Abstract
Antagonistic selection—where alleles at a locus have opposing effects on male and female fitness (“sexual antagonism”) or between components of fitness (“antagonistic pleiotropy”)—might play an important role in maintaining population genetic variation and in driving phylogenetic and genomic patterns of sexual dimorphism and life-history evolution. While prior theory has thoroughly characterized the conditions necessary for antagonistic balancing selection to operate, we currently know little about the evolutionary interactions between antagonistic selection, recurrent mutation, and genetic drift, which should collectively shape empirical patterns of genetic variation. To fill this void, we developed and analyzed a series of population genetic models that simultaneously incorporate these processes. Our models identify two general properties of antagonistically selected loci. First, antagonistic selection inflates heterozygosity and fitness variance across a broad parameter range—a result that applies to alleles maintained by balancing selection and by recurrent mutation. Second, effective population size and genetic drift profoundly affect the statistical frequency distributions of antagonistically selected alleles. The “efficacy” of antagonistic selection (i.e., its tendency to dominate over genetic drift) is extremely weak relative to classical models, such as directional selection and overdominance. Alleles meeting traditional criteria for strong selection (Nes >> 1, where Ne is the effective population size, and s is a selection coefficient for a given sex or fitness component) may nevertheless evolve as if neutral. The effects of mutation and demography may generate population differences in overall levels of antagonistic fitness variation, as well as molecular population genetic signatures of balancing selection.
PLEIOTROPY may impose widespread evolutionary genetic constraints against adaptation (Fisher 1958, Orr 1998, Otto 2004). Allocation tradeoffs between fitness components, coupled with a shared genetic basis between them, may limit evolutionary opportunities to simultaneously maximize individual components of fitness (Houle 1991). Conflicting selection may emerge at underlying loci, so that alleles improving one fitness component reduce others—a scenario referred to as “antagonistic pleiotropy” (Rose 1982; Curtsinger et al. 1994). A similar scenario arises when patterns of selection differ between males and females. Opposing, sex-specific selection, or “sexual antagonism,” arises when mutations favorable within one sex are costly when expressed in the other (Rice and Chippindale 2001; Bonduriansky and Chenoweth 2009; van Doorn 2009).
Antagonistic selection may play an important role in maintaining genetic variation related to fitness and driving patterns of genome evolution. Direct estimates of sex- and context-specific selection on quantitative traits confirm that pleiotropic and sexually antagonistic fitness tradeoffs are ubiquitous (Curtsinger et al. 1994; Roff 1996; Delph 2007; Cox and Calsbeek 2009; Poissant and Coltman 2009; Poissant et al. 2010). Emerging data from the field of genomics suggest that processes of antagonistic selection promote the evolutionary accumulation of gene duplicates and genes with sex-biased transcription patterns (Ellegren and Parsch 2007; Mank 2009). Antagonistically selected alleles also comprise an important component of genetic variation for life-history traits and fitness (Charlesworth and Hughes 1999; Chippindale et al. 2001; Rice and Chippindale 2001; Bonduriansky and Chenoweth 2009; van Doorn 2009; Innocenti and Morrow 2010).
Despite accumulating empirical support for antagonistic selection and genetic variation, we still lack a general population genetic framework for characterizing the evolutionary dynamics and equilibrium states at antagonistically selected loci. This may seem surprising given the intensity of interest (e.g., Bonduriansky and Chenoweth 2009; van Doorn 2009; Stewart et al. 2010) and the considerable theoretical tradition regarding antagonistic selection and variation (e.g., Owen 1953; Bennett 1957; Parsons 1961; Haldane 1962; Haldane and Jayakar 1964; Li 1967; Mérat 1969; Mandel 1971; Kidwell et al. 1977; Pamilo 1979; Curtsinger 1980; Rose 1982, 1985; Rice 1984; Curtsinger et al. 1994; Babcock and Asmussen 1996; Hedrick 1999, 2007; Prout 2000; Gavrilets and Rice 2006; Unckless and Herren 2009; Patten and Haig 2009a,b; Fry 2010; Patten et al. 2010; Úbeda et al. 2011; Arnqvist 2011; Jordan and Charlesworth 2011). Prior theory has thoroughly described the requisite parameter criteria for balancing selection, yet it has neglected the impact of recurrent mutation and finite population size on genetic variability. This strong emphasis on balancing selection and deterministic evolutionary dynamics has led to two questionable conclusions, based on the current theory:
The generally restrictive parameter space for antagonistic balancing selection is thought to imply that conditions for maintaining antagonistic genetic variation are also restrictive (e.g., Curtsinger et al. 1994; Merilä and Sheldon 1999; Hedrick 1999). However, such criteria for maintaining genetic variation are overly conservative because recurrent mutation also maintains genetic variation in the absence of balancing selection (see Lynch and Walsh 1998, pp. 655–656; Radwan 2008). Assessing the potential impact of antagonistic selection on patterns of genetic variation requires a theoretical analysis of the full range of allele frequency dynamics and equilibria within balancing and nonbalancing selection domains of parameter space. Such an approach should facilitate effective contrasts between the theoretical predictions of antagonistic selection and those of the classic null model: unequivocally deleterious genetic variation maintained by recurrent mutation (e.g., Mukai et al. 1974; Charlesworth 1987).
Deterministic models are assumed to reasonably describe the evolutionary dynamics of antagonistic alleles within (real) populations of finite size. As with any evolutionary dynamic, deterministic models can be used to describe properties of finite populations as long as directional evolutionary forces (e.g., mutation, migration, selection) are strong relative to stochastic forces like genetic drift (Rice 2004; Charlesworth and Charlesworth 2010). In most selection models, this criterion is easily met because the expected rate of change due to selection is typically much greater than that due to drift. However, under antagonistic selection, the expected frequency change per generation reflects the net outcome of opposing forces, leading to a very slow overall rate of directional change (Livingstone 1992). Deterministic results may often prove misleading if antagonistically selected loci are particularly sensitive to genetic drift and factors that influence effective population size. This possibility has yet to be considered theoretically.
With these issues in mind, we developed general population genetic models of sexual antagonism and antagonistic pleiotropy and examined how interactions between antagonistic selection, recurrent mutation, and genetic drift combine to influence equilibrium patterns of heterozygosity and fitness variation at single loci. The analysis leads to two primary results. First, a substantial parameter range of antagonistic selection can inflate both heterozygosity and fitness variance compared to simple models of directional selection (i.e., those without antagonism). This result does not require that conditions for antagonistic balancing selection be permissive. Second, antagonistic selection is often ineffective in the face of genetic drift, with the impact of drift being particularly pronounced at or near the parameter domain for balancing selection. For those loci that meet the necessary parameter conditions, antagonistic balancing selection may nevertheless fail to stably maintain balanced polymorphisms. Such loci should therefore fail to exhibit molecular population genetic signatures of balancing selection (e.g., Charlesworth and Charlesworth 2010), despite its occurrence.
Model
Following prior models of sexual antagonism and antagonistic pleiotropy, genotypic fitness for sex-linked or autosomal loci are parameterized as shown in Table 1. In analyzing the allele frequency dynamics at each locus, we make four simplifying assumptions.
Table 1. Relationship between genotype and fitness under models of sexual antagonism and antagonistic pleiotropy.
| Sexually antagonistic selection | |||
|---|---|---|---|
| Genotype | Af, AfAf | AfAm | Am, AmAm |
| Female fitness | 1 | 1 – sfhf | 1 – sf |
| Male fitness (autosome) | 1 – sm | 1 – smhm | 1 |
| Male fitness (X linked) | 1 – sm | — | 1 |
| Antagonistic pleiotropy | |||
| Genotype | A1A1 | A1A2 | A2A2 |
| Context 1 fitness | 1 – s1 | 1 – s1h1 | 1 |
| Context 2 fitness | 1 | 1 – s2h2 | 1 – s2 |
| Net fitness | 1 – s1 | (1 – s1h1)(1 – s2h2) | 1 – s2 |
First, selection coefficients are assumed to be small (0 < sm, sf ≪ 1 under sexual antagonism; 0 < s1, s2 ≪ 1 under antagonistic pleiotropy), as likely applies to the vast majority of loci within a genome. This assumption greatly increases model tractability by facilitating the analytical treatment of multiple evolutionary processes, e.g., selection, recurrent mutation, and drift. It also justifies our use of diffusion approximations for describing the finite population allele frequency distributions.
