Abstract
In higher eukaryotes many genes encode protein isoforms whose properties and biological roles are often poorly characterized. Here we describe systematic approaches for detection of either distinct isoforms, or separate pools of the same isoform, with differential biological properties. Using information from ion intensities we have estimated protein abundance levels and using rates of change in stable isotope labeling with amino acids in cell culture isotope ratios we measured turnover rates and subcellular distribution for the HeLa cell proteome. Protein isoforms were detected using three data analysis strategies that evaluate differences between stable isotope labeling with amino acids in cell culture isotope ratios for specific groups of peptides within the total set of peptides assigned to a protein. The candidate approach compares stable isotope labeling with amino acids in cell culture isotope ratios for predicted isoform-specific peptides, with ratio values for peptides shared by all the isoforms. The rule of thirds approach compares the mean isotope ratio values for all peptides in each of three equal segments along the linear length of the protein, assessing differences between segment values. The three in a row approach compares mean isotope ratio values for each sequential group of three adjacent peptides, assessing differences with the mean value for all peptides assigned to the protein. Protein isoforms were also detected and their properties evaluated by fractionating cell extracts on one-dimensional SDS-PAGE prior to trypsin digestion and MS analysis and independently evaluating isotope ratio values for the same peptides isolated from different gel slices. The effect of protein phosphorylation on turnover rates was analyzed by comparing mean turnover values calculated for all peptides assigned to a protein, either including, or excluding, values for cognate phosphopeptides. Collectively, these experimental and analytical approaches provide a framework for expanding the functional annotation of the genome.
Biological regulatory mechanisms and cellular responses are predominantly mediated by proteins and multi-protein complexes. The structures and properties of these proteins are crucial for their function and can vary greatly. For example, protein expression levels in mammalian cells vary over a large dynamic range of 106 or more (1), whereas subcellular localization patterns, post-translational modifications, rates of synthesis, and degradation and interactions with partner proteins are also variable properties (2). Furthermore, all of these properties not only vary between proteins, they are also dynamic and can vary for the same protein either at different times, or in different subcellular locations, depending on parameters such as cell cycle progression, growth rate, and signaling events.
In higher eukaryotes, many genes encode two or more separate protein isoforms (3, 4). Even minor structural differences between isoforms can alter their biological properties and result in distinct pools of related proteins whose subcellular location, function, and interactions vary (5, 6). Furthermore, even apart from isoforms, single polypeptides can partition into two or more distinct functional pools within the cell that have different roles. For example, a single isoform of protein phosphatase I can interact with numerous different interaction partners to create different phosphatase enzymes that target different substrates (7). Proteomes are thus inherently complex and their properties in constant flux. This presents a major challenge for proteomic studies, as we aspire not only to identify which proteins are expressed in a cell or organelle, but also to characterize their properties and quantify how these change in response to different perturbations and cell cycle stages etc (8).
Alternative splicing of pre-mRNA transcripts is commonplace and this can generate multiple mRNAs from the same gene and hence multiple different proteins (9, 10). Shoemaker et al. have reported that over 73% of all human genes are alternatively spliced (11). Such isoforms can vary in length, share common exons, include variable exons, and even have very different amino acid sequences because splicing events can alter the translational reading frame of the differentially spliced mRNAs. It is estimated that 15% of all point mutations causing human genetic disease result in an mRNA splicing defect (12). Isoforms can also arise from differential post-translational processing and modification (6, 13) of a polypeptide encoded by a single mRNA. In other cases gene duplication results in expression of closely related protein paralogs that share extensive sequence identity and may thus be hard to distinguish by MS depending on the number and structures of peptides that encode the variance between these paralogs.
The structural and functional diversity of the expressed proteome in multicellular eukaryotes is thus generated by a combination of alternative splicing, together with other processes such as the use of alternative transcription start sites, alternative polyadenylation, RNA editing, SNPs, as well as complex patterns of post-translational modification and cleavage events (14). Although the use of mass spectrometry has now revolutionized the efficient and sensitive detection and quantitation of cell proteins, a complication of interpreting the results of protein identification and quantification using mass spectrometry is that proteins are typically extracted from cells and digested into peptides before MS analysis. This affects the interpretation of the resulting data because the same peptide sequence can be present in either multiple different proteins, or protein isoforms (3). As noted above, the same peptides may even belong in vivo to functionally distinct pools of the same protein. Such shared peptides therefore can lead to ambiguities, both in determining the identities of proteins and in reliably measuring their functional properties. It will therefore be helpful to develop methods that can help to distinguish between different isoforms and functionally distinct protein pools when interpreting MS data.
Some of the current approaches used to identify protein isoforms include deep sequencing, tiling arrays, protein processing through identification of new N-terminal peptides, SNP detection, alignment of identified peptides to the genome combined with target analysis of predicted peptides and Expressed Sequence Tags (15). A recent study by Alm et al. reported the identification of isoforms via alignment of mass spectra of spots on two-dimensional gels by use of extracted peak lists and hierarchical clustering (16). Methods for de novo sequencing and identification of post-translational modifications have also been developed, which operate independent of sequence databases (17). Combining transcriptome data with MS-based proteomics in specific forms of cancer cells has enabled identification of novel protein isoforms and splicing variants (18). Bioinformatics approaches have made use of Expressed Sequence Tag and RNA and genomic sequence data to match new splice forms with peptides revealed in MS spectra. Nonetheless, these strategies are limited by the availability and often incompleteness and fragmentation of relevant gene expression data (19).
The functional annotation of genome expression will be improved if it is possible to take into account and measure expression levels, structures, properties, and biological roles of separate protein isoforms and protein pools. In general this information on isoforms and protein pools is not available in most large-scale proteomic analyses (20). It will aid the biological interpretation of proteomics experiments to decide whether all peptides identified and quantified that are mapped to a specific gene are encoded either in a single polypeptide, or in two or more isoforms, and whether the peptides belong to a polypeptide that behaves within the cell as one or more functional pools with respect to its properties, such as subcellular localization and/or turnover rate. For example, when studying subcellular localization, the averaged value for all peptides mapped to a specific gene may indicate that the protein is present in both the cytoplasm and the nucleus, when in fact they belong to two isoforms, with one isoform predominantly cytoplasmic and the other predominantly nuclear. This is likely to be of general importance for annotating the genome because a recent comparative study of subcellular protein localization in three human cell lines detected ∼40% of the 4000 genes analyzed localizing to multiple subcellular compartments (21).
