Abstract
Genomic selection can increase genetic gain per generation through early selection. Genomic selection is expected to be particularly valuable for traits that are costly to phenotype and expressed late in the life cycle of long-lived species. Alternative approaches to genomic selection prediction models may perform differently for traits with distinct genetic properties. Here the performance of four different original methods of genomic selection that differ with respect to assumptions regarding distribution of marker effects, including (i) ridge regression–best linear unbiased prediction (RR–BLUP), (ii) Bayes A, (iii) Bayes Cπ, and (iv) Bayesian LASSO are presented. In addition, a modified RR–BLUP (RR–BLUP B) that utilizes a selected subset of markers was evaluated. The accuracy of these methods was compared across 17 traits with distinct heritabilities and genetic architectures, including growth, development, and disease-resistance properties, measured in a Pinus taeda (loblolly pine) training population of 951 individuals genotyped with 4853 SNPs. The predictive ability of the methods was evaluated using a 10-fold, cross-validation approach, and differed only marginally for most method/trait combinations. Interestingly, for fusiform rust disease-resistance traits, Bayes Cπ, Bayes A, and RR–BLUB B had higher predictive ability than RR–BLUP and Bayesian LASSO. Fusiform rust is controlled by few genes of large effect. A limitation of RR–BLUP is the assumption of equal contribution of all markers to the observed variation. However, RR-BLUP B performed equally well as the Bayesian approaches.The genotypic and phenotypic data used in this study are publically available for comparative analysis of genomic selection prediction models.
P LANT and animal breeders have effectively used phenotypic selection to increase the mean performance in selected populations. For many traits, phenotypic selection is costly and time consuming, especially so for traits expressed late in the life cycle of long-lived species. Genome-wide selection (GWS) (Meuwissen et al. 2001) was proposed as an approach to accelerating the breeding cycle. In GWS, trait-specific models predict phenotypes using dense molecular markers from a base population. These predictions are applied to genotypic information in subsequent generations to estimate direct genetic values (DGV).
Several analytical approaches have been proposed for genome-based prediction of genetic values, and these differ with respect to assumptions about the marker effects (de los Campos et al. 2009a; Habier et al. 2011; Meuwissen et al. 2001). For example, ridge regression–best linear unbiased prediction (RR–BLUP) assumes that all marker effects are normally distributed and that these marker effects have identical variance (Meuwissen et al. 2001). In Bayes A, markers are assumed to have different variances and are modeled as following a scaled inverse χ2 distribution (Meuwissen et al. 2001). The prior in Bayes B (Meuwissen et al. 2001) assumes the variance of markers to equal zero with probability π, and the complement with probability (1 – π) follows an inverse χ2 distribution, with v degree of freedom and scale parameter S. The definition of the probability π depends on the genetic architecture of the trait, suggesting an improvement to the Bayes B model, known as Bayes Cπ. In Bayes Cπ, the mixture probability π has a prior uniform distribution (Habier et al. 2011). A drawback of Bayesian methods is the need for the definition of priors. The requirement of a prior for the parameter π is circumvented in the Bayesian LASSO method, which needs less information (de los Campos et al. 2009b; Legarra et al. 2011b). Methods for genomic prediction of genetic values may perform differently for different phenotypes (Meuwissen et al. 2001; Usai et al. 2009; Habier et al. 2011) and results may diverge because of differences in genetic architecture among traits (Hayes et al. 2009; Grattapaglia and Resende 2011). Therefore, it is valuable to compare performance among methods with real data and identify those that provide more accurate predictions.
Recently, GWS was applied to agricultural crops (Crossa et al. 2010) and trees (Resende et al. 2011). Here we report, for an experimental breeding population of the tree species loblolly pine (Pinus taeda L.), a comparison of GWS predictive models for 17 traits with different heritabilities and predicted genetic architectures. Genome-wide selection models included RR–BLUP, Bayes A, Bayes Cπ, and the Bayesian LASSO. In addition, we evaluated a modified RR–BLUP method that utilizes a subset of selected markers, RR–BLUP B. We show that, for most traits, there is limited difference among these four original methods in their ability to predict GBV. Bayes Cπ performed better for fusiform rust resistance—a disease-resistance trait shown previously to be controlled in part by major genes—and the proposed method RR–BLUP B was similar to or better than Bayes Cπ when a subsample of markers was fitted to the model.