Second, we assume that mutation rates are symmetrical and the same in males and females—i.e., that the mutation rate per locus is u, per gamete, regardless of the allelic state of the locus or the sex within which it resides. This assumption is fairly trivial and serves to simplify presentation of the results. Incorporating unequal forward and backward mutation rates, or sex-biased mutation, requires only minor modification of each model.
Third, we constrain our analysis to alleles with additive to recessive fitness costs within heterozygotes (hi ≤ 1/2), as this represents a particularly interesting and biologically plausible range of dominance (see Fry 2010; Connallon and Clark 2010; Jordan and Charlesworth 2011). We focus on this critical parameter range by imposing symmetry and analyzing cases where hm = hf ≤ 1/2 and h1 = h2 ≤ 1/2. Additive models arise when hi = hj = 1/2, and dominance reversals arise when hi = hj < 1/2 (dominance reversal refers to the fact that the deleterious allele in each sex, or in each context of selection, is recessive to the beneficial allele when hi, hj < 1/2). Additional results for constant dominance between contexts (e.g., hf = 1 – hm) are presented in the supporting information, File S1; such scenarios behave similarly to the additive case.
Finally, in results for models of sex-linked inheritance, we assume that the heterogametic sex is hemizygous, which applies to species with degenerate Y or W chromosomes that do not carry functional homologs of X-linked or Z-linked genes (for recent studies of balancing selection at sex-linked loci with nondegenerate copies on the Y or W, see Kirkpatrick et al. 2010, and Jordan and Charlesworth 2011). X-linked results are presented below and are equivalent to Z linkage with the sexes reversed.
Deterministic allele frequency dynamics
Sexually antagonistic (SA) selection:
Consider the population frequency of the male-beneficial allele, Am. Given the assumption of small sex-specific selection coefficients and weak mutation (u, sm, sf ≪ 1), the frequency of Am (denoted q = [Am] = 1 – [Af]) will be approximately equal in each sex. Under autosomal linkage, the allele frequency change due to selection in males and selection in females (respectively) will be
| (1a) |
and
| (1b) |
Incorporating mutation, the overall frequency change per generation will be Δq ≈ Δqf/2 + Δqm/2 + u(1 – 2q).
Under X-linked inheritance, the allele frequency change due to selection in males is
| (1c) |
The change due to selection in females remains the same as in the autosomal model. Since each X is carried by females with two-thirds probability and by males with one-third probability, the overall change in frequency of an X-linked, male-beneficial allele is given by Δq = 2Δqf/3 + Δqm/3 + u(1 – 2q).
Antagonistic pleiotropy:
Under antagonistic pleiotropy, the frequency of the A2 allele (in Table 1) is q = [A2] = 1 – [A1]. Under symmetrical dominance (letting h = h1 = h2; see above), the allele frequency trajectory on an autosome is
| (2) |
X linkage is slightly less conducive to polymorphism under antagonistic pleiotropy, although allele frequency behavior is similar between the X and autosomes. Results for autosomal inheritance are presented below. Additional results under X-linked inheritance can be found in File S1.
Extension to finite populations
Following Wright (1945), we obtain equilibrium allele frequency distributions under mutation, selection, and drift using the relation
| (3) |
where C is a constant to ensure that , M is the expected frequency change due to selection and mutation (M = Δq, from the deterministic models), and V is the variance of allele frequencies due to binomial sampling in a Wright–Fisher population. VA = q(1 – q)/NA for autosomal loci, and VX = q(1 – q)/NX for X-linked loci, where NA and NX represent the effective population size relevant to each autosomal and X-linked locus. For example, with equal numbers of breeding males and females, NA = 2Ne and NX = 3Ne/2, although a wider range of values for NA/NX is biologically plausible (see Charlesworth 2009; Ellegren 2009). Stochastic simulations in a Wright–Fisher population confirm that the stationary distribution approximation provides an excellent characterization of the antagonistic allele frequency distribution at mutation–selection–drift equilibrium (T. Connallon and A. G. Clark, unpublished results).
All numerical integration was performed using the R statistical software package (R Development Core Team 2005).
Results
To evaluate the interaction between antagonistic selection and recurrent mutation, we first describe evolutionary dynamics and equilibrium properties under deterministic conditions (i.e., without genetic drift). We subsequently consider the impact of finite population size by analyzing the statistical distributions of antagonistically selected alleles evolving by recurrent mutation, selection, and genetic drift, in a population of constant size.
We present results for four general scenarios that have each received attention from prior theory (e.g., Kidwell et al. 1977; Pamilo 1979; Rice 1984; Curtsinger 1994; Patten and Haig 2009; Fry 2010):
Autosomal sexually antagonistic selection with additive fitness effects (hm = hf = 0.5). Although we focus on the additive case for simplicity, similar results emerge under models of sexual antagonism with constant dominance between the sexes, i.e., hf = 1 – hm (see File S1).
Autosomal sexually antagonistic selection with reversed dominance (hm = hf < 0.5).
X-linked sexually antagonistic selection with additive to recessive expression of male-beneficial alleles in females (hf ≤ 0.5).
Antagonistic pleiotropy under additive to reversed dominance (h2 = h2 ≤ 0.5).
Although our modeling approach differs somewhat from previous theory regarding sexual antagonism and antagonistic pleiotropy, we arrive at complementary results in many cases where direct comparisons are possible. In particular, prior work has heavily emphasized the set of parameter conditions giving rise to balancing selection (these are generally based on an analysis of evolutionary stability at the boundary equilibria, q = 0 and q = 1, using systems of exact recursion equations). Our results correspond closely to these (T. Connallon and A. G. Clark, unpublished results).
Allele frequency dynamics and equilibria under selection and recurrent mutation
In each of the antagonistic selection scenarios considered, allele frequency trajectories follow the same basic form: , where f(si, sj, h) is a function of the selection and dominance parameters and depicts the mode of selection acting at the locus (see below for model-specific definitions). Net directional selection emerges when (selection favors elimination of Am and A2) or when (selection favors fixation of Am and A2). Balancing selection arises when , in which case, selection favors the maintenance of both alleles in the population. Under balancing selection, corresponds to the interior equilibrium allele frequency in the absence of recurrent mutation.
Autosomal, additive SA selection:
For the case of autosomal sexual antagonism with additive allelic effects (hm = hf = 1/2), the frequency trajectory of a male-beneficial allele reduces to
| (4a) |
where . Under conditions necessary for balancing selection (and given the assumption of small selection coefficients), takes a much simpler form compared to the exact equilibrium results originally derived by Kidwell et al. (1977, p. 174). When sm2 ≫ sm – sf, the sexually antagonistic equilibrium (in the absence of subsequent mutations) further simplifies to , which has a form reminiscent of the overdominant selection model (for example, overdominance involving two alleles, i and j, with genotypic fitness wii = 1 – si, wij = 1, and wjj = 1 – sj, gives rise to an equilibrium frequency of the form si/(si + sj) for allele j and sj/(si + sj) for allele i).
By incorporating recurrent mutation, and assuming weak mutation compared to selection (16u ≪ sm2 + sf2), the equilibrium frequency of a male-beneficial allele becomes
| (4b) |
These and subsequent approximations compare well with exact numerical evaluation of the relevant roots of the recurrence equation at equilibrium (see File S1). Because the term 16u/(sm2 + sf2) is small, qeq ≈ under most conditions of balancing selection, with exceptions when is close to zero or one. Under most conditions of net directional selection, qeq ≈ 4u/[sf – sm(1 + sm)], when < 0 and 2 ≫ 16u/(sm2 + sf2); qeq ≈ 1 – 4u/[sm – sf(1 + sf)] for > 1 and (1 – )2 ≫ 16u/(sm2 + sf2).
