Abstract
A substantial proportion of the world's living species, including one-third of the reef-building corals, are threatened with extinction and in pressing need of conservation action. In order to reduce biodiversity loss, it is important to consider species' contribution to evolutionary diversity along with their risk of extinction for the purpose of setting conservation priorities. Here I reconstruct the most comprehensive tree of life for the order Scleractinia (1,293 species) that includes all 837 living reef species, and employ a composite measure of phylogenetic distinctiveness and extinction risk to identify the most endangered lineages that would not be given top priority on the basis of risk alone. The preservation of these lineages, not just the threatened species, is vital for safeguarding evolutionary diversity. Tests for phylogeny-associated patterns show that corals facing elevated extinction risk are not clustered on the tree, but species that are susceptible, resistant or resilient to impacts such as bleaching and disease tend to be close relatives. Intensification of these threats or extirpation of the endangered lineages could therefore result in disproportionate pruning of the coral tree of life.
Introduction
Worldwide, ocean-scale effects of sea surface warming and acidification are subjecting reef corals to severe stresses, resulting in intensified bleaching and disease, as well as declining calcification rates [1]–[6]. Local anthropogenic impacts such as overfishing and pollution have also forced coral reefs through regime shifts toward macroalgal domination [4], [7]–[10]. Alarmingly, 32.8% of all zooxanthellate reef-building coral species are considered to be threatened with global extinction [11] (see also [12]).
Limited resources constrain scientists and managers to focus their efforts on a subset of the world's coral reefs to minimise extinction risk [13]. Consequently, the decision-making process associated with assignment of funds and manpower has become a major research focus in conservation biology [14]–[17]. One of the most widely-used frameworks for assessing threats to species and setting conservation priorities is the International Union for Conservation of Nature (IUCN) Categories and Criteria [18], [19]. Indeed, the identification and design of protected areas are often guided by the distribution of species with the highest risk of extinction, and in particular, the most threatened species of the IUCN Red List [20]–[22].
Extinction probabilities aside, species are not equal. Rather, evolutionary processes render each species unique with a characteristic history that can be quantified for the purpose of conservation prioritisation [14], [23]–[26]. Assessments that integrate phylogenetic distinctiveness and extinction threat have been performed mainly for mammalian groups, drawing attention to extraordinary species from lesser known localities and lineages (i.e. lines of ancestry and descent [27]) [28]–[33]. The dire situation of reef corals necessitates an equivalent treatment.
The utility of phylogenetic trees extends beyond the recognition of distinct lineages that are at risk. Due to the hierarchical nature of phylogenies, random losses of species rarely perturb the branches of evolutionary history [34], but concentration of threatened species or risk factors in particular parts of the phylogeny can imperil entire clades [35]–[38]. Threats to reef corals have traditionally been generalised based on species' taxonomic memberships [39], [40]. The family Faviidae, for instance, is reputed to be resilient to environmental disturbances [41], but the extreme polyphyly of the group has called into question such inferences [42] (see also [43]). Considering evolutionary history in the analysis of extinction risk will certainly aid in the development of informed conservation strategies against threats facing corals of the world today.
The aim of this study is to apply the phylogenetic approach on all reef corals of the order Scleractinia to identify, first, the most endangered coral lineages, and second, evolutionary patterns associated with extinction probability and various threats. To rank corals according to both distinctiveness and imperilment, I use the EDGE (evolutionarily distinct and globally endangered) metric [29], which combines a unique measure of phylogenetic diversity [44] with the conservation status of each species. Data for the latter are based on the IUCN Red List that includes 827 reef-building scleractinians assessed by the world's leading coral experts in 2006 and 2007 [11]. Of the 688 species not deemed Data Deficient (DD), 32.7% are considered threatened. These comprise―in decreasing likelihood of extinction―four Critically Endangered (CR), 23 Endangered (EN) and 198 Vulnerable (VU) corals. The remaining species are categorised as Near Threatened (NT; 174 species) or of Least Concern (LC; 289 species).
Methods
Phylogenetic data and analyses
To reconstruct the scleractinian phylogeny, 827 species from the IUCN Red List dataset [11], five previously omitted corals, five new species described since the assessment [45]–[47], and 65% of non-reef corals [48] were included in the analysis (Table S1). The supertree approach [49], [50] was used to combine data from molecular, morphologic and taxonomic sources. Unlike Kerr [51], the last published Scleractinia supertree, I reanalysed the molecular data rather than use available phylogenies because several DNA markers were utilised repeatedly in different studies (e.g. [52] and [53]). Using these phylogenies as source trees would result in data duplication [54], [55].
Mitochondrial DNA markers each with coverage of >40 species were obtained from GenBank to assemble a 463-species dataset (365 reef, 98 non-reef). The seven markers used were 12S small subunit ribosomal RNA (12S), 16S ribosomal RNA (16S), ATP synthase F0 subunit 6 (AT6), cytochrome c oxidase subunit I (COI), control region (CTR), cytochrome b (CYB) and NADH dehydrogenase subunit 5 (ND5) (Table S1). Corallimorphs Discosoma and Ricordea florida were included as outgroups. Matrices were aligned with MAFFT 6.8 [56], [57] and concatenated for analysis under the maximum likelihood criterion, using RAxML 7.2.8 [58], [59] implemented at the Cyberinfrastructure for Phylogenetic Research (http://www.phylo.org) [60]. Tree searches were carried out in 1000 alternate runs from distinct parsimony starting trees, utilising the partitioned GTRGAMMA model. Nodal supports were assessed via 1000 bootstrap replicates.
Thirteen morphological datasets were used to obtain source trees for the supertree reconstruction [61]–[69] (Table 1). All except one [61] were included in Kerr's [51] study. Congeners were assumed monophyletic unless otherwise shown in recent phylogenies (see remarks, Table S1). Maximum parsimony analyses were performed in PAUP* 4.0b10 [70] using the branch-and-bound algorithm for matrices with ≤25 terminals and heuristic searches (105 random additions with a rearrangement limit of 107 per replicate) for larger datasets. Nodal supports were determined with 1000 bootstrap replicates (100 random additions per replicate for heuristic searches). For 145 reef species with no available data, a source tree was used to represent likely sister relationships based on a review of literature, favouring the more recent hypotheses in cases of conflict [71]–[97] (see remarks, Table S1).
Table 1. Morphological data used as source matrices for supertree reconstruction.
| Taxon | No. of genera | No. of species | Analysis parameters | Reference |
| Faviina | 11 | 26 | equal weights; unordered | [61] |
| Turbinoliidae | 22 | 57 | characters weighted; one character ordered | [62] |
| Dendrophylliidae | 20 | 164 | characters weighted; two characters ordered | [63] |
| Scleractinia | 29 | 440 | equal weights; unordered | [64] |
| Fungiidae | 15 | 40 | equal weights; unordered | [65] |
| Pleuractis | 1 | 6 | equal weights; unordered | [66] |
| Mussidae | 12 | 44 | characters weighted; Lundberg rooting | [67] |
| Lobophyllia+Symphyllia | 2 | 10 | characters weighted | [67] |
| Siderastreidae | 6 | 29 | characters weighted; Lundberg rooting | [67] |
| Coscinaraea+Psammocora | 2 | 14 | characters weighted | [67] |
| Scleractinia+Corallimorpharia | 38 | 47 | includes two outgroups | [68] |
| Acroporidae | 6 | 291 | equal weights; unordered | [69] |
| Acropora+Isopora | 2 | 139 | 10 sister species grafted post-analysis | [69] |
Numbers in bold represent the taxonomic levels of analyses performed in the original studies.
Including the molecular phylogeny, 1293 scleractinian species (837 reef, 456 non-reef) were analysed. All source trees were coded into bootstrap percentage-weighted matrix representation with parsimony using SuperMRP 1.2.1 [98]. To ensure that analyses were driven primarily by data, weights of nodes derived from taxonomic information were each set at one. Maximum parsimony analysis of the 792-character dataset was carried out as above (rearrangement limit of 108 per replicate) to obtain 18978 minimum length trees.
The molecular data were then fitted to the strict consensus supertree using RAxML (1000 replicate runs) to derive the best branch length estimates [99]. Polytomies in the supertree were randomly resolved to generate 1000 different resolutions. Species with no available DNA sequence data were assigned a terminal branch length of zero, though still represented by their ancestral branches based on topology. This procedure yielded estimates for the lower limit of distinctiveness, a conservative approach given the lack of data. Calculations that followed were carried out for each of the 1000 resolutions; reported results are means over all randomly resolved trees.