Materials and Methods
Training population and genotypic data
The loblolly pine population used in this analysis is derived from 32 parents representing a wide range of accessions from the Atlantic coastal plain, Florida, and lower Gulf of the United States. Parents were crossed in a circular mating design with additional off-diagonal crosses, resulting in 70 full-sib families with an average of 13.5 individuals per family (Baltunis et al. 2007a). This population is referred to hereafter as CCLONES (comparing clonal lines on experimental sites). A subset of the CCLONES population, composed of 951 individuals from 61 families (mean, 15; standard deviation, 2.2) was genotyped using an Illumina Infinium assay (Illumina, San Diego, CA; Eckert et al. 2010) with 7216 SNP, each representing a unique pine EST contig. A subset of 4853 SNPs were polymorphic in this population and were used in this study. None of the markers were excluded on the basis of minimum allele frequency. Genotypic data and pedigree information are available in the Supporting Information, File S1 and File S2.
Phenotypic data
The CCLONES population was phenotyped for growth, developmental, and disease-resistance traits in three replicated studies. The first was a field study established using single-tree plots in eight replicates (one ramet of each individual is represented in each replicate) that utilized a resolvable alpha-incomplete block design (Williams et al. 2002). In that field trial, four replicates were grown under a high-intensity and four were grown under a standard silvicultural intensity regime. The traits stem diameter (DBH, cm), total stem height (HT, cm), and total height to the base of the live crown (HTLC, cm) were measured in the eight replicates at years 6, 6, and 4, respectively. At year 6, crown width across the planting beds (CWAC, cm), crown width along the planting beds (CWAL, cm), basal height of the live crown (BLC, cm), branch angle average (BA, degrees), and average branch diameter (BD, cm) were measured only in the high-intensity silvicultural treatment. Phenotypic traits tree stiffness (Stiffness, km2/sec2), lignin content (Lignin), latewood percentage at year 4 (LateWood), wood specific gravity (Density), and 5- and 6-carbon sugar content (C5C6) were measured only in two repetitions, in the high-intensity culture (Baltunis et al. 2007a; Emhart et al. 2007; Li et al. 2007; Sykes et al. 2009).
The second study was a greenhouse disease-resistance screen. The experimental design was a randomized complete block with single-tree plots arranged in an alpha lattice with an incomplete block (tray container). Fusiform rust (Cronartium quercuum Berk. Miyable ex Shirai f. sp. fusiforme) susceptibility was assessed as gall volume (Rust_gall_vol) and presence or absence of rust (Rust_bin) (Kayihan et al. 2005; Kayihan et al. 2010).
Finally, in the third study the rooting ability of cuttings was investigated in an incomplete block design (tray container) with four complete repetitions, in a controlled greenhouse environment. Root number (Rootnum) and presence or absence of roots (Rootnum_bin) were quantified (Baltunis et al. 2005; Baltunis et al. 2007b).
Breeding value prediction
Analyses were carried out using ASReml v.2 (Gilmour et al. 2006) with the following mixed linear model,
where y is the phenotypic measure of the trait being analyzed, b is a vector of the fixed effects, i is a vector of the random incomplete block effects within replication ∼N(0, Iσ2iblk), a is a vector of random additive effects of clones, ∼N(0, Aσ2a), c is a vector of random nonadditive effects of clones ∼N(0, Iσ2c), f is a vector of random family effects ∼N(0, Iσ2f), d1 and d2 are described below, e is the vector of random residual effects ∼N(0, DIAGσ2e), X and Z1–Z6 are incidence matrices, and I, A, and DIAG are the identity, numerator relationship, and block diagonal matrices, respectively. For traits measured in the field study under both high and standard culture intensities, the model also included d1, a vector of the random additive × culture type interaction ∼N(0, DIAGσ2d1), and d2, a vector of the random family × culture type interaction ∼N(0, DIAGσ2d2). Narrow-sense heritability was calculated as the ratio of the additive variance σ2a to the total or phenotypic variance (e.g., for the field experiment total variance was σ2a + σ2n + σ2f + σ2d1 + σ2d2 + σ2e). Prior to use in GWS modeling, the estimated breeding values were deregressed into phenotypes (DP) following the approach described in Garrick et al. (2009), to remove parental average effects from each individual. Breeding values and deregressed phenotypes are available in File S3 and File S4.