Autosomal SA selection: dominance reversal:
Under dominance reversal conditions, with h = hm = hf < 1/2, and sm2, sf2 ≪ 1 – 2h, the deterministic allele frequency trajectory simplifies to
| (5a) |
where . In the absence of recurrent mutation, values of under balancing selection correspond to previously described internal equilibrium results (Connallon and Clark 2010, 2011). Adding recurrent mutation to the model leads to the equilibrium:
| (5b) |
X-linked SA selection:
Under X-linked inheritance, with h = hf ≤ 1/2, the frequency trajectory of an X-linked, male-beneficial allele is
| (6a) |
where . Under conditions of balancing selection and in the absence of recurrent mutation, is similar to previous interior equilibrium results (see the appendix of Albert and Otto 2005; Connallon and Clark 2010, 2011; both provide results for the case in which 1 – 2h ≫ sf2, sm2). Adding recurrent mutation, the equilibrium is
| (6b) |
Antagonistic pleiotropy:
For the case of antagonistic pleiotropy with h = h1 = h2 ≤ 1/2, the allele frequency change of A2 is given by
| (7a) |
with . Under the additive case (h = 1/2), the internal equilibrium under balancing selection (without recurrent mutation) reduces to the pleasing = 1/2 + (s1 – s2)/(s1s2), which is similar in form to the balanced polymorphic equilibrium under additive sexual antagonism. Incorporating recurrent mutation (and assuming s1s2 ≫ 8u), the equilibrium is
| (7b) |
Equilibrium heterozygosity and fitness variance
It is useful to express these results in terms of population heterozygosity, H = 2q(1 – q), as this metric is intimately related to patterns of molecular genetic and phenotypic variation. From the molecular population genetics standpoint, heterozygosity can be estimated directly from population samples and is often useful for inferring the strength of selection acting on putatively functional sites within a genome (e.g., Loewe et al. 2006; Loewe and Charlesworth 2006; Eyre-Walker and Keightley 2007). Genetic variation of phenotypes, including fitness, is also a positive function of heterozygosity, typically being maximized near 2q(1 – q) = 1/2 (Falconer and Mackay 1996).
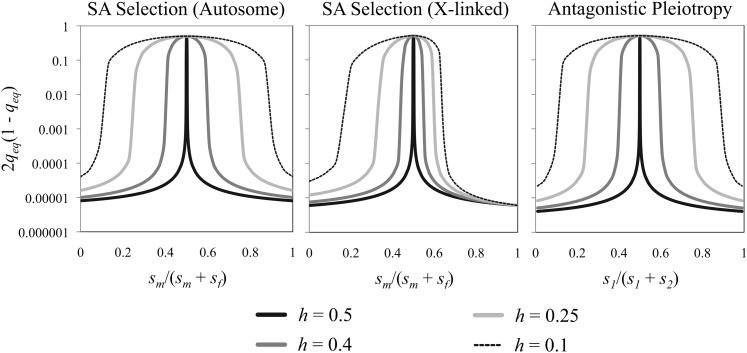
Under antagonistic selection with recurrent mutation, equilibrium heterozygosity is always positive and reaches a zenith of high heterozygosity as the two selection coefficients approach each other in magnitude (i.e., as si approaches sj; Figure 1). The spikes in heterozygosity in Figure 1 represent the regions of parameter space corresponding to balancing selection. Low-heterozygosity regions largely reflect parameter combinations leading to net directional selection, where mutation–selection balance equilibria prevail. As in prior models, dominance can substantially affect equilibrium patterns of heterozygosity. When alleles have additive fitness effects (hi = hj = 1/2), all models produce a narrow spike of high heterozygosity near the point where si = sj. When deleterious fitness effects are recessive (hi, hj < 1/2), regions of high heterozygosity become broader due to expanded parameter space for balancing selection.
Figure 1.
Equilibrium heterozygosity under antagonistic selection and recurrent mutation. Results are based on equations (4-7), with u = 10−8, max(si, sj) = 0.01. For autosomal models, h = hi = hj; for the X-linked model, h = hf.
The critical question is whether (and when) antagonistic selection is expected to inflate population variability compared to nonantagonistic models of evolution, such as the classic null of unequivocally deleterious alleles at mutation–selection balance. Alleles maintained by balancing selection will clearly generate greater heterozygosity than deleterious alleles maintained at mutation–selection balance. However, the parameter space for balancing selection is often restrictive, making it unclear whether genetic variation should typically be inflated by antagonistic selection across the broader parameter space.
To directly compare equilibrium genetic variation between antagonistic and nonantagonistic models, consider the following hypothetical scenario. Suppose within “population A” there is a single, autosomal locus evolving via mutation and sexually antagonistic selection; assume for simplicity that the alleles have additive fitness effects per sex. In population A, equilibrium fitness variation in males and females is given by expressions Vm(A) = sm2qeq(1 – qeq)/2 and Vf(A) = sf2qeq(1 – qeq)/2, respectively, with values of qeq from Equation 4b. Suppose that a second population (“population B”) has resolved the antagonistic selection by way of gene duplication and sex-specific cooption of the paralogs (e.g., Connallon and Clark 2011). Following duplication, and supposing that the sex-specific selection coefficients do not systematically change, male-deleterious alleles will evolve to a frequency qm ≈ 4u/sm and female-deleterious alleles evolve to a frequency qf ≈ 4u/sf. Equilibrium fitness variation in population B will be Vm(B) = sm2qm(1 – qm)/2 and Vf(B) = sf2qf(1 – qf)/2.
Comparing the two populations, unresolved sexual antagonism leads to inflated fitness variation in both sexes as long as qeq(1 – qeq) > max{qm(1 – qm), qf(1 – qf)}, which requires that the following conditions are both true:
| (8a) |
| (8b) |
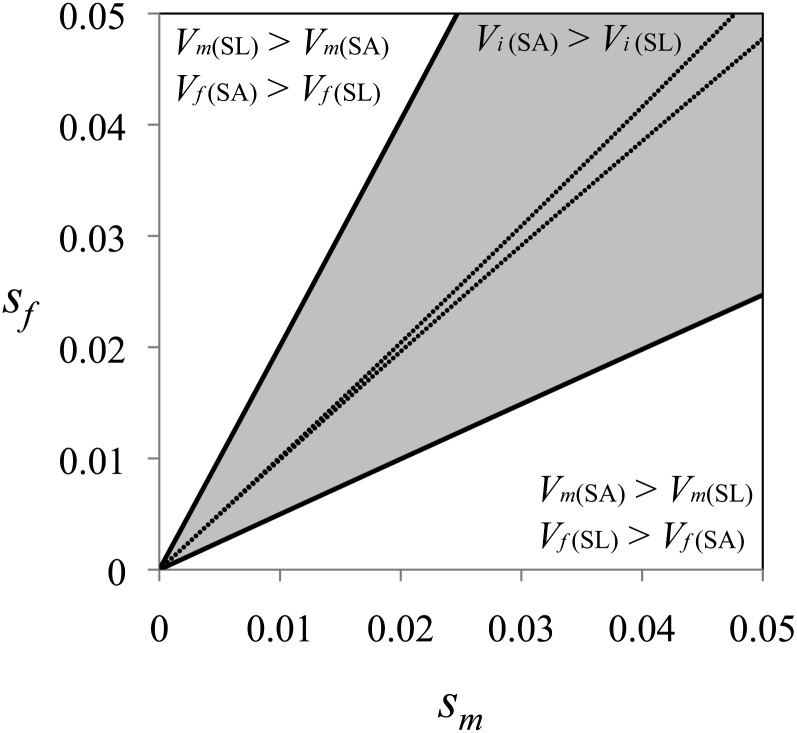
(Note that these results also apply when contrasting heterozygosity in population A vs. B. A locus under sexual antagonism will have higher heterozygosity than either paralog when both conditions in (8) hold.) When Equations 8a and 8b fail, sexual antagonism will inflate fitness variation in only one sex, so that min{qm(1 – qm), qf(1 – qf)} < qeq(1 – qeq) < max{qm(1 – qm), qf(1 – qf)}. Figure 2 shows the parameter conditions where sex-specific fitness variance is inflated by sexual antagonism (i.e., where variance in both sexes is greater in population A compared to population B). These conditions are quite broad, particularly when compared to those required for balancing selection (Figure 2). Models involving dominance reversals, X-linkage, and antagonistic pleiotropy generate similar to more exaggerated results.