Determining species priorities
For each reef species in the Scleractinia supertree, Tuatara 1.01 [100] was used to evaluate evolutionary distinctiveness (ED) by summing the terminal branch length and its species-weighted allocation of ancestral branches. ED was then multiplied by extinction probability (PE) to obtain the EDGE score, a measure of expected loss of evolutionary history [29], [101]. PE was calculated based on the IUCN100 transformation of the Red List categories [102]. LC species' PE was set at 0.001, assuming that at most about one of the 289 LC corals would go extinct in 100 years; NT corals were given an intermediate PE of 0.01. For the 149 DD species, a PE value between the lowest Red List categories (LC and NT) was assigned [33]. The ‘Isaac’ and ‘Pessimistic’ transformations of Mooers et al. [102] led to an LC species consistently achieving the top two highest scores, an overly conservative result that is not discussed (available in Table S1). Species were ranked according to their EDGE scores. Analyses repeated exclusively for the reef species show that incomplete sampling of Scleractinia (i.e. the non-reef corals) had minimal effect (mean rank variation: top 30 species = 1.5, all 837 species = 12.8).
Testing for phylogenetic signal
Phylogenetic signal of PE was tested using a randomisation procedure [103] in R package Picante 1.3 [104] that determined whether the actual phylogeny better fits a set of continuous data relative to data that had been randomly permuted across the tips of the tree (1000 replicates per supertree; K = 0 for random traits). For binary traits, Fritz & Purvis' [105] D was computed in CAIC 1.0.4 [106]. This metric was based on the trait's sum of sister-clade disparities on the tree (D = 0 for clumped traits, D = 1 for random traits). The phylogenetic patterns of three extinction risk levels, EN and above, VU and above and at least NT, were determined. In addition, eight species-specific binary traits assessed by Carpenter et al. [11] were tested for phylogenetic signal (Table 2).
Table 2. Phylogenetic signal of IUCN Red List categories and traits of reef corals.
| Category/trait | Proportion of species | D | P for H0: D = 0 | P for H0: D = 1 |
| Endangered and above | 0.032 | 1.096±0.063 | <0.001 | 0.780 |
| Vulnerable and above | 0.269 | 0.960±0.023 | <0.001 | 0.167 |
| Near Threatened and above | 0.477 | 0.853±0.018 | <0.001 | <0.001 |
| moderately or highly susceptible to bleaching | 0.419 | 0.229±0.010 | <0.001 | <0.001 |
| moderately or highly resistant to bleaching | 0.116 | 0.300±0.023 | 0.001 | <0.001 |
| moderately or highly susceptible to disease | 0.310 | 0.124±0.012 | 0.024 | <0.001 |
| moderately or highly resistant to disease | 0.058 | −0.172±0.015 | 0.887 | <0.001 |
| recovers quickly from bleaching or disease | 0.134 | 0.125±0.013 | 0.068 | <0.001 |
| moderately or highly susceptible to crown-of-thorns seastar predation | 0.273 | 0.052±0.011 | 0.180 | <0.001 |
| restricted or highly fragmented range | 0.124 | 1.136±0.037 | <0.001 | 0.973 |
| reported collection of > 1000 pieces per year | 0.157 | 0.630±0.021 | <0.001 | <0.001 |
Results based on D, a measure of total sister-clade disparities on the phylogeny (± SD; 0 for clumped traits, 1 for random traits). Numbers in bold denote non-significant results (i.e. not different from 0 or 1).
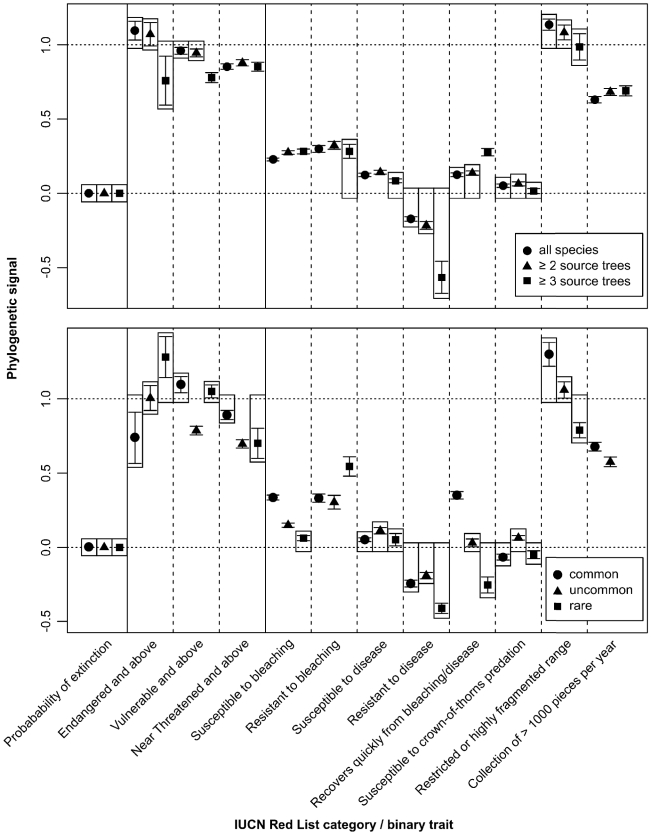
Two potential confounding factors associated with the above analyses were investigated. First, species assembled in the supertree differ in the degree of representation among source trees. It may be argued that poorly-sampled species are generally placed, unresolved, outside of clades with well-sampled species, leading to bias in calculations. The 1000 random resolutions of the strict consensus supertree should circumvent this problem, but to be sure, the tests were repeated for two reduced datasets with species present in at least two and three source trees respectively. Second, the level of phylogenetic signal inferred for each trait may be influenced by variation in species abundances, hence the analyses were also performed separately for species that are considered common (including one abundant taxon), uncommon and rare (data from [11]). Phylogenetic signal of the trait ‘reported collection of >1000 pieces per year’ for the ‘rare’ dataset could not be computed as it is represented by just two species.
Carpenter et al. [11] also found that several taxa that are susceptible to bleaching also appear to be heavily impacted by disease and predation by the crown-of-thorns seastar, Acanthaster planci. To ascertain if this relationship holds with the incorporation of phylogenetic information, I tested for correlation among traits associated with coral bleaching, disease and predation using phylogenetically independent contrasts [107]. This was implemented in APE 2.7 [108], with statistical significance evaluated based on fit to a linear model.
Finally, I determined whether the decrease in phylogenetic diversity (PD) [44] under various extinction scenarios was different from a null model of random extinction. PD was compared between rarefied trees based on threat status (EN and above, VU and above, NT and above) and 1000 randomly pruned trees with the same species richness, using the one-sample t-test [109]. This analysis was also carried out for 30 species with the highest EDGE scores.
Results
Integrating the diverse data types using a supertree approach yields a 1293-species phylogeny of Scleractinia that includes all 837 reef-building corals (Figures 1, 2, 3). Despite the vast increase in taxon sampling over previous phylogenies [42], [82], the present analysis recovers a highly similar topology. In particular, all 21 clades recognised by Fukami et al. [42] (labelled I to XXI) are present in the supertree.
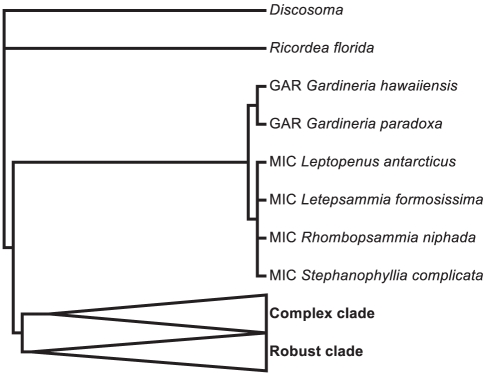
Figure 1. Supertree of Scleractinia with corallimorph outgroups Discosoma and Ricordea florida.
Cladogram of 1293 corals inferred by maximum parsimony analysis of the 792-character dataset assembled using 15 source trees (13 morphological, one molecular and one taxonomic). Complex and robust clades shown in Figures 2 and 3 respectively. GAR: Gardineriidae, MIC: Micrabaciidae.
