Abstract
MS-based proteomics generates rapidly increasing amounts of precise and quantitative information. Analysis of individual proteomic experiments has made great strides, but the crucial ability to compare and store information across different proteome measurements still presents many challenges. For example, it has been difficult to avoid contamination of databases with low quality peptide identifications, to control for the inflation in false positive identifications when combining data sets, and to integrate quantitative data. Although, for example, the contamination with low quality identifications has been addressed by joint analysis of deposited raw data in some public repositories, we reasoned that there should be a role for a database specifically designed for high resolution and quantitative data. Here we describe a novel database termed MaxQB that stores and displays collections of large proteomics projects and allows joint analysis and comparison. We demonstrate the analysis tools of MaxQB using proteome data of 11 different human cell lines and 28 mouse tissues. The database-wide false discovery rate is controlled by adjusting the project specific cutoff scores for the combined data sets. The 11 cell line proteomes together identify proteins expressed from more than half of all human genes. For each protein of interest, expression levels estimated by label-free quantification can be visualized across the cell lines. Similarly, the expression rank order and estimated amount of each protein within each proteome are plotted. We used MaxQB to calculate the signal reproducibility of the detected peptides for the same proteins across different proteomes. Spearman rank correlation between peptide intensity and detection probability of identified proteins was greater than 0.8 for 64% of the proteome, whereas a minority of proteins have negative correlation. This information can be used to pinpoint false protein identifications, independently of peptide database scores. The information contained in MaxQB, including high resolution fragment spectra, is accessible to the community via a user-friendly web interface at http://www.biochem.mpg.de/maxqb.
Bottom-up proteomics consists of the MS analysis of enzymatically digested proteomes. During the last few years, measurements have increasingly been performed in a high resolution, quantitative format (1–3). Each proteomic experiment typically generates large amounts of raw MS and MS/MS data, which should be made available with each experiment (4). Computational proteomics is then used to extract high confidence peptide and protein identifications and relative ratios between conditions, as well as to distill biological implications from the data (5–8). Apart from the analysis of individual projects, several repositories for proteomic experiments have been developed, each with different purposes in mind. The Global Proteome Machine (9) and PeptideAtlas (10, 11) are two of the earliest such collections, with the primary goal of providing a collection of peptide identifications. These collections can, for example, be mined for the design of multiple reaction monitoring experiments in targeted proteomics (12). In contrast, TRANCHE (proteomecommons.org/tranche) is a repository for the raw mass spectrometric data (13). PRoteomics IDEntifications database (PRIDE) is a large effort at the European Bioinformatics Institute, which has collected peptide and protein identification data from more than 10,000 experiments (14, 15). PRIDE, PeptideAtlas, and TRANCHE are also part of the ProteomeXchange consortium, whose objective is to provide a single point of submission for MS-based proteomics data (www.proteomexchange.org). Many dedicated databases for specific organelles or organisms also exist (see for example Refs. 16 and 17).
Most of these databases accept data from heterogeneous sources, which presents a challenge in analysis. For instance, data acquired with different proteomics technologies, different computational pipelines and different quantification strategies may be combined in the database. Although these problems have been addressed to some degree by open standards and joint analysis of deposited raw data, we reasoned that there should be a role for a database designed for homogeneous, quantitative, high resolution data, which nevertheless covers a large part of diverse proteomes. Here we describe the construction of the MaxQB database, which is meant to address the above challenges, allow novel types of analyses, and serve as a public resource via a versatile web interface. We illustrate MaxQB with deep proteome data generated in an accompanying paper (18). In that study, the proteomes of 11 widely used cell lines were mapped in depth with high resolution MS and MS/MS data. We describe analysis and visualization tools of MaxQB, a solution to the problem of inflated false positive protein identifications, and examine the reproducibility of peptide intensity rank order for each protein in different proteomes.
EXPERIMENTAL PROCEDURES
Database Implementation
MaxQB is structured as a classical three-tiered application consisting of data, application logic, and presentation. The data tier is a relational database managed by Oracle Standard Edition Database 11g (Oracle, Redwood Shores, CA). Because only standard SQL features are used, it is in principle possible to port the database to other relational database management systems like the free and open source MySQL database (Oracle). The application logic tier is implemented in Java 1.6 (Oracle) and Groovy (http://groovy.codehaus.org) using the Grails web application framework version 1.3.3 (http://grails.org). The web application runs on a Tomcat 7 web server (http://tomcat.apache.org). Finally, the presentation tier is comprised of dynamically generated html pages and JavaScript.