Statistical methods
The SNP effects were estimated on the basis of five different statistical methods: RR–BLUP, Bayes A (Meuwissen et al. 2001), Bayes Cπ (Habier et al. 2011), the Improved Bayesian LASSO (BLASSO) approach proposed by Legarra et al. (2011b), and RR–BLUP B (a modified RR–BLUP). In all cases the genotypic information was fitted using the model
where DP is the vector of phenotypes deregressed from the additive genetic values (Garrick et al. 2009), β is the overall mean fitted as a fixed effect, m is the vector of random marker effects, and ε is the vector of random error effects, 1 is a vector of ones, and Z is the incidence matrix m, constructed from covariates based on the genotypes. No additional information, such as marker location, polygenic effects, or pedigree was used in those models.
Once the marker effects were estimated using one of the methods, the predicted DGV of individual j for that method was given by
where i is the specific allele of the ith marker on individual j and n is the total number of markers.
Random regression–best linear unbiased predictor
The RR–BLUP assumed that the SNP effects, m, were random (Meuwissen et al. 2001). The variance parameters were assumed to be unknown and were estimated by restricted maximum likelihood (REML), which is equivalent to Bayesian inference using an uninformative, flat prior. The first and second moments for this model are
where is the variance common to each marker effect and is the residual variance.
The mixed model equation for the prediction of m is equivalent to:
where refers to the total additive variance of the trait and η, due to standardization of the Z matrix, refers to the total number of markers (Meuwissen et al. 2009). The matrix Z was parameterized and standardized to have a mean of zero and variance of one as previously described (Resende et al. 2010; Resende et al. 2011). The analyses were performed in the software R (available at CRAN, http://cran.r-project.org/) and the script is available in File S5.
Bayes A
The Bayes A method proposed by Meuwissen et al. (2001) assumes the conditional distribution of each effect (given its variance) to follow a normal distribution. The variances are assumed to follow a scaled inversed χ2 distribution with degrees of freedom νa and scale parameter . The unconditional distribution of the marker effects can be shown to follow a t-distribution with mean zero (Sorensen and Gianola 2002). Bayes A differs from RR–BLUP in that each SNP has its own variance. In this study, νa was assigned the value 4, and was calculated from the additive variance according to Habier et al. (2011) as
where
and pk is the allele frequency of the kth SNP, is the variance of a given marker and is the additive genetic variance explained by the SNPs.
Bayes Cπ
Bayes Cπ was proposed by Habier et al. (2011). In this method, the SNP effects have a common variance, which follows a scaled inverse χ2 prior with parameters νa, . As a result, the effect of a SNP fitted with probability (1 − π) follows a mixture of multivariate Student's t-distributions, t(0, νa, I), where π is the probability of a marker having zero effect. Parameters νa and were chosen as described for Bayes A. The π parameter is treated as unknown with a uniform (0,1) prior distribution.
Bayes A and Bayes Cπ were performed using the software GenSel (Fernando and Garrick 2008); available at http://bigs.ansci.iastate.edu/bigsgui/) for which an R package is available in File S6. The marker input file was coded as −10, 0, and 10 for marker genotypes 0, 1, and 2, respectively. A total of 50,000 iterations were used, with the first 2000 excluded as the burn-in.
Bayesian LASSO
The Bayesian LASSO method was performed as proposed by Legarra et al. (2011b), using the same model equation used previously for the estimation of the markers effects. However, in this case:
where MNV represents a multivariate normal distribution and λ is the “sharpness” parameter.
Using a formulation in terms of an augmented hierarchical model including an extra variance component associated with each marker locus, we have:
Therefore, .
The prior distribution for was an inverted χ2 distribution with 4 degrees of freedom and expectations equal to the value used in regular genetic evaluation for . Analyses were performed using the software GS3 (Legarra et al. 2011a), available in http://snp.toulouse.inra.fr/∼alegarra/. The chain length was 100,000 iterations, with the first 2000 excluded as the burn-in and a thinning interval of 100.
RR–BLUP B
We also evaluated a modified, two-step RR–BLUP method that reduces the number of marker effects estimated. In this case, the DGV for each trait was generated on the basis of a reduced subset of markers. To define the number of markers in the subset, the marker effects from the RR–BLUP were ranked in decreasing order by their absolute values and grouped in multiples of 10 (10, 20, 30, …, 4800). Each group was used, with their original effects, to estimate DGV. The size, q, of the subset that maximized the predictive ability was selected as the optimum number of marker effects to be used in subsequent analyses. Next, markers effects were reestimated in a second RR–BLUP, using only the selected q markers within each training partition (see below). The estimated effects derived from this analysis were used to predict the merit of the individuals in the validation partition that were not present in the training partition. This process was repeated for different allocations of the data into training and validation partitions. In each validation, a different subset of markers was selected, on the basis of the highest absolute effects within that training partition. Therefore, the only restriction applied to the second analysis was related to q, the number of markers to be included in each data set. The same approach was performed with two additional subsets of markers of the same size as a control: the first subset contained randomly selected markers and the second subset contained markers with the smallest absolute effect values.