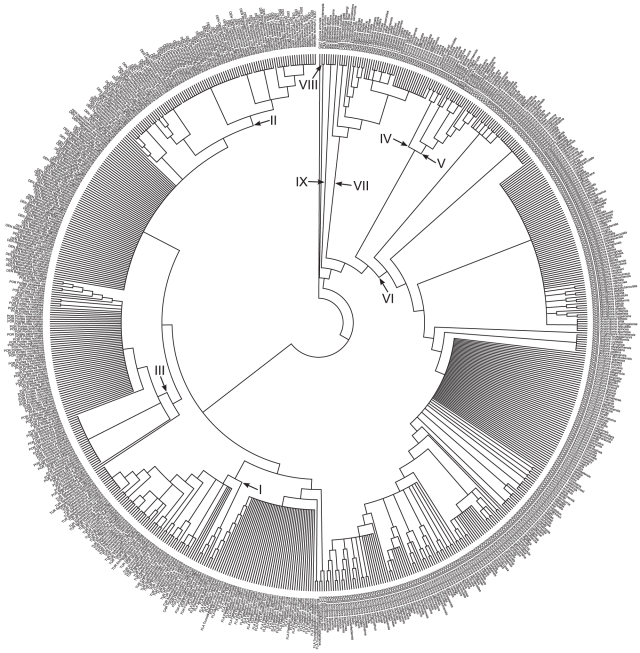
Figure 2. Cladogram of scleractinian corals in the complex clade.
A total of 735 corals, including 462 reef species, are represented on this maximum parsimony cladogram that is part of the scleractinian supertree (Figure 1). Roman numerals denote clades based on the phylogeny in Fukami et al. [42]. ACR: Acroporidae, AGA: Agariciidae, AST: Astrocoeniidae, CAR: Caryophylliidae, DEN: Dendrophylliidae, EUP: Euphylliidae, FLA: Flabellidae, FUA: Fungiacyathidae, GUY: Guyniidae, MEA: Meandrinidae, OCU: Oculinidae, POR: Poritidae, SID: Siderastreidae, TUR: Turbinoliidae.
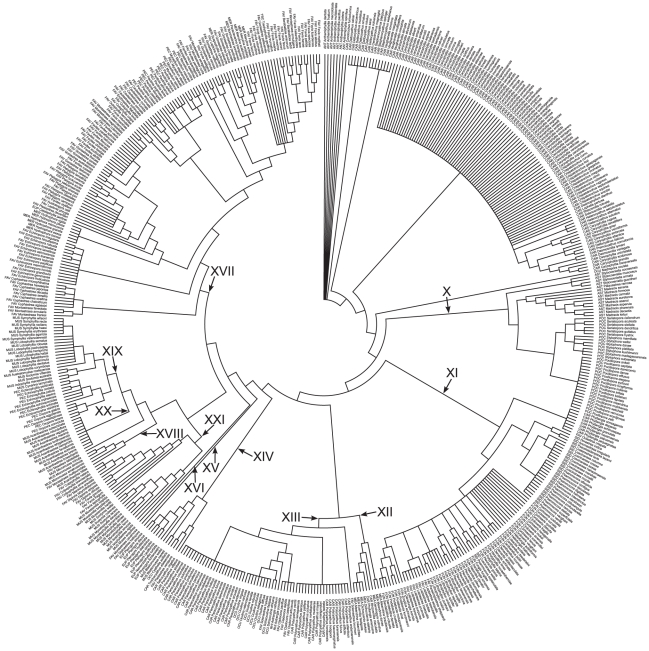
Figure 3. Cladogram of scleractinian corals in the robust clade.
A total of 552 corals, including 375 reef species, are represented on this maximum parsimony cladogram that is part of the scleractinian supertree (Figure 1). Roman numerals denote clades based on the phylogeny in Fukami et al. [42]. ANT: Anthemiphyllidae, AST: Astrocoeniidae, CAR: Caryophylliidae, EUP: Euphylliidae, FAV: Faviidae, FUN: Fungiidae, MEA: Meandrinidae, MER: Merulinidae, MUS: Mussidae, OCU: Oculinidae, PEC: Pectiniidae, POC: Pocilloporidae, RHI: Rhizangiidae, SID: Siderastreidae, STE: Stenocyathidae, TRC: Trachyphylliidae.
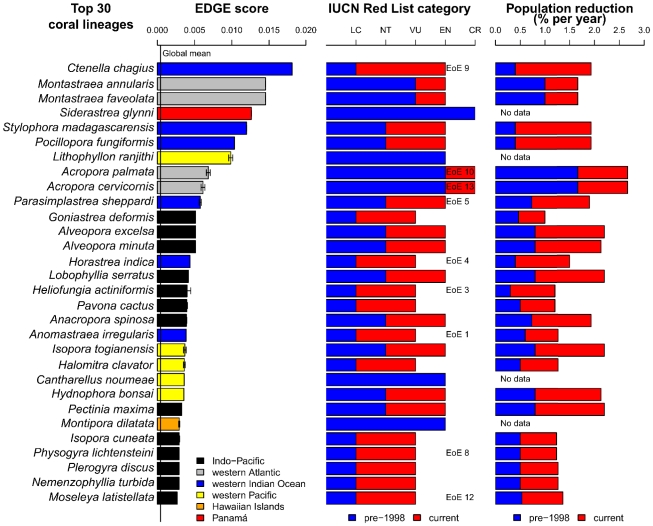
The analysis of EDGE scores has produced a priority list of reef-building corals that are both phylogenetically unique and facing elevated extinction risk (Figure 4; for full ranking, see Table S1). Conservation of these endangered lineages is critical for the preservation of evolutionary diversity. The priority scores of the top 30 species exceed the mean of all reef corals by at least an order of magnitude, and a significantly greater than random loss of phylogenetic diversity would occur should these species go extinct (P<0.001).
Figure 4. Top 30 reef corals ranked according to EDGE scores.
List of corals representing high evolutionary distinctiveness and extinction risk. Left panel shows the EDGE score for each species. Global mean score for all 837 reef corals denoted by vertical line through bars, which are coloured to indicate respective geographic ranges. Error bars represent standard deviation. Middle panel shows pre-1998 and present IUCN Red List categories, as well as ranks according to the EDGE of Existence (EoE) programme. Right panel shows pre-1998 and present rates of global population reduction. IUCN Red List and population reduction data derived from Carpenter et al. [11].
Extinction probability of corals exhibits negligible phylogenetic signal since the hypothesis that there is no signal cannot be rejected given the data, i.e. non-zero K values are only non-zero by chance (P = 0.745, K = 1.584×10−11). Threatened species (EN and above, and VU and above) are randomly distributed on the phylogeny, while species given a minimum status of NT are only slightly more clumped than random (Figure 5, Table 2). The datasets comprising species with increased source tree sampling and fixed abundances show very similar patterns, indicating that these factors have limited influence on phylogenetic signal strength (Figure 6). Gains in statistical significance (more clumped than random) are recorded for VU and above corals that are present in ≥3 source trees, as well as for taxa considered at least VU and NT for the uncommon species, but values of D remain close to one (random). Simulated extinction scenarios of reef corals based solely on threat status result in smaller than random losses of PD (P<0.001, EN and above, VU and above, NT and above, all significantly less than random loss).
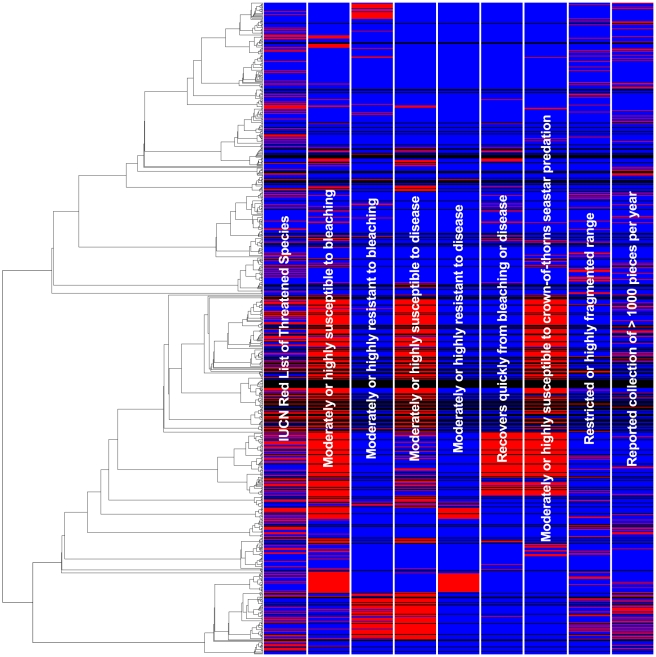
Figure 5. Cladogram of reef corals illustrating phylogenetic signal of traits.
This tree represents the first of 1000 random resolutions of the strict consensus supertree. Vertical bars illustrate, in red, degrees of clumping among species classified as Vulnerable (VU) and above, susceptible and/or resistant to specific threats, and those recovering quickly from bleaching and disease. Taxa absent for the above traits are in blue. Data Deficient (DD) species, which are not phylogenetically clumped, are in black.