Protein Index and Mapping to Genome
Several human proteome databases were uploaded to MaxQB to build a comprehensive protein index: Uniprot version 09/2011 (including variants), Ensembl build 64, and International Protein Index (IPI) version 3.87. Identical entries were collapsed to a single logical protein entry where identity of entries is defined by strict sequence identity. For example, the entry for the human protein CDK2 refers to the Uniprot accession number P24941, the Ensembl protein accession number ENSP00000266970, and the IPI accession number IPI00031681. All three database entries have identical sequences. The sequences were first transformed to a hash key using the Secure Hash Algorithm, which dramatically increased the speed of mapping identical sequences. The locations of the genes on the chromosomes were obtained from Ensembl (19), and the ortholog pairs of proteins in different organisms were obtained from InParanoid eukaryotic ortholog database (20).
Cell Line Data
MaxQB already serves as a general repository for experiments performed in our laboratory. Therefore, it will contain an increasing number of deep proteome mapping experiments of human, mouse, and other cell types and species in the future. The data analyzed here are mainly from a proteome profiling experiment of 11 cell lines described in the accompanying paper (18). Briefly, A549, GAMG, HEK293, HeLa, HepG2, Jurkat, K562, LnCap, MCF7, RKO, and U2OS cell lines were grown in standard conditions, lysed, and prepared according to the Filter Aided Sample Preparation method (21) and fractionated by pipette-based strong anion exchange into six fractions. Resulting peptide mixtures were analyzed on-line by LC-MS/MS on a linear ion trap Orbitrap (VELOS, Thermo Fisher Scientific) in higher energy collisional dissociation mode (22). Each proteome measurement—consisting of six 200-min gradients—was repeated in triplicate. Analysis of the results was performed in MaxQuant (23) using the Andromeda search engine (24). The results that are presented here and are accessible in MaxQB are based on data processed with the “match between runs” feature enabled. However, the increase of identifications for additional analyzed cell lines (see Fig. 2) and the correlation analysis (see Fig. 7) is based on data processed with this feature disabled. For details see Ref. 18. MaxQB and the results of the 11-cell line proteome can be accessed freely upon publication at http://www.biochem.mpg.de/maxqb.
Fig. 2.
Number of proteins (red bars) and peptides (blue bars) identified in increasing number of cell lines. In total, 10,183 non-redundant proteins and 103,869 non-redundant peptides were identified (see text for details).
Fig. 7.
A, distribution of correlation values. For each protein group with two or more peptides identified, the Spearman correlation between the intensities of the peptides and the detection probability were calculated. B and C, examples of proteins with high correlation (0.92): Q8NFI3-ENGASE (B) and low correlation (0.27): Q92918-MAP4K1 (C).
Mouse Tissue Data
In addition to the cell line data, we also analyze data from a proteome profiling experiment of 28 mouse tissues (18). Briefly, 28 tissues were dissected from C57BL/6 mice and snap frozen in liquid nitrogen. The tissues were homogenized, lysed, and mixed with a SILAC1 spike-in standard. Protein digestion was performed with endoprotease Lys-C, followed by peptide fractionation by isoelectric focusing. The resulting peptide mixtures were analyzed on-line by LC-MS/MS on a linear ion trap Orbitrap (XL, Thermo Fisher Scientific) in CID mode. Each proteome measurement—consisting of twelve 100-min gradients—was repeated in triplicate. Analysis of the results was performed in MaxQuant (23) using the Andromeda search engine (24).
RESULTS AND DISCUSSION
Database Architecture
MaxQB serves as a generic repository and analysis platform for high resolution MS-based proteomics experiments. As such, it stores details about protein and peptide identifications together with the corresponding high or low resolution fragment spectra and quantitative information, such as SILAC ratios or label-free intensities. To enable smooth upload of data, MaxQB is tightly integrated with MaxQuant (23) (Fig. 1). At the end of data processing, the user of MaxQuant is asked whether she wants to upload the data to the database. In this case, the data is submitted by calling a simple object-based protocol (SOAP)-based web service. Alternatively, the data can be manually uploaded through the user interface of MaxQB. In either case, the user is asked to enter additional meta information, such as the project name, experiment name, and workflow parameters. All of the data are stored in a relational SQL database running on an Oracle relational database management system. The user can browse, search, and retrieve the data through a web interface. Furthermore, the data can be accessed either through SQL queries or preferably through SOAP web services from visualization and data analysis tools like the Perseus module for bioinformatic analysis in MaxQuant, R (www.r-project.org), Matlab (The Mathworks, Natick, MA), or Spotfire (TIBCO, Palo Alto, CA).
Fig. 1.
Database architecture and interfaces to other applications.
Protein Index and Cell Line Data
To demonstrate the general concepts and features of MaxQB, data from the proteome profiling of 11 cancer cell lines described in the accompanying paper (18) were uploaded to the database. The combined data were searched against the IPI database 3.68 using MaxQuant version 1.2.0.34. A frequent problem of proteomics experiments is the difficulty of matching protein accession numbers between experiments that were searched against different databases or even just against different versions of the same database (25). Here, we sought to solve this problem by building a protein index that matches the accession numbers of various popular protein sequence databases to a logical protein entry. For the human species, these databases are Uniprot (including variants), IPI, and Ensembl. In brief, sequence database entries that refer to identical sequence and species are mapped to a unique protein index entry (see “Experimental Procedures” for more details). The protein index contains 19,515 human entries with identical sequences in IPI and Ensembl (see Table I). This nonredundant set was the basis for further analysis. From these proteins, we calculated the number of tryptic peptides readily observable and identifiable by MS (mass between 600 and 4,000 Da; no missed cleavages). There are 536,593 such peptides, and interestingly only 6% of them are shared between two or more proteins.
Table I. Number of identified proteins (Ensembl genes with identical IPI sequence) and peptides in comparison with respective numbers in the ENSEMBL human database.
Observable peptides are in silico digested peptides with masses between 0.6 and 4 kDa and no missed cleavages.
| Human proteome | Identified in 11 cell lines | |
|---|---|---|
| Proteins | 19,515 | 10,183 (52.2%) |
| Observable peptides | 536,593 | |
| Observable, sequence-unique peptides | 506,080 | |
| Observable peptides from identified proteins | 338,496 | 109,862 (32.5%) |
| Observable, sequence-unique peptides from identified proteins | 316,585 | 103,869 (32.8%) |
Triplicate analysis of one cell line alone identified 7,337 proteins, each subsequently added cell line contributed a decreasing number of new proteins, and analysis of all 11 cell line proteomes together identified 10,183 nonredundant proteins (Fig. 2 and Table I). In silico digest of the identified proteins generated 338,496 observable tryptic peptides, of which 32.5% were identified in the cell line data set at a false discovery rate of 1%. For each of these peptides, the database contains the corresponding database identification score, the posterior error probability, individual evidences for the peptide identification, and the corresponding fragment spectra. At this point, proteins encoded by more than half of all human genes and a large proportion of all their possible, unmodified tryptic peptides are identified in the database.
Apart from the 11-cell line project, MaxQB contains a number of large scale experiments on human proteomes. Interestingly, these experiments together already account for proteins encoded by 64% of all human genes and 39% of their possible, unmodified tryptic peptides. This suggests that improving technology will soon make it possible to obtain reference spectra for a large part of the proteome from homogeneous data sources given a supply of diverse proteomes in which all human proteins are expressed.
Use Cases
To illustrate practical use of MaxQB, we next describe three “use cases” dealing with diverse types of questions that can be addressed by this novel database. As a first use case, we assume that the user is interested in members of a specific protein family—here DNA polymerase epsilon subunits (POLE)—and wants to investigate their expression across the different cell lines and additionally across mouse tissues. The user can query the database by various fields, specifically by gene name, organism, and source database. The query terms can be combined by Boolean logic and grouped using parentheses. Alternatively, the query builder can be used if one is not familiar with the query syntax. In this example, the user searches for all human Uniprot entries that have a gene name beginning with “POLE” (Fig. 3A). The query returns four subunits. By clicking on one of the hits (POLE), the user obtains additional details (Fig. 3B). In particular, this resulting page specifies the entries in the databases IPI and Ensembl with identical sequence. On the protein expression tab, a bar chart visualizes the protein expression across the 11 human cell lines. Expression of POLE varies by more than 2 orders of magnitude between LnCap (lowest expression) and HEK293 (highest expression) calculated by label-free quantification in MaxQuant (26) (Fig. 3C). In addition to estimating expression of the same protein between proteomes, MaxQB can also display expression within any of the proteomes, compared with all other quantified proteins in that proteome. Here, the expression of the protein is estimated by the sum of its peptide signals, after normalization of the total proteome signals to each other in MaxQuant. The iBAQ algorithm (27) is now implemented into MaxQuant and can also be used to estimate protein amounts. In Fig. 3D, selection of the HEK293 proteome brings up a distribution plot comparing the expression of the protein of interest with all other proteins in this cell line. This reveals that POLE is among the highly expressed proteins in these cells (within the top 15-percentile). The sequence coverage tab for the corresponding protein group shows the distribution of identified peptides along the sequence of POLE and across the 11 cell lines and their biological replicates (Fig. 4). Additionally, the in silico digested peptides with masses between 0.6 and 4 kDa and the known domains as retrieved from Uniprot (28) are displayed.
Fig. 3.
A, query proteins for human DNA polymerase epsilon subunits. B, select POLE and show details on this protein. C, histogram of protein expression across 11 cell lines. D, expression of POLE compared with expression of all other detected proteins in HEK293 cells. E, expression of the mouse ortholog across 28 mouse tissues.
Fig. 4.
Sequence coverage of POLE. The blue boxes are two c4-type domains. The gray boxes are in silico digested peptides with masses between 0.6 and 4 kDa. Detected peptides are colored by their label-free intensities across the 11 tested cell lines with three replicates each.
The user may also be interested in the expression of POLE in other organisms. The InParanoid eukaryotic ortholog database contains pairwise orthologs of 100 organisms (20). MaxQB integrates this information to allow the user to jump directly to the proteomes of other organisms. For example, Fig. 3B lists two ortholog proteins in yeast and mouse. Clicking on the mouse ortholog (E9QKW1), the user obtains additional information on the mouse protein. As an example of how MaxQB can integrate data from various studies, Fig. 3E shows the expression of POLE in 28 mouse tissues (data not published). POLE was identified in embryonic tissue, jejunum, olfactory bulb, spleen, and white fat.
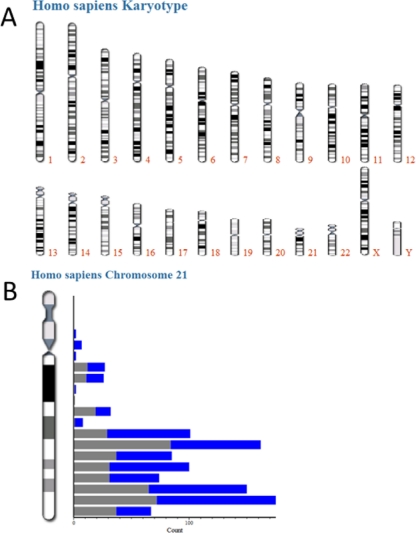
Recently, several projects aiming to identify all proteins encoded on specific chromosomes have been started under the auspice of the Human Proteome Organization (29). In a second use case, we ask how many proteins have been identified for a certain chromosome and whether there are any regions with low identification rates. MaxQB lists all human (or mouse or yeast) chromosomes and allows the user to select one of them for further analysis (Fig. 5A). In the case of chromosome 21, for example, this results in the distribution of protein coding genes and the respective protein identifications in the cell line proteomes shown in Fig. 5B. By clicking on one of the bars, the user can drill down to the list of proteins encoded in the corresponding region of the chromosome as well as the underlying peptide information. As expected, ∼50% of all annotated genes on chromosome 21 are associated with high confidence protein identification information, and the distribution across the chromosome appears to be uniform.
Fig. 5.
A, human karyotype. B, histogram of proteins identified by MS in the 11 cell line project (gray) and annotated proteins (blue) on chromosome 21.
A popular use of proteomics repositories is the selection of peptides suitable for targeted methods such as multiple reaction monitoring. In the third use case, a user is interested in establishing an multiple reaction monitoring assay for the cell cycle protein CDK2 and starts by searching for all peptides that are unique for CDK2, have an Andromeda identification score larger than 80, and have no missed cleavages. As in the search for proteins described above, query terms can be combined by Boolean logic (Fig. 6A). The query returns seven peptides fulfilling these criteria. The user selects the peptide AFGVPVR and displays the fragment spectrum for the best identification evidence for this peptide (Fig. 6B). The user can now export the list of peaks together with the masses and annotations and use this as a basis for creating multiple reaction monitoring transitions. A particular advantage of using MaxQB for this use case is the fact that this database contains high resolution fragmentation spectra that are obtained by the higher energy collisional dissociation method, which produces very similar transitions to those that would be observed in triple quadrupole methods (30).
Fig. 6.
A, query for unique peptides for CDK2 with a score greater 80 and no missed cleavages. B, the fragment spectrum with the best evidence for peptide AFGVPVR.
Inflation of False Identifications
It is a common strategy in MS-based proteomics to control the proportion of false identifications by searching against a combined forward and decoy sequence database and then adjusting the cutoff score to a value, such that the proportion of identified decoy hits is equal to a given false discovery rate (FDR) (31, 32). Although decoy database search is a robust method to control FDR in single projects, an additional challenge arises when combining the results of many different experiments covering the same proteome. In this case the number of true identifications saturates because the same “true” proteome is sampled repeatedly (see for example Fig. 2). However, the false identifications are largely independent of each other and therefore accumulate, leading to an inflation of false identifications. A related but different problem arises when combining the search scores from multiple search engines obtained for the same data set (33, 34). Here, we instead investigate the combination of identifications from multiple proteomes that have been analyzed with the same search engine.
Although generally known this issue has to our knowledge not been quantified with experimental data. We investigated the severity of this problem by analyzing the effects of successively adding data sets to a database instead of analyzing all data together as described above. For this purpose, the raw data of the proteome profiling experiment of 11 cell lines described in the accompanying paper (18) were arbitrarily partitioned into three sets, each consisting of four or three cell lines and their corresponding biological replicates. These three sets were reprocessed by MaxQuant with a fixed FDR for protein and peptide identification of 1%. Each set resulted in ∼10,000 total protein identifications and 100 decoy hits (Table II). If these sets were successively added to a single database, the number of true identifications would increase by 28%, whereas the number of decoy hits would increase by 225% compared with the average number in the individual sets. The resulting FDR would now be 1.82% instead of the desired 1%. Clearly, the more proteomics data sets are added to a database, the larger the inflation of false identifications. Often the underlying data are not available for reprocessing, or reprocessing for each added data set would be impractical. For these cases, we propose to solve the issue by adjusting the cutoff score to a more stringent database-wide level. The adjustment is performed such that the ratio between the number of unique decoy database hits and the total number of unique identifications on a protein and peptide level is equal to the desired FDR. MaxQB follows this proposal by calculating a local FDR (q value) for each peptide and each protein identification. This q value for a protein is essentially the ratio between the number of decoy hits and the total number of identifications with scores smaller or equal to the score of that protein (and the peptide q value is calculated analogously). The identifications with q values below the preset FDR of 1% are filtered out when the user is analyzing data from the whole database rather than data from a single experiment. This strategy is possible because all projects in MaxQB are analyzed with the same search engine (Andromeda) and therefore use the same type of identification score.
Table II. Number of identified proteins if raw files are processed in three disjointed sets.
Set 1 includes A549, HEK293, GAMG, and HeLa. Set 2 includes HepG2, Jurkat, K562, and MCF7. Set 3 includes RKO, LNCap, and U2OS. Union refers to the union of the proteins identified in the individual sets. True is the number of forward hits, decoy the number of decoy hits, and FDR is the corresponding false discovery rate.
| Set 1 | Set 2 | Set 3 | Union | |
|---|---|---|---|---|
| True | 9,922 | 9,690 | 9,054 | 12,211 |
| Decoy | 103 | 105 | 93 | 226 |
| FDR | 1.03% | 1.07% | 1.02% | 1.82% |
Reproducibility of Identified Peptides
As can be seen in the sequence coverage plot of POLE (Fig. 4), the identified peptides are not random between different proteomes but follow certain patterns. As one would expect, the cell line with the highest expression level (HEK293) also shows the largest number of identified peptides. Furthermore, a few POLE peptides are identified in almost all samples (e.g. positions 60–77, 1520–1540, and 2132–2145), and these peptides are also the ones with the highest label-free intensity as indicated by the color code. These observations motivated us to investigate possible general relationships between the probability of peptide identification and peptide intensity. For each protein having at least two peptides, we calculated the Spearman rank correlation between the sum of label-free peptide intensities and the number of experiments in which the peptide was detected. The histogram of the correlation values for each protein shows a strong accumulation of proteins with high correlation values (Fig. 7A). A total of 64% of the proteins had Spearman rank correlations of more than 0.8. Fig. 7B shows the example of ENGASE, a protein with a high correlation value (0.92). Here, the peptides detected many times are also the most intense ones and vice versa. A few proteins have small or even negative correlations and were therefore investigated in detail. For example, MAP4K1 has a small correlation of 0.27 between peptide intensities and peptide detection probabilities. Whereas the peptides detected in the three Jurkat samples show a high overlap, the peptide VSGDLVALK starting at position 38 was only detected in one replicate of A549, HeLa, HepG2, K562, LNCap, and MCF7, respectively, and it was also the only peptide detected for this protein in these cell lines. We speculate that this peptide is a false identification in the non-Jurkat cell lines, which is further supported by a relatively high posterior error probability. Our analysis clearly suggests that rank order statistics of identified peptides for each protein are sufficiently high to pinpoint false protein identifications, independently of peptide database scores. Therefore a comprehensive catalog of protein and peptide identifications compiled from high quality data, such as those in MaxQB, could be used to improve protein identification in proteomics experiments. Furthermore, public databases could incorporate such algorithms to judge the quality of submitted data sets. If peptide rank correlation of the new data set to established data sets is low, this may indicate problems with the newly submitted data.
Conclusions and Outlook
We have described MaxQB, a resource for high resolution and quantitative MS-based proteomics data. MaxQB draws on a homogenous set of proteome measurements, which allows types of analyses that are difficult to perform in many other public repositories. Here, the capabilities of MaxQB have been illustrated using deep proteome measurements of 11 different cell lines. These data already cover more than half of the human proteome, and for these proteins any researcher can visualize expression patterns across cell lines as well as estimated expression levels within each of them. The expression data may be used, for example, to select a cell line or tissue that highly expresses the protein of interest. We plan to add even deeper and more diverse data sets in the near future. As an example, the expression levels of an ortholog protein in 28 different mouse tissues can be visualized in MaxQB. Although these efforts may not lead to complete coverage of the proteome, because a number of proteins may not be expressed in readily available sources, we predict that the large majority of proteins and peptides typically observable in proteomics experiments will soon be represented. As in other repositories, the peptide information can be mined for establishing targeted proteomics assays. However, in this regard MaxQB has the advantage of drawing on a relatively focused set of experiments that are strictly controlled for overall false discovery rate. As technology advances, increasingly accurate proteome measurements will be feasible within short measurement times. We envision that the data in MaxQB will periodically be replaced with these superior data (while keeping access to the old data), something that is difficult in broad data repositories that cannot discriminate between data submitted at different stages of technology development. Although MaxQB currently contains proteome data of human cancer cell lines and a set of 26 mouse tissues, we envision that proteomes of additional cell types and species will be added in the future. Furthermore, MaxQB can serve as a repository for more specialized data, for example, proteome changes after treatment with drugs or data on post-translational modifications. All of these data will have in common that they contain high resolution identifications and are produced with a homogenous set of technologies. We plan to implement an automatic submission of the experiments in MaxQB to PRIDE, so that MaxQB data are also available in the databases that are part of ProteomeXchange.
MaxQB also allowed us to investigate the reproducibility of peptide identifications for each protein across proteome experiments. We found a high correlation of peptide rank order, sufficient to highlight false positive protein identifications independently of peptide identification score. This suggests that the peptide rank order can be used as a component of a protein identification score. As MaxQB contains more and more of the typically identifiable proteins and peptides, it will be interesting to investigate whether these data can contribute to better proteome characterization.
Additionally, MaxQB features a number of analysis tools that are not currently present in other databases. For example, we here introduced a procedure to adjust the required cutoff scores to keep the overall false positive rate constant when incremental proteome projects are added. These analysis tools can be used in MaxQB, but they could also be incorporated into other proteome databases.
Acknowledgments
We thank Mario Oroshi for helpful discussions.
Footnotes
* This work was funded by European Commission's 7th Framework Program PROSPECTS Grant HEALTH-F4-2008-201648. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- FDR
- false discovery rate
- SOAP
- simple object access protocol
- IPI
- International Protein Index.
REFERENCES
- 1. Mallick P., Kuster B. (2010) Proteomics: A pragmatic perspective. Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 2. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 3. Domon B., Aebersold R. (2010) Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 4. Olsen J. V., Mann M. (2011) Effective representation and storage of mass spectrometry-based proteomic data sets for the scientific community. Sci. Signal. 4, pe7. [DOI] [PubMed] [Google Scholar]
- 5. Taylor C. F., Paton N. W., Garwood K. L., Kirby P. D., Stead D. A., Yin Z., Deutsch E. W., Selway L., Walker J., Riba-Garcia I., Mohammed S., Deery M. J., Howard J. A., Dunkley T., Aebersold R., Kell D. B., Lilley K. S., Roepstorff P., Yates J. R., 3rd, Brass A., Brown A. J., Cash P., Gaskell S. J., Hubbard S. J., Oliver S. G. (2003) A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 21, 247–254 [DOI] [PubMed] [Google Scholar]
- 6. Kumar C., Mann M. (2009) Bioinformatics analysis of mass spectrometry-based proteomics data sets. FEBS Lett. 583, 1703–1712 [DOI] [PubMed] [Google Scholar]
- 7. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaab C. (2011) Analysis of phosphoproteomics data. Methods Mol. Biol. 696, 41–57 [DOI] [PubMed] [Google Scholar]
- 9. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 10. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Reports 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hüttenhain R., Malmström J., Picotti P., Aebersold R. (2009) Perspectives of targeted mass spectrometry for protein biomarker verification. Curr. Opin. Chem. Biol. 13, 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill J. A., Smith B. E., Papoulias P. G., Andrews P. C. (2010) ProteomeCommons.org collaborative annotation and project management resource integrated with the Tranche repository. J. Proteome Res. 9, 2809–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Côté R., Reisinger F., Martens L., Barsnes H., Vizcaino J. A., Hermjakob H. (2010) The Ontology Lookup Service: Bigger and better. Nucleic Acids Res. 38, W155–W160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: The proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 16. Ahmad Y., Boisvert F. M., Gregor P., Cobley A., Lamond A. I. (2009) NOPdb: Nucleolar Proteome Database: 2008 update. Nucleic Acids Res. 37, D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gnad F., Oroshi M., Birney E., Mann M. (2009) MAPU 2.0: High-accuracy proteomes mapped to genomes. Nucleic Acids Res. 37, D902–D906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 10.1074/mcp.M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S., Gordon L., Hendrix M., Hourlier T., Johnson N., Kähäri A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Larsson P., Longden I., McLaren W., Overduin B., Pritchard B., Riat H. S., Rios D., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Spudich G., Tang Y. A., Trevanion S., Vandrovcova J., Vilella A. J., White S., Wilder S. P., Zadissa A., Zamora J., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernández-Suarez X. M., Herrero J., Hubbard T. J., Parker A., Proctor G., Vogel J., Searle S. M. (2011) Ensembl 2011. Nucleic Acids Res. 39, D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ostlund G., Schmitt T., Forslund K., Köstler T., Messina D. N., Roopra S., Frings O., Sonnhammer E. L. (2010) InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38, D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 22. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 25. Griss J., Cote R. G., Gerner C., Hermjakob H., Vizcaino J. A. (2011) Published and Perished? The influence of the searched protein database on the long-term storage of proteomics data. Mol. Cell. Proteomics 10: M111.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
- 27. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 28. Consortium U. (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C. H., Corthals G. L., Costello C. E., Deutsch E. W., Domon B., Hancock W., He F., Hochstrasser D., Marko-Varga G., Salekdeh G. H., Sechi S., Snyder M., Srivastava S., Uhlen M., Wu C. H., Yamamoto T., Paik Y. K., Omenn G. S. (2011) The Human Proteome Project: Current state and future direction. Mol. Cell. Proteomics 10: M111.009993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Graaf E. L., Altelaar A. F., van Breukelen B., Mohammed S., Heck A. J. (2011) Improving SRM assay development: A global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res. 10, 4334–4341 [DOI] [PubMed] [Google Scholar]
- 31. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 32. Nesvizhskii A. I. (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10: M111.007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwon T., Choi H., Vogel C., Nesvizhskii A. I., Marcotte E. M. (2011) MSblender: A probabilistic approach for integrating peptide identifications from multiple database search engines. J. Proteome Res. 10, 2949–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]