Validation of the models
Two cross-validation schemes were tested in the RR–BLUP method: 10-fold and leave-one-out. For the 10-fold cross-validation approach a random subsampling partitioning, fixed for all methods, was used (Kohavi 1995). Briefly, the data for each trait were partitioned into two subsets. The first one was composed of the majority of the individuals (90%) and was used to estimate the marker effects. The second one, the validation partition (10%), had their phenotypes predicted on the basis of the marker effects estimated in the training set. Randomly taken samples of N = (9/10) × NT individuals were used as training sets, while the remaining individuals were used for validation (NT is the total number of individuals in the population). The process was repeated 10 times, each time with a different set of individuals as the validation partition, until all individuals had their phenotypes predicted (Legarra et al. 2008; Usai et al. 2009; Verbyla et al. 2010). In the leave-one-out approach, a model was constructed using NT − 1 individuals in the training population and validated in a single individual that was not used in the training set. This was repeated NT times, such that each individual in the sample was used once as the validation individual. This method maximized the training population size.
Accuracy of the models
The correlation between the DGV and the DP was estimated using the software ASReml, v. 2 (Gilmour et al. 2006), from a bivariate analysis, including the validation groups as fixed effects since each validation group had DGV estimated from a different prediction equation and might have had a different mean. This correlation represented the predictive ability of GS to predict phenotypes and was theoretically represented (Resende et al. 2010) by
where was the accuracy of GS and h was the square root of the heritability of adjusted phenotypes, which is associated to Mendelian sampling effects and is given by
where n was the number of ramets used in each study. To remove the influence of the heritability upon the predictive ability and thus estimate the accuracy, the following formula was applied
In addition, for each method and trait, the slope coefficient for the regression of DP on DGV was calculated as a measurement of the bias of the DGV. Unbiased models are expected to have a slope coefficient of 1, whereas values greater than 1 indicate a biased underestimation in the DGV prediction and values smaller than 1 indicate a biased overestimation of the DGV.
Results
Cross-validation method
Testing the effect of cross-validation using two methods, 10-fold and leave-one-out (N-fold), showed that their predictive ability was not significantly different (Table S1). The largest difference was detected for the trait CWAC, where the leave-one-out method outperformed the 10-fold cross-validation by 0.02 (standard error, 0.03). Likewise, no significant differences were found for bias of the regressions (slope) in both methods (Table S2). Thus, the 10-fold approach was selected and used for comparing all methods.
Predictive ability of the methods
Four well-established genome-wide selection methods were compared in 17 traits with heritabilities ranging from 0.07 to 0.45. Overall, the ability to predict phenotype ranged from 0.17 for Lignin to 0.51 for BA (Table 1). Although the methods differ in a priori assumptions about marker effects, their predictive ability was similar—no significant differences were detected for any of the 17 traits. The standard errors for each method and trait are described in Table S3.
Table 1.
Predictive ability of genomic selection models using four different methods
| Methods |
||||||
| Trait category | Trait | h2 | RR–BLUP | BLASSO | Bayes A | Bayes Cπ |
| Growth | HT | 0.31 | 0.39 | 0.38 | 0.38 | 0.38 |
| HTLC | 0.22 | 0.45 | 0.44 | 0.44 | 0.44 | |
| BHLC | 0.35 | 0.49 | 0.49 | 0.49 | 0.49 | |
| DBH | 0.31 | 0.46 | 0.46 | 0.46 | 0.46 | |
| Development | CWAL | 0.27 | 0.38 | 0.36 | 0.36 | 0.36 |
| CWAC | 0.45 | 0.48 | 0.46 | 0.47 | 0.47 | |
| BD | 0.15 | 0.27 | 0.25 | 0.27 | 0.27 | |
| BA | 0.33 | 0.51 | 0.51 | 0.51 | 0.51 | |
| Rootnum_bin | 0.10 | 0.28 | 0.28 | 0.27 | 0.28 | |
| Rootnum | 0.07 | 0.24 | 0.26 | 0.25 | 0.24 | |
| Disease resistance | Rust_bin | 0.21 | 0.29 | 0.28 | 0.34 | 0.34 |
| Rust_gall_vol | 0.12 | 0.23 | 0.24 | 0.28 | 0.29 | |
| Wood quality | Stiffness | 0.37 | 0.43 | 0.39 | 0.42 | 0.42 |
| Lignin | 0.11 | 0.17 | 0.17 | 0.17 | 0.17 | |
| LateWood | 0.17 | 0.24 | 0.24 | 0.23 | 0.24 | |
| Density | 0.09 | 0.20 | 0.22 | 0.23 | 0.22 | |
| C5C6 | 0.14 | 0.26 | 0.25 | 0.25 | 0.25 | |
h2 is the narrow-sense heritability of the trait.