Figure 6. Species' source tree representation and abundances show limited effect on phylogenetic signal strength.
Measure of phylogenetic signal based on K for probability of extinction (K = 0 for random continuous traits) and D for all other traits (D = 0 for clumped and D = 1 for random binary traits). Upper and lower panels show levels of phylogenetic signal for datasets with varying degrees of source tree representation and abundance respectively. Error bars represent standard deviation. Means not significantly different from zero or one are enclosed by boxes with those values.
The tests for phylogenetic signal show that species susceptible to bleaching, disease, and predation by Acanthaster planci, as well as those resistant to and recovering quickly from bleaching and disease (i.e. resilient [110]) are at least moderately clumped on the coral tree (Figure 5, Table 2; see [105]). Species' source tree representation and abundances have negligible effects on these inferences (Figure 6). In fact, phylogenetic signal increases among taxa represented by at least three source trees for the traits ‘resistant to bleaching’, ‘susceptible to disease’ and ‘resistant to disease’. It should be noted that in the dataset comprising only rare corals, species resistant to bleaching display relatively low signal (D = 0.545±SD 0.065), but are still significantly more clustered than random on the phylogeny (P = 0.016).
Among lineages, correlations are evident between susceptibilities to bleaching events and disease (P = 0.001), as well as susceptibilities to bleaching and predation (P<0.001). Negative linear relationships are present between susceptibility and resistance for both bleaching (P<0.001) and disease (P<0.001), although there is a positive correlation between susceptibility to disease and quick recovery from bleaching/disease (P = 0.025).
Discussion
Using the most comprehensive coral phylogeny to date, this study has quantified the expected loss of evolutionary history for reef species based on the EDGE (evolutionarily distinct and globally endangered) measure. The ranking provided here, the first of its kind for corals, has been successful in identifying distinct lineages that warrant the highest conservation attention.
The top-30 list captures three of four CR species and 16 of the 23 EN species, the majority of which have restricted ranges (Figure 4). In particular, the most endangered lineage represented by Ctenella chagius is known only from the Chagos Archipelago, Mauritius and La Réunion, while Siderastrea glynni, fourth on the list, is endemic to Panamá in the tropical eastern Pacific [92]. The remaining 11 species are of VU status and could be accorded lower conservation priority based upon extinction risk alone. Five of these, Horastrea indica, Heliofungia actiniformis, Anomastraea irregularis, Physogyra lichtensteini and Moseleya latistellata have only recently been highlighted by the EDGE of Existence programme (http://www.edgeofexistence.org/coral_reef) that aims to identify evolutionarily distinct and globally endangered species. Yet it has failed to recognise 21 of the 30 corals shown here to be of top priority; neither the ‘Isaac’ nor ‘Pessimistic’ transformation increases its representation of high EDGE-scoring species (22 and 24 species overlooked respectively). The programme's methodology remains unknown, but likely utilisation of an incomplete phylogeny may have precluded a comprehensive listing (see also materials and methods in [29]).
Distinctiveness metrics such as ED often account for a greater proportion of total PD than expected [111]. Recent evidence also suggests that evolutionarily distinct species and high PD represent a broader distribution of ecological diversity and higher ecosystem function than expected [112]–[117] (but see [109]). If the preservation of biological diversity is a goal of reef conservation, then such phylogenetically-informed rankings would shore up priority setting efforts that currently focus on species richness, rarity and connectivity [13], [118]–[121].
Despite the heightened risk in a larger fraction of corals relative to birds and mammals [11], groups that exhibit phylogenetic clustering of threat status [105], [122], extinction probability and threatened species of corals show negligible signal associated with phylogeny (Figure 5). That species facing elevated extinction risk are not concentrated in particular parts of the phylogeny is no cause for optimism, however, as recent simulations have shown that other factors are involved in determining the magnitude of PD loss during extinctions [123]. In particular, trees derived from real data generally have asymmetric topologies [124]–[128]; the coral supertree is no exception (P<0.001, Colless' [129] index significantly greater than predicted by the Yule model). Under this circumstance, even random exterminations of species can lead to disproportionate losses of PD [34], [123], [130]. High average extinction probability among reef corals [11] may also exacerbate this pattern [123]. Indeed, random extinction scenarios of coral species lead to larger declines in PD compared to extinctions based on IUCN Red List threat status. In other words, while none of the major clades of reef corals are in immediate danger of complete obliteration, the unbalanced phylogeny and high mean extinction risk suggest that any extinction event can substantially reduce overall PD.
Bleaching, disease, and predation by A. planci are three of the most serious stressors affecting coral health today [131], [132]. Tests for phylogenetic signal show that species susceptible to these threats, as well as those resistant and resilient to bleaching and disease are clustered on the tree, indicating that the aggravation of these risk factors can result in disproportionately large PD declines. More worrying is the finding that lineages vulnerable to bleaching events are also more likely to be susceptible to disease and predation. These threats often impact similar sets of species [11], [133]–[136], yet this relationship holds even after controlling for effects of shared common ancestry.
The value of investigating extinction risk in the phylogenetic context has been emphasised in considerable detail elsewhere [26], [29], [32], [38], [101], [137], [138]. Specifically for corals, confusion surrounding traditional taxonomy makes it difficult to accurately generalise traits exhibited by species to higher level taxa [42]. For instance, following the massive bleaching event in 1998, the family Faviidae, including Leptastrea purpurea and L. transversa, has been declared a ‘winner’ in the recovery process at Sesoko Island, Japan [39], [40]. Yet phylogenies inferred in the last 15 years have unequivocally demonstrated that Leptastrea is more closely related to members of Fungiidae rather than Faviidae [42], [52], [53], [82], [139] (see also [140], [141]), recovered within clade X with corals that are resistant to or recover quickly from bleaching (Figures 3, 5). Results here suggest that these traits are conserved on the evolutionary tree, irrespective of species' taxonomic affiliations.
Vulnerabilities of reef corals to bleaching and disease appear to be mediated by the same physiological mechanisms, and immune responses against these threats tend to be similar among close relatives, with Acroporidae and Porites (Poritidae) possessing the lowest and highest immunity levels respectively [142]. Consequently, the enhanced susceptibility of Alveopora to bleaching [11] is better understood in the context of recent phylogenies that show the genus being placed within Acroporidae (clade VI) rather than, traditionally, Poritidae (clade III) [42], [82]. It is clear that, conventional taxonomy notwithstanding, close relatives are likely to share similar levels of susceptibility, resistance and resilience to various risk factors, underscoring the utility of phylogenetic approaches in understanding specific responses of corals to environmental perturbations.
Subsequent analyses will utilise these results in distinguishing reef regions that make the greatest contribution to evolutionary history, in comparison to the most species-rich areas [143]. A biogeographically-weighted evolutionary distinctiveness (ED) metric has the potential for regional prioritisation [144], but a probabilistic approach that accounts for future extinctions of related species may be more suitable than the static allocation of conservation value afforded by the ED measure [32], [145], [146].
Analyses demonstrating phylogenetic clustering of susceptibilities, resistance and resilience to various risk factors rely on accurate and precise species-specific data. The conservation status of Data Deficient species clearly needs to be assessed while regular updates are necessary for all corals [147], [148]. Increasingly, recent research is revealing a wider range of species responses to environmental threats than before [149]–[152]. Given that these threats exhibit considerable phylogenetic signal, the coral tree of life will prove an excellent framework for investigating these variabilities.
Supporting Information
Reef and non-reef coral species included in the phylogenetic analysis of Scleractinia. For each species, the IUCN Red List category and ranks according to the EDGE of Existence (EoE) programme and the present study are shown where appropriate. Species not assessed are indicated as N/A. GenBank accession numbers are provided for DNA sequences (see text for names of markers).
(PDF)
Acknowledgments
I thank Gregory Rouse and Kaustuv Roy for ideas, discussion and critical reading of the manuscript. Nicholas Holland, Arne Mooers, Mikhail Matz and five anonymous reviewers helped improve the initial versions of the paper. I am grateful to Marcelo Kitahara for sharing unpublished information, and Nancy Knowlton and Florian Maderspacher for providing references. This work is dedicated to the late conservation biologist Navjot Sodhi.
Footnotes
Competing Interests: The author has declared that no competing interests exist.