Mass spectrometry-based proteomics has become the technology of choice for the direct identification and characterization of proteins (22). In combination with quantitative approaches, such as SILAC (stable isotope labeling with amino acids in cell culture)1, mass spectrometry can not only identify proteins and post-translational modifications, but also measure how relative protein levels change in cells under different conditions (23, 24). This provides a flexible assay format for proteomic studies that evaluate differences between two or more cell states, each defined by metabolic labeling of proteins with amino acids that have different combinations of isotopes incorporated into selected amino acids. Subsequent isolation of proteins and enzyme cleavage results in mixtures of isotopically labeled peptides where the relative levels of each isotopic form can be resolved and quantified by mass spectrometry. The peptide isotope ratios are then mapped back to the genome sequences encoding the cognate proteins and used to infer whether either the levels, or properties, of these proteins have been changed. The SILAC strategy has been used for quantitative studies of cell and organelle proteomes and for comparative studies of protein modifications, and interactions (22) and to identify proteins isolated from mitotic chromosomes (25). It has also been used in combination with cell fractionation to generate “isotope-encoded” subcellular compartments allowing subcellular protein localization to be evaluated on a system-wide level (26, 27). By examining incorporation rates of isotope-labeled amino acids into proteins, pulse-labeling SILAC has been employed to measure protein turnover in cells and organelles (28–32). We have recently reported a global analysis of protein properties in human cells using a combined pulse-labeling, spatial proteomics and data analysis strategy to characterize the expression, localization, synthesis, degradation and turnover rates of endogenously expressed, untagged human proteins in different subcellular compartments (33). Mass spectrometry combined with pulsed incorporation of stable isotopes of arginine and lysine were used to perform quantitative analyses of the rates of synthesis, degradation, and turnover of HeLa cell proteins. Cells were pulsed for 0.5, 4, 7, 11, 27, and 48 h before being fractionated into cytoplasmic, nucleoplasmic, and nucleolar fractions. Proteins from each of the respective subcellular fractions and time points were further fractionated by 1-D SDS-PAGE and each of 16 gel slices trypsin digested. The resulting peptides were analyzed by liquid chromatography (LC)-tandem MS (MS/MS) and ratios between light, medium, and heavy isotopic forms for each peptide quantified using MaxQuant and the data managed and analyzed using PepTracker. A total of 80,098 peptides from an estimated 8041 HeLa proteins were quantified, and their spatial distribution between the cytoplasm, nucleus, and nucleolus determined as described in the related paper (33). Using information from ion intensities and rates of change in isotope ratios, protein abundance levels and protein synthesis, degradation, and turnover rates were calculated for the whole cell and for the respective cytoplasmic, nuclear, and nucleolar compartments.
Here we analyze this same HeLa proteomics data set (33) using systematic approaches for the detection of protein isoforms and protein pools with differential biological properties. We evaluate methods that can identify human protein isoforms whose turnover and/or subcellular localization properties vary and analyze phosphorylated peptides that are correlated with altered rates of protein turnover in the separate cytoplasmic, nuclear, and nucleolar compartments. The methods described here maximize the opportunity of using empirically measured protein properties to identify functionally distinct pools of proteins and protein isoforms.
EXPERIMENTAL PROCEDURES
Cell Culture, Translation Inhibition, and Transfection
HeLa cells were cultured as adherent cells in DMEM (Dulbeccos's modified Eagle medium; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin and 2 mm l-Glutamine. For translation inhibition, HeLa cells were plated on 6-well dishes at 250,000 cells per well. Cells were either mock treated, or treated with 100 μg/ml cycloheximide (Sigma) for 0.5, 1, 4, 7, and 24 h. HeLa cells were transfected using Effectene Transfection Reagent (Qiagen, Dorking, Surrey, UK) as per the manufacturer's protocol.
Antibodies and Western Blotting
For protein blot analysis cells were lysed by mixing with loading sample buffer (Invitrogen), then boiled for 10 min and separated by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex mini-gel, Invitrogen) and transferred to nitrocellulose (iBlot, Invitrogen) prior to Western blotting. Antibodies from the Human Protein Atlas (34), HBS1L (HPA02729), CTSD (HPA003001), and RPRD1A (HPA040602) were used at a 1:1000 dilution for Western blotting. The anti-GFP mouse monoclonal antibody (Roche Diagnostics) was used at a dilution of 1:3000.
Plasmid Construction and Stable Cell Lines
For the creation of the GFP-NudCD1 isoform expressing HeLa cell lines, each isoform of NudCD1 was PCR amplified from pENTRY plasmids (Open Biosystems, Thermo Scientific) and inserted into pDONR221 vector using BP Clonase II (Invitrogen) and subsequently transferred into pG-LAP1 (N-terminal GFP-Lap tag) using LR Clonase II (Invitrogen) and integrated into HeLa Flp-In cells (Invitrogen) (35). Cells were selected for Hygromycin and Blasticidine resistance and expression of the GFP-fusion proteins was induced by adding Doxycycline at a concentration of 2 μg/ml for 48 h.
Immunofluorescence
HeLa cells were grown on glass coverslips and fixed with 1% paraformaldehyde in PBS for 10 min. Cells were then permeabilized in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 for 10 min, and then labeled with antibodies recognizing GFP (Roche Diagnostic). After washing with PBS containing 0.1% Triton X-100 and PBS, cells were then labeled with a secondary antibody coupled to Alexa 546 (Molecular Probes, Eugene, OR) and mounted on slides with Vectashield (Vector Laboratories Inc., Burlingame, CA) containing DAPI. Fluorescence imaging was performed on a DeltaVision Spectris widefield deconvolution microscope (Applied Precision) (Washington, United States) equipped with a CoolMax charge-coupled device camera (Roper Scientific, Trenton, NJ). Cells were imaged using a 60x NA 1.4 Plan-Apochromat objective (Olympus) and the appropriate filter sets (Chroma Technology Corp., Brattleboro, VT), with 20 optical sections of 0.5 μm each acquired. SoftWorX software (Applied Precision) was used for both acquisition and deconvolution.
Quantification and Bioinformatic Analysis
The methods used for preparation of SILAC labeled HeLa proteins from nuclear, nucleolar, and cytoplasmic fractions, protein chromatography by SDS-PAGE, trypsin digestion, and mass spectrometry were described previously (33). Peptide identification, quantitation, and phosphopeptide analysis was performed using MaxQuant version 1.1.1.14 (36, 37). The derived peak list was searched using the in-built Andromeda database search engine in MaxQuant for peptide identifications against the International Protein Index (IPI) human protein database version 3.68 containing 89,422 proteins, to which 175 commonly observed contaminants and all the reversed sequences had been added. The initial mass tolerance was set to 7 p.p.m. and MS/MS mass tolerance was 0.5 Da. Enzyme was set to trypsin/p with two missed cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, whereas N-acetyl protein, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine were searched as variable modifications. Identification was set to a false discovery rate of 1%. To achieve reliable identifications, all proteins were accepted based on the criteria that the number of forward hits in the database was at least 100-fold higher than the number of reverse database hits, thus resulting in a false discovery rate of less than 1%. A minimum of two peptides were quantified for each protein. Data analysis was performed using the PepTracker™ software environment. Clustering analysis was performed using the software Cluster with complete linkage clustering and visualized using Treeview (http://rana.lbl.gov/EisenSoftware.htm) (38).
To maximize the identification of potential novel protein isoforms, prior annotation of known proteins was removed to ensure the analysis was carried out as unbiased as possible. Hence, the predicted protein groups generated by MaxQuant were discarded and instead the evidences of individual peptides were used to build protein profiles based on data corresponding to our empirical measurements of protein properties. Using these measured behavioral properties it was then possible to identify novel pools of proteins that were functionally distinct. By performing the analysis in this way, it was possible to link the existence of predicted isoforms with changes in their measured properties.