Bayesian approaches performed better for traits in the disease-resistance category. For Rust_bin, the methods Bayes A and Bayes Cπ were 0.05 superior than RR–BLUP and 0.06 superior to BLASSO. For Rust_gall_vol, Bayes Cπ was 0.05 superior to RR–BLUP and BLASSO. The accuracy () for each genome-wide prediction method was also estimated and varied from 0.37 to 0.77 (Table S4).
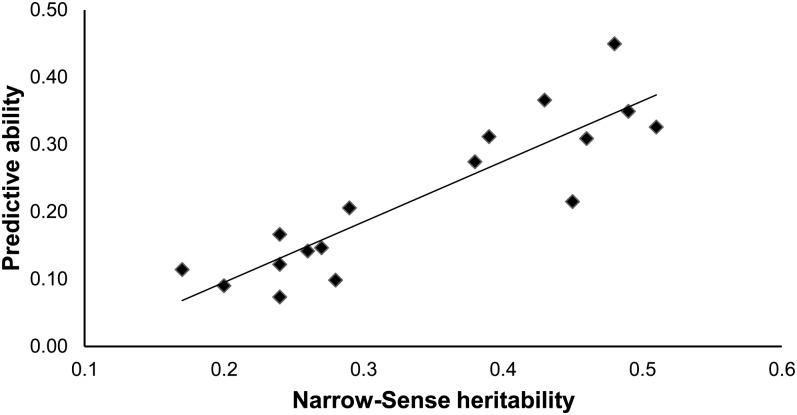
For all methods, the ability to predict phenotypes was linearly correlated with trait heritability. The strongest correlation (0.79) was observed for RR–BLUP (Figure 1). The correlation is expected, as traits with lower heritability have phenotypes less reflective of their genetic content, and are expected to be less predictable through genomic selection.
Figure 1 .
Regression of RR–BLUP predictive ability on narrow-sense heritability for 17 traits (trend line is shown, R2 = 0.79).
Bias of the methods
The coefficient of regression (slope) of DP on DGV was calculated as a measurement of the bias of each method. Ideally, a value of β equal to one indicates no bias in the prediction. For all traits, the slopes of all the models were not significantly different than one, indicating no significant bias in the prediction. In addition, no significant differences among the methods were detected (Table S5). Although no evidence of significant bias was detected, the value of β derived from RR–BLUP was slightly higher for all traits (average across traits equal to 1.18).
Markers Subset and RR–BLUP B
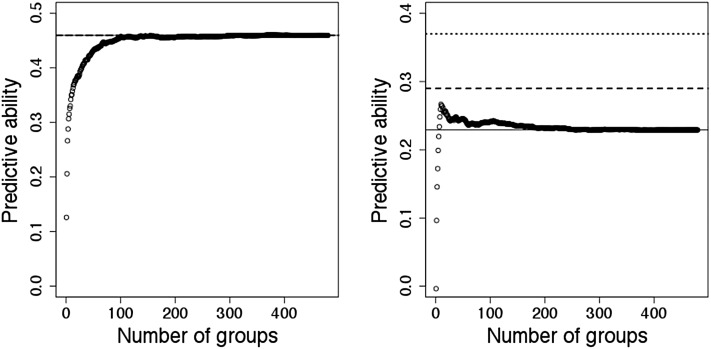
Prediction of phenotype was also performed with RR–BLUP, but adding increasingly larger marker subsets, until all markers were used jointly in the prediction. The predictive ability was plotted against the size of the subset of markers (Figure 2). The pattern of the prediction accuracy was similar for 13 out 17 traits (Figure 2A), where differences were mainly found in the rate with which the correlation reached the asymptote. In these cases, the size of the subset ranged from 820 to 4790 markers. However, a distinct pattern was detected for disease-resistance-related traits, density, and CWAL (Figure 2B). In these cases, maximum predictive ability was reached with smaller marker subsets (110–590 markers) and decreased with the addition of more markers. An additional RR–BLUP was performed using as covariates only the marker subset for which maximum predictive ability was obtained. For traits where a large number of markers (>600) explain the phenotypic variability, RR–BLUP B was similar to RR–BLUP or Bayesian methods (Table S6). However, for traits where the maximum predictive ability (Density, Rust_bin, Rust_gall_vol) was reached with a smaller number of marker (<600), RR–BLUP B performed significantly better than RR–BLUP. For example, the predictive ability of the trait Rust_gall_vol was 61% higher using RR–BLUP B (0.37) compared to the traditional RR–BLUP (0.23) and also improved relative to BLASSO (0.24), Bayes A (0.28 and Bayes Cπ (0.29).
Figure 2 .
Example of the two patterns of predictive ability observed among traits, as an increasing number of markers is added to the model. Each marker group is represented by a set of 10 markers. (Left) For DBH, the maximum predictive ability was detected when 380 groups of markers (3800 markers) were included in the model. (Right) For the trait Rust_gall_vol, predictive ability pattern reached a maximum when only 10 groups (100 markers) were added. Lines indicate the predictive ability of RR–BLUP (solid line), Bayes Cπ (dashed line), and RR–BLUP B (dotted line) as reported in Table 1 and Table S6.
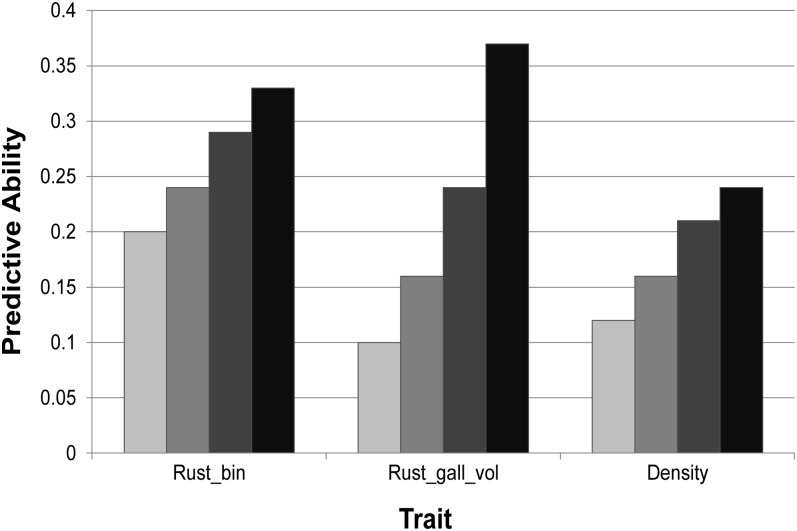
We also contrasted these results with the predictive ability using a subset of markers of similar size, but selected either randomly or to include those markers with lower effects. As expected, for the three traits the predictive ability was larger for the subset selected by RR–BLUP B over the subsets with lower effects and random effects (Figure 3). A significant difference over the lower and random subsets was found for rust-resistance-related traits (Rust_bin, Rust_gall_vol), while for Density the markers selected by RR–BLUP B were only significantly different than the lower marker subset but not different than the random marker subset.
Figure 3 .
Predictive ability for subsets of 310 markers for Rust_bin, 110 markers for Rust_gall_vol, and 240 markers for Density. Subsets were generated by selecting markers with the lowest absolute effects (light shading), with random values (medium shading), including all markers (dark shading), and including only those markers with largest absolute effects (solid).
Discussion
We characterized the performance of RR–BLUP/RR–BLUP B, Bayes A, Bayesian LASSO regression, and Bayes Cπ for GWS of growth, developmental, disease resistance, and biomass quality traits in common data set generated from an experimental population of the conifer loblolly pine. In general, the methods evaluated differed only modestly in their predictive ability (defined by the correlation between the DGV and DP).
The suitability of different methods of developing GWS predictive models is expected to be trait dependent, conditional on the genetic architecture of the characteristic. RR–BLUP differs from the other approaches used in this study in that the unconditional variance of marker effects is normally distributed, with the same variance for all markers (Meuwissen et al. 2001). This assumption may be suitable when considering an infinitesimal model (Fisher 1918), in which the characters are determined by an infinite number of unlinked and nonepistatic loci, with small effect. Not surprisingly, BLUP-based methods underperformed relative to Bayesian approaches for oligogenic traits. For instance, Verbyla et al. (2009) showed that BLUP-based GWS had lower accuracy, compared to Bayesian methods, in prediction of fat percentage in a population in which a single gene explains ∼50% of the genetic variation. Similarly, our observation that Bayes A and Bayes Cπ were more accurate in predicting fusiform rust-resistance traits, compared to RR–BLUP, may reflect a simpler genetic architecture, with a few loci of large effect. While the causative genes that regulate fusiform rust resistance have not yet been uncovered, several genetic studies support the role of few major genes in the trait variation. For example, the Fr1 locus confers resistance to specific fungus aeciospore isolates (Wilcox et al. 1996), and at least five families within the CCLONES population segregate for this locus (Kayihan et al. 2010).
The underperformance of RR–BLUP for predicting oligogenic traits is a consequence of fitting a large number of markers to model variation at a trait controlled by few major loci, leading to model overparameterization. In Bayes A and Bayes Cπ, the shrinkage of effects is marker specific, while in BLUP all markers are penalized equally. To address this limitation, we proposed an alternative, RR–BLUP B, to Bayesian and the traditional RR–BLUP approaches, aimed at reducing the number of parameters. In RR–BLUP B, marker effects are initially estimated and ranked using RR–BLUP. Next, increasing markers subsets that include initially those with larger effect are used to estimate DGV. The number of markers that maximizes the predictive ability is then defined and used in a second RR–BLUP model. For rust disease resistance and wood-density traits, the modified RR–BLUP B approach performed better that traditional RR–BLUP and as well as the Bayesian methods. Previous studies using simulated data have shown that improvements in predictive ability could be obtained by using an approach similar to the one proposed here (Zhang et al. 2010, Zhang et al. 2011), although with a different strategy of marker selection. While RR–BLUP B may add an additional step to the development of predictive models (i.e., initial marker selection), it is overall simpler and computationally less expensive than Bayesian approaches. Therefore, it may provide a suitable alternative to the use of BLUP-based methods for traits that do not fit an infinitesimal model and are rather regulated by few major loci.
Supplementary Material
Acknowledgments
The authors thank members of the Forest Biology Research Cooperative (FBRC) for their support in establishing, maintaining, and measuring field trials used in this study. The work was supported by the National Science Foundation Plant Genome Research Program (award no. 0501763 to G.F.P., J.M.D., and M.K.) and the U.S. Department of Agriculture National Institute of Food and Agriculture Plant Breeding and Education Program (award no. 2010-85117-20569 to M.K., G.F.P., and J.M.D.). We also acknowledge the valuable input from two anonymous reviewers.
Footnotes
Edited by Dirk-Jan de Koning and Lauren M. McIntyre
Literature Cited
- Baltunis B. S., Huber D. A., White T. L., Goldfarb B., Stelzer H. E., 2005. Genetic effects of rooting loblolly pine stem cuttings from a partial diallel mating design. Can. J. For. Res. 35: 1098–1108 [Google Scholar]
- Baltunis B. S., Huber D. A., White T. L., Goldfarb B., Stelzer H. E., 2007a. Genetic analysis of early field growth of loblolly pine clones and seedlings from the same full-sib families. Can. J. For. Res. 37: 195–205 [Google Scholar]
- Baltunis B. S., Huber D. A., White T. L., Goldfarb B., Stelzer H. E., 2007b. Genetic gain from selection for rooting ability and early growth in vegetatively propagated clones of loblolly pine. Tree Genet. Genomes 3: 227–238 [Google Scholar]
- Crossa J., de los Campos G., Perez P., Gianola D., Burgueno J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Rosa G. J. M., 2009a. Reproducing kernel Hilbert spaces regression: a general framework for genetic evaluation. J. Anim. Sci. 87: 1883–1887 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Naya H., Gianola D., Crossa J., Legarra A., et al. , 2009b. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert A. J., van Heerwaarden J., Wegrzyn J. L., Nelson C. D., Ross-Ibarra J., et al. , 2010. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185: 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emhart V. I., Martin T. A., White T. L., Huber D. A., 2007. Clonal variation in crown structure, absorbed photosynthetically active radiation and growth of loblolly pine and slash pine. Tree Physiol. 27: 421–430 [DOI] [PubMed] [Google Scholar]
- Fernando R., Garrick D. J., 2008. GenSel: User Manual for a Portfolio of Genomic Selection Related Analyses. Animal Breeding and Genetics, Iowa State University, Ames, IA. Available at http://bigs.ansci.iastate.edu/bigsgui; accessed January 2, 2012
- Fisher R. A., 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52: 34 [Google Scholar]
- Garrick D. J., Taylor J. F., Fernando R. L., 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R., 2006. ASReml User Guide Release 2.0. VSN International, Hemel Hempstead, UK [Google Scholar]
- Grattapaglia D., Resende M. D. V., 2011. Genomic selection in forest tree breeding. Tree Genet. Genomes 7: 241–255 [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Goddard M. E., 2009. Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92: 433–443 [DOI] [PubMed] [Google Scholar]
- Kayihan G. C., Huber D. A., Morse A. M., White T. L., Davis J. M., 2005. Genetic dissection of fusiform rust and pitch canker disease traits in loblolly pine. Theor. Appl. Genet. 110: 948–958 [DOI] [PubMed] [Google Scholar]
- Kayihan G. C., Nelson C. D., Huber D. A., Amerson H. V., White T. L., et al. , 2010. Clonal evaluation for fusiform rust disease resistance: effects of pathogen virulence and disease escape. Can. J. For. Res. 40: 1042–1050 [Google Scholar]
- Kohavi R., 1995. A study of cross-validation and bootstrap for accuracy estimation and model selection, pp. 1137–1143 in Proceedings of the Fourteenth International Joint Conference on Artificial Intelligence, edited by C. S. Mellish. Morgan Kaufmann, San Francisco [Google Scholar]
- Legarra A., Robert-Granie C., Manfredi E., Elsen J. M., 2008. Performance of genomic selection in mice. Genetics 180: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., Ricardi A., Filangi O., 2011a. GS3: Genomic Selection, Gibbs Sampling, Gauss-Seidel (and BayesCπ). http://snp.toulouse.inra.fr/∼alegarra/
- Legarra A., Robert-Granie C., Croiseau P., Guillaume F., Fritz S., 2011b. Improved LASSO for genomic selection. Genet. Res. 93: 77–87 [DOI] [PubMed] [Google Scholar]
- Li X. B., Huber D. A., Powell G. L., White T. L., Peter G. F., 2007. Breeding for improved growth and juvenile corewood stiffness in slash pine. Can. J. For. Res. 37: 1886–1893 [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Solberg T. R., Shepherd R., Woolliams J. A., 2009. A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet. Sel. Evol. 41: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende M. D., Resende M. F. R., Jr, Aguiar A. M., Abad J. I. M., Missiaggia A. A., et al. , 2010. Computação da Seleção Genômica Ampla (GWS). Embrapa Florestas, Colombo [Google Scholar]
- Resende M. F. R., Jr, Munoz P., Acosta J. J., Peter G. F., Davis J. M., et al. , 2011. Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol. 10.1111/j.1469–8137.2011.03895.x [DOI] [PubMed] [Google Scholar]
- Sorensen D., Gianola D., 2002. Likelihood, Bayesian and MCMC Methods in Quantitative Genetics. Springer-Verlag, New York [Google Scholar]
- Sykes R., Yung M., Novaes E., Kirst M., Peter G. F., et al. , 2009. High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectrometry, pp. 169–183 Biofuels: Methods and Protocols, edited by Mielenz J. R. Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- Usai M. G., Goddard M. E., Hayes B. J., 2009. LASSO with cross-validation for genomic selection. Genet. Res. 91: 427–436 [DOI] [PubMed] [Google Scholar]
- Verbyla K. L., Hayes B. J., Bowman P. J., Goddard M. E., 2009. Accuracy of genomic selection using stochastic search variable selection in Australian Holstein Friesian dairy cattle. Genet. Res. 91: 307–311 [DOI] [PubMed] [Google Scholar]
- Verbyla K. L., Calus M. P. L., Mulder H. A., de Haas Y., Veerkamp R. F., 2010. Predicting energy balance for dairy cows using high-density single nucleotide polymorphism information. J. Dairy Sci. 93: 2757–2764 [DOI] [PubMed] [Google Scholar]
- Wilcox P. L., Amerson H. V., Kuhlman E. G., Liu B. H., Omalley D. M., et al. , 1996. Detection of a major gene for resistance to fusiform rust disease in loblolly pine by genomic mapping. Proc. Natl. Acad. Sci. USA 93: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. R., Matheson A. C., Harwood C. E., 2002. Experimental Design and Analysis for Tree Improvement. Commonwealth Scientific and Industrial Research Organization, Melbourne [Google Scholar]
- Zhang Z., Liu J., Ding X., Bijma P., de Koning D.-J., et al. , 2010. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS ONE 5: e12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ding X., Liu J., Zhang Q., de Koning D.-J., 2011. Accuracy of genomic prediction using low-density marker panels. J. Dairy Sci. 94: 3642–3650 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.