Funding: This work was supported by the Ah Meng Memorial Conservation Fund (Wildlife Reserves Singapore) and the Friends of the International Center Scholarship (University of California San Diego). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hughes TP, Baird AH, Bellwood DR, Card M, Connolly SR, et al. Climate change, human impacts, and the resilience of coral reefs. Science. 2003;301:929–933. doi: 10.1126/science.1085046. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- 2.Bruno JF, Selig ER, Casey KS, Page CA, Willis BL, et al. Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 2007;5:e124. doi: 10.1371/journal.pbio.0050124. doi: 10.1371/journal.pbio.0050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- 4.Knowlton N, Jackson JBC. Shifting baselines, local impacts, and global change on coral reefs. PLoS Biol. 2008;6:e54. doi: 10.1371/journal.pbio.0060054. doi: 10.1371/journal.pbio.0060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De'ath G, Lough JM, Fabricius KE. Declining coral calcification on the Great Barrier Reef. Science. 2009;323:116–119. doi: 10.1126/science.1165283. doi: 10.1126/science.1165283. [DOI] [PubMed] [Google Scholar]
- 6.Veron JEN, Hoegh-Guldberg O, Lenton TM, Lough JM, Obura DO, et al. The coral reef crisis: the critical importance of <350 ppm CO2. Mar Poll Bull. 2009;58:1428–1436. doi: 10.1016/j.marpolbul.2009.09.009. doi: 10.1016/j.marpolbul.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Hughes TP. Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science. 1994;265:1547–1551. doi: 10.1126/science.265.5178.1547. doi: 10.1126/science.265.5178.1547. [DOI] [PubMed] [Google Scholar]
- 8.McCook LJ. Macroalgae, nutrients and phase shifts on coral reefs: scientific issues and management consequences for the Great Barrier Reef. Coral Reefs. 1999;18:357–367. doi: 10.1007/s003380050213. [Google Scholar]
- 9.Pandolfi JM, Jackson JBC, Baron N, Bradbury RH, Guzman HM, et al. Are U.S. coral reefs on the slippery slope to slime? Science. 2005;308:1742–1743. doi: 10.1126/science.1104258. doi: 10.1126/science.1104258. [DOI] [PubMed] [Google Scholar]
- 10.Hughes TP, Rodrigues MJ, Bellwood DR, Ceccarelli D, Hoegh-Guldberg O, et al. Phase shifts, herbivory, and the resilience of coral reefs to climate change. Curr Biol. 2007;17:360–365. doi: 10.1016/j.cub.2006.12.049. doi: 10.1016/j.cub.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter KE, Abrar M, Aeby GS, Aronson RB, Banks S, et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science. 2008;321:560–563. doi: 10.1126/science.1159196. doi: 10.1126/science.1159196. [DOI] [PubMed] [Google Scholar]
- 12.Polidoro BA, Elfes CT, Sanciangco JC, Pippard H, Carpenter KE. Conservation status of marine biodiversity in Oceania: an analysis of marine species on the IUCN Red List of Threatened Species. J Mar Biol. 2011;2011:247030. doi: 10.1155/2011/247030. [Google Scholar]
- 13.Roberts CM, McClean CJ, Veron JEN, Hawkins JP, Allen GR, et al. Marine biodiversity hotspots and conservation priorities for tropical reefs. Science. 2002;295:1280–1284. doi: 10.1126/science.1067728. doi: 10.1126/science.1067728. [DOI] [PubMed] [Google Scholar]
- 14.Vane-Wright RI, Humphries CJ, Williams PH. What to protect?-Systematics and the agony of choice. Biol Conserv. 1991;55:235–254. doi: 10.1016/0006-3207(91)90030-D. [Google Scholar]
- 15.Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- 16.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 17.Murdoch W, Polasky S, Wilson KA, Possingham HP, Kareiva P, et al. Maximizing return on investment in conservation. Biol Conserv. 2007;139:375–388. doi: 10.1016/j.biocon.2007.07.011. [Google Scholar]
- 18.IUCN. IUCN Red List Categories and Criteria: Version 3.1. Gland, Switzerland and Cambridge, UK: IUCN; 2001. 30 [Google Scholar]
- 19.Mace GM, Collar NJ, Gaston KJ, Hilton-Taylor C, Akçakaya HR, et al. Quantification of extinction risk: IUCN's system for classifying threatened species. Conserv Biol. 2008;22:1424–1442. doi: 10.1111/j.1523-1739.2008.01044.x. doi: 10.1111/j.1523-1739.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues ASL, Akçakaya HR, Andelman SJ, Bakarr MI, Boitani L, et al. Global gap analysis: priority regions for expanding the global protected-area network. Bioscience. 2004;54:1092–1100. doi: 10.1641/0006-3568(2004)054[1092:GGAPRF]2.0.CO;2. [Google Scholar]
- 21.Rodrigues ASL, Pilgrim JD, Lamoreux JF, Hoffman M, Brooks TM. The value of the IUCN Red List for conservation. Trends Ecol Evol. 2006;21:71–76. doi: 10.1016/j.tree.2005.10.010. doi: 10.1016/j.tree.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann M, Brooks TM, da Fonseca GAB, Gascon C, Hawkins AFA, et al. Conservation planning and the IUCN Red List. Endang Species Res. 2008;6:113–125. doi: 10.3354/esr00087. [Google Scholar]
- 23.May RM. Taxonomy as destiny. Nature. 1990;347:129–130. doi: 10.1038/347129a0. [Google Scholar]
- 24.Altschul SF, Lipman DL. Equal animals. Nature. 1990;348:493–494. doi: 10.1038/348493c0. doi: 10.1038/348493c0. [DOI] [PubMed] [Google Scholar]
- 25.Witting L, Loeschcke V. The optimization of biodiversity conservation. Biol Conserv. 1995;71:205–207. doi: 10.1016/0006-3207(94)00041-N. [Google Scholar]
- 26.Mace GM, Gittleman JL, Purvis A. Preserving the tree of life. Science. 2003;300:1707–1709. doi: 10.1126/science.1085510. doi: 10.1126/science.1085510. [DOI] [PubMed] [Google Scholar]
- 27.de Queiroz K. Branches in the lines of descent: Charles Darwin and the evolution of the species concept. Biol J Linn Soc. 2011;103:19–35. doi: 10.1111/j.1095-8312.2011.01634.x. [Google Scholar]
- 28.Pavoine S, Ollier S, Dufour AB. Is the originality of a species measurable? Ecol Lett. 2005;8:579–586. doi: 10.1111/j.1461-0248.2005.00752.x. [Google Scholar]
- 29.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE. 2007;2:e296. doi: 10.1371/journal.pone.0000296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agnarsson I, Kuntner M, May-Collado LJ. Dogs, cats, and kin: a molecular species-level phylogeny of Carnivora. Mol Phylogenet Evol. 2010;54:726–745. doi: 10.1016/j.ympev.2009.10.033. doi: 10.1016/j.ympev.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 31.Kuntner M, May-Collado LJ, Agnarsson I. Phylogeny and conservation priorities of afrotherian mammals (Afrotheria, Mammalia). Zool Scr. 2011;40:1–15. doi: 10.1111/j.1463-6409.2010.00452.x. [Google Scholar]
- 32.Collen B, Turvey ST, Waterman C, Meredith HMR, Kuhn TS, et al. Investing in evolutionary history: implementing a phylogenetic approach for mammal conservation. Philos Trans R Soc B-Biol Sci. 2011;366:2611–2622. doi: 10.1098/rstb.2011.0109. doi: 10.1098/rstb.2011.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May-Collado LJ, Agnarsson I. Phylogenetic analysis of conservation priorities for aquatic mammals and their terrestrial relatives, with a comparison of methods. PLoS ONE. 2011;6:e22562. doi: 10.1371/journal.pone.0022562. doi: 10.1371/journal.pone.0022562.t005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nee S, May RM. Extinction and the loss of evolutionary history. Science. 1997;278:692–694. doi: 10.1126/science.278.5338.692. doi: 10.1126/science.278.5338.692. [DOI] [PubMed] [Google Scholar]
- 35.McKinney ML. Extinction vulnerability and selectivity: combining ecological and paleontological views. Annu Rev Ecol Syst. 1997;28:495–516. doi: 10.1146/annurev.ecolsys.28.1.495. [Google Scholar]
- 36.Russell GJ, Brooks TM, McKinney MM, Anderson CG. Present and future taxonomic selectivity in bird and mammal extinctions. Conserv Biol. 1998;12:1365–1376. doi: 10.1111/j.1523-1739.1998.96332.x. [Google Scholar]