Figure 2.
Antagonistic selection inflates fitness variation with or without balanced polymorphism. Models of sexually antagonistic selection (SA, corresponding to the hypothetical population A in the text) are contrasted with a model of sex-limited evolution (SL, with conflict completely resolved; corresponding to population B in the text). Shown is the case of autosomal sexual antagonism with additive allelic effects. The regions of parameter space are divided into those that increase fitness variation in both sexes (shaded) and those that increase variation in only one sex (open). The parameter space necessary for balancing selection is bounded by the dotted lines.
Allele frequency distributions under selection, recurrent mutation, and genetic drift
The deterministic equations for allele frequency change (above) give the expected change per generation in a finite population. Thus, for each scenario of antagonistic selection, the expected change takes the form: . In a Wright–Fisher population, the variance in allele frequency change per generation is VA = q(1 – q)/NA for autosomal loci, and VX = q(1 – q)/NX for X-linked loci, where NA and NX refer to the effective population size per autosomal and X-linked locus, respectively. Using these expressions of M and V in evaluating Equation 3 gives the equilibrium allele frequency distribution for models of antagonistic selection, mutation, and genetic drift,
| (9) |
where L = {A, X}, and specific values of f(si, sj, h) and are presented above, for each model of selection and linkage. With this simple result, we can quantify the impact of population size on equilibrium patterns of heterozygosity and fitness variance at antagonistically selected loci.
The efficacy of net directional selection:
Under sexual antagonism and antagonistic pleiotropy, many parameter sets generate directional selection to eliminate one of the alleles from the population. Recall that net directional selection arises when < 0 (selection favors elimination of Am and A2) or > 1 (selection favors elimination of Af and A1). For each model, the effective strength of directional selection depends on the magnitude of the term NLf(si, sj, h) in Equation 9. For simplicity, we provide results below for the case of net directional selection against allele Am or A2 ( < 0). Given the symmetry of these models, analysis of positive selection for Am and A2 leads to a similar set of conclusions.
Under strong net directional selection against Am or A2 (i.e., |NLf(si, sj, h)2| ≫ 1 and 2|| ≫ 0), Equation 9 can be approximated with the gamma distribution
| (10) |
(see Nei 1968; Orr and Unckless 2008). In this case, mean heterozygosity is , which is approximately equal to heterozygosity under the deterministic models (based on some manipulation of Equations 4–7). Thus, when |NLf(si, sj, h)2| ≫ 1 and 2|| ≫ 0, mean heterozygosity in a finite population is pseudodeterministic: 2E[q(1 – q)] ≈ 2qeq(1 – qeq).
Although the above approximations apply to much of the parameter range of net directional selection, they break down for parameter sets approaching the boundary between directional and balancing selection (i.e., near ≈ 0). In this case, there are two potential evolutionary outcomes. If the selection term is sufficiently small (i.e., NLf(si, sj, h) → 0), Equation 9 converges to a beta distribution with parameters α = β = 2NLu, which is the same as a neutral locus under symmetrical mutation. Assuming weak mutation (NLu ≪ 1), mean heterozygosity at the locus will be 2E[q(1 – q)] ≈ 2NLu. If instead the selection term NLf(si, sj, h) remains large (but || is near zero), Equation 9 takes a form that is very similar to models of selection against completely recessive deleterious alleles (see Wright 1937; Nei 1968, 1969; Hedrick 2002). Under strong mutation (NLu > 1), heterozygosity is similar to the deterministic case: E[2q(1 – q)] ≈ . When mutation is weak (NLu ≪ 1), mean heterozygosity is approximately , which is lower than in the deterministic case.
The efficacy of balancing selection:
For parameter conditions that generate balancing selection (those parameter sets leading to 0 < < 1), the efficacy with which selection maintains both alleles at a locus is extremely sensitive to the location of the deterministic equilibrium for a pair of alleles () and population size. This is best depicted visually by plotting allele frequency distributions under weak mutation (NLu ≪ 1). Effective selection will generate a tight probability distribution with a center of mass surrounding . Inefficient selection results in a distribution spanning a greater range of q, with high probability density near one or both boundary states (q = 0 and/or q = 1). In exaggerated cases, a locus under balancing selection can resemble the U-shaped distribution characteristic of a neutrally evolving locus.
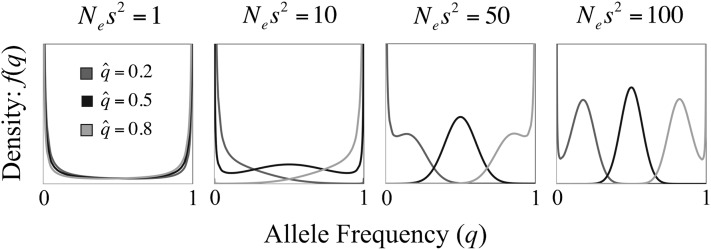
Under models of sexual antagonism and antagonistic pleiotropy, the efficacy of balancing selection is generally very weak, particularly when alleles have additive effects on fitness (h ≈ 1/2) and/or the deterministic equilibrium associated with a given parameter set is close to zero or one (Figures 3 and 4). For the case of strictly additive allelic effects, the strength of balancing selection is determined by the compound parameters Ne(sm2 + sf2) or Nes1s2, under sexual antagonism and antagonistic pleiotropy, respectively. Interestingly, large values of Nesi for each sex or context of selection—the standard criteria for effectively strong selection—may nevertheless yield sufficiently small values of Nesi2 or Nes1s2 (i.e., on order of 1; Figure 3), so that antagonistically selected alleles evolve as if they are neutral. For example, in species like Drosophila with population size on the order of Ne = 106, Nesi = 1000 corresponds to Nesi2 = 1. Although Nesi = 1000 would represent very strong selection under models of directional or overdominant selection, it corresponds to effectively weak selection under antagonistic pleiotropy or sexual antagonism. For the specific case mentioned, allele frequency spectra would be almost indistinguishable from those of a neutrally evolving locus (see Figure 3, left).
Figure 3.
The efficacy of balancing selection as a function of Nes2. The four graphs plot the allele frequency distributions corresponding to deterministic balanced polymorphic equilibria () under sexually antagonistic selection, with autosomal inheritance and additive allelic effects (hf = hm = 0.5). For each case NA = 2Ne = 20,000 and s = max(sm, sf) = 0.01. Results are based on Equation 9, with weak recurrent mutation (Neu → 0). Qualitatively similar results emerge in additive models of antagonistic pleiotropy and X-linked sexual antagonism.
Figure 4.
The efficacy of balancing selection is enhanced by recessive sexually antagonistic fitness costs. Results are based on Equation 9, ignoring recurrent mutation (Neu → 0). For each case NA = 4NX/3 = 2Ne = 10,000, Nes2 = 1, and Nes = 100, where s = max(sm, sf) = 0.01. Models of antagonistic pleiotropy with dominance reversals produce results similar to that of autosomal sexual antagonism.
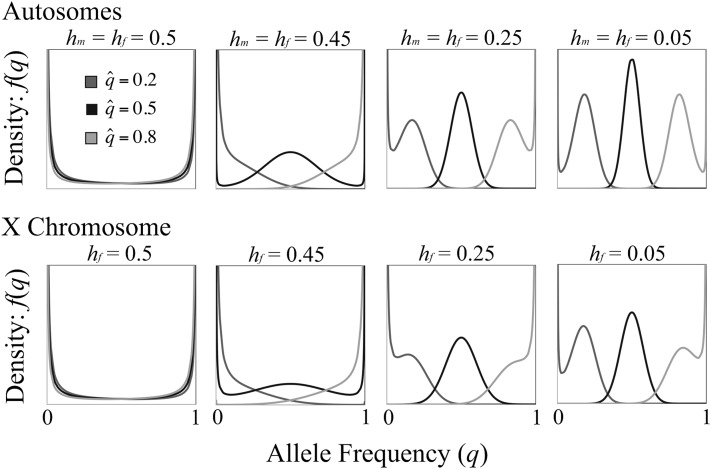
The efficacy of balancing selection increases drastically when the fitness costs of antagonistic alleles are at least partially recessive (Figure 4; hi, hj < 1/2 for the autosome model; hf < 1/2 for the X-linked sexual antagonism model). Partial masking in heterozygotes of conditionally deleterious alleles (i.e., dominance reversals) can generate net overdominance for fitness, averaged across each context of selection. Dominance reversals increase the parameter range favoring polymorphism (as previously shown; e.g., Kidwell et al. 1977; Curtsinger et al. 1994; Fry 2010) and also increase the efficacy with which both alleles are maintained. Under dominance reversal conditions, the effective strength of balancing selection is on order 2Nesi(1 – 2hi), which may be orders of magnitude greater than Nesi2 (Figure 4).
