 15 (FM predictions), computing gt from Equation 7b.
Finally, we simulate the evolution of mean viability under EFC. In this case, the shrunk population consisted each generation of N/2 full-sib families. For each family, offspring were randomly sampled and tested for survival, until a surviving male and female were obtained to form the panmictic breeding pool. Results were compared to IP and FM predictions (gt computed from Equation 18 substituted into the linear one-locus versions of Equation 21 and of Equation 19 bounded with Equation 20, respectively), and to neutral predictions obtained using fEt instead of gEt into the linear version of Equation 21.
Multilocus case
The genome-wide predictions were also checked against simulation results published by Pérez-Figueroa et al. (2009) and against the EFC–NM ones published by Fernández and Caballero (2001). These results were obtained using simulation programs that basically conform to the model analyzed here, although deleterious mutation was simulated as recurrent by Fernández and Caballero and as nonrecurrent by Pérez-Figueroa et al. In both cases, the homozygous deleterious effect s was assumed to be gamma distributed, and h was uniformly distributed between 0 and exp[ –ks], with k chosen to produce the desired expected value (E(h)). Loci were unlinked, fitness was multiplicative across loci, and only nonepistatic results are used from Pérez-Figueroa et al.
To compute overall predictions, we sample large numbers of mutational effects (s, h) from the appropriate distribution, compute predictions for each, and average them over the sample. For each (s, h) pair, the ancestral population was assumed to be at the MSD balance for the appropriate effective population size (103 in the case of Perez-Figueroa et al. results, sizes specified by García-Dorado (2008) in the case of Fernández and Caballero results). Under NM, IP predictions are obtained from Equation 4 and FM ones from Equation 14, with gt from Equation 7a. Neutral predictions were obtained from Equation 4 using ft instead of gt. Under EFC, a similar procedure was followed for viability, for which gEt was computed from Equation 18, IP predictions were obtained from Equation 21, FM predictions from Equation 19 were bounded by Equation 20, and neutral predictions were obtained using fEt instead of gEt into Equation 21.
Results
One-locus results
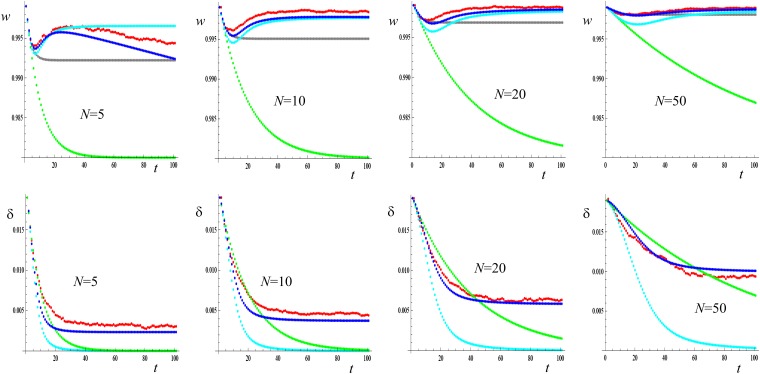
Figure 1 shows results over 100 generations after the effective size of a large population shrinks to a smaller stable N value. They correspond to a single locus where deleterious mutations of constant effect (s = 0.4, h = 0, d = 0.2) continuously occur at a rate μ = 0.001. Mean fitness initially decays at a slowing rate, subsequently recovering to values that, except for very small N, are very close to the ancestral average.
Figure 1 .
One-locus results after reduction of the effective size to different stable values N starting from a population at the MS balance. s = 0.4, h = 0, μ=0.001. Upper plates: Evolution of mean fitness. Lower plates: Evolution of the inbreeding depression rate. Red: simulation results; gray: predictions accounting for inbreeding and purge according to García-Dorado 2008; light blue: IP predictions; deep blue: FM predictions; green: neutral predictions computed ignoring new mutation.
Mean fitness predicted using the IP approach fits quite closely the simulated results although, as expected, it is conservatively smaller until fixation of new deleterious mutations becomes relevant. Analogous purge-inbreeding predictions from the older approach (García-Dorado 2008) also provide a reasonable approximation in the short term or for large N values, but they fail to account for the fitness recovery. Therefore, they can induce substantial bias in the medium-long term for small populations. Both approaches predict that purge can be important and that it is more efficient for larger populations where, of course, both inbreeding and purge proceed more slowly.
The accuracy of FM predictions improves that of IP ones. In the short term, FM produces a smaller downward bias than IP, because it takes into account the fraction of the inbreeding depression rate that is canceled out by standard selection. In the long term, FM predictions reduce the upward bias that can arise for IP predictions due to fixation of new deleterious mutations. In our case, this decline is relevant just for the smallest size considered. In that particular case (N = 5), FM predicts a too-large long-term decline, probably because we assume nonrecurrent mutation to predict Dm, while mutation is recurrent in this one-locus simulation program. Apart from that, for these cases, FM seems to account for all major genetic processes determining the evolution of mean fitness, even for very small populations.
The genealogical approach gives means for the standard inbreeding coefficient (ft) that are virtually identical to those expected from N, showing that selection upon a single locus starting at the MS balance is too weak to affect genealogies. The averages of the purged inbreeding coefficient (gt) estimated from the genealogies of simulation results are also virtually identical to those expected from N, and the corresponding predictions for the evolution of mean fitness are indistinguishable at the graphical level.
Similar results are obtained for other values of s and h (not shown), as far as Nd ≥ 1. For Nd < 1, however, the smaller Nd, the more similar the fitness reduction to the corresponding neutral prediction. If Ns is also small, the main determinant of fitness decline in the long term (say, t > N) is the rate of fixation of new deleterious mutation.
Figure 1 also shows that, due to purging and despite new mutation, δt initially declines faster than predicted from the increase in ft. Equation 11 provides reasonable prediction for δt, which approaches the δ* value corresponding to the new MSD balance (Equation 12). The larger N, the smaller (δ − δ*), so that the maximum depression predicted by IP becomes considerably larger than observed or than predicted by FM.
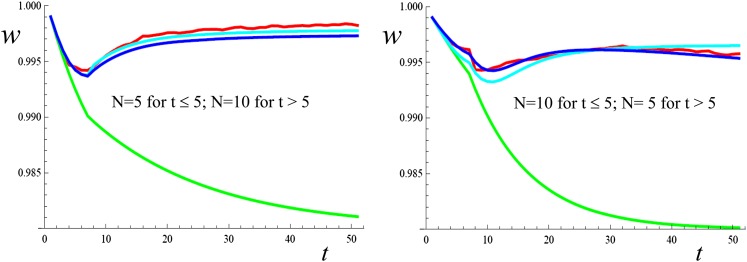
Figure 2 gives similar results for two cases where, at t = 5, N changes to a new value, also much smaller than the ancestral one. These results reasonably fit the corresponding predictions. However, the conditions for the reliability of predictions under variable N should be further explored.
Figure 2 .
One-locus mean fitness evolution after reduction of the effective sizes to a variable Nt value starting from a population at the MS balance. s = 0.4, h = 0, μ=0.001. Red: simulation results; green: neutral predictions computed ignoring new mutation; light blue: IP predictions; deep blue: FM predictions.
Multilocus results
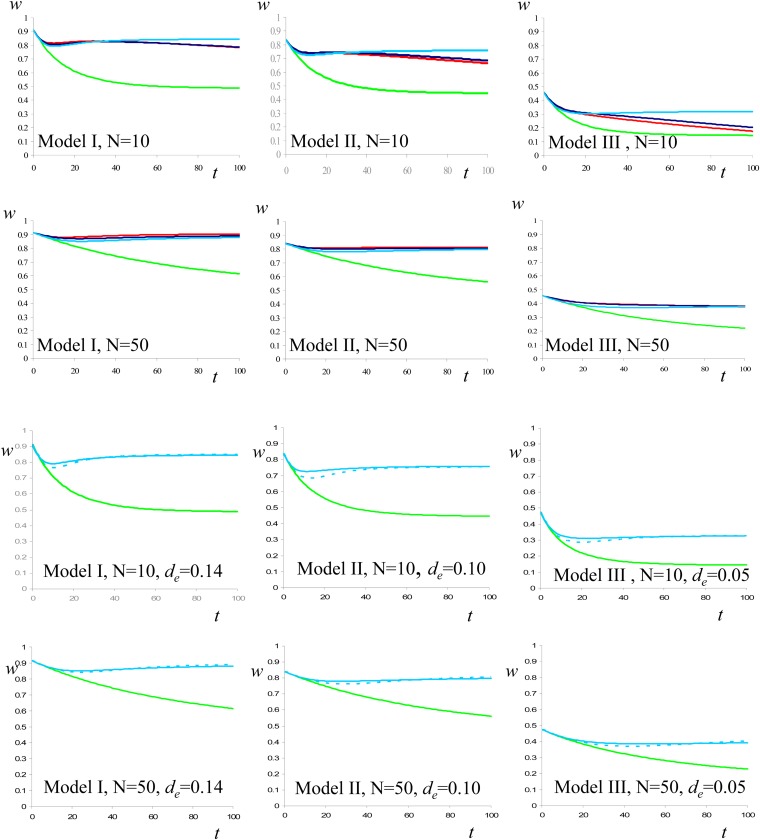
Figure 3 (top six graphs) gives the evolution of mean fitness after the effective size of a large population shrinks to a smaller stable value (N = 10 or N = 50). Three very different mutational models are considered, always assuming nonrecurrent mutation over the genome, with randomly variable deleterious effect (Perez-Figueroa et al. 2009). It shows a pattern qualitatively similar to the one described for the one-locus model, i.e., some early decline of fitness, which later recovers to a considerable extent.
Figure 3 .
Evolution of mean fitness through 100 generations after the reduction of population size from the 103 value of the ancestral MSD balance population to two different N values (10 or 50) under the mutational models I, II and III used in Pérez-Figueroa et al. (2009). Six upper plates: Red: simulation results given by Pérez-Figueroa et al. (2009); green: neutral predictions computed ignoring new mutation; light blue: IP predictions; deep blue: FM predictions. Six lower plates: Green: neutral predictions computed ignoring new mutation; light blue solid line: IP predictions averaged over the joint distributions of s and h; light blue dashed line: IP predictions computed using de.