- 37.Purvis A, Agapow PM, Gittleman JL, Mace GM. Nonrandom extinction and the loss of evolutionary history. Science. 2000;288:328–330. doi: 10.1126/science.288.5464.328. doi: 10.1126/science.288.5464.328. [DOI] [PubMed] [Google Scholar]
- 38.Purvis A. Phylogenetic approaches to the study of extinction. Annu Rev Ecol Evol Syst. 2008;39:301–319. doi: 10.1146/annurev-ecolsys-063008-102010. [Google Scholar]
- 39.Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, et al. Coral bleaching: the winners and the losers. Ecol Lett. 2001;4:122–131. doi: 10.1046/j.1461-0248.2001.00203.x. [Google Scholar]
- 40.van Woesik R, Sakai K, Ganase A, Loya Y. Revisiting the winners and the losers a decade after coral bleaching. Mar Ecol-Prog Ser. 2011;434:67–76. doi: 10.3354/meps09203. [Google Scholar]
- 41.Bellwood DR, Hughes TP. Regional-scale assembly rules and biodiversity of coral reefs. Science. 2001;292:1532–1534. doi: 10.1126/science.1058635. doi: 10.1126/science.1058635. [DOI] [PubMed] [Google Scholar]
- 42.Fukami H, Chen CA, Budd AF, Collins A, Wallace CC, et al. Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PLoS ONE. 2008;3:e3222. doi: 10.1371/journal.pone.0003222. doi: 10.1371/journal.pone.0003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Díaz M, Madin J. Macroecological relationships between coral species' traits and disease potential. Coral Reefs. 2011;30:73–84. doi: 10.1007/s00338-010-0668-4. [Google Scholar]
- 44.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. doi: 10.1016/0006-3207(92)91201-3. [Google Scholar]
- 45.Forsman ZH, Birkeland C. Porites randalli: a new coral species (Scleractinia, Poritidae) from American Samoa. Zootaxa. 2009;2244:51–59. [Google Scholar]
- 46.Hoeksema BW. Attached mushroom corals (Scleractinia: Fungiidae) in sediment-stressed reef conditions at Singapore, including a new species and a new record. Raffles Bull Zool. 2009;S22:81–90. [Google Scholar]
- 47.Wallace CC, Turak E, DeVantier LM. Novel characters in a conservative coral genus: three new species of Astreopora (Scleractinia: Acroporidae) from West Papua. J Nat Hist. 2011;45:1905–1924. doi: 10.1080/00222933.2011.573098. [Google Scholar]
- 48.Cairns SD. Phylogenetic list of 722 valid Recent azooxanthellate scleractinian species, with their junior synonyms and depth ranges. In: Roberts JM, Wheeler A, Freiwald A, Cairns SD, editors. Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats. New York: Cambridge University Press; 2009. Available: http://www.lophelia.org/online-appendices. Accessed 2011 Jun 6. [Google Scholar]
- 49.Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. doi: 10.2307/1222480. [Google Scholar]
- 50.Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. doi: 10.1016/1055-7903(92)90035-F. [DOI] [PubMed] [Google Scholar]
- 51.Kerr AM. Molecular and morphological supertree of stony corals (Anthozoa: Scleractinia) using matrix representation parsimony. Biol Rev. 2005;80:543–558. doi: 10.1017/S1464793105006780. doi: 10.1017/S1464793105006780. [DOI] [PubMed] [Google Scholar]
- 52.Romano SL, Palumbi SR. Evolution of scleractinian corals inferred from molecular systematics. Science. 1996;271:640–642. doi: 10.1126/science.271.5249.640. [Google Scholar]
- 53.Romano SL, Cairns SD. Molecular phylogenetic hypotheses for the evolution of scleractinian corals. Bull Mar Sci. 2000;67:1043–1068. [Google Scholar]
- 54.Bininda-Emonds ORP, Gittleman JL, Steel MA. The (super)tree of life: procedures, problems, and prospects. Annu Rev Ecol Syst. 2002;33:265–289. doi: 10.1146/annurex.ecolysis.33.010802.150511. [Google Scholar]
- 55.Bininda-Emonds ORP. The evolution of supertrees. Trends Ecol Evol. 2004;19:315–322. doi: 10.1016/j.tree.2004.03.015. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 59.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 60.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010. pp. 1–8. Proceedings of the Gateway Computing Environments Workshop. New Orleans. doi: 10.1109/GCE.2010.5676129.
- 61.Budd AF, Smith ND. Diversification of a new Atlantic clade of scleractinian reef corals: insights from phylogenetic analysis of morphologic and molecular data. Paleontol Soc Pap. 2005;11:103–128. [Google Scholar]
- 62.Cairns SD. A generic revision and phylogenetic analysis of the Turbinoliidae (Cnidaria: Scleractinia). Smithsonian Contrib Zool. 1997;591:1–55. [Google Scholar]
- 63.Cairns SD. A generic revision and phylogenetic analysis of the Dendrophylliidae (Cnidaria: Scleractinia). Smithsonian Contrib Zool. 2001;615:1–75. [Google Scholar]
- 64.Daly M, Fautin DG, Cappola VA. Systematics of the Hexacorallia (Cnidaria: Anthozoa). Zool J Linn Soc. 2003;139:419–437. doi: 10.1046/j.1096-3642.2003.00084.x. [Google Scholar]
- 65.Hoeksema BW. Taxonomy, phylogeny and biogeography of mushroom corals (Scleractinia: Fungiidae). Zool Verh Leiden. 1989;254:1–295. [Google Scholar]
- 66.Hoeksema BW. Historical biogeography of Fungia (Pleuractis) spp. (Scleractinia: Fungiidae), including a new species from the Seychelles. Zool Meded Leiden. 1993;67:639–654. [Google Scholar]
- 67.Pandolfi JM. Successive isolation rather than evolutionary centres for the origination of Indo-Pacific reef corals. J Biogeogr. 1992;19:593–609. doi: 10.2307/2845703. [Google Scholar]
- 68.Pires DO, Castro CB. Scleractinia and Corallimorpharia: An analysis of cnidae affinity. Proc 8th Int Coral Reef Symp. 1997;2:1581–1586. [Google Scholar]
- 69.Wallace CC. Staghorn Corals of the World: A Revision of the Coral Genus Acropora. Collingwood: CSIRO Publishing; 1999. 421 [Google Scholar]
- 70.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland: Sinauer Associates; 2003. [Google Scholar]
- 71.Benzoni F, Stefani F, Stolarski J, Pichon M, Mitta G, et al. Debating phylogenetic relationships of the scleractinian Psammocora: molecular and morphological evidences. Contrib Zool. 2007;76:35–54. [Google Scholar]
- 72.Benzoni F, Stefani F, Pichon M, Galli P. The name game: morpho-molecular species boundaries in the genus Psammocora (Cnidaria, Scleractinia). Zool J Linn Soc. 2010;160:421–456. doi: 10.1111/j.1096-3642.2010.00622.x. [Google Scholar]
- 73.Best MB, Hoeksema BW. New observations on scleractinian corals from Indonesia: 1. Free-living species belonging to the Faviina. Zool Meded Leiden. 1987;61:387–403. [Google Scholar]
- 74.Chevalier J-P. Les scléractiniaires de la Mélanésie Francaise (Nouvelle Calédonie, Iles Chesterfield, Iles Loyauté, Nouvelles Hébrides). Première partie. Expéd Française Récifs Coralliens Nouvelle Calédonie. 1971;5:1–307. [Google Scholar]
- 75.Claereboudt MR. Galaxea paucisepta nom. nov. (for G. pauciradiata), rediscovery and redescription of a poorly known scleractinian species (Oculinidae). Galaxea. 1990;9:1–8. [Google Scholar]
- 76.Claereboudt MR, Al-Amri IS. Calathiscus tantillus, a new genus and new species of scleractinian coral (Scleractinia, Poritidae) from the Gulf of Oman. Zootaxa. 2004;532:1–8. [Google Scholar]
- 77.Ditlev H. New scleractinian corals (Cnidaria: Anthozoa) from Sabah, North Borneo. Description of one new genus and eight new species, with notes on their taxonomy and ecology. Zool Meded Leiden. 2003;77:193–219. [Google Scholar]
- 78.Fenner DP. Species distinctions among several Caribbean stony corals. Bull Mar Sci. 1993;53:1099–1116. [Google Scholar]
- 79.Gittenberger A, Reijnen BT, Hoeksema BW. A molecularly based phylogeny reconstruction of mushroom corals (Scleractinia: Fungiidae) with taxonomic consequences and evolutionary implications for life history traits. Contrib Zool. 2011;80:107–132. [Google Scholar]