RESULTS
Protein Isoform Analysis at the Peptide Level: Candidate Approach
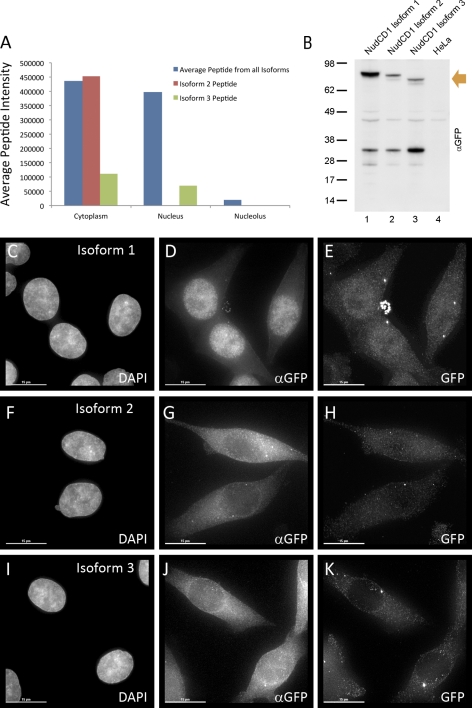
We have analyzed HeLa cell SILAC data describing global protein abundance, localization, and turnover (33), using three approaches to detect protein isoforms that have differential properties (Fig. 1). First, a candidate approach was used. For genes encoding known isoforms, average intensity values were compared for peptides shared between all isoforms with candidate, isoform-specific peptides (Fig. 2). This is illustrated for the NudCD1 protein, which has three reported isoforms (39). Using average values for all peptides detected that are common to the three isoforms (blue), there is similar average peptide intensity in the cytoplasm and nucleus, with little signal in the nucleolus. However, although analysis of a peptide predicted to be specific to isoform 3 showed intensity in both cytoplasmic and nuclear compartments (green, ∼3:2 cytoplasmic/nuclear), a peptide predicted to be specific for isoform 2 (red), instead showed exclusively cytoplasmic signal (Fig. 2A). We were unable to detect reliably any peptides that were unique to isoform 1. However, as there is strong overall peptide signal in the nucleus that cannot be accounted for by the intensities of either the isoform 2, or isoform 3-specific peptides, we infer that isoform 1 is likely to be enriched in the nucleus.
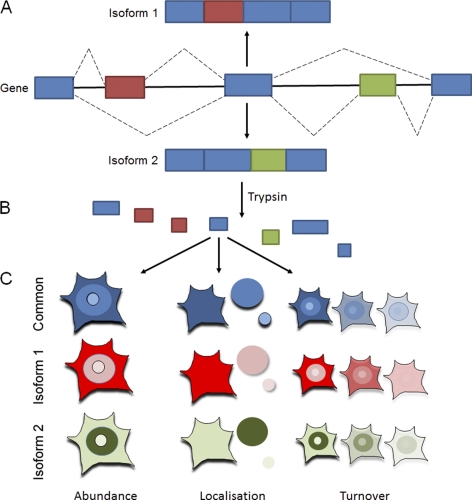
Fig. 1.
Alternative splicing leading to protein isoforms. A, A single gene can encode multiple proteins because of alternative splicing. After transcription of a gene, exons of the resultant RNA can be reconnected in multiple ways during RNA splicing, resulting in the translation of protein isoforms, with red and green areas signifying differences between the isoforms. B, Protein isoforms represent several different forms of a protein and have a largely shared sequence, however small differences occur. Mass spectrometery can be used to identify both common peptides shared by all isoforms and specific peptides (shown in red and green) that are unique to each isoform. C, Using these identified peptides it is possible to extract abundance, localization, and turnover information for a protein by using either shared peptides or isoform specific peptides.
Fig. 2.
NUDCD1 protein isoform identification and localization from peptide intensities. Identification of NudCD1, NudC domain-containing protein 1. A, Chart showing intensity (y axis) in different cellular compartments (x axis) for three peptides. The blue series provides average intensity of common peptides shared between isoforms of NudCD1. Remaining series show intensity of MLYLQGWSMPAVAEVK (isoform 2 peptide - red) and YNQDTALGKPR (isoform 3 peptide - green). The isoform 3 specific peptide has intensity in both cytoplasmic and nuclear compartments (∼3:2 cytoplasmic/nuclear), and the isoform 2 peptide shows exclusively cytoplasmic signal. B, HeLa cells expressing GFP used to detect isoforms of NudCD1. Upper bands on the Western blot (orange arrow) show three protein isoforms recognized, migrating at the predicted molecular weights of NudCD1 isoforms. Fluorescence microscopy analysis of the HeLa cells expressing the respective GFP-fusion proteins was performed using both antibody to GFP (panels D, G, and J), and direct GFP fluorescence (panels E, H, and K). NudCD1 Isoform 1 shows nuclear accumulation in panels D and E, NudCD1 isoform 2 shows predominantly cytoplasmic accumulation in panels G and H and both cytoplasmic and nuclear accumulation is shown of NudCD1 isoform 3 in panels J and K.
As no suitable isoform-specific antibodies for NudCD1 were available, the localization patterns of the three NudCD1 isoforms were next compared by immunofluorescence microscopy analysis of HeLa cells expressing GFP fused at the N terminus to isoform-specific cDNAs (Figs. 2B–2K). All three GFP-NudCD1 isoform fusions were used to establish stable HeLa cell lines where expression of the fusion protein is under the control of a tetracycline-regulated promoter (see Experimental Procedures). All three stable HeLa cell lines produced proteins of the expected sizes when induced by addition of tetracycline and analyzed by protein blotting, detected using an anti-GFP antibody (Fig. 2B). Fluorescence microscopy analysis of HeLa cells expressing the respective GFP-fusion proteins was performed, using both an antibody to GFP (Figs. 2D, 2G, and 2J), and direct GFP fluorescence (Figs. 2E, 2H, and 2K), to determine their localization patterns. In agreement with the spatial proteomics data, this showed predominantly nuclear accumulation of NudCD1 isoform 1 (panels 2D and 2E), predominantly cytoplasmic accumulation of NudCD1 isoform 2 (panels 2G and 2H) and both cytoplasmic and nuclear accumulation of NudCD1 isoform 3 (panels 2J and 2K). None of the three GFP-NudCD1 isoform fusions accumulated in nucleoli.
These data analyzing NudCD1 isoforms illustrate the validity of the candidate approach but also highlight its limitations. It relies upon prior annotation to predict the existence of isoforms and the detection of unique peptides that are isoform-specific. As seen for isoform 1, it is not always possible to detect isoform-specific peptides. Even when isoform-specific peptides can be detected, as with isoforms 2 and 3, they are usually few in number (often only one) and this reduces the accuracy of the overall quantitation. Nonetheless, the NudCD1 data show clearly that analysis of a key protein property, such as subcellular localization, can be misleading when values for all peptides are averaged without taking into account the existence of distinct pools of protein with differential localization phenotypes.
Rule of Thirds Approach
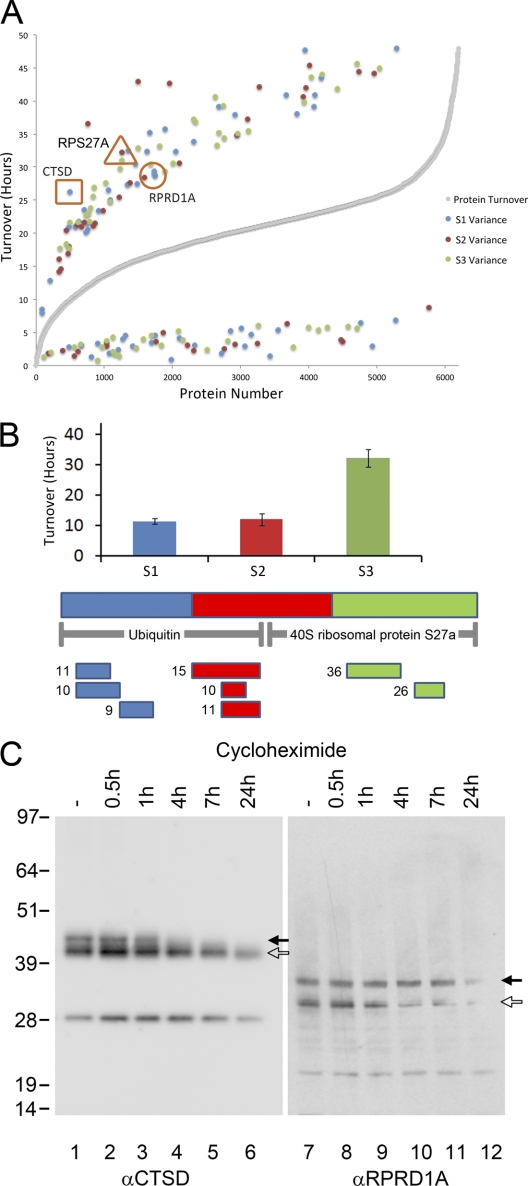
Next, we used two methods to systematically evaluate whether the mean value of all peptides quantitated for a given protein included clusters of adjacent peptides with significantly different mean values. First, a “rule of thirds” approach was used to search the data for examples in which the mean values of peptides from the amino-terminal (S1, blue), central (S2, red), or carboxyl terminal (S3, green) segments of the protein differed by at least one standard deviation from an adjacent segment (supplemental Table S1). This was evaluated for over 6000 HeLa proteins, where at least two peptides had been quantitated within each segment of the protein sequence. The mean turnover rate for each segmented third of every protein was plotted on the y axis against total proteins, ranked on the x axis by the mean turnover value derived from all peptides assigned to that protein (Fig. 3A). Examples where the turnover value of any one third segment of a given protein differed by more than 70% from the overall turnover value for the same protein, i.e. the mean of all the peptides assigned to that protein, are highlighted and color coded in blue, red, and green for segments S1–S3, respectively (Fig. 3A).
Fig. 3.
Protein isoform identification from protein sequence segmentation. A, Graph showing all proteins (x axis) (ordered by average protein turnover) plotted against turnover in hours (y axis). Gray series shows average turnover for each protein. Blue, red, and green series highlight segments of proteins that have a variance of 70% or more compared with average protein turnover. B, Chart shows the three equal segments of RPS27A, Ubiquitin-40S ribosomal protein S27a, plotted on x axis against average turnover on y axis. Peptides for each segment (examples shown underneath) have significantly different turnover in carboxyl terminal segment 3 compared with segments 1 and 2. RPS27A protein is expressed as a precursor that is processed to yield ubiquitin. C, Further proteins tested were, CTSD - Cathepsin d- (shown in orange square in A) and RPRD1A - Regulation of nuclear pre-mRNA domain-containing protein 1A (shown in orange circle in A). Cycloheximide inhibition experiments were performed on HeLa cells to block protein synthesis and thus measure the rate of protein degradation. The Western blot for CTSD shows a band at predicted molecular weight of 44.55 kDa (white arrow). Another, higher, band is visible (black arrow), which shows a faster turnover across the five timepoints (lanes 2–6). The second protein tested, RPRD1A, shows two bands on the Western blot, which correlate with the expected molecular weight of the known isoforms of RPRD1A (isoform 1 at 35.72kDa, isoform 2 at 32.92kDa and 31.63kDa). The upper band (black arrow, isoform 1) shows slower degradation over the timecourse (lanes 8–12) compared with the lower band (white arrow) (isoforms 2 and 3).
The validity of the rule of thirds approach was confirmed by its unbiased identification of RPS27A as one of the proteins with a segment showing differential turnover to the mean value for the whole protein (Fig. 3B). In this case the mean turnover value of peptides from the carboxyl terminal segment (green, ∼31 h), was ∼threefold higher than the mean turnover values for the peptides in either of the other two segments (blue ∼11 h and red ∼14 h) and ∼twofold higher than the mean of all the peptides in this protein (∼15 h). Interestingly, the full length RPS27A protein is expressed as a precursor that is subsequently processed to yield ubiquitin, which accounts for ∼70% of the sequence, and a carboxyl terminal segment of ∼30% that corresponds to the mature ribosomal small subunit protein S27A (40). As ubiquitin is subsequently conjugated to proteins as a post-translational modification that can promote proteasome-mediated degradation, whereas ribosomal proteins are typically stable after incorporation into ribosome subunits, it is not surprising that these two products of the original RPS27A polypeptide exhibit different turnover values.
We selected two other examples from the group of highlighted proteins for further analysis, for which antibodies were available, corresponding to Cathepsin D (CTSD) and Regulation of nuclear pre-mRNA domain-containing protein 1A (RPRD1A) (Fig. 3C). A cycloheximide inhibition experiment was performed on HeLa cells to block protein synthesis and thus measure the rate of protein degradation. Both CTSD (lanes 1–6) and RPRD1A (lanes 7–12) were detected by immunoblotting, using specific antibodies generated by the Human Protein Atlas Project (see Experimental Procedures). In both cases the blotting experiments revealed two bands for each protein that decay at different rates following cycloheximide treatment (Fig. 3C, arrows). These data support the prediction from the rule of thirds analysis that the CTSD and RPRD1A proteins are expressed as distinct polypeptides with different turnover values.
Three in a Row Approach
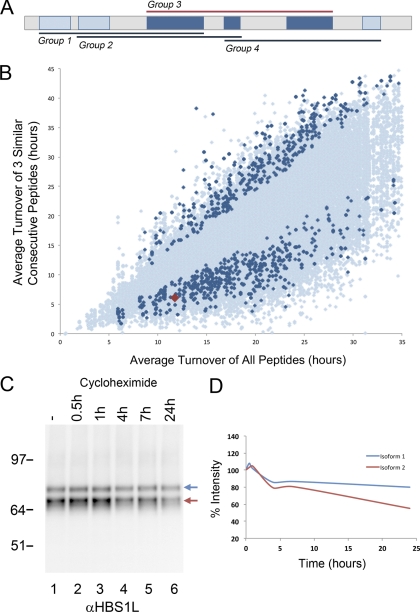
A limitation with the rule of thirds approach is that not all isoforms will have structures that are separable based on analysis of equal third regions of the protein. The available peptide coverage is also often not evenly distributed between each of these three equal segments. To provide a more general approach for predicting isoform expression, based on local clusters of peptide values, we therefore turned to a “three in a row” method. Here, mean turnover values were calculated for each set of three consecutive peptides within the total set of peptides assigned to a given protein, moving along one peptide at a time from the amino to carboxyl terminus (Fig. 4A). The resulting mean turnover values for every group of three consecutive peptides were then plotted on the y axis, against the corresponding mean turnover value on the x axis calculated using all peptides mapped to each protein (Fig. 4B). In this plot each triple peptide mean value is shown either in light blue (default), or in dark blue if two conditions are met. Thus, dark blue indicates that both the turnover value for that group of three consecutive peptides differs by 20% or more, (either higher or lower), than the mean value of all of the peptides assigned to that protein and that all three peptides in the group have similar values, i.e. all three are either higher, or lower, than the protein mean (see Supp. Table S2).