In all the cases shown, the FM prediction satisfactorily fits the simulation results. Perez-Figueroa et al. (2009) provided, in their Figure 3, predictions from the García-Dorado (2008) early full approach (i.e., accounting for new mutation, as well as for inbreeding and purge), the fit being worse than that shown here for the new FM approach. For N = 2, drift renders purging quite inefficient, preventing fitness recovery so that none of the two approaches provides reliable predictions (results not shown), and accumulation of newly arisen mutation is the main cause of fitness decline in the medium-long term.
Furthermore, the simple IP approach gives very reasonable predictions. For small effective population sizes (N = 10, which implies small δ*), it gives good approximation during at least 2N generations under the three mutational models, until the fitness decline caused by fixation of new deleterious mutations becomes relevant. For larger populations (N = 50), purging becomes so efficient that gt is always small, so that using δ instead of (δ − δ*) induces just a slight underestimate of fitness in the short term and is irrelevant thereafter. In this case, since Dm is negligible, IP predictions roughly hold during long periods.
The six bottom graphs in Figure 3 compare the evolution of fitness computed by averaging IP predictions over the joint distribution of s and h with that computed using a single “effective purging coefficient” (de), chosen “ad hoc” to predict the same long-term fitness as by averaging predictions over individual mutations. Using de usually gives good approximations, even for Nde = 0.5, although tends to predict too conspicuous minima. This de value depends upon the distribution of true d values, ranging from 0.05 for model III up to 0.14 for model I.
Results for populations under EFC
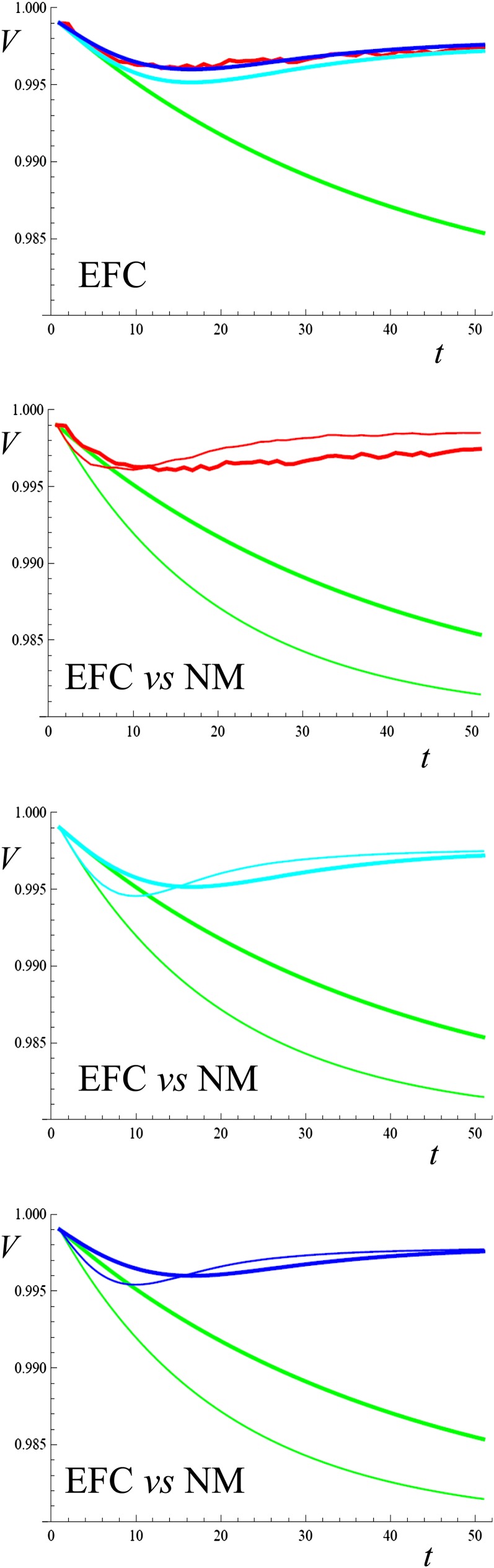
Figure 4 presents a comparison of single-locus analytical predictions and simulation results for mean viability after the effective size of a large population shrinks to a smaller stable value (N = 10) and the population starts breeding under EFC (results for fecundity were discussed in García-Dorado 2008). At this locus, deleterious mutations with s = 0.4 and h = 0 continuously occur at a rate μ=10−3. In the top graph, both simulation results and IP predictions show that purge upon viability is efficient under EFC, maintaining the average viability well above the neutral prediction. It also shows that FM predictions are very accurate. The fact that, for h = 0, FM does not account for the increase of the expressed segregating load Ls ascribed to EFC (ΔLs) is here irrelevant, since this load is expected to be negligible for this single-locus model.
Figure 4 .
Evolution of mean viability under NM or EFC through 50 generations after a reduction of population size to N = 10. Ancestral population at the MS balance for one locus with s = 0.4, h = 0, μ=0.001. Upper plate: results when the population is maintained under EFC while N = 10. Remaining plates: EFC results (thick line) vs. NM results (thin lines). Red: simulation results; green: neutral predictions computed ignoring new mutation; light blue: IP predictions; deep blue: FM predictions.
The three bottom graphs compare the evolution of mean viability under EFC and under NM. Simulation results show that EFC slows both inbreeding and purge, leading to a minimum for mean viability that is similar to that observed under NM, but that is attained later, and also to a subsequent slower recovery. Therefore, EFC produces some advantage for viability in the short term, and some disadvantage in the medium term. Both IP and FM predictions qualitatively fit this observed pattern. On the contrary, under the former approach published by the author (García-Dorado 2008), purging did not account for this medium-term disadvantage. The reason is that the former approach predicted no fitness recovery, neither under NM nor under EFC. Although the FM prediction was more accurate than the simpler IP one, both produce, in this case, the same qualitative picture regarding the merits of EFC vs. NM.
Table 1 gives simulation results for viability from Fernandez and Caballero (2001) assuming mutation over the genome with randomly variable deleterious effect for a large variety of mutational models, both under EFC and under NM. On the whole, under EFC, FM predictions improve those obtained in García-Dorado (2008), which were quite reasonable but gave a mean squared error more than fourfold that of the FM approach.
Table 1. Simulation results and analytical predictions for viability after 50 generations of no management or equal family contribution.
| Equal family contributions |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No management:Mean viability |
Mean viability |
|||||||||||||||
| Case | λ | E(s) | Shape | E(h) | δ | N | δ* | Neutral | IPb | FMc | Sd | Neutral | IPb | FMc | Sd | ΔLs at t = 50a |
| a | 2 | 0.01 | 0.07 | 0.35 | 3.75 | 25 | 0.55 | 0.09 | 0.25 | 0.28 | 0.24 | 0.09 | 0.33 | 0.38 | 0.38 | 0.003 |
| b | 1 | 0.025 | 0.25 | 0.35 | 3.82 | 25 | 0.53 | 0.08 | 0.32 | 0.35 | 0.34 | 0.09 | 0.37 | 0.38 | 0.40 | 0.079 |
| c | 0.5 | 0.05 | 1.0 | 0.35 | 1.65 | 25 | 0.29 | 0.35 | 0.56 | 0.57 | 0.51 | 0.35 | 0.62 | 0.52 | 0.52 | 0.215 |
| d | 0.1 | 0.10 | 0.5 | 0.35 | 0.74 | 25 | 0.10 | 0.64 | 0.88 | 0.88 | 0.88 | 0.62 | 0.89 | 0.85 | 0.86 | 0.066 |
| e | 0.03 | 0.264 | 2.3 | 0.35 | 0.19 | 25 | 0.04 | 0.89 | 0.98 | 0.98 | 0.98 | 0.89 | 0.97 | 0.95 | 0.95 | 0.024 (0.092) |
| h | 0.03 | 0.264 | 2.3 | 0.20 | 0.51 | 25 | 0.08 | 0.74 | 0.96 | 0.97 | 0.97 | 0.72 | 0.95 | 0.93 | 0.95 | 0.024 (0.042) |
| i | 0.5 | 0.05 | 1.0 | 0.35 | 1.66 | 100 | 0.62 | 0.69 | 0.76 | 0.83 | 0.84 | 0.69 | 0.84 | 0.67 | 0.69 | 0.255 |
| j | 0.03 | 0.264 | 2.3 | 0.20 | 0.50 | 100 | 0.14 | 0.90 | 0.96 | 0.98 | 0.98 | 0.89 | 0.97 | 0.95 | 0.96 | 0.024 (0.050) |
| Mean squared error for FM | 0.00070 | 0.00017 | ||||||||||||||
| Mean squared error for García-Dorado 2008 “full” predictions | 0.00089 | 0.00079 | ||||||||||||||
Results are given for different cases, each characterized by the effective sizes after population shrinkage (N), the deleterious mutation rate (λ), the mean and shape parameter of the gamma distribution for s (E(s) and “Shape”) and the average dominance coefficient (E(h). δ and δ* are, respectively, the inbreeding depression rates at the ancestral an the new MSD balance under no management.
Increase of the relative expressed segregating load ascribed to EFC. Regular font: bounded prediction computed from Equation B2; Italic font: unbounded prediction computed from Equation B1a, given only when larger than the MS bound.
Inbreeding-Purge predictions.
Full-Model predictions.
Simulation results published by Fernández and Caballero (2001).
Under NM, the overall viability decline from fixation of new deleterious mutation, predicted from diffusion theory, was considerable for cases a, b, and c (∼0.07), and was always smaller than 0.01 in the remaining cases. Predictions under NM show a slight upward bias in some cases, where a relevant fraction of δ is due to mildly deleterious mutations so that purging becomes inefficient against drift (Nd < 1).
Under EFC, the above upward bias is never observed, because selection, despite being slower than under NM, is more efficient against drift. Furthermore, the overall viability decline from fixation of new deleterious mutation was always small (∼0.01 for cases a, b, and c, ∼10−3 or smaller in the remaining cases). In addition, the accuracy of the FM approach supports the use of the bounded prediction for the increase in expressed segregating load induced by EFC (ΔLs, computed for Equation B2 in Appendix B). A detailed interpretation of the conditions determining large ΔLs values is given in Appendix B. Since the prediction of purging is reliable and the overall viability decline from fixation of new deleterious mutations is negligible, IP predictions are accurate as long as ΔLs remains small.
Discussion
This work considers a large population that undergoes a reduction in size and studies the different causes operating upon both standing and newly arisen deleterious mutations and determining the decline of mean fitness.
In the first place, we assume that the reduction in size is so drastic that the evolution of mean fitness in the short-medium term depends only on the consequences of inbreeding and purge upon the ancestral genetic variation. We find that, in agreement with a previous heuristic approach proposed by the author in 2008, the purged inbreeding depression can be predicted from the ancestral concealed load (δ) using a purged inbreeding coefficient (gt).
This purged inbreeding coefficient takes into account the consequences of natural selection upon the 2d increase of the deleterious effect in the homozygotes that are induced by inbreeding. For each deleterious allele, gt is the expected value of the product of Wright’s inbreeding coefficient by the ratio of its frequency at generation t to the ancestral frequency; i.e., gt = E(ft qt/q0). Then, simple expressions that predict to a good approximation the evolution of gt and its asymptotic value are derived, which are functions of d and of the reduced effective population size N (Equations 7 and 8). These expressions improve a former intuitive approach (García-Dorado 2008), quantifying how purge acts upon previously accumulated inbreeding so that its consequences are somewhat delayed with respect to those of inbreeding. Then, the evolution of fitness expected from inbreeding and purge (IP predictions) can be computed using gt, which predicts a minimum and a later a recovery for mean fitness, in agreement with empirical observations and simulation results (Crnokrak and Barrett 2002; Pérez-Figueroa et al. 2009; Larsen et al. 2011; Sánchez-Molano and García-Dorado 2011). This minimum is achieved earlier as larger d and smaller N values are considered.
In practice, d values vary for different mutations, so that predictions should be averaged over the distribution of d (Pérez-Figueroa et al. 2009). However, good approximations can be computed using an effective purging coefficient (de) that could be intended to roughly account for overall long-term purging. Nevertheless, since mutations with different (larger) d values lead to minimum fitness at different (earlier) generations, the evolution of overall fitness tends to show a minimum that is less conspicuous than predicted using de.
These IP predictions have been checked through simulation and, when the reduction in population size is so drastic that purging becomes the only relevant selective force, they are reliable in the short-medium term, until the fitness loss from fixation of new deleterious mutation becomes relevant. Reliability requires Nd ≥ 1 since, for Nd < 1, purging becomes inefficient against drift. However, under a wide variety of mutational models explored by Pérez-Figueroa et al. (2009), predictions were reliable even for N = 10.
It should be stressed that these predictions apply only to the inbreeding depression caused by partially recessive deleterious alleles, because depression caused by overdominance cannot be purged. However, since segregating recessive deleterious mutations are ubiquitous, the concealed load they produce is the most parsimonious explanation for inbreeding depression and, in fact, seems to be usually its main source (Charlesworth and Charlesworth 1999).
This approach also provides expressions for variable N (Equations 15 and 16), giving reasonable predictions under similar conditions (i.e., N values drastically below the ancestral size, and Nd > 1). In addition, it can be translated into an algorithm that uses genealogical information and that provides accurate prediction for both the expected gt and the expected fitness, at least in panmictic populations. However, these predictions will be reliable only if Ned d > 1, where Ned is the drift effective population size. If genealogies are drawn at random from a panmictic population, Ned equals the inbreeding effective population size N, which can be inferred from the genealogies. In principle, deliberately mating individuals that are more related than randomly expected, as under systematic inbreeding (Glemin 2003) or restricted panmixia (Ávila et al. 2010), could enhance the efficiency of purging by inducing inbreeding with little reduction of Ned. However, the reliability of this genealogical algorithm should still be carefully studied for genealogies obtained without panmixia, both using simulation and inbreeding depression from pedigree data (Boakes et al. 2007; Gulisija and Crow 2007). Furthermore, the variability of individual fitness around its expected value should be assayed. In any case, deliberate inbreeding should be considered with caution, as it will cause some initial fitness loss that might be unbearable for endangered populations.
In the second place, we consider a FM that applies after an arbitrary reduction in size and considers that the population shifts from an ancestral MSD equilibrium to that corresponding to its new N value. This approach also predicts the evolution of the inbreeding depression rate (δt, Equation 11) as it approaches the value corresponding the new MSD balance (δ*, Equation 12). During the shift, standard selection cancels out the inbreeding depression ascribed to δ*, implying that purged inbreeding operates on the remaining ancestral concealed load (δ − δ*). This FM approach accounts for purged inbreeding and standard selection and for the fixation of continuously arising deleterious mutations (Equations 13 and 14; Equations 15 and 16 when N varies through generations). We check these FM predictions against published simulation results for largely different mutational models and found that they are very accurate, as far as the new size N is large enough that most of that ancestral δ is contributed by alleles with Nd ≥ 1. We also find that, if N is relatively large, standard selection through the process, as well as δ*, can be relevant.
The accuracy of the FM predictions supports the underlying model as one providing a comprehensive description of the main processes that determine the evolution of fitness after a reduction of population size. However, due to our limited knowledge of the parameters involved in FM, the IP approach can be much more useful. Fortunately, this IP approach has shown to be quite reliable, even when based on the effective purging coefficient (de). This confers remarkable practical importance to this predictive method, because de can be empirically estimated. Basically, if δ can be estimated in the ancestral population, the gt value achieved after t generations with reduced effective size N can be inferred from the corresponding fitness decline (Equation 4). Then, de can be estimated as the value leading to this gt value by recurrently applying Equation 7, or by using Equation 8 in case a stable fitness value has been achieved. Although this is methodologically feasible, obtaining reliable estimates can be empirically elusive, as it relays on the evaluation of temporal changes in fitness, which can be obscured by adaptive processes, and of effective population sizes.
The reduction of fitness when population size shrinks plays an important role on the evolution of different biological mechanisms that can allow populations to deal with their load, as diploidy, sexual reproduction, recombination, or breeding population structure (Charlesworth and Charlesworth 1987; Kondrashov and Crow 1991). Therefore, the analytical model presented here, in addition to contributing to the understanding of the causes of fitness decline, can be useful in the study of the evolution of those biological mechanisms.
Furthermore, these predictions can provide useful insight into the surviving prospects of endangered populations and into the consequences of the strategies that can be implemented in conservation and animal breeding programs (Hedrick and Kalinowski 2000). Here we have considered a particularly simple strategy consisting in equating family contributions to the breeding pool (EFC), which is often recommended to slow the loss of genetic diversity and to reduce inbreeding in conservation programs (Frankham et al. 2002). Under this strategy, no natural selection occurs upon fecundity, which can be readily predicted (Equation 28 in García-Dorado 2007 and Equation 17 in this article). Regarding viability, the evolution of the purged inbreeding coefficient under EFC can be computed from a straightforward modification (Equation 18) of the general expression (Equation 7a) and, then, used to predict, to a good approximation, the joint consequences of inbreeding and purge (IP). However, EFC and NM strategies not only differ regarding inbreeding and purge: the rate of fixation for viability deleterious alleles is lower under EFC, while the segregating viability load is larger under EFC. And it should be noted that these two effects depend on the whole set of deleterious mutations, whether or not they contribute to inbreeding depression. The consequences of the lower rate of deleterious fixation under EFC are usually irrelevant, at least in the short-medium term under plausible mutational models. Regarding the excess of segregating load induced by EFC (ΔLs), which is not accounted for by IP predictions, it increases with N ftE, as well as with hs and λ, and is bounded by the segregating load expected under EFC at the mutation–selection balance. In principle, assuming large deleterious mutation rates, ΔLs might determine an important disadvantage for EFC. However, empirical evidence suggests that, excluding very small deleterious effects, the rate of spontaneous deleterious mutation is usually small (García-Dorado et al. 2004). Furthermore, the ΔLs caused by deleterious mutations with very small effect is mainly constrained by drift in small populations and accumulates very slowly in large ones. Therefore, ΔLs is not expected to cause important disadvantage for mean viability under EFC in most practical circumstances. In addition, it must be noted that ΔLs should be reversible after the EFC breeding strategy is discontinued.
Therefore, taking into account that IP operates upon the same inbreeding depression rate (δ − δ*) under both EFC and NM breeding strategies (Appendix B), the simple IP expressions provide an insight on the relative merits of the two strategies for mean viability. Simulation results show that EFC slows both inbreeding and purging and suggest that it may cause some short-term viability advantage due to slowed inbreeding and some later viability disadvantage due to slowed purging. However, since EFC increases the purge/drift ratio (i.e., the effective Ns product), the later disadvantage is expected to disappear in the long term.
I have presented a simple but powerful and versatile approach to the prediction of the consequences of purge and nonpurging (standard) selection upon the evolution of fitness after population shrinkage, which can be applied in different evolutionary and conservation contexts. After drastic shrinkage, fitness evolution can be predicted as a function of the ancestral inbreeding depression rate δ and of a purged inbreeding coefficient gt. The latter can be approached in terms of an effective purging coefficient de that accounts for overall purging through the whole genome and that should be experimentally estimated. The reliability of this predictive method should be further explored for different practical situations, depending on historical population sizes, on the breeding structure, or on the availability of genealogical information.
Supplementary Material
Acknowledgments
I am grateful to Carlos López-Fanjul for helpful suggestions. This work was supported by grant CGL2011-25096 from the Ministerio de Ciencia e Innovación.
Appendix A
We consider a single locus where a deleterious allele m segregates with low frequency q0 in a large ancestral panmictic population. The fitness of the wild homozygous (++), the heterozygous (+m), and the mutant homozygous (mm) genotypes are 1, 1 − hs, and 1 − s, respectively. Therefore, the average fitness for this one-locus model in that population is w0 = 1 − 2p0q0hs − q02s. At generation t = 0, the effective size of this population reduces to N, so that its expected properties can be derived under the classical Wright’s model of the “subdivided population.” After t generations of subdivision, the expected reduction in average fitness can be written as
where E denotes expected values over subpopulations. The overall probability of mm homozyotes is E(qt2) = E(qt ft) + ζt, where ζt is the probability of being homozygous for m and not being homozygous by descent. Therefore,
Since we assume that, due to natural selection, the larger sh is, the smaller the expected value for q0 is, we can neglect hsq0 (and, therefore, hs[q0 – E(qt)]), as well as (− ζt)), and we obtain w0 – wt ≈ 2dE(qt ft), so that
| (A1) |
where gt is the purged inbreeding coefficient defined by Equation 2 in the main text.
The evolution of gt can be predicted from those of ft and qt, which are due to inbreeding and natural selection. With random mating, if the frequency of a deleterious allele in a given subpopulation i at generation t before natural selection acts is qit, then, the frequency expected at the same generation after selection can be written as
| (A2) |
Since natural selection is operating through the whole process, we assume that qit is small enough that, for the one-locus model, wit ≈1 and pit qit2 ≈ qit2. Therefore, the expected frequency at generation t after natural selection is q’it ≈ qit – [sh pitqit + 2dqit2]. For this Inbreeding-Purging approach (IP), we assume that q0 was close to zero and that the population size reduces so drastically that all relevant selection occurs due to the homozygosis for m induced by inbreeding. In other words, we assume that q’it ≈ qit – 2dqit2, and that ζt can be neglected in E(qt2) = E(qt ft) + ζ t, and we obtain
| (A3) |
Thus, Equation A3 predicts the reduction of qt expected from purging, ignoring the selection expected on the basis of the heterozygotic disadvantage hs, as well as that upon the 2d excess of deleterious effect of the homozygotes ζ that are not due to inbreeding.
Then, a new generation t+1 is produced through panmictic mating, where, for each line, fi t+1 = (1 – 1/2N) fit + 1/2N. According to the definition of gt (Equation 2), gt+1 = E(ft+1 qt+1)/q0 . Substituting qi t+1 with q’it from Equation A3, this gives
To simplify the first term in this expression, we neglect the covariance between (1 − 2d ft) and , obtaining
If the covariance (covt(f, qf)) between f and the proportion of homozygous by descent for m at generation t is positive, the above approximation is a conservative one.
Furthermore, the second term can be rewritten as
which equals
where decreases as purging progresses, and where the scaled covariance between f and q at generation t caused by purging is negative. Therefore, by omitting in the above expression we overestimate the effect of purge against the increase in gt that would be expected at generation t due to a 1/2N increase of ft if purging were relaxed at that point (i.e., we overestimate purging against Δgt = ). On the contrary, by dropping we overestimate Δgt. As far as the consequences of the later overestimate overcome those of the former, the approximation
would also be conservative.
Therefore, using the above approximations for the terms in and in , we obtain Equation 7a.
Finally, the inbreeding depression rate within subpopulations that is expected at generation t due to deleterious alleles that were segregating in the ancestral population is
where the sum is over loci for which partially recessive deleterious alleles were segregating at the ancestral population. Therefore, according to Equation 2,
Furthermore, assuming E(f t q t) ≈ E(qt)ft implies that E(qt)≈ q0 gt / ft, so that, for ft > 0,
and, since δ0 = ∑ 2d q0, we obtain
| (A4) |
Appendix B
Consider a nonmanaged ancestral population (NM) with effective size equal to the actual population size N0, which is at the MSD balance with viability inbreeding depression rate δ, and where the viability decline from deleterious fixation can be neglected. At that population, the per generation rate of viability decline expected from inbreeding (δ/2N0) is cancelled out by standard selection acting upon the viability additive variance. Then, EFC is established at generation t = 0, so that the population roughly doubles its effective size. Therefore, the per-generation viability decline expected from inbreeding upon the δ load concealed at the MSD balance in the ancestral nonmanaged population, is halved. However, it is still cancelled out by natural selection acting upon half the ancestral additive variance for viability (Falconer and Mackay 1996, p. 232). By analogy, we assume that, if EFC is accompanied by the reduction of the population size to a new value N, standard selection cancels out the inbreeding depression corresponding to load δ* that would be concealed in a nonmanaged population of size N at the MSD balance. Thus, under this assumption, both inbreeding and purge occur at the expense of (δ − δ*).
Now, to inquire into the consequences of the increase in segregating load under EFC, we consider a large population at the MS balance that begins to breed under EFC. Then, selection upon just half the additive variance compensates for just half the 2λhs viability decline from new mutation, so that viability initially decays at a rate λhs. Therefore, the overall frequency of deleterious alleles slowly increases until a new MS balance is attained, where the expressed segregating load is larger than in the ancestral balance. To account for this process in a small population, we assume that, for h > 0, the expressed segregating viability load Ls increases at a rate λhs per generation in the 1-fEt “ancestral” (or panmictic) fraction. Then, after this load accumulates during t generations, it reduces average fitness by a factor = exp[−4NλhsfEt] (García-Dorado 2007, 2008), leading to Equation 19. Thus, the increase of the relative segregating load (ΔLs) in a small population after t generations of EFC can be predicted as:
| (B1a) |
However, if the new population size is relatively large (Ns >> 1), the approach to the new equilibrium is faster than the approach to fEt=1, and the accumulated increase of segregating load is up-bounded by the value expected at the MS balance. Under NM, assuming h>0, the segregating load reduces mean viability by a factor exp[-2λ] at the MS balance (Crow 1970, p. 147). Under EFC, it can be similarly shown to reduce it by a factor . Thus, at the MS balance, EFC reduces the mean viability by a factor compared to NM, due to increased segregating load. This leads to Equation 20. In other words, in an infinite population, the excess in (relative) segregating load induced by EFC will have a MS equilibrium value
| (B1b) |
where the hat denotes equilibrium. This settles an upper bound to the equilibrium ΔLs value in a finite population at the MSD balance under EFC, where mean viability is expected to be reduced due to the increased segregating load by, at most, a factor .
Using the above approximations, we will predict the increase of segregating load through the period of reduced population size under EFC as the smaller of the two expressions derived above, i.e.,
Table 1 (see main text for explanation) illustrates how ΔLs depends on N and on the distribution of mutational effects. For cases a, b and c (and i), the deleterious mutation rate is so high that ΔLs would be expected to amount 0.81, 0.56 and 0.34 at the MS balance, respectively (Equation B1b). However, for cases a and b, the table gives much smaller predictions. The reason is that, in these cases, ΔLs is mainly due to tiny deleterious effects, so that, for N = 25, it is strongly constrained by drift. Thus, as inbreeding increases, ΔLs approaches a much smaller value than expected at the MS balance (Equation B1a gives 0.007 and 0.189, respectively, for fEt=1 and N=25). Furthermore, even for this small N, the equilibrium ΔLs values had not been attained by generation 50. On the contrary, for case c, most deleterious effects are larger than for cases a and b (say, mild instead of tiny), so that, at the new equilibrium, the relevant constraint upon ΔLs is natural selection, and the large ΔLs value predicted for the MS balance (0.34) is expected to be asymptotically attained (the prediction obtained from Equation B1a for fEt=1 and N=25 is larger, amounting 0.450). It is interesting to note that, in this case (as well as in case i, where N = 100), the ΔLs value at generation 50 is relatively close to the MS bound but can still be predicted by the Equation B1a, as shown by the agreement between FM predictions and simulation results. In cases e, h and j, deleterious effects are usually large, so that the observed ΔLs is constrained by the load corresponding to the MS balance.
Footnotes
Communicating editor: H. G. Spencer
Literature Cited
- Ávila V., Amador C., García-Dorado A., 2010. The purge of genetic load through restricted panmixia in a Drosophila experiment. J. Evol. Biol. 23: 1937–1946 [DOI] [PubMed] [Google Scholar]
- Boakes E., Wang J., Amos W., 2007. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity 98: 172–182 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1987. Inbreeding depression and and its evolutionary consequences. Ann. Rev. Ecol. System. 14: 237–268
- Charlesworth B., Charlesworth D., 1999. The genetic basis of inbreeding depression. Genet. Res. Camb. 74: 329–340 [DOI] [PubMed] [Google Scholar]
- Crnokrak P., Barrett S. C. H., 2002. Purging the genetic load: a review of the experimental evidence. Evolution 56: 2347–2358 [DOI] [PubMed] [Google Scholar]
- Crow J. F., 1970. Genetic loads and the cost of natural selection, pp. 128–177 Mathematical Topics in Population Genetics, edited by Kojima K. Springer-Verlag, New York [Google Scholar]
- Crow J. F., 2008. Mid-century controversies in population genetics. Annu. Rev. Genet. 42: 1–16 [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York [Google Scholar]
- Falconer D. S., Mackay T., 1996. Introduction to Quantitative Genetics, Ed. 4 Longman, Essex, England [Google Scholar]
- Fernández J., Caballero A., 2001. Accumulation of deleterious mutations and equalization of parental contributions in the conservation of genetic resources. Heredity 86: 480–488 [DOI] [PubMed] [Google Scholar]
- Frankham R., Ballou J. D., Briscoe D. A., 2002. Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK [Google Scholar]
- García-Dorado A., 2003. Tolerant vs. sensitive genomes: the impact of deleterious mutation on fitness and conservation. Conserv. Genet. 4: 311–324 [Google Scholar]
- García-Dorado A., 2007. Shortcut predictions for fitness properties at the MSD balance and for its build-up after size reduction under different management strategies. Genetics 176: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorado A., 2008. A simple method to account for natural selection when predicting inbreeding depression. Genetics 180: 1559–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorado A., López-Fanjul C., Caballero A., 2004. Rates and effects of deleterious mutations and their evolutionary consequences, pp. 20–32 Evolution: From Molecules to Ecosystems, edited by Moya A., Font E. Oxford University Press, Oxford [Google Scholar]
- Glémin S., 2003. How are deleterious mutations purged?: drift vs. nonrandom mating. Evolution 57: 2678–2687 [DOI] [PubMed] [Google Scholar]
- Gulisija D., Crow J. F., 2007. Inferring the purging from pedigree data. Evolution 61: 1043–1051 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., Kalinowski S. T., 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31: 139–162 [Google Scholar]
- Keller L. F., Waller D. M., 2002. Inbreeding effects in wild populations. Trends in Ecol. Evol. 17: 230–241 [Google Scholar]
- Kimura M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A. S., Crow J. F., 1991. Haploidy or diploidy: Which is better? Nature 351: 314–315 [DOI] [PubMed] [Google Scholar]
- Larsen L. K., Pélabon C., Bolstad G. H., Viken A., Fleming I. A., et al. , 2011. Temporal change in inbreeding depresion in life-history traits in captive populations of guppy (Poecilia reticulata): evidence for purging? J. Evol. Biol. 24: 823–834 [DOI] [PubMed] [Google Scholar]
- Leberg P. L., Firmin B. D., 2007. Role of inbreeding depression and purging in captive breeding and restoration programmes. Mol. Ecol. 17: 334–343 [DOI] [PubMed] [Google Scholar]
- Pérez-Figueroa A., Caballero A., García-Dorado A., López-Fanjul C., 2009. The action of purifying selection, mutation and drift on fitness epistatic systems. Genetics 183: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Molano E., García-Dorado A., 2011. The consequences on fitness of equating family contributions: inferences from a Drosophila experiment. Conserv. Genet. 12: 343–353 [Google Scholar]
- Wright S., 1938. Size of population and breeding structure in relation to evolution. Science 87: 430–431 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.