- 80.Head SM. An undescribed species of Merulina and a new genus and species of siderastreid coral from the Red Sea. J Nat Hist. 1983;17:419–435. doi: 10.1080/00222938300770281. [Google Scholar]
- 81.Huang D, Licuanan WY, Baird AH, Fukami H. Cleaning up the “Bigmessidae”: molecular phylogeny of scleractinian corals from Faviidae, Merulinidae, Pectiniidae and Trachyphylliidae. BMC Evol Biol. 2011;11:37. doi: 10.1186/1471-2148-11-37. doi: 10.1186/1471-2148-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS ONE. 2010;5:e11490. doi: 10.1371/journal.pone.0011490. doi: 10.1371/journal.pone.0011490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin M-F, Luzon KS, Licuanan WY, Ablan-Lagman MC, Chen CA. Seventy-four universal primers for characterizing the complete mitochondrial genomes of scleractinian corals (Cnidaria; Anthozoa). Zool Stud. 2011;50:513–524. [Google Scholar]
- 84.Locke JM, Weil E, Coates KA. A newly documented species of Madracis (Scleractinia: Pocilloporidae) from the Caribbean. Proc Biol Soc Wash. 2007;120:214–226. doi: 10.2988/0006-324X(2007)120[214:ANDSOM]2.0.CO;2. [Google Scholar]
- 85.Moll H, Best MB. New scleractinian corals (Anthozoa: Scleractinia) from the Spermonde Archipelago, South Sulawesi, Indonesia. Zool Meded Leiden. 1984;58:47–58. [Google Scholar]
- 86.Scheer G, Pillai CSG. Report on the stony corals from the Red Sea. Zoologica. 1983;131:1–198. [Google Scholar]
- 87.Vaughan TW. The stony corals of the Porto Rican waters. Bull US Fish Commiss. 1901;1900 2:289–320. [Google Scholar]
- 88.Veron JEN, Pichon M, Wijsman-Best M. Scleractinia of Eastern Australia. Part II. Families Faviidae, Trachyphylliidae. Townsville: Australian Institute of Marine Science; 1977. 233 [Google Scholar]
- 89.Veron JEN, Pichon M. Scleractinia of Eastern Australia. Part III. Families Agariciidae, Siderastreidae, Fungiidae, Oculinidae, Merulinidae, Mussidae, Pectiniidae, Caryophylliidae, Dendrophylliidae. Townsville: Australian Institute of Marine Science; 1980. 422 [Google Scholar]
- 90.Veron JEN. Corals of Australia and the Indo-Pacific. Sydney: Angus & Robertson; 1986. 644 [Google Scholar]
- 91.Veron JEN. New Scleractinia from Japan and other Indo-West Pacific countries. Galaxea. 1990;9:95–173. [Google Scholar]
- 92.Veron JEN. Corals of the World. Townsville: Australian Institute of Marine Science; 2000. 1382 [Google Scholar]
- 93.Veron JEN. New Species Described in Corals of the World. Townsville: Australian Institute of Marine Science; 2002. 209 [Google Scholar]
- 94.Wallace CC, Chen CA, Fukami H, Muir PR. Recognition of separate genera within Acropora based on new morphological, reproductive and genetic evidence from Acropora togianensis, and elevation of the subgenus Isopora Studer, 1878 to genus (Scleractinia: Astrocoeniidae; Acroporidae). Coral Reefs. 2007;26:231–239. doi: 10.1007/s00338-007-0203-4. [Google Scholar]
- 95.Wells JW. New genera of Mesozoic and Cenozoic corals. J Paleontol. 1937;11:73–77. [Google Scholar]
- 96.Wijsman-Best M. Systematics and ecology of New Caledonian Faviinae (Coelenterata – Scleractinia). Contrib Zool. 1972;42:3–90. [Google Scholar]
- 97.Yabe H, Sugiyama T. Recent reef-building corals from Japan and the south sea islands under the Japanese mandate. II. Sci Rep Tôhoku Imp Univ 2nd Ser (Geol) Spec Vol. 1941;2:67–91. [Google Scholar]
- 98.Bininda-Emonds ORP, Beck RMD, Purvis A. Getting to the roots of matrix representation. Syst Biol. 2005;54:668–672. doi: 10.1080/10635150590947113. doi: 10.1080/10635150590947113. [DOI] [PubMed] [Google Scholar]
- 99.Jones KE, Bininda-Emonds ORP, Gittleman JL. Bats, clocks, and rocks: diversification patterns in Chiroptera. Evolution. 2005;59:2243–2255. doi: 10.1111/j.0014-3820.2005.tb00932.x. [PubMed] [Google Scholar]
- 100.Maddison WP, Mooers AØ. Tuatara: Conservation Priority in a Phylogenetic Context. Version 1.0. 2007. Available: http://mesquiteproject.org/packages/tuatara. Accessed 2011 Jun 20.
- 101.Redding DW, Mooers AØ. Incorporating evolutionary measures into conservation prioritization. Conserv Biol. 2006;20:1670–1678. doi: 10.1111/j.1523-1739.2006.00555.x. doi: 10.1111/j.1523-1739.2006.00555.x. [DOI] [PubMed] [Google Scholar]
- 102.Mooers AØ, Faith DP, Maddison WP. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE. 2008;3:e3700. doi: 10.1371/journal.pone.0003700. doi: 10.1371/journal.pone.0003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 104.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 105.Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv Biol. 2010;24:1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 106.Orme D, Freckleton RP, Thomas G, Petzoldt T, Fritz SA. CAIC: Comparative Analyses Using Independent Contrasts. R Package Version 1.0.4-94. 2008. Available: http://r-forge.r-project.org/projects/caic. Accessed 2011 Jun 20.
- 107.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [Google Scholar]
- 108.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 109.Fritz SA, Purvis A. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc R Soc B-Biol Sci. 2010;277:2435–2441. doi: 10.1098/rspb.2010.0030. doi: 10.1098/rspb.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nyström M, Graham NAJ, Lokrantz J, Norström AV. Capturing the cornerstones of coral reef resilience: linking theory to practice. Coral Reefs. 2008;27:795–809. doi: 10.1007/s00338-008-0426-z. [Google Scholar]
- 111.Redding DW, Hartmann K, Mimoto A, Bokal D, DeVos M, et al. Evolutionarily distinctive species often capture more phylogenetic diversity than expected. J Theor Biol. 2008;251:606–615. doi: 10.1016/j.jtbi.2007.12.006. doi: 10.1016/j.jtbi.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 112.Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- 113.Cadotte MW, Cardinale BJ, Oakley TH. Evolutionary history and the effect of biodiversity on plant productivity. Proc Natl Acad Sci USA. 2008;105:17012–17017. doi: 10.1073/pnas.0805962105. doi: 10.1073/pnas.0805962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE. 2009;4:e5695. doi: 10.1371/journal.pone.0005695. doi: 10.1371/journal.pone.0005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Redding DW, DeWolff CV, Mooers AØ. Evolutionary distinctiveness, threat status, and ecological oddity in primates. Conserv Biol. 2010;24:1052–1058. doi: 10.1111/j.1523-1739.2010.01532.x. doi: 10.1111/j.1523-1739.2010.01532.x. [DOI] [PubMed] [Google Scholar]
- 116.Connolly J, Cadotte MW, Brophy C, Dooley Á, Finn J, et al. Phylogenetically diverse grasslands are associated with pairwise interspecific processes that increase biomass. Ecology. 2011;92:1385–1392. doi: 10.1890/10-2270.1. doi: 10.1890/10-2270.1. [DOI] [PubMed] [Google Scholar]
- 117.Flynn DFB, Mirotchnick N, Jain M, Palmer MI, Naeem S. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem function relationships. Ecology. 2011;92:1573–1581. doi: 10.1890/10-1245.1. doi: 10.1890/10-1245.1. [DOI] [PubMed] [Google Scholar]
- 118.Hughes TP, Bellwood DR, Connolly SR. Biodiversity hotspots, centres of endemicity, and the conservation of coral reefs. Ecol Lett. 2002;5:775–784. doi: 10.1046/j.1461-0248.2002.00383.x. [Google Scholar]
- 119.Beger M, Jones GP, Munday PL. Conservation of coral reef biodiversity: a comparison of reserve selection procedures for corals and fishes. Biol Conserv. 2003;111:53–62. doi: 10.1016/S0006-3207(02)00249-5. [Google Scholar]