Fig. 4.
Protein isoform identification from consecutive peptide analysis. A, Groups of three consecutive peptides were identified and average turnover value of each group calculated. A linear representation of protein is shown, with highlighted regions indicating identified peptides. Any group showing a 20% variance in turnover from the average protein turnover is labeled as interesting (Group 3). B, Graph showing average protein turnover from all peptides (x axis) versus average turnover of three consecutive peptides from the protein (y-axis). Data points highlighted in blue indicate three consecutive peptides that all have a turnover that varies by 20% greater or less than the average protein turnover. Highlighted in red, is protein HBS1L, HBS1-like protein. C, Cycloheximide experiment was carried out to measure the degradation rate of HBS1L. Antibody for HBS1L detected two bands, consistent with expression of two isoforms (blue and red arrows). These bands correlate to the known isoforms of HBS1L (isoform 1 at 75.5 kDa (blue arrow), isoforms 2 and 3 at 70.13 kDa and 70.63 kDa (red arrow)). D, Quantitation of the two bands at multiple time points from 0.5–24 h (lanes 2–6) is shown on the graph. The percentage intensity is plotted on the y axis, across the timecourse on the x axis. Graph shows that the two putative isoforms of HBS1L differ in their degradation rates.
For the whole cell protein turnover data set, analysis of 178,509 groups of three consecutive peptides identified 1790 groups (∼1%) that met these criteria and hence are shaded dark blue (Fig. 4B). To validate this approach we selected one of the highlighted proteins for which specific antibodies were available, i.e. HBS1L (red diamond in Fig. 4B). A cycloheximide experiment was carried out to measure independently the degradation rate of HBS1L (Figs. 4C and 4D). An antibody from the Human Protein Atlas Project specific for HBS1L detected two bands on an immunoblot, consistent with expression of two isoforms (Fig. 4C). Quantitation of the two bands at multiple time points from 0.5–24 h following cycloheximide treatment showed that the two putative isoforms of HBS1L differed in their degradation rates (Fig. 4D). We conclude that the three in a row approach can help to detect proteins expressed as isoforms with differential properties.
Isoform Analysis by Combined Protein Fractionation and Peptide MS
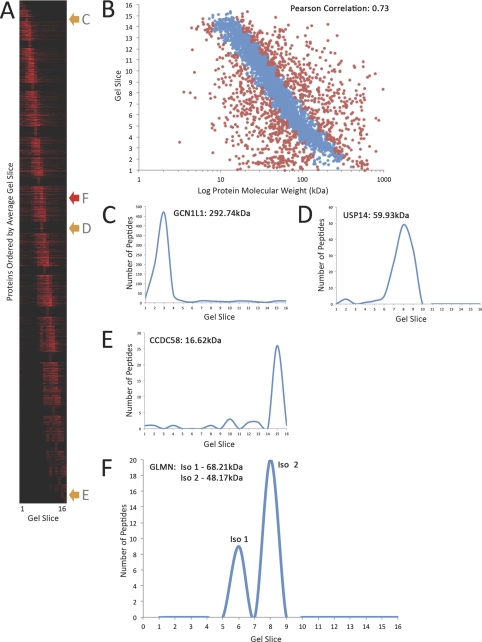
Protein isoforms that differ in size can be separated by chromatography prior to enzyme cleavage and MS identification of peptides. We have therefore incorporated into our analysis information derived from fractionation of HeLa cell proteins by one-dimensional SDS-PAGE (Fig. 5). HeLa cell extracts were separated on a 4–12% SDS-PAGE gel, which was then cut into 16 slices, numbered from the top (slice 1, largest proteins) to the bottom (slice 16, smallest proteins) of the gel (Fig. 5A). Proteins in each gel slice were digested with trypsin and the resulting peptides eluted and analyzed by MS (33), with the resulting data plotted on a graph showing gel slice on the y axis and Log predicted molecular weight of each identified protein, based on genome sequence annotation, on the x axis (Fig. 5B). These empirical data demonstrate that, as expected, the position of protein migration on SDS-PAGE is positively correlated with predicted molecular weight, (Pearson correlation coefficient 0.73). In this gel system, that correlation holds true at least within the size range from ∼10–180 kDa. Using the MS identification information the approximate size range of proteins migrating in each gel slice can thus be estimated. Based upon a best linear fit within the 10–180 kDa size range, the majority of proteins (∼78%), migrate at their predicted molecular weight ±40% (Fig. 5B, blue dots). Nonetheless, a substantial number of proteins identified by MS (>20%), migrate anomalously with respect to predicted molecular weight (Fig. 5B, red dots). Reasons for apparently anomalous migration is likely to include the expression of novel protein isoforms and processed polypeptides, as well as effects of post-translational modifications on migration behavior.
Fig. 5.
Protein migration study on gel fractionation. A, Heat map of the 16 gel slices (horizontally) is shown with every protein identified (vertically) ordered by the average gel slice the protein was found in. B, Graph showing predicted log protein molecular weight (x axis) against gel slice (y axis), indicating that proteins predictably migrated across the gel based on their molecular weights (Pearson Correlation: 0.73). Graphs C, D, and E highlight three examples of proteins, GCN1L1 (Translational activator GCN1), USP14 (Ubiquitin carboxyl-terminal hydrolase 14) and CCDC58 (Coiled-coil domain-containing protein 58) respectively, where the peptide count (y axis) is plotted against gel slice (x axis). These graphs show that the proteins migrate at different gel slices consistent with their molecular weight. F, Graph showing peptide count (y axis) plotted versus gel slice and molecular weight (x axis) for protein GLMN (Glomulin). The gel fractionation data indicate that this protein migrates at two gel slices, 6 and 8. GLMN, has in fact two known isoforms, isoform 1 at 68.21 kDa and isoform 2 at 48.17 kDa.
Examination of the number of unique peptide identifications assigned to a given protein in each gel slice reveals the migration profile of that protein in SDS-PAGE (Figs. 5C–5E). For representative large (Fig. 5C, GCN1L1, 293 kDa), medium (Fig. 5D, USP14, 60 kDa), and small (Fig. 5E, CCDC58, 17 kDa) proteins, the number of unique peptides identified shows a clear single peak across the respective gel slices. The breadth of the unique peptide abundance peak is positively correlated with protein abundance (supplemental Fig. S1), such that the most abundant proteins show broad horizontal lines in the heat map (Fig. 5A). The unique peptide count per gel slice also helps to identify distinct protein isoforms. As shown for the protein Glomulin (GLMN), which has two known isoforms of 48 kDa and 68 kDa, respectively. Two peaks of GLMN peptides are detected, centered on different gel slices (Fig. 5F). Thus, combined protein chromatography on SDS-PAGE, together with peptide MS analysis, can detect the presence of protein isoforms and combined with ion intensity values provide information concerning protein expression levels (see Supp. Tables S3–S6). Importantly, this approach can aid detection of previously unknown isoforms and/or processed and modified pools of proteins, which may have different biological properties, without prior knowledge of isoform-specific peptides or the availability of specific antibodies.
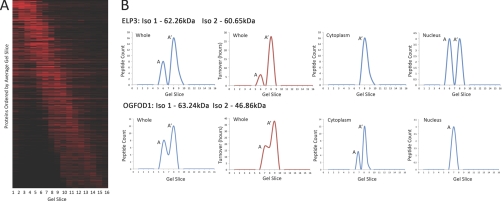
Next, correlation analyses were performed to examine potential differences in subcellular localization and protein turnover properties for examples of protein isoforms predicted from the combined SDS-PAGE and peptide MS data (Fig. 6). By independently evaluating the SILAC data reflecting subcellular protein localization and turnover (33) for the separate sets of unique peptides found in different gel slices, we can thus predict whether the different protein isoforms/processed forms differ in their properties. This is illustrated for proteins Elongator complex protein 3 (ELP3) and 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1 (OGFOD1), both of which are detected in two peaks of unique peptide abundance in SDS-PAGE (Fig. 6B). In the case of ELP3, the larger (A) isoform has a turnover value of ∼5 h and is detected specifically in the nucleus. In contrast, the smaller (A′) isoform has an apparent turnover more than fivefold slower (∼27 h) and is detected equally in the nucleus and cytoplasm. In the case of protein OGFOD1, the two isoforms detected also differ in both turnover and in subcellular distribution. The larger (A) OGFOD1 isoform has a ∼50% faster turnover than the smaller (A′) isoform, (∼18 h and ∼37 h, respectively). The two isoforms are differentially distributed, with the larger A isoform detected in both the cytoplasm and nucleus, and the smaller A′ isoform concentrated specifically in the cytoplasm. We conclude that this prechromatography approach can reveal the presence of protein isoforms with differential properties.
Fig. 6.
Protein isoform identification from gel fractionation. A, Heat map showing the 16 gel slices and their corresponding molecular weights (horizontally) for every protein identified (vertically) ordered by the average gel slice the protein was found in. This heat map was filtered to show only proteins that migrate at multiple gel slices. B, Two example proteins are shown, ELP3 (Elongator complex protein 3) and OGFOD1 (2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1). The graphs on the left show peptide count (y axis) versus gel slice (x axis) as an aggregate for the whole cell, indicating that both ELP3 and OGFOD1 migrate at two separate gel slices. The cytoplasmic graph (top-middle-right) and nuclear graph (top-right) for ELP3 indicate that only one isoform is present in the Cytoplasm (A′), whereas both isoforms are detected in the Nucleus (A and A′). The turnover graph (top-middle-left), showing the turnover values detected (y axis), in each gel slice (x axis), indicates that both isoforms of ELP3 have a different turnover, i.e. 6 h (A) and 28 h (A′) respectively. In relation to OGFOD, the cytoplasmic graph (bottom-middle-right) and nuclear graph (bottom-right) show the isoform A is found in both the cytoplasm and nucleus, however isoform A′ is only found in the cytoplasm. The turnover graph (bottom-middle-left), showing the turnover values detected (y axis) in each gel slice (x axis), indicates that the two forms of the OGFOD1 protein at the different gel slices have different turnovers, i.e. 18.18 h (A) and 37.49 h (A′).
Correlating Post-translational Modification with Protein Properties
Finally, we investigated the potential relationship between post-translational modifications and the properties of subcellular localization, turnover and abundance we have measured for HeLa proteins using SILAC. In this study we analyzed the effect of phosphorylation on either serine, threonine or tyrosine residues on rates of protein turnover in each of the cytoplasmic, nuclear, and nucleolar compartments (Fig. 7) (see Supp. Table S7). Phosphopeptides were detected and quantitated for the HeLa protein localization and turnover SILAC data set using MaxQuant (see Experimental Procedures). Overall, 2444 phosphopeptides were detected and quantitated in this analysis, identifying phosphorylated residues in ∼46% of the HeLa proteins (supplemental Fig. S2). A comparison of protein abundance levels with the detection of phosphorylated peptides showed only a weak positive correlation. This indicates that the phosphopeptides studied are not reflecting the properties of only the most abundant proteins. The majority (53%) of phosphoproteins were identified with a single phosphorylated residue, although 23% had two phosphorylated peptides and 24% had three or more (supplemental Fig. S2).
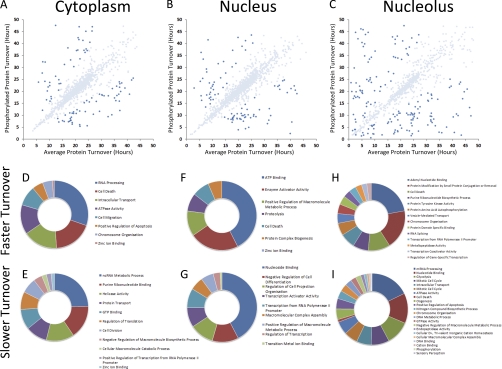
Fig. 7.
Correlation analysis of phosphopeptides with turnover. Graphs A, B, and C show the average protein turnover using nonphosphorylated peptides (x axis) against average protein turnover using both phosphorylated and nonphosphorylated peptides in each of the cytoplasmic, nuclear, and nucleolar compartments. Highlighted in blue are phosphorylated proteins that show a 1.5-fold change compared with the nonphosphorylated form of the protein. Comparison of graphs A (cytoplasm), B (nucleus), and C (nucleolus) show that the nucleolus has the greatest number of phosphorylated proteins compared with the cytoplasm and nucleus. The pie charts D, E, and F show the gene ontology analysis of the phosphorylate proteins that have a slower turnover in comparison with phosphorylated from and, similarly, pie charts G, H, and I show the gene ontology analysis of the phosphorylated proteins that have a faster turnover in comparison with the nonphosphorylated form.
For proteins identified in the respective cytoplasmic, nuclear and nucleolar fractions, the mean turnover value for all peptides assigned to each protein, including all phosphopeptides detected, was plotted on the y axis against the corresponding mean turnover value for all peptides assigned to the same protein, but excluding phosphopeptides, plotted on the x axis (Figs. 7A–7C). In the graphs any protein where the presence of phosphopeptides either increases, or decreases, the mean turnover value by 1.5-fold, or greater, is colored dark blue. The data show that for the phosphosites detected, in most HeLa proteins the presence of one or more phosphorylated residues has little or no effect on mean turnover rate. However, a subset of proteins showed changes in turnover rate when phosphopeptides are present. Interestingly, a larger fraction of nucleolar proteins showed effects of phosphorylation on turnover rates (Fig. 7C), as opposed to either cytoplasmic, or nuclear proteins (Fig. 7A and 7B). This is not correlated with the subset of highest abundance nucleolar proteins, such as ribosomal proteins and nucleophosmin, suggesting that there is a broader effect of phosphorylation on modulating nucleolar protein turnover rates.
Gene ontology analysis was carried out to categorize the phosphorylated proteins showing the greatest increase (Figs. 7D, 7F, and 7H) and greatest decrease in turnover (Figs. 7E, 7G, and 7I), for the cytoplasmic (Fig. 7D and 7E), nuclear (Fig. 7F and 7G) and nucleolar (Fig. 7H and 7I) compartments, respectively. This shows specific groups of proteins whose turnover rates are most affected by phosphorylation at the sites identified. This includes ATP and nucleotide binding proteins, multiple cell cycle regulated proteins, and proteins involved in apoptosis and cell death response mechanisms.
DISCUSSION
This study has investigated multiple data analysis approaches that can be used to identify the expression of protein isoforms that exhibit differential localization and/or turnover properties. We have also identified examples of protein phosphorylation correlating with altered turnover rates in different subcellular compartments. These analyses are performed on SILAC-based quantitative mass spectrometry data from fractionated HeLa cells, where changes in isotope ratios are used to measure turnover rates in the separate cytoplasmic, nuclear and nucleolar compartments (33). We have shown that combining cell fractionation and the separation of intact proteins by chromatography, prior to enzyme digestion and peptide identification by mass spectrometry, can be effectively coupled with SILAC analysis of changes in peptide isotope ratios to identify distinct protein pools and isoforms and assess whether they have different properties. Collectively, these experimental procedures and data analysis approaches provide a new framework for the systematic detection and analysis of protein pools and isoforms that can be correlated with biological properties and hence used to expand the functional annotation of the genome.
Differential proteomic analysis using SILAC involves measuring differences in the ratio of separate isotopic forms of the same peptide, which in turn is related to a specific biological property according to the experimental design. Thus, differences in isotope ratios can be used, inter alia, to measure changes in protein expression levels following drug treatment, to discriminate between specific and nonspecific protein interaction partners or to compare subcellular protein localization. Typically, mean values are calculated for the different isotopic forms of all of the peptides detected that map to a given protein, as deduced from genomic sequence information. A potential limitation with this strategy however is that it usually does not discriminate between peptides arising from functionally distinct pools of protein encoded by the same gene. Thus, ensemble measurements are generated that can average the separate properties, or responses, either of two or more protein isoforms, or of distinct pools of the same protein. We show here that, at least in part, it is possible to circumvent these limitations and to identify protein isoforms and pools and compare their properties, both using information provided by detailed analysis of isotope ratios for separate peptides assigned to the same protein group and by incorporating information from cell fractionation and protein chromatography prior to enzyme digestion and MS analysis.
For each approach presented here, we have independently validated the results by examining randomly selected examples of proteins highlighted to be functionally distinct by MS using alternative methods such as fluorescence microscopy and cycloheximide-SDS-PAGE analyses. The candidates were chosen primarily because specific antibodies were available to assist the follow-up validation experiments. In each case the analysis of the selected proteins successfully confirmed that the MS techniques described here are valid. However, it is accepted that there could also be false positives and/or negatives with these techniques, and it has not been possible because of issues of scale to systematically test all of the many proteins identified as showing differential properties. We anticipate that the reliability of the analysis techniques will also improve in future as more detailed MS analysis provides higher sequence coverage and more accurate measurement of ion intensities and SILAC ratios.
The candidate peptide approach is conceptually simple and can be effective, as demonstrated here for the protein NudCD1 (Fig. 2). However, it is often not possible either to identify, or to reliably quantitate, isoform-specific peptides. This restricts the use of the candidate peptide approach to the analysis of protein isoforms whose structures are already characterized and where one or more isoform-specific peptides have been identified. Even in these cases, quantitation of the isoform response is often derived from analysis of only one or two specific peptides, which can reduce the overall reliability of the measurements. We show that instead the systematic evaluation of mean isotope ratio values for groups of peptides within the total set of peptides mapped to a specific gene provide promising general approaches for detecting isoforms and comparing their properties. Importantly, with both the rule of thirds and three peptides in a row approaches, analysis of the SILAC data can predict the potential existence of either isoforms, or processed forms of proteins, as well as compare their properties, without prior knowledge of either isoform structures, or expression. In each case, the mean isotope ratio values of subgroups of peptides can be evaluated with respect to the mean value, either for all the peptides in the protein, or for values for neighboring groups of peptides, or both. Objective statistical criteria can be applied to these comparisons that will aid the reliable detection of isoforms and thereby help to annotate the functional expression of the genome.
This study has validated the effectiveness of data analysis strategies involving statistical comparisons of isotope ratio values for local clusters of peptides within a protein. When comparing the strategies described, 131 proteins were identified using all three approaches (7% of total proteins identified by the three methods, see supplemental Fig. S3). This highlights an interesting group of proteins for future analysis but also indicates that each approach has strengths and weaknesses. With the degree of peptide coverage currently available we argue for the use of multiple approaches, rather than relying entirely on one method. We also envisage several ways in which these approaches can be enhanced further in future. For example, using filters that compare more closely variations in values between peptides in a group and by defining peptide groups with reference to 3-D crystal structure information on proteins.
Whatever future improvements are made to the data analysis procedures, it is clear that a critical point is having a high quality data set for the proteome under study and in particular having as wide a peptide coverage as possible for each protein. The HeLa SILAC data set studied here included over 80,000 peptides from ∼8000 proteins, with an average coverage of ∼10 peptides per protein identified. Our recent analyses indicate that this is already a large enough sample of the expressed HeLa proteome to be highly representative of the general behavior of cell proteins (33). In future, therefore, we will seek to expand the number of peptides analyzed, not primarily to increase the total number of proteins identified, but rather seeking to enhance the peptide coverage for each protein. We anticipate this will aid the unbiased detection of protein isoforms and their properties that can in turn be related to biological mechanisms and responses.
In most cases, differences in structure between protein isoforms alters their size and/or charge, which in turn provides an opportunity to separate them by chromatography, as demonstrated here using one-dimensional SDS-PAGE. Our results show that independently evaluating the differences in peptide isotope ratios for the same peptides migrating in different chromatographic fractions (in the present case different gel slices), provides a powerful approach for detecting protein isoforms and assessing differences in their properties. Combining fractionation of protein extracts with downstream enzyme cleavage and MS analysis thus provides important information that is lost in procedures in which entire extracts are digested without pre-fractionation and peptides analyzed en masse. The isoform information is similarly lost if extract fractionation is performed at the peptide, rather than protein level. To provide higher resolution separation of isoforms, therefore, we plan in future to increase the degree of protein fractionation prior to MS analysis. For example, using two-dimensional fractionation of extracts, combining ion exchange and gel filtration chromatography. We anticipate that such two-dimensional protein fractionation strategies, combined with increased peptide coverage, will further enhance the efficiency of detecting isoforms and characterizing their properties.
We have shown previously that the subcellular distribution of the proteome can be measured using a SILAC strategy where different cell compartments and organelles are isotope-encoded (26, 27, 33). We showed also that system-wide changes in protein localization could be measured in response to drug treatment and in cells with different genotypes. Here we have extended this “spatial proteomics” approach to detect protein isoforms that are differentially localized within the cell and to analyze differential effects of protein phosphorylation on turnover in different subcellular compartments. This can be developed further in future in several ways. First, a higher resolution map of proteome localization can be derived by more extensive cell fractionation prior to protein chromatography and MS analysis. For example, the cytoplasmic compartment can be further subfractionated into cytosol and organelle fractions and work is underway to implement this. Second, many other post-translational modifications in addition to phosphorylation can be analyzed and their potential effects on the properties of specific protein families and protein isoforms evaluated and compared in different cellular compartments. Enrichment strategies can also be used to increase the efficiency of detecting phosphorylation sites and other modified residues on peptides. Third, our analyses to date have analyzed mixtures of cells at different cell cycle stages. However, it is already known for specific proteins that their expression levels and properties, including localization and PTMs, can change during different stages of interphase and mitosis. We therefore plan to expand our future studies to encompass system-wide, quantitative analysis of the properties of protein isoforms both in multiple subcellular locations and at different cell cycle stages. The resulting data are likely to provide a useful source of information that can reveal unexpected and novel molecular relationships and potential regulatory mechanisms for future investigation.
Footnotes
* This work was supported in part by the European Commission's FP7 (GA HEALTH-F4–2008-201648/PROSPECTS) (www.prospects-fp7.eu/), by RASOR (Radical Solutions for Researching the Proteome) and by a Wellcome Trust program grant to AIL (073980/Z/03/Z). The work performed as part of the Human Protein Atlas project was funded by the Knut and Alice Wallenberg foundation. AIL is a Wellcome Trust Principal Research Fellow. Yasmeen Ahmad is supported by a BBSRC PhD studentship.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S7.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acids in cell culture
- DMEM
- Dulbeccos's modified Eagle medium
- GFP
- green fluorescent protein
- IPI
- International Protein Index.
REFERENCES
- 1. Corthals G. L., Wasinger V. C., Hochstrasser D. F., Sanchez J. C. (2000) The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis 21, 1104–1115 [DOI] [PubMed] [Google Scholar]
- 2. Hinkson I. V., Elias J. E. (2011) The dynamic state of protein turnover: It's about time. Trends Cell. Biol. 21, 293–303 [DOI] [PubMed] [Google Scholar]
- 3. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of Shotgun Proteomic Data. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 4. Rappsilber J., Mann M. (2002) What does it mean to identify a protein in proteomics? Trends Biochem. Sci. 27, 74–78 [DOI] [PubMed] [Google Scholar]
- 5. Godovac-Zimmermann J., Kleiner O., Brown L. R., Drukier A. K. (2005) Perspectives in spicing up proteomics with splicing. Proteomics 5, 699–709 [DOI] [PubMed] [Google Scholar]
- 6. Lubec G., Afjehi-Sadat L. (2007) Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 107, 3568–3584 [DOI] [PubMed] [Google Scholar]
- 7. Cohen P. (1989) The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 58, 453–508 [DOI] [PubMed] [Google Scholar]
- 8. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell. Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 9. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 10. Matlin A. J., Clark F., Smith C. W. (2005) Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 6, 386–398 [DOI] [PubMed] [Google Scholar]
- 11. Johnson J. M., Castle J., Garrett-Engele P., Kan Z., Loerch P. M., Armour C. D., Santos R., Schadt E. E., Stoughton R., Shoemaker D. D. (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 [DOI] [PubMed] [Google Scholar]
- 12. Krawczak M., Reiss J., Cooper D. N. (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90, 41–54 [DOI] [PubMed] [Google Scholar]
- 13. Mann M., Jensen O. N. (2003) Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 [DOI] [PubMed] [Google Scholar]
- 14. Nilsen T. W., Graveley B. R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lisacek F. C., Traini M. D., Sexton D., Harry J. L., Wilkins M. R. (2001) Strategy for protein isoform identification from expressed sequence tags and its application to peptide mass fingerprinting. Proteomics 1, 186–193 [DOI] [PubMed] [Google Scholar]
- 16. Alm R., Johansson P., Hjerno K., Emanuelsson C., Ringnér M., Häkkinen J. (2006) Detection and identification of protein isoforms using cluster analysis of MALDI-MS mass spectra. J. Proteome Res. 5, 785–792 [DOI] [PubMed] [Google Scholar]
- 17. Potthast F., Ocenasek J., Rutishauser D., Pelikan M., Schlapbach R. (2005) Database independent detection of isotopically labeled MS/MS spectrum peptide pairs. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 817, 225–230 [DOI] [PubMed] [Google Scholar]
- 18. Hatakeyama K., Ohshima K., Fukuda Y., Ogura S., Terashima M., Yamaguchi K., Mochizuki T. (2011) Identification of a novel protein isoform derived from cancer-related splicing variants using combined analysis of transcriptome and proteome. Proteomics 11, 2275–2282 [DOI] [PubMed] [Google Scholar]
- 19. Modrek B., Lee C. (2002) A genomic view of alternative splicing. Nat. Genet. 30, 13–19 [DOI] [PubMed] [Google Scholar]
- 20. Jungblut P. R., Holzhutter H. G., Apweiler R., Schlüter H. (2008) The speciation of the proteome. Chem. Cent. J 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fagerberg L., Stadler C., Skogs M., Hjelmare M., Jonasson K., Wiking M., Abergh A., Uhlén M., Lundberg E. (2011) Mapping the Subcellular Protein Distribution in Three Human Cell Lines. J. Proteome Res. 10, 3766–3777 [DOI] [PubMed] [Google Scholar]
- 22. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell. Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 24. Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 25. Ohta S., Bukowski-Wills J. C., Sanchez-Pulido L., Alves Fde L., Wood L., Chen Z. A., Platani M., Fischer L., Hudson D. F., Ponting C. P., Fukagawa T., Earnshaw W. C., Rappsilber J. (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boisvert F. M., Lamond A. I. (2010) p53-Dependent subcellular proteome localization following DNA damage. Proteomics 10, 4087–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doherty M. K., Whitehead C., McCormack H., Gaskell S. J., Beynon R. J. (2005) Proteome dynamics in complex organisms: using stable isotopes to monitor individual protein turnover rates. Proteomics 5, 522–533 [DOI] [PubMed] [Google Scholar]
- 29. Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 30. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwanhäusser B., Gossen M., Dittmar G., Selbach M. (2009) Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9, 205–209 [DOI] [PubMed] [Google Scholar]
- 32. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 33. Boisvert F. M., Ahmad Y., Gierlinski M., Charriere F., Lamond D., Scott M., Barton G., Lamond A. I. (2011) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 35. Torres J. Z., Miller J. J., Jackson P. K. (2009) High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9, 2888–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 37. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 38. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan Y., Phan L., Yang F., Talpaz M., Yang Y., Xiong Z., Ng B., Timchenko N. A., Wu C. J., Ritz J., Wang H., Yang X. F. (2004) A novel mechanism of alternative promoter and splicing regulates the epitope generation of tumor antigen CML66-L. J. Immunol. 172, 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan Y. L., Suzuki K., Wool I. G. (1995) The carboxyl extensions of two rat ubiquitin fusion proteins are ribosomal proteins S27a and L40. Biochem. Biophys. Res. Commun. 215, 682–690 [DOI] [PubMed] [Google Scholar]