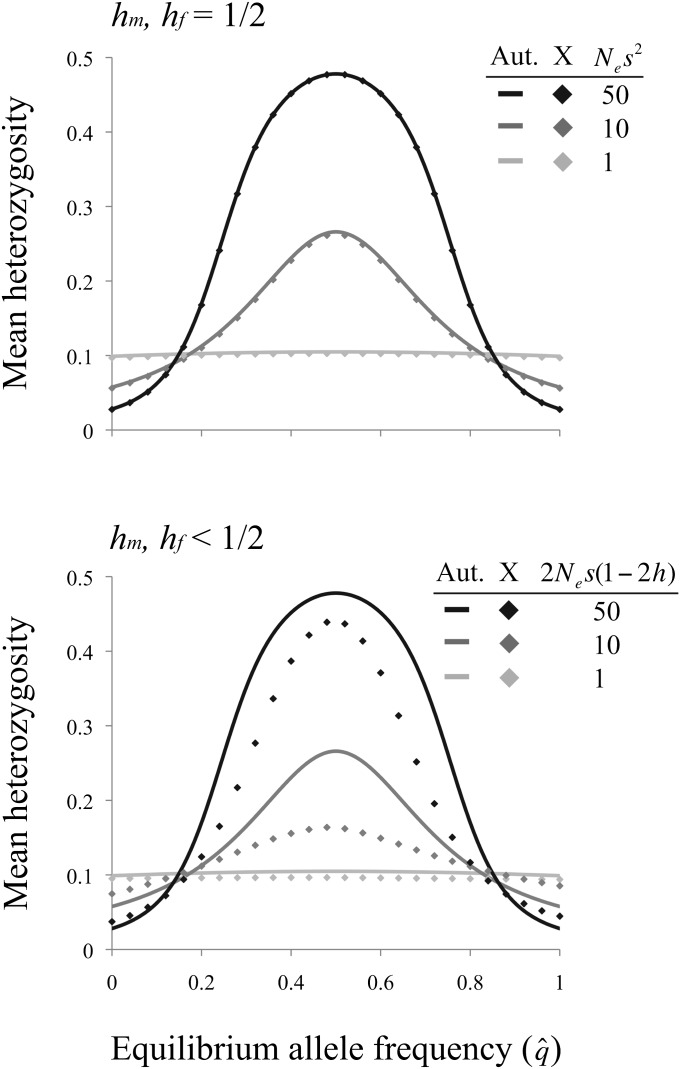
The inherently weak efficacy of antagonistic balancing selection may drastically influence equilibrium heterozygosity in populations of variable sizes (since fitness variance is a function of heterozygosity, the following results also apply to the relative fitness variance of small vs. large populations). For the most extreme case of balancing selection with Nesi2 < 1 and additive fitness effects (hi = hj = 0.5), the allele frequency spectrum approaches that of a beta distribution with parameters α = β = 2NLu. Assuming weak mutation per locus (NLu ≪ 1), heterozygosity is simply a function of the population mutation rate: 2E[q(1 – q)] ≈ 2NLu. In contrast, heterozygosity in a large (effectively deterministic) population is 2qeq(1 – qeq), with qeq defined entirely by the set of selection and dominance parameters (see above). Deterministic heterozygosity falls within the range 0 < 2qeq(1 – qeq) < 1/2. From these results, we might expect heterozygosity to be greater in a small population when the deterministic balanced polymorphic equilibrium is close to zero or one and greater in a large population when the deterministic equilibrium is at intermediate frequency. Such intuition is supported by numerical analysis of Equation 9 across a range of parameter space (Figure 5). Under models of antagonistic selection at an autosomal locus, the efficacy of balancing selection increases with Nesi2 (under the additive model) or Nesi(1 – 2h) (under dominance reversal conditions), yielding reduced heterozygosity near = 0 and = 1, and increased heterozygosity across the remaining range of . Qualitatively similar results emerge under X-linked sexual antagonism.
Figure 5.
Mean heterozygosity at sexually antagonistic loci under balancing selection. Results condition on both alleles segregating in the population. In other words, mean heterozygosity is , with f(q) given in Equation 9. Results are shown for NA = 4NX/3 = 2Ne, s = max(sm, sf), and Neu = 0.01. For the case of X linkage with dominance (hm = hf < 1/2, bottom), results are shown for hf = 1/4; lower values of hf increase the disparity between the X and autosomes. Models of antagonistic pleiotropy produce results similar to that of autosomal sexual antagonism.
Discussion
Antagonistic selection can potentially emerge as a consequence of evolutionary tradeoffs between fitness components, or from sexually dimorphic fitness landscapes. The basic environmental and genetic conditions that give rise to antagonistic selection—discordant selection and genetic correlations between traits or between the sexes—appear to be common in natural populations (Curtsinger et al. 1994; Roff 1996; Delph 2007; Cox and Calsbeek 2009; Poissant and Coltman 2009; Poissant et al. 2010), yet we currently know very little of the underlying population genetic details of antagonistically selected genes. The critical question going forward is whether antagonistic selection gives rise to distinct and readily measurable patterns of variation, which might be used to address hypotheses about the role of antagonism in maintaining population or quantitative genetic variation.
Previous theory overwhelmingly focuses on defining the parameter conditions for balancing selection at antagonistically selected loci (see Introduction), and this has inspired two main conclusions about the relationship between genetic variation and antagonistic selection. First, because the necessary parameter conditions for balancing selection are often highly restrictive, the contribution of antagonistically selected mutations to population or quantitative genetic variation is often predicted to be minor (e.g., Curtsinger et al. 1994; Hedrick 1999). This interpretation has been criticized, as it ignores the important interaction between selection and recurrent mutation that likely mediates a large proportion of molecular and fitness variation, genome-wide (Lynch and Walsh 1998; Radwan 2008). Nevertheless, few theoretical studies explicitly consider mutation–selection equilibrium variation for antagonistically selected loci and how this might compare to a null model of variation maintained at nonantagonistically selected loci. Second, by focusing on deterministic evolutionary dynamics, prior theory tacitly assumes that the parameter conditions generating balancing selection are sufficient for the maintenance of stable, balanced polymorphisms (but see Hedrick 1999). Yet, one might expect the role of genetic drift and population size to be particularly important during the evolution of antagonistically selected loci. Genetic drift becomes an increasingly important process when the rate of directional change at a locus is slow (e.g., Wright 1945; Rice 2004). While slow evolutionary dynamics should often characterize antagonistically selected loci (Livingstone 1992), the connection between sexual antagonism, antagonistic pleiotropy, and genetic drift has not been previously considered.
The results presented here are motivated by the need for a theory of antagonistic selection that takes into account the interactions between mutation, selection, and genetic drift in a finite population. Given the growing interest in antagonistic selection and its evolutionary genomic consequences, our major theoretical results are applied to two important—and measurable—quantities in evolutionary genetics: genetic variance for fitness (or its components) and molecular population genetic variation. Below, we summarize our major conclusions and discuss their relevance to empirical evolutionary genetics and genomics.
The maintenance of fitness variation by antagonistic selection
Loci evolving under antagonistic selection may evolve under net directional (averaged across contexts of selection) or balancing selection, and as a result, allele frequency equilibria may evolve at a balance between the opposing forces of mutation and net directional selection or by the active maintenance of polymorphism by selection. By characterizing equilibrium allele frequencies across the full parameter range of sexual antagonism and antagonistic pleiotropy, we show that fitness variance should generally be inflated by antagonistic selection, whether or not conditions for balanced polymorphism are met. This finding is in line with prior verbal models, which posit that mutations with opposing fitness effects between contexts of selection should disproportionately contribute to population variation because they are slowly removed from the population relative to unequivocally deleterious or beneficial mutations (Lynch and Walsh 1998, p. 656).
We show that the proportional rate of directional change under antagonism may be as low as the sum of squared selection coefficients (under sexual antagonism: sm2 + sf2) or the product of context-specific selection coefficients (under antagonistic pleiotropy: s1s2). In contrast, directional selection against unequivocally deleterious (or beneficial) alleles predicts a rate of change proportional to the selection coefficient for each allele. If selection coefficients are small, as is generally expected, then the rate of directional change under antagonism may typically be orders of magnitude slower than the evolutionary rate under nonantagonistic directional selection. Under antagonism, net directional selection is effectively weaker, despite nontrivial fitness effects on individual components of fitness. This naturally leads to a higher mean heterozygosity under antagonistic selection, once recurrent mutation is accounted for.
The slow rate of evolutionary change expected under antagonistic selection also generates severe sensitivity of equilibrium allele frequency distributions to the effective size of a population. For loci that approach parameter conditions for balancing selection, genetic drift may often dominate over selection, even in populations that are quite large. These results are reminiscent of classic models of overdominant selection in a finite population (Robertson 1962; Rice 2004; albeit the importance of drift is more drastic under antagonistic selection) and are conceptually similar to idealized cases of spatial and temporal variation, such as Levene’s (1953) model of a panmictic population with two equally abundant niches and Haldane and Jayakar’s (1963) model of opposing selection across generations (see Prout 2000 for an overview of deterministic opposing selection models). As in the case of antagonistic pleiotropy and sexual antagonism, the rate of directional allele frequency change under environmental heterogeneity may be relatively slow if there is opposing selection between environmental contexts, which should lead to strong interactions between recurrent mutation, finite population size, and environment-specific directional selection (for related theory, see Avery 1978; Gillespie 1978; Hedrick 1974, 1976, 1978). Because the efficacy of antagonistic balancing selection is so sensitive to genetic drift, reducing population size may drastically decrease the mean heterozygosity and mean contribution to fitness variance of antagonistically selected loci. To the extent that phenotypic variation is influenced by loci under balancing selection, we expect that the relative contribution of sexually antagonistic or antagonistic pleiotropic alleles to total population variance should increase with the effective size of a population.
Sexually antagonistic variation in fitness and life-history traits has been detected in a variety of vertebrate and insect populations (e.g., Chippindale et al. 2001; Gibson et al. 2002; Calsbeek and Sinervo 2004; Fedorka and Mousseau 2004; Pischedda and Chippindale 2006; Brommer et al. 2007; Foerster et al. 2007; Long and Rice 2007; Calsbeek and Bonneaud 2008; Connallon and Jakubowski 2009; Delcourt et al. 2009; Cox and Calsbeek 2010; Innocenti and Morrow 2010; Mokkonen et al. 2011), which suggests that such variation makes an important contribution to total genetic variance for fitness. While observations of sexually antagonistic variation are often interpreted through the lens of balancing selection models, it is also plausible that much of this antagonistic variation is maintained by a combination of recurrent mutation, antagonistic selection, and genetic drift. If so, the genetic basis of antagonistically selected traits may therefore differ between subpopulations of a species, and the specific sexually antagonistic alleles in a population may be transiently rather than stably maintained.
Interpretations of the genomic distribution of antagonistic variation—e.g., between the X or Z and autosomes—also commonly assume that balancing selection, rather than mutation, is the most important factor maintaining sexually antagonistic variation. To the extent that antagonistic variation is mutation driven, we might expect considerable lineage-specific differences in the relative proportion of antagonistic variation that is sex linked. Such lineage differences may be driven, in part, by variation in the ratio of effective population size between sex-linked and autosomal loci, which can be influenced by mating system, recombination and hitchhiking, and population demography (e.g., Charlesworth 2001, 2009; Betancourt et al. 2004; Pool and Nielsen 2008; Ellegren 2009). Sex-biased mutation may also affect equilibrium variation between chromosomes, by inflating the sex-linked mutation rate in species with a Z chromosome or the autosomal mutation rate in species with an X (Ellegren 2007; Hedrick 2007). Finally, some methods for detecting sexually antagonistic variation are biased toward finding large sex-linked effects, even when levels of genetic diversity are similar between chromosomes (Connallon 2010). This array of contributing factors may complicate attempts to explain why and how sex chromosomes might disproportionately contribute to the maintenance of genetic variation for fitness.
Inferences of selection from molecular population genomic data
Molecular genetic variation is influenced by natural selection in the wild. Population and comparative genomic data provide a partial record of past and ongoing adaptation, which has proven extremely useful for characterizing the statistical properties of selection on individual loci or mutations (Nielsen 2005; Eyre-Walker 2006; Eyre-Walker and Keightley 2007; Wright and Andolfatto 2008). To this end, measures of DNA sequence variation and evolutionary constraint are often used to estimate the proportion of a genome that is functionally relevant to the fitness of organisms, to quantify details of the fitness effect distribution of new mutations and segregating genetic variation, or to locate DNA linkage blocks whose genealogical structure indicates recent or ongoing selection.
Inferences of directional selection are typically based on measures of evolutionary constraint between species, taken to represent the proportion of sites that are functionally important for fitness (Wright and Andolfatto 2008), or by model fitting, where the observed allele frequency distribution across many sites in a genome is compared to predicted distributions under a model of directional selection per site and a hypothetical distribution of selection coefficients among sites (reviewed in Eyre-Walker and Keightley 2007). Under the assumption of directional selection, these methods can be used to recover aspects of the distribution of Nes—the effective strength of directional selection against deleterious mutations. If directional selection is instead context dependent, leading to cases of antagonism between fitness components or between the sexes, then these studies will generally interpret the genome as being less functionally constrained than it actually is. Some regions of parameter space under antagonistic selection can generate patterns that are indistinguishable from a neutral model. The parameter regions outside of this pseudoneutral range also shift the site frequency spectrum toward more intermediate frequency alleles (compared to directional selection), which would be interpreted as weak selection under standard assumptions.
Balancing selection, when effective, can indefinitely maintain polymorphism at a locus. Supposing that some balancing selection occurs within a genome, one might locate candidate genes by identifying shared alleles between closely related species (Asthana et al. 2005), by finding gene genealogies with exceptionally long internal branches compared to the rest of the genome (Kaplan et al. 1988; Hudson and Kaplan 1988; Charlesworth and Charlesworth 2010), or by finding genomic regions with an excess abundance of sites with intermediate allele frequency (Andrés et al. 2009, 2010). To date, such loci appear to be exceedingly rare, which is taken to mean that: (i) balancing selection rarely occurs, or (ii) balancing selection is too temporally fleeting to leave a recognizable pattern in the molecular data. A third plausible option emerges from our analysis: if antagonism is the primary driver of balancing selection, it might be largely ineffective at stably maintaining variation at a locus. Supposing that antagonistic selection affects many loci in a genome, a subset of these may meet conditions for balancing selection. However, such balancing selection may typically be weak, either because the deterministic equilibrium frequency for one of the alleles is close to zero (as would apply also to overdominant selection models; Robertson 1962), or instead because the efficacy of selection under antagonism is inherently weak, even in very large populations. This possibility will be particularly relevant under additive conditions, where the efficacy of balancing selection is proportional to Ne(sm2 + sf2) or Nes1s2, under sexual antagonism and antagonistic pleiotropy, respectively. Thus, related species may not share polymorphism even when these are selected for in both lineages. The genealogical method also depends on alleles being stably maintained for a long time, causing genomic scans for loci under balancing selection to often fail, despite their presence.
Implications for the resolution of antagonistic selection
Antagonistic selection should favor the evolution of dimorphic gene expression patterns between different tissues, life-history stages, or between the sexes. If mutations with sex- or tissue-limited effects are not available to evolution, then evolutionary transitions to dimorphic gene expression may require the fixation of “complex substitutions,” such that pairs of epistatically interacting loci (for fitness) coevolve during population transitions to dimorphic expression. As suggested previously, the resolution of antagonistic selection potentially involves coevolution between antagonistic loci and cis- or trans-modifiers, gene duplication and specialization of paralogs, or genomic imprinting (Rice 1984; Day and Bonduriansky 2004; Proulx and Phillips 2006; Ellegren and Parsch 2007; Innan and Kondrashov 2010).
Such scenarios of adaptation predict that the strength and/or speed with which evolution can resolve adaptive conflict depends heavily on the initial frequencies of segregating alleles at the loci subject to antagonistic selection (e.g., Connallon and Clark 2010, 2011). For example, if antagonistic alleles segregate at intermediate frequencies, modifier alleles and gene duplicates are readily selected for, whereas when variation is low or absent at an antagonistic locus, the mean waiting time until the resolution of conflict at the locus is typically extended by several orders of magnitude. A well-known result from theoretical population genetics is that the waiting time until establishment of a favored allele is a decreasing function of the effective population size (Wright 1949). Larger populations are expected to respond genetically to selection more rapidly than small populations. This qualitative difference between large and small populations should become amplified under complex substitution models, because the rate of response at a interacting locus depends on allele frequencies at the original, antagonistic locus. As we show above, there may be little opportunity in small populations for stable balanced polymorphism at antagonistic loci, in which case, selection for modifier alleles, new gene duplicates, or genomic imprinting may be sufficiently weak as to render these evolutionary pathways largely inaccessible. Lineages with relatively small effective population size may instead resolve antagonism by either waiting for mutations with context- or sex-specific effects to arise and become fixed (e.g., Darwin 1871; Rhen 2000; Coyne et al. 2008) or for the environment to change and thereby eliminate antagonistic selection (such environmental changes may include mating system divergence: see Seger and Trivers 1986; Albert and Otto 2005).
Supplementary Material
Acknowledgments
This work benefitted from discussions with J. R. Arguello, G. Arnqvist, M. Cardoso-Moreira, R. P. Meisel, E. H. Morrow, A. Uesugi, and R. L. Unckless and from comments by two anonymous reviewers. This work was supported by National Institutes of Health grant R01 GM064590 to A. G. Clark and A. B. Carvalho.
Footnotes
Communicating editor: W. Stephan
Literature Cited
- Albert A. Y. K., Otto S. P., 2005. Sexual selection can resolve sex-linked sexual antagonism. Science 310: 119–121 [DOI] [PubMed] [Google Scholar]
- Andrés A. M., Hubisz M. J., Indap A., Torgerson D. G., Degenhardt J. D., et al. , 2009. Targets of balancing selection in the human genome. Mol. Biol. Evol. 26: 2755–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S. Q., et al. , 2010. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6: e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G., 2011. Assortative mating by fitness and sexually antagonistic genetic variation. Evolution 65: 2111–2116 [DOI] [PubMed] [Google Scholar]
- Asthana S., Schmidt S., Sunyaev S., 2005. A limited role for balancing selection. Trends Genet. 21: 30–32 [DOI] [PubMed] [Google Scholar]
- Avery P. J., 1978. Selection effects in a model of two intermigrating colonies of finite size. Theor. Popul. Biol. 13: 24–39 [DOI] [PubMed] [Google Scholar]
- Babcock C. S., Asmussen M. A., 1996. Effects of differential selection in the sexes on cytonuclear polymorphism and disequilibria. Genetics 144: 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. H., 1957. Selectively balanced polymorphism at a sex-linked locus. Nature 180: 1363–1364 [DOI] [PubMed] [Google Scholar]
- Betancourt A. J., Kim Y., Orr H. A., 2004. A pseudohitchhiking model of X vs. autosomal diversity. Genetics 168: 2261–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky R., Chenoweth S. F., 2009. Intralocus sexual conflict. Trends Ecol. Evol. 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Brommer J. E., Kirkpatrick M., Qvarnström A., Gustafsson L., 2007. The intersexual genetic correlation for lifetime fitness in the wild and its implications for sexual selection. PLoS ONE 2: e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsbeek R., Bonneaud C., 2008. Postcopulatory fertilization bias as a form of cryptic sexual selection. Evolution 62: 1137–1148 [DOI] [PubMed] [Google Scholar]
- Calsbeek R., Sinervo B., 2004. Within-clutch variation in offspring sex determined by differences in sire body size: cryptic mate choice in the wild. J. Evol. Biol. 17: 464–470 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1987. The heritability of fitness, pp. 21–40 Sexual Selection: Testing the Alternatives, edited by Bradbury J. W., Andersson M. B. Wiley, New York [Google Scholar]
- Charlesworth B., 2001. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 77: 153–166 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2009. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts, Greenwood Village, CO [Google Scholar]
- Charlesworth B., Hughes K. A., 1999. The maintenance of genetic variation in life history traits, pp. 369–392 Evolutionary Genetics: From Molecules to Morphology, edited by Singh R. S., Krimbas C. B. Cambridge University Press, Cambridge, UK [Google Scholar]
- Chippindale A. K., Gibson J. R., Rice W. R., 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., 2010. Genic capture, sex linkage, and the heritability of fitness. Am. Nat. 175: 564–576 [DOI] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64: 3417–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2011. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T., Jakubowski E., 2009. Association between sex ratio distortion and sexually antagonistic fitness consequences of female choice. Evolution 63: 2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. M., Calsbeek R., 2009. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am. Nat. 173: 176–187 [DOI] [PubMed] [Google Scholar]
- Cox R. M., Calsbeek R., 2010. Cryptic sex ratio bias provides indirect genetic benefits despite sexual conflict. Science 328: 92–94 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Kay E. H., Pruett-Jones S., 2008. The genetic basis of sexual dimorphism in birds. Evolution 62: 214–219 [DOI] [PubMed] [Google Scholar]
- Curtsinger J. W., 1980. On the opportunity for polymorphism with sex-linkage or haplodiploidy. Genetics 96: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger J. W., Service P. M., Prout T., 1994. Antagonistic pleiotropy, reversal of dominance, and genetic polymorphism. Am. Nat. 144: 210–228 [Google Scholar]
- Darwin C., 1871. The Descent of Man and Selection in Relation to Sex. Murray, London [Google Scholar]
- Day T., Bonduriansky R., 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167: 1537–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt M., Blows M. W., Rundle H. D., 2009. Sexually antagonistic genetic variance in an ancestral and a novel environment. Proc. Biol. Sci. 276: 2009–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph L. F., 2007. The genetic integration of sexually dimorphic traits in the dioecious plant, Silene latifolia, pp. 115–123 in Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism, edited by Fairbairn D. J., Blanckenhorn W. U. Oxford University Press, New York [Google Scholar]
- Ellegren H., 2007. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc. Biol. Sci. 274: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., 2009. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 25: 278–284 [DOI] [PubMed] [Google Scholar]
- Ellegren H., Parsch J., 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8: 689–698 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. 21: 569–575 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Pearson, Essex, UK [Google Scholar]
- Fedorka K. M., Mousseau T. A., 2004. Female mating bias results in conflicting sex-specific offspring fitness. Nature 429: 65–67 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1958. The Genetical Theory of Natural Selection, Ed. 2 Dover, New York [Google Scholar]
- Foerster K., Coulson T., Sheldon B. C., Pemberton J. M., Clutton-Brock T. H., et al. , 2007. Sexually antagonistic genetic variation for fitness in red deer. Nature 447: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Fry J. D., 2010. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution 64: 1510–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S., Rice W. R., 2006. Genetic models of homosexuality: generating testable predictions. Proc. Biol. Sci. 273: 3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. R., Chippindale A. K., Rice W. R., 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. Biol. Sci. 269: 499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1978. A general model to account for enzyme variation in natural populations. V. The SAS-CFF model. Theor. Popul. Biol. 14: 1–45 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1962. Conditions for stable polymorphism at an autosomal locus. Nature 193: 1108. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., Jayakar S. D., 1963. Polymorphism due to selection of varying direction. J. Genet. 58: 237–242 [Google Scholar]
- Haldane J. B. S., Jayakar S. D., 1964. Equilibria under natural selection at a sex-linked locus. J. Genet. 59: 29–36 [Google Scholar]
- Hedrick P. W., 1974. Genetic variation in a heterogeneous environment. I. Temporal heterogeneity and the absolute dominance model. Genetics 78: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 1976. Genetic variation in a heterogeneous environment. II. Temporal heterogeneity and directional selection. Genetics 84: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 1978. Genetic variation in a heterogeneous environment. V. Spatial heterogeneity in finite populations. Genetics 89: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 1999. Antagonistic pleiotropy and genetic polymorphism: a perspective. Heredity 82: 126–133 [Google Scholar]
- Hedrick P. W., 2002. Lethals in finite populations. Evolution 56: 654–657 [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 2007. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution 61: 2750–2771 [DOI] [PubMed] [Google Scholar]
- Houle D., 1991. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution 45: 630–648 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kondrashov F., 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Innocenti P., Morrow E. H., 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan C. Y., Charlesworth D., 2011. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66: 505–516 [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Darden T., Hudson R. R., 1988. The coalescent process in models with selection. Genetics 120: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell J. F., Clegg M. T., Stewart F. M., Prout T., 1977. Regions of stable equilibria for models of differential selection in the two sexes under random mating. Genetics 85: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Guerrero R. F., Scarpino S. V., 2010. Patterns of neutral genetic variation on recombining sex chromosomes. Genetics 184: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H., 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87: 331–333 [Google Scholar]
- Li C. C., 1967. Genetic equilibrium under selection. Biometrics 23: 397–484 [PubMed] [Google Scholar]
- Livingstone F. B., 1992. Polymorphism and differential selection for the sexes. Hum. Biol. 64: 649–657 [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., Bartolomé C., Nöel V., 2005. Estimating selection on nonsynonymous mutations. Genetics 172: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T. A. F., Rice W. R., 2007. Adult locomotory activity mediates intralocus sexual conflict in a laboratory-adapted population of Drosophila melanogaster. Proc. Biol. Sci. 274: 3105–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh J. B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mandel S. P. H., 1971. Owen’s model of a genetical system with differential viability between sexes. Heredity 26: 49–63 [DOI] [PubMed] [Google Scholar]
- Mank J. E., 2009. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am. Nat. 173: 141–150 [DOI] [PubMed] [Google Scholar]
- Mérat P., 1969. Different selection in the two sexes: general discussion of the opportunities for balance for an autosomal locus with two alleles. Ann. Genet. Sel. Anim. 1: 49–65 (in French) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilä J., Sheldon B. C., 1999. Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity 83: 103–109 [DOI] [PubMed] [Google Scholar]
- Mokkonen M., Kokko H., Koskela E., Lehtonen J., Mappes T., et al. , 2011. Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science 334: 972–974 [DOI] [PubMed] [Google Scholar]
- Mukai T., Cardellino R. A., Watanabe T. K., Crow J. F., 1974. The genetic variance for viability and its components in a local population of Drosophila melanogaster. Genetics 78: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1968. The frequency distribution of lethal chromosomes in finite populations. Proc. Natl. Acad. Sci. USA 60: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1969. Heterozygous effects and frequency changes of lethal genes in populations. Genetics 63: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Orr H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949 [DOI] [PubMed] [Google Scholar]
- Orr H. A., Unckless R. L., 2008. Population extinction and the genetics of adaptation. Am. Nat. 172: 160–169 [DOI] [PubMed] [Google Scholar]
- Otto S. P., 2004. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. Biol. Sci. 271: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen A. R. G., 1953. A genetical system admitting of two distinct stable equilibria under natural selection. Heredity 7: 97–102 [Google Scholar]
- Pamilo P., 1979. Genetic variation at sex-linked loci: quantification of regular selection models. Hereditas 91: 129–133 [DOI] [PubMed] [Google Scholar]
- Parsons P. A., 1961. The initial progress of new genes with viability differences between the sexes and with sex linkage. Heredity 16: 103–107 [Google Scholar]
- Patten M. M., Haig D., 2009a Maintenance or loss of genetic variation under sexual and parental antagonism at a sex-linked locus. Evolution 63: 2888–2895 [DOI] [PubMed] [Google Scholar]
- Patten M. M., Haig D., 2009b Parental sex discrimination and intralocus sexual conflict. Biol. Lett. 5: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten M. M., Haig D., Úbeda F., 2010. Fitness variation due to sexual antagonism and linkage disequilibrium. Evolution 64: 3638–3642 [DOI] [PubMed] [Google Scholar]
- Pischedda A., Chippindale A. K., 2006. Intralocus sexual conflict diminishes the benefits of sexual selection. PLoS Biol. 4: 2099–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J., Coltman D. W., 2009. The ontogeny of cross-sex genetic correlations: an analysis of patterns. J. Evol. Biol. 22: 2558–2562 [DOI] [PubMed] [Google Scholar]
- Poissant J., Wilson A. J., Coltman D. W., 2010. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution 64: 97–107 [DOI] [PubMed] [Google Scholar]
- Pool J. E., Nielsen R., 2008. The impact of founder events on chromosomal variability in multiply mating species. Mol. Biol. Evol. 25: 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx S. R., Phillips P. C., 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60: 881–892 [PubMed] [Google Scholar]
- Prout T., 2000. How well does opposing selection maintain variation? pp. 157–181 Evolutionary Genetics: From Molecules to Morphology, edited by Singh R. S., Krimbas C. B. Cambridge University Press, Cambridge, UK [Google Scholar]
- R Development Core Team , 2005. R: A Language and Environment for Statistical Computing, Reference Index v. 2.2.1 R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Radwan J., 2008. Maintenance of genetic variation in sexual ornaments: a review of the mechanisms. Genetica 134: 113–127 [DOI] [PubMed] [Google Scholar]
- Rhen T., 2000. Sex-limited mutations and the evolution of sexual dimorphism. Evolution 54: 37–43 [DOI] [PubMed] [Google Scholar]
- Rice S. H., 2004. Evolutionary Theory: Mathematical and Conceptual Foundations. Sinauer Associates, Sunderland, MA [Google Scholar]
- Rice W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742 [DOI] [PubMed] [Google Scholar]
- Rice W. R., Chippindale A. K., 2001. Intersexual ontogentic conflict. J. Evol. Biol. 14: 685–693 [Google Scholar]
- Robertson A., 1962. Selection for heterozygotes in small populations. Genetics 47: 1291–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. A., 1996. The evolution of genetic correlations: an analysis of patterns. Evolution 50: 1392–1403 [DOI] [PubMed] [Google Scholar]
- Rose M. R., 1982. Antagonistic pleiotropy, dominance, and genetic variation. Heredity 48: 63–78 [Google Scholar]
- Rose M. R., 1985. Life history evolution with antagonistic pleiotropy and overlapping generations. Theor. Popul. Biol. 28: 342–358 [Google Scholar]
- Seger J., Trivers R., 1986. Asymmetry in the evolution of female mating preferences. Nature 319: 771–773 [Google Scholar]
- Stewart A. D., Pischedda A., Rice W. R., 2010. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J. Hered. 101: S94–S99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda F., Haig D., Patten M. M., 2011. Stable linkage disequilibrium owing to sexual antagonism. Proc. Biol. Sci. 278: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Herren J. K., 2009. Population genetics of sexually antagonistic mitochondrial mutants under inbreeding. J. Theor. Biol. 260: 132–136 [DOI] [PubMed] [Google Scholar]
- van Doorn G. S., 2009. Intralocus sexual conflict. Ann. N. Y. Acad. Sci. 1168: 52–71 [DOI] [PubMed] [Google Scholar]
- Wright S., 1937. The distributions of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1945. The differential equation of the distribution of gene frequencies. Proc. Natl. Acad. Sci. USA 31: 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1949. Adaptation and selection, pp. 365–389 in Genetics, Paleontology and Evolution, edited by Jepsen G. L., Mayr E., Simpson G. G. Princeton University Press, Princeton [Google Scholar]
- Wright S. I., Andolfatto P., 2008. The impact of selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 39: 193–213 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.