- 120.Almany GR, Connolly SR, Heath DD, Hogan JD, Jones GP, et al. Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs. 2009;28:339–351. doi: 10.1007/s00338-009-0484-x. [Google Scholar]
- 121.Carpenter KE, Barber PH, Crandall ED, Ablan-Lagman MC, Ambariyanto, et al. Comparative phylogeography of the Coral Triangle and implications for marine management. J Mar Biol. 2011;2011:396982. doi: 10.1155/2011/396982. [Google Scholar]
- 122.Davies TJ, Fritz SA, Grenyer R, Orme CDL, Bielby J, et al. Phylogenetic trees and the future of mammalian biodiversity. Proc Natl Acad Sci USA. 2008;105:11556–11563. doi: 10.1073/pnas.0801917105. doi: 10.1073/pnas.0801917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parhar RK, Mooers AØ. Phylogenetically clustered extinction risks do not substantially prune the Tree of Life. PLoS ONE. 2011;6:e23528. doi: 10.1371/journal.pone.0023528. doi: 10.1371/journal.pone.0023528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mooers AØ. Tree balance and tree completeness. Evolution. 1995;49:379–384. doi: 10.1111/j.1558-5646.1995.tb02251.x. doi: 10.2307/2410349. [DOI] [PubMed] [Google Scholar]
- 125.Mooers AØ, Heard SB. Inferring evolutionary process from phylogenetic tree shape. Q Rev Biol. 1997;72:31–54. doi: 10.1086/419657. [Google Scholar]
- 126.Purvis A, Agapow PM. Phylogeny imbalance: taxonomic level matters. Syst Biol. 2002;51:855–854. doi: 10.1080/10635150290102546. doi: 10.1080/10635150215871. [DOI] [PubMed] [Google Scholar]
- 127.Blum MGB, François O. Which random processes describe the Tree of Life? A large-scale study of phylogenetic tree imbalance. Syst Biol. 2006;55:685–691. doi: 10.1080/10635150600889625. doi: 10.1080/10635150600889625. [DOI] [PubMed] [Google Scholar]
- 128.Purvis A, Fritz SA, Rodríguez J, Harvey PH, Grenyer R. The shape of mammalian phylogeny: patterns, processes and scales. Philos Trans R Soc B-Biol Sci. 2011;366:2462–2477. doi: 10.1098/rstb.2011.0025. doi: 10.1098/rstb.2011.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Colless DH. Review of “Phylogenetics: The Theory and Practice of Phylogenetic Systematics.”. Syst Zool. 1982;31:100–104. [Google Scholar]
- 130.Heard SB, Mooers AØ. Phylogenetically patterned speciation rates and extinction risks change the loss of evolutionary history during extinctions. Proc R Soc Lond B-Biol Sci. 2000;267:613–620. doi: 10.1098/rspb.2000.1046. doi: 10.1098/rspb.2000.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bellwood DR, Hughes TP, Folke C, Nyström M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- 132.Bruno JF, Selig ER. Regional decline of coral cover in the Indo-Pacific: timing, extent, and subregional comparisons. PLoS ONE. 2007;2:e711. doi: 10.1371/journal.pone.0000711. doi: 10.1371/journal.pone.0000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marshall PA, Baird AH. Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs. 2000;19:155–163. doi: 10.1007/s003380000086. [Google Scholar]
- 134.Miller J, Waara R, Muller E, Rogers C. Coral bleaching and disease combine to cause extensive mortality on reefs in US Virgin Islands. Coral Reefs. 2006;25:418. doi: 10.1007/s00338-006-0125-6. [Google Scholar]
- 135.Brandt ME, McManus JW. Disease incidence is related to bleaching extent in reef-building corals. Ecology. 2009;90:2859–2867. doi: 10.1890/08-0445.1. doi: 10.1890/08-0445.1. [DOI] [PubMed] [Google Scholar]
- 136.Yee SH, Santavy DL, Barron MG. Assessing the effects of disease and bleaching on Florida Keys corals by fitting population models to data. Ecol Model. 2011;222:1323–1332. doi: 10.1016/j.ecolmodel.2011.01.009. [Google Scholar]
- 137.Weitzman ML. The Noah's Ark problem. Econometrica. 1998;66:1279–1298. doi: 10.2307/2999617. [Google Scholar]
- 138.Faith DP, Magallón S, Hendry AP, Conti E, Yahara T, et al. Evosystem services: an evolutionary perspective on the links between biodiversity and human well-being. Curr Opin Environ Sustain. 2010;2:66–74. doi: 10.1016/j.cosust.2010.04.002. [Google Scholar]
- 139.Romano SL, Palumbi SR. Molecular evolution of a portion of the mitochondrial 16S ribosomal gene region in scleractinian corals. J Mol Evol. 1997;45:397–411. doi: 10.1007/pl00006245. doi: 10.1007/PL00006245. [DOI] [PubMed] [Google Scholar]
- 140.Fukami H, Budd AF, Paulay G, Sole-Cava AM, Chen CA, et al. Conventional taxonomy obscures deep divergence between Pacific and Atlantic corals. Nature. 2004;427:832–835. doi: 10.1038/nature02339. doi: 10.1038/nature02339. [DOI] [PubMed] [Google Scholar]
- 141.Budd AF, Romano SL, Smith ND, Barbeitos MS. Rethinking the phylogeny of scleractinian corals: a review of morphological and molecular data. Integr Comp Biol. 2010;50:411–427. doi: 10.1093/icb/icq062. doi: 10.1093/icb/icq062. [DOI] [PubMed] [Google Scholar]
- 142.Palmer CV, Bythell JC, Willis BL. Levels of immunity parameters underpin bleaching and disease susceptibility of reef corals. FASEB J. 2010;24:1935–1946. doi: 10.1096/fj.09-152447. doi: 10.1096/fj.09-152447. [DOI] [PubMed] [Google Scholar]
- 143.Veron JEN, DeVantier LM, Turak E, Green AL, Kininmonth S, et al. Delineating the Coral Triangle. Galaxea. 2009;11:91–100. doi: 10.3755/galaxea.11.91. [Google Scholar]
- 144.Cadotte MW, Davies TJ. Rarest of the rare: advances in combining evolutionary distinctiveness and scarcity to inform conservation at biogeographical scales. Divers Distrib. 2010;16:376–385. doi: 10.1111/j.1472-4642.2010.00650.x. [Google Scholar]
- 145.Steel M, Mimoto A, Mooers AØ. Hedging our bets: the expected contribution of species to future phylogenetic diversity. Evol Bioinform. 2007;3:237–244. [PMC free article] [PubMed] [Google Scholar]
- 146.Faith DP. Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv Biol. 2008;22:1461–1470. doi: 10.1111/j.1523-1739.2008.01068.x. doi: 10.1111/j.1523-1739.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- 147.Knowlton N, Nunes F. Atlantic corals—least of our concerns? Science E- Letter. 2008. Available: http://www.sciencemag.org/content/321/5888/560.full/reply#sci_el_11734. Accessed 2011 Sep 5.
- 148.Carpenter KE, Polidoro BA, Livingstone SR, Aronson RB, Precht WF. Response to N. Knowlton and F. Nunes' E-Letter. Science E-Letter. 2008. Available: http://www.sciencemag.org/content/321/5888/560.full/reply#sci_el_11734. Accessed 2011 Sep 5.
- 149.Maynard JA, Baird AH, Pratchett MS. Revisiting the Cassandra syndrome; the changing climate of coral reef research. Coral Reefs. 2008;27:745–749. doi: 10.1007/s00338-008-0432-1. [Google Scholar]
- 150.Fabricius KE, Langdon C, Uthicke S, Humphrey C, Noonan S, et al. Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat Clim Change. 2011;1:165–169. doi: 10.1038/nclimate1122. [Google Scholar]
- 151.Pandolfi JM, Connolly SR, Marshall DJ, Cohen AL. Projecting coral reef futures under global warming and ocean acidification. Science. 2011;333:418–422. doi: 10.1126/science.1204794. doi: 10.1126/science.1204794. [DOI] [PubMed] [Google Scholar]
- 152.Rodolfo-Metalpa R, Houlbrèque F, Tambutté E, Boisson F, Baggini C, et al. Coral and mollusc resistance to ocean acidification adversely affected by warming. Nat Clim Change. 2011;1:308–312. doi: 10.1038/nclimate1200. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reef and non-reef coral species included in the phylogenetic analysis of Scleractinia. For each species, the IUCN Red List category and ranks according to the EDGE of Existence (EoE) programme and the present study are shown where appropriate. Species not assessed are indicated as N/A. GenBank accession numbers are provided for DNA sequences (see text for names of markers).
(PDF)