Abstract
Chemical cross-linking in combination with mass spectrometric analysis offers the potential to obtain low-resolution structural information from proteins and protein complexes. Identification of peptides connected by a cross-link provides direct evidence for the physical interaction of amino acid side chains, information that can be used for computational modeling purposes. Despite impressive advances that were made in recent years, the number of experimentally observed cross-links still falls below the number of possible contacts of cross-linkable side chains within the span of the cross-linker. Here, we propose two complementary experimental strategies to expand cross-linking data sets. First, enrichment of cross-linked peptides by size exclusion chromatography selects cross-linked peptides based on their higher molecular mass, thereby depleting the majority of unmodified peptides present in proteolytic digests of cross-linked samples. Second, we demonstrate that the use of proteases in addition to trypsin, such as Asp-N, can additionally boost the number of observable cross-linking sites. The benefits of both SEC enrichment and multiprotease digests are demonstrated on a set of model proteins and the improved workflow is applied to the characterization of the 20S proteasome from rabbit and Schizosaccharomyces pombe.
The analysis of the three-dimensional structure of proteins and the spatial arrangement of subunits within protein complexes is of great importance to study their biological function (1). Historically, methods such as x-ray crystallography and NMR spectroscopy, together with a variety of other spectroscopic techniques have been used, each with their own drawbacks. Obtaining diffracting crystals still remains a challenge, especially for large protein assemblies, and the application of NMR is predominantly limited to smaller proteins. Lower resolution methods based on electron microscopy have revealed interesting insight into the structure of large macromolecular assemblies. However, their use for smaller complexes remains limited. In recent years, mass spectrometry has contributed a number of emerging approaches that deliver low-resolution structural information on proteins that can provide complementary data to the above-mentioned methods. Among the most established of these are hydrogen/deuterium exchange (2), oxidative footprinting (3), and—for the analysis of intact protein complexes—native MS techniques (4).
In addition, various forms of cross-linking have been employed to study protein structure and protein interactions in combination with mass spectrometry (5–7). In its simplest form, noncovalent interactions in protein complexes can be stabilized, for example using the rather unselective formaldehyde as reagent. In such studies, proteins co-isolated using affinity purification are cross-linked during the isolation step and interaction partners are identified without deriving dedicated information about the exact cross-linking sites. Different forms of cross-linking strategies may be employed that are either based on the introduction of photoreacive amino acids at specific sites into target proteins or, alternatively, cross-linking reagents containing photoreactive groups are used. In both cases, covalent links with interactors are formed upon UV irradiation.
In contrast to these studies where relatively unspecific cross-linking reagents have been used to stabilize complexes, studies attempting the precise localization of cross-linking sites by mass spectrometry—using chemically selective cross-linkers—have proven more challenging, but also more rewarding (5–7). The thus obtained “distance restraints” provide direct information about the spatial proximity of the reactive sites, taking into account the spacer length of the reagent and the length of the amino acid side chains. Such restraints can be used for computational purposes or in combination with other low-resolution techniques such as electron microscopy and small angle x-ray scattering. For structural studies, chemical cross-linking reagents that target specific functional groups within the protein(s) of interest are used, most commonly amine-reactive succinimide esters such as disuccinimidyl suberate (DSS)1 or bis(sulfosuccinimidyl) suberate, which cross-link primary amino groups on lysine residues and protein N termini.
Recently, the concept of chemical cross-linking/mass spectrometry has seen a noticeable increase in attention and is becoming more widely adopted. New cross-linking reagents (6) and software tools (8) for data analysis have been introduced, and advances in MS instrumentation have enabled lower detection limits and improved mass accuracy. However, fundamental limitations in the workflow remain. The yield from a cross-linking experiment is typically low. In the case of succinimide esters, hydrolysis of the cross-linking reagent in aqueous solution is a competitive reaction, and using a large excess of reagent might impede further sample processing, e.g. because of the reduced efficiency of enzymatic digestion in highly cross-linked samples. Therefore, enrichment of cross-linked peptides is essential when more complex samples are analyzed.
To this end, various strategies have been proposed. They include the use of reagents containing affinity tags (9–11) or the use of strong cation exchange chromatography (SCX) (12–14). Although affinity tagged reagents have been used successfully to some extent, most notably by the Bruce laboratory in the Protein Interaction Reporter concept (15–17), their synthesis is challenging and many studies reported in the literature remain proof-of-principle only. SCX enrichment makes use of the fact that when two peptides are connected via a cross-link, they are more highly charged in solution than their linear counterparts because of the presence of a higher number of protonation sites (usually N- and C termini when proteases such as trypsin or Lys-C are used). This strategy, originally introduced by Rinner et al. (12), has recently been applied to larger complexes such as RNA polymerase II (13) or the ribosome (18).
Most of the cross-linking studies to date have used trypsin as the proteolytic enzyme. This protease is widely used in proteomic research because of its robustness and its tendency to produce peptides with advantageous properties (length, charge, and fragmentation behavior) for MS and tandem MS (MS/MS) analysis. However, this may not necessarily be the case for the analysis of cross-linked peptides, because in this case it is required that both connected regions of the protein(s) yield peptides of suitable length. Cross-links with short peptides typically yield only few informative fragment ions, whereas excessively long peptides might cause several other problems—such peptides begin to deviate from the common fragmentation model where amide bonds within the peptide backbone are more or less randomly cleaved, leading to incomplete fragmentation. This issue appears to be further exacerbated in cross-linked peptides, where little general information about their fragmentation behavior is known (19).
To overcome suboptimal fragmentation in conventional collision-induced dissociation, alternative strategies such as the use of electron transfer dissociation (20) or gas-phase cleavable reagents (15–17, 21) have been introduced, although both techniques come with their own drawbacks such as low fragmentation efficiency and reduced scan speeds because of the requirement of performing MS3 scans.
Moreover, long peptides, especially when present in cross-links, are difficult to separate efficiently by reversed-phase chromatography, leading to peak broadening because of poor mass transfer in solution, and might also exceed the optimal mass range for MS analysis, causing a decrease in sensitivity. Therefore, the use of multiple proteases might be advantageous to enhance the yield of cross-linking data. Up to now, no systematic study of the use of different proteases for cross-linking/mass spectrometry has been reported. For conventional proteomics approaches, however, Swaney et al. have recently found a clear benefit in the use of additional proteases to increase both proteome and protein sequence coverage (22).
We have shown previously that computational modeling approaches require a considerable amount of low-resolution restraints (such as those from cross-linking experiments) to provide a noticeable benefit (5). To restrain the conformation of interaction partners in protein complexes, the yield of cross-linking experiments should therefore be maximized. To achieve this, the development of more sophisticated and powerful sample preparation strategies is one of the most promising strategies. We introduce peptide size exclusion chromatography (SEC) as a novel chromatographic technique for enriching cross-linked peptides, and thereby expand the yield of structural information from cross-linking experiments. Although not very frequently used for proteomics applications, peptide-level SEC has recently been employed by Quadroni and coworkers to select large tryptic peptides for secondary digestions using complementary enzymes (23). Although most of the peptides resulting from an enzymatic digest have a molecular mass below 2000 Da, the majority of cross-link identifications resulting from the combination of two peptides plus the cross-linker mass result from precursor masses above this level. Therefore even a relatively crude chromatographic separation—as can be expected from a low-efficiency technique such as SEC—should result in appreciable depletion of unmodified peptides and peptides carrying the partially hydrolyzed cross-linker as a modification (designated as “mono-link” according to our nomenclature (5)).
Here, we show that SEC can indeed be used to fractionate digests of cross-linked proteins and complexes and that the technique enriches for cross-linked peptides in higher mass fractions. The use of multiple proteases (Asp-N, Glu-C, Lys-C, and Lys-N in addition to trypsin) provides complementary distance restraints, therefore extending the amount of information that can be used for computational purposes. The methods are first discussed in detail using a set of model proteins and applied to the cross-linking of the 20S proteasome as an example for a protein complex of high biological relevance. For the model proteins, a total of 240 nonredundant Lys-Lys contacts were covered by combining data from five enzymes. The concept was also found to be applicable to small sample amounts as demonstrated by the proteasome data where the number of cross-links was expanded by 20–50% with the use of a second enzyme (Asp-N) for digestion.
EXPERIMENTAL PROCEDURES
Cross-linking of Model Proteins
Individual stock solutions of the eight model proteins (bovine catalase, rabbit creatine kinase, rabbit fructose-bisphosphate aldolase, bovine lactoferrin, chicken ovotransferrin, rabbit pyruvate kinase, bovine serotransferrin, bovine serum albumin; all obtained from Sigma-Aldrich Buchs, Switzerland) were prepared at concentrations of 5–10 mg ml−1 in 20 mm HEPES/KOH buffer (pH 8.2). Samples were diluted for each protein separately to a final concentration of 2 mg ml−1 in the same buffer, and 4 μl of the cross-linker solution (25 mm each of DSS-d0 and DSS-d12 (Creative Molecules, Canada) in anhydrous DMF) were added per 100 μl protein solution. Samples were incubated for 30 min at 37 °C in an Eppendorf Thermomixer (mixing speed 750 rpm). Remaining cross-linking reagent was quenched by adding aqueous NH4HCO3 solution to a final concentration of 50 mm, followed by incubation for further 20 min. Aliquots of the individually cross-linked protein solutions were then combined and evaporated to dryness in a vacuum centrifuge before further processing.
Cross-linking of 20S Proteasome Samples
The 20S proteasome from rabbit (Oryctolagus cuniculus) was obtained from Sigma-Aldrich and cross-linked at a concentration of ∼0.8 mg ml−1. Schizosaccharomyces pombe proteasome was prepared following a protocol from Saeki et al. (24) with modifications as described in (25). The sample was concentrated by ultrafiltration to a final concentration of 0.2 to 0.3 mg ml−1. The cross-linking reaction was carried out as described above for model proteins, with the amounts of cross-linking reagent adjusted according to the lower protein concentrations. Cross-linked samples were split in half and digested with trypsin and Asp-N in both cases, as described below.
Enzymatic Digestions
Dried cross-linked samples were resuspended in 8 m urea solution to a final concentration of 1 mg ml−1 (0.2 mg ml-1 for S. pombe proteasome). Five microliters of a 50 mm tris(carboxyethyl) phosphine stock solution in water were added and the samples were incubated at 37 °C for 30 min. Subsequently, 5 μl of a 100 mm aqueous iodoacetamide stock solution were added and the samples were incubated for 20 min at room temperature and protected from light.
Trypsin
Following reduction and alkylation, the sample was diluted with 50 mm NH4HCO3 to 1 m urea and trypsin (proteomics grade; Promega, Charbonnières, France) was added at an enzyme-to-substrate ratio of 1:50. The solution was incubated at 37 °C overnight.
Lys-C
Following reduction and alkylation, the sample was diluted with 150 mm NH4HCO3 to 4 m urea and lysyl endopeptidase (mass spectrometry grade; Wako Chemicals, Richmond, VA) was added at an enzyme-to-substrate ratio of 1:100. The solution was incubated at 37 °C overnight.
Lys-N
Following reduction and alkylation, the sample was diluted with 150 mm NH4HCO3 to 6 m urea and endoproteinase Lys-N (gift from Albert J. R. Heck, University of Utrecht) was added at an enzyme-to-substrate ratio of 1:100. The solution was incubated at 37 °C overnight.
Glu-C
Following reduction and alkylation, the sample was diluted with 50 mm NH4HCO3 to 2 m urea and endoproteinase Glu-C (sequencing grade, Sigma-Aldrich) was added at an enzyme-to-substrate ratio of 1:100. The solution was incubated at 25 °C overnight.
Asp-N
Following reduction and alkylation, the sample was diluted with 50 mm sodium phosphate buffer, pH 8, to 1.6 m urea and endoproteinase Asp-N (sequencing grade, Roche Diagnostics Rotkreuz, Switzerland) was added at an enzyme-to-substrate ratio of 1:100. The solution was incubated at 37 °C overnight.
After overnight digestion, all samples were acidified to 2% formic acid and purified by solid-phase extraction using 50 mg Sep-Pak tC18 cartridges (Waters, Milford, MA). The eluate (water/acetonitrile/formic acid, 50:50:0.1) was evaporated to dryness in a vacuum centrifuge.
Fractionation of Cross-Linked Peptides by Size Exclusion Chromatography
Purified samples were reconstituted in 20 μl of SEC mobile phase (water/acetonitrile/trifluoroacetic acid, 70:30:0.1). 15 μl were injected on a GE Healthcare Äkta micro system consisting of autosampler, binary pump, UV/pH/conductivity detectors and fraction collector. This corresponded to total protein amounts of 200 μg of standard proteins, 50 μg of rabbit proteasome, and 10 μg of S. pombe proteasome, respectively. Peptides were separated on a Superdex Peptide PC 3.2/30 column (300 × 3.2 mm) at a flow rate of 50 μl min−1 using the SEC mobile phase. The separation was monitored by UV absorption at 215, 254, and 280 nm. Two-minute fractions (100 μl) were collected into 96-well plates over a separation window of one column volume (2.4 ml = 48 min). For analysis by liquid chromatography (LC)-MS/MS, fractions of interest (retention volumes 0.9–1.4 ml) were removed and evaporated to dryness. For the proteasome samples, only the two main fractions (1.0–1.1 and 1.1–1.2 ml) were analyzed.
Liquid Chromatography-Tandem Mass Spectrometry
LC-MS/MS analysis was carried out on an Eksigent 1D-NanoLC-Ultra system connected to a Thermo LTQ Orbitrap XL mass spectrometer equipped with a standard nanoelectrospray source. SEC fractions were reconstituted in mobile phase A (water/acetonitrile/formic acid, 97:3:0.1). The injection volume was chosen according to the 215 nm UV absorption signal from the SEC separation.
A fraction corresponding to an estimated 1 μg (if available) of the total recovered peptide amount was injected onto a 11 cm × 0.075 mm I.D. column packed in house with Michrom Magic C18 material (3 μm particle size, 200 Å pore size). Peptides were separated at a flow rate of 300 nl min−1 using the following gradient: 0–5 min = 5% B, 5–95 min = 5–35% B, 95–97 min = 35–95% B, and 97–107 min = 95% B, where B = (acetonitrile/water/formic acid, 97:3:0.1).
The mass spectrometer was operated in data-dependent mode, selecting up to five precursors from a MS1 scan (resolution = 60,000) in the range of m/z 350–1600 for collision-induced dissociation. An intensity threshold of 150 counts was chosen for triggering fragmentation, and singly and doubly charged precursor ions and precursors of unknown charge states were excluded from fragmentation. Collision-induced dissociation was performed for 30 ms using 35% normalized collision energy and an activation q of 0.25. Dynamic exclusion was activated with a repeat count of 1, exclusion duration of 30 s, list size of 300, and a mass window of ±50 ppm. Ion target values were 1,000,000 (or maximum 500 ms fill time) for full scans and 10,000 (or maximum 200 ms fill time) for MS/MS scans, respectively. Fragment ions were detected at low resolution in the linear ion trap.
Data Analysis
For data analysis, Thermo Xcalibur .raw files were converted into the open mzXML format using ReAdW, version 4.0.2, using the default settings. mzXML files were directly used as input for xQuest searches, while they were further converted into the .mgf (Mascot generic file) format using the tool MzXML2Search, part of the Trans-Proteomics Pipeline (26). MzXML2Search was executed with the option “-T10000” to export precursors with a mass above the default value of 4200 Da.
Mascot Search
Unmodified peptides from the eight-protein mix were identified by database search using an in-house Mascot (27) server, version 2.3.0, against the Uniprot/SwissProt database (version 51.6, 257964 entries). Search parameters were as follows: Maximum number of missed cleavages = 2, taxonomy = chordata, fixed modifications = carbamidomethyl-Cys, variable modification = Met oxidation, MS1 tolerance = 15 ppm, MS2 tolerance = 0.6 Da, instrument type = ESI-TRAP, decoy mode = on. For validation, the peptide probability was set to p < 0.05, additional filters used were require bold red = yes and peptide score = > 20.
xQuest Search
Cross-linked peptides and peptide mono-links were identified using an in-house version of the dedicated search engine, xQuest, using the same scoring model as described in (12). Tandem mass spectra of precursors differing in their mass by 12.07532 Da (difference between DSS-d0 and DSS-d12) were paired if they had a charge state of 3+ to 8+ and were triggered within 2.5 min of each other. These spectra were then searched against a preprocessed .fasta database as described in the following.
For the eight-protein mixture, the database contained the UniProt/SwissProt entries of the target proteins. Two separate entries were created for the two isoenzymes of pyruvate kinase, and known signal peptides as annotated in UniProt were removed from the primary sequence. Rabbit proteasome data was searched against all 35 human 26S proteasome subunits retrieved from UniProt because only two probable rabbit subunit sequences are available. S. pombe data was searched against a database of all 32 S. pombe 26S subunits in UniProt/SwissProt supplemented with the sequence of rabbit creatine kinase (P00563) and YLK1_SCHPO (Q9P7H8), two known contaminants. (No cross-links from contaminants were identified.)
xQuest search parameters were as follows: Maximum number of missed cleavages (excluding the cross-linking site) = 2, peptide length = 4–40 amino acids, fixed modifications = carbamidomethyl-Cys (mass shift = 57.02146 Da), mass shift of the light cross-linker = 138.06808 Da, mass shift of mono-links = 156.07864 and 155.09643 Da, MS1 tolerance = 15 ppm, MS2 tolerance = 0.2 Da for common ions and 0.3 for cross-link ions, search in enumeration mode (exhaustive search). Search results were filtered according to the following criteria: MS1 mass tolerance window = –3 to +7 ppm (–4 to +7 ppm for proteasome samples), %TIC explained ≥ 0.1, xQuest score ≥ 16 for trypsin, Lys-C and Lys-N and ≥ 18 for Glu-C and Asp-N. Finally, all spectra were then manually validated. Identifications were only considered for the final result list when both peptides had at least four bond cleavages in total or three adjacent ones, respectively, and a minimum length of six amino acids (see also Results and Discussion section).
RESULTS AND DISCUSSION
Design of the Study
To evaluate the use of SEC for enrichment purposes and of multiple proteases for obtaining complementary digestion patterns, we first optimized the method on a well-defined set of model proteins (Table I). They range in size from ∼40 kDa to 80 kDa, thereby offering sufficient potential cross-linking sites, and are quite diverse in their amino acid composition. The model proteins were cross-linked individually using a mixture of two differentially isotope-coded forms of the amine reactive disuccinimidyl suberate, d0- and d12-DSS, before mixing (12). This way, all observed interprotein cross-links can be easily assigned as false positive identifications acting as a control for the estimation of false discovery rates. Cross-linked samples were then digested in parallel by five different proteases as described in the Experimental procedures, and analyzed by liquid chromatography-tandem mass spectrometry. Following the assessment of the data from this pilot study, we then applied the optimized workflow to the proteasome, a protein complex currently extensively studied by our group.
Table I. Model proteins used in the 8-protein mix. Shown are UniProt/SwissProt accession numbers, molecular mass (excluding modifications), and the relative content of relevant amino acids, Lys as potential cross-linking and cleavage site and Arg, Asp and Glu as cleavage sites. All parameters were calculated from the processed forms of the proteins, after cleavage of initiator methionines and signal peptides as assigned in UniProt.
| Protein name | Accession number | Molecular mass (kDa) | % Lys | % Arg | % Asp | % Glu |
|---|---|---|---|---|---|---|
| Catalase, bovine | P00432 | 59.8 | 5.3 | 6.1 | 7.0 | 4.9 |
| Creatine kinase, rabbit | P00563 | 43.1 | 8.9 | 4.7 | 7.3 | 7.1 |
| Fructose-bisphosphate aldolase A, rabbit | P00883 | 39.2 | 7.2 | 4.1 | 3.9 | 6.6 |
| Lactoferrin, bovine | P24627 | 76.1 | 7.8 | 5.4 | 5.2 | 5.8 |
| Ovotransferrin, chicken | P02789 | 75.8 | 8.6 | 4.4 | 6.7 | 6.6 |
| Pyruvate kinase, rabbit (isoenzyme 1) | P11974 | 57.9 | 7.0 | 5.8 | 5.7 | 7.0 |
| Serotransferrin, bovine | Q29443 | 75.8 | 9.3 | 3.4 | 6.9 | 6.4 |
| Serum albumin, bovine | P02769 | 66.4 | 10.1 | 3.9 | 6.9 | 10.1 |
Establishing Peptide Size Exclusion Chromatography for the Fractionation of Digests of Cross-link Samples
We used a polymeric FPLC size-exclusion column suitable for a separation range of 1000 to 7000 Da, according to the manufacturer's specifications. We first evaluated the efficiency of the SEC column by analyzing a peptide mixture consisting of insulin (5.7 kDa), oxidized insulin A chain (2.5 kDa), and angiotensin II (1.0 kDa). Careful optimization of the mobile phase was required as symmetric peaks were only observed in the presence of an acidic aqueous/organic mobile phase. The use of 30% acetonitrile and 0.1% trifluoroacetic acid resulted in acceptable separation of the three analytes, particularly in the range of 1–3 kDa that is most relevant for cross-linked peptides, as shown in Fig. 1A. This volatile mobile phase composition also ensured direct compatibility with downstream LC-MS analysis, requiring only an evaporation step and, in contrast to SCX fractionation, no further sample clean-up.
Fig. 1.
Peptide separations by size exclusion chromatography. UV traces at 215 nm are shown. A, Separation of a model peptide mixture (1 μg per peptide injected) consisting of insulin (1; 5.7 kDa), oxidized insulin A chain (2; 2.5 kDa), and angiotensin II (3; 1.0 kDa). B, Separation of the eight-protein mix cross-linked with DSS and digested with trypsin as the protease (100 μg total protein digest injected). The fractions collected for LC-MS analysis are highlighted. Elution profiles using other proteases are shown in the supplemental Material Fig. S1.
We next used the optimized conditions to analyze a tryptic digest of the eight-protein mixture cross-linked with DSS. The resulting UV chromatogram is shown in Fig. 1B. Based on the elution profile, we collected five individual 100 μl-fractions (corresponding to 2 min windows) as shown (elution volumes 0.9–1.4 ml). Preliminary experiments showed that the fraction from 1.1 to 1.2 ml gave the highest number of cross-link identifications and was labeled fraction “0.” Higher mass fractions were termed “+1” and “+2” and lower mass fractions “–1” and “–2,” respectively, to label their positions relative to the main fraction.
To assess the distribution of different types of peptides over the SEC elution, we analyzed the individual fractions by LC-MS/MS on a linear ion trap-Orbitrap hybrid instrument. MS/MS spectra were analyzed using two software platforms. Unmodified peptides were identified with the widely used search engine, Mascot (27), whereas cross-linked peptides and single peptide chains carrying a hydrolyzed cross-linker modification (mono-links) were assigned with the dedicated cross-linking software, xQuest (12). For this purpose, modified peptides can easily be discerned because they appear as doublets in the MS1 spectrum, separated by 12/z Da, corresponding to the mass shift of the cross-linking reagent.
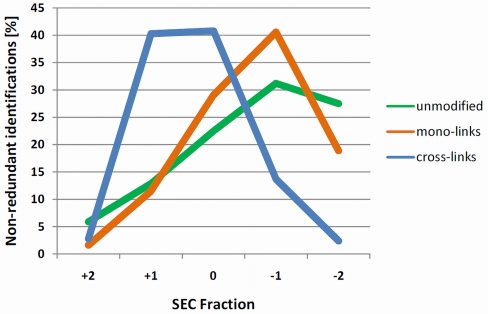
We assumed that because of the increase in molecular mass upon cross-linking, cross-linked peptides would appear in earlier fractions and can therefore be enriched to some degree. Fig. 2 shows the distribution of three classes of peptides over the five SEC fractions that were examined. As expected, the maximum number of cross-link identifications is shifted to higher mass fractions compared with unmodified peptides and peptides carrying mono-links. In particular, most of the cross-link identifications were observed in only two fractions (0 and +1). The majority of the linear peptides, unmodified peptides or mono-links, eluted in later fractions (–1 and –2). The actual numbers of unmodified peptides are expected to be even higher as data was obtained from samples where only precursors of charge states three and higher were selected for fragmentation. This means that likely a considerable number of peptides in the lower molecular weight fraction were excluded from sequencing because they were only present in lower charge states. According to a rough estimate based on UV absorption, these low molecular weight fractions cover ∼80% of the total peak area, corresponding to substantial depletion of peptides that are not directly relevant to cross-linking studies. On the other end of the elution window, the number of identifications in the highest MW fractions was quite low. This is most likely a result of the combination of lower abundance and unfavorable analytical properties in this region.
Fig. 2.
Relative distributions of three classes of peptides (unmodified peptides, green; mono-links, orange; and cross-links, blue) among the SEC fractions from a trypsin digest of the eight-protein mix. Data points are normalized so that for each peptide class, the sum of identifications in all five fractions is set to 100%. Distributions for other proteases are shown in supplemental material Fig. S2.
These very promising initial results led us to conclude that an acceptable degree of separation between the subpopulation of cross-linked peptides and the rest of the peptide pool was possible.
Using Different Proteases for the Digestion of Cross-link Samples
To further expand the number of cross-links that can be recovered in combination with SEC fractionation, we evaluated other commonly used proteases for enzymatic cleavage in addition to trypsin: Asp-N and Glu-C for cleavage at acidic residues and Lys-C and Lys-N for exclusive cleavage at lysine residues.
Evaluating Enzyme Specificity
Preliminary studies were carried out on noncross-linked peptides to evaluate the specificity of the proteases under typical conditions. This was important to keep the search space to a minimum while at the same time using realistic cleavage settings for xQuest. Both Lys-C and Lys-N were found to be highly specific, exhibiting negligible unspecific cleavage at other residues. In contrast, the endoproteinases Asp-N and Glu-C were not exclusively specific for their expected cleavage site: Asp-N was also found to cleave on the N-terminal side of Glu residues, and Glu-C exhibited cleavage also C-terminal to Asp. Specificity was found to be slightly higher for Glu-C, but we decided to consider cleavages at aspartic and glutamic acid for xQuest analysis in both cases, because both peptides connected in a cross-link need to adhere to the defined specificity, and thus even a single deviation in the required four cleavage events would preclude identification of the cross-link.
SEC Fractionation of Cross-linked Samples
Sample preparation for the four additional proteases was carried out as for the trypsin treatment. Proteins individually cross-linked with DSS (the same samples as for the trypsin data set) were digested using procedures as recommended by the manufacturers or, in the case of Lys-N, according to a protocol supplied by the Heck laboratory. SEC fractionation was performed as described above and the same five fractions were collected and analyzed by LC-MS/MS. As shown in supplemental Fig. S1, elution profiles were highly similar for Lys-C, Lys-N, and Glu-C, whereas the Asp-N profile resembled the trypsin chromatogram. The first three enzymes also exhibited a significant peak at the void volume (∼0.3 column volumes), pointing to the generation of very large peptides and/or incomplete digestion under the conditions used.
Redundancy and Orthogonality of the Cross-linking Data Sets for Standard Proteins
We next set out to compare the cross-linking identifications from the fractionated digests of the five proteases and to assess the benefit of using additional proteases. In order to comprehensively profile the sample, we first analyzed all five SEC fractions per protease in duplicate. Thus, in total, 50 LC-MS/MS runs were performed for the model protein samples as part of this study. Using the xQuest software pipeline, scan pairs corresponding to light/heavy pairs with a mass shift of 12.07532 Da were detected. Depending on the enzyme used, up to more than 2000 of such scan pairs were detected in a single fraction and submitted to the database search. Results from the technical replicates were then combined before further analysis and manually validated.
Fig. 3 gives an overview of the distribution of nonredundant cross-link identifications in each SEC fraction for the five proteases investigated; and supplemental Fig. S2 compares the distribution of different classes of peptides as shown for trypsin in Fig. 2 above. In all cases, the majority of cross-link identifications are again confined to two fractions; additional fractions yielded much lower numbers of identifications that were also partially overlapping with the set from the two main fractions. Mono-links and unmodified peptides show an elution pattern that is shifted by at least one SEC fraction, which is comparable to the trypsin data set. The separation of cross-linked peptides appears to be somewhat less pronounced for the Asp-N and Glu-C samples, which can be attributed to the fact the cross-linking reaction does not result in missed cleavage sites for these enzymes.
Fig. 3.
Distribution of cross-link identifications in different SEC fractions. Shown are nonredundant cross-linked peptides in five individual SEC fractions per enzyme, with the two main fractions highlighted in red. (A) Trypsin, (B) Asp-N, (C) Glu-C, (D) Lys-C, (E) Lys-N.
Focusing on the identification of cross-linked peptides, when collapsing all identifications from the five SEC fractions per enzyme, trypsin yielded by far the highest number of nonredundant identifications (Fig. 4). In total, 150 different intraprotein cross-links were identified from this digest, which is a substantial improvement compared with previously reported methods. For comparison, we also analyzed in duplicate an unfractionated sample of the tryptic digest directly by LC-MS/MS. In this case, only 44 intraprotein cross-links were identified. The difference can be explained both by the reduction in sample complexity and the proportional increase in loading for cross-linked peptides because of the partial enrichment.
Fig. 4.
Comparison of cross-link identifications with five different proteases. Shown are nonredundant cross-linked peptides combined over all SEC fractions (in blue) and found in the two main fractions (in red), respectively.
The second highest number of cross-link identifications, 95, was observed for Asp-N, with Glu-C, Lys-C, and Lys-N following. Fig. 4 shows also the contribution of the two SEC fractions richest in cross-links, demonstrating that the majority of cross-links identifiable with the current approach (88–100%) fall within two fractions for all proteases.
To obtain this comprehensive data set, preliminary results were initially filtered according to the achievable mass tolerance (± 5 ppm, an asymmetric search window was used as no recalibration of the raw data was carried out) and multiple identifications corresponding to the same peptide sequences and the same cross-linking sites (“cross-link topologies”) within the peptides were collapsed into single hits. Only the highest scoring identifications were kept. As explained above, because of the experimental design, all putative interprotein cross-links can be confidently classified as false positives. Despite the stringent filtering criteria, the number of these false identifications was still considerable (above 10% in some cases) at the level of unique nonredundant cross-links. Using interprotein cross-link assignments as a guide, we performed additional filtering steps to reduce the number of random hits. We excluded cross-link identifications that contained a peptide shorter than six amino acids as they were found to contain a disproportionally high number of false positives. Furthermore, the acceptable xQuest score threshold needed to be raised from 16 to 18 for the Glu-C and Asp-N data sets, because in these cases, the search space is more than three times larger than for trypsin as a consequence of the higher abundance of acidic residues as cleavage sites. Finally, the remaining spectra were all manually examined for the number of observed bond cleavages in each peptide. We found that many of the assigned interprotein cross-links had only poor sequence coverage in one peptide. Similar to the classification scheme reported by Rappsilber and coworkers (13), we excluded all identifications with less than four bond cleavages in total or three consecutive bond cleavages in both peptides. The final refined data sets are expected to yield FDRs of below 5%, which was also confirmed by structural validation (see below). During preparation of the manuscript, Lauber and Reilly reported similar observations and developed their own filtering criteria for xQuest results (18).
These results already clearly demonstrate that SEC is a powerful tool for cross-linking analysis. However, we cannot exclude at this stage that different instrumental set-ups and refined software would facilitate the identification of additional cross-links, especially in the higher molecular mass fractions. This could especially be the case for enzymes such as Lys-C and Lys-N, which cleave less frequently and yielded comparably fewer identifications.
Improvement in Cross-link Coverage on the Protein Level
Breaking down the identifications according to the individual proteins presents a very interesting picture. Table II lists the identified unique cross-linking sites observed with the different enzymes for all eight proteins. Here, cross-linking topologies that were identified in different peptides because of the use of different enzyme cleavage specificities or to the presence of missed cleavage sites were combined as they all provide the same spatial information. Although all proteins are relatively comparable in size, the number of distance restraints differs by nearly an order of magnitude (compare serum albumin, 66 kDa, and catalase, 60 kDa). Most apparent is a trend that the number of cross-links observed for a given protein in the trypsin data set is proportional to its lysine content. Although this may not come as a surprise because a lysine-specific cross-linking reagent was used in the study, the connection is still relevant. It shows that for proteins with a disproportionally high lysine content (e.g. 10.1% for BSA), unusually high numbers of cross-links are achievable whereas the information that is recoverable for proteins with average lysine content is probably less extensive. Such trends are not as apparent for the enzymes that yield smaller numbers of identifications, such as Lys-C and Lys-N. Interestingly, Glu-C and Asp-N cross-links differ substantially for some proteins (lactoferrin, serum albumin) despite an overall quite similar content of acidic residues for all proteins. This may be connected to the dominant cleavage specificity for the individual proteases.
Table II. Non-redundant Lys-Lys contacts at the individual protein level for each single protease and combined. Numbers in parentheses show contacts not observed in the trypsin data set.
| Protein name | Trypsin | Asp-N | Glu-C | Lys-C | Lys-N | Total |
|---|---|---|---|---|---|---|
| Catalase, bovine | 3 | 2 (2) | 0 | 3 (1) | 0 | 6 |
| Creatine kinase, rabbit | 12 | 6 (4) | 4 (4) | 3 (2) | 1 (1) | 19 |
| Fructose-bisphosphate aldolase A, rabbit | 12 | 6 (4) | 2 (1) | 7 (2) | 1 (0) | 19 |
| Lactoferrin, bovine | 21 | 11 (8) | 1 (1) | 2 (1) | 1 (1) | 31 |
| Ovotransferrin, chicken | 19 | 13 (12) | 2 (2) | 9 (4) | 4 (2) | 38 |
| Pyruvate kinase, rabbit (isoenzyme 1) | 8 | 7 (6) | 2 (2) | 1 (1) | 1 (1) | 15 |
| Serotransferrin, bovine | 28 | 21 (14) | 13 (12) | 6 (3) | 4 (2) | 57 |
| Serum albumin, bovine | 34 | 12 (9) | 20 (10) | 9 (4) | 7 (4) | 55 |
| TOTAL | 137 | 78 (59) | 44 (32) | 40 (18) | 19 (10) | 240 |
Finally, these identifications were cumulated to provide a list of nonredundant distance restraints (Lys-Lys contacts) for the whole data set (supplemental Tables S1 and S2). The complete data set provided a total of 240 restraints, with numbers for individual proteins ranging from 6 (catalase) to 57 (bovine transferrin). Trypsin-derived data alone accounted for a total of 137 restraints from these eight proteins. Although all other enzymes contributed less information, Asp-N provided the largest amount of complementary restraints, with 59 intramolecular connections out of a total of 78 not covered by trypsin (Table II). Again, Glu-C, Lys-C and Lys-N followed in the same order as for the total nonredundant identifications, contributing 32, 18, and 10 contacts not found using trypsin.
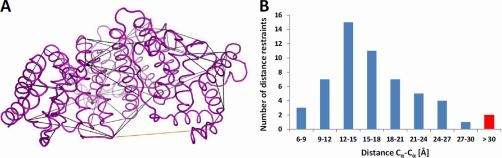
To illustrate the extensive cross-link coverage obtained for BSA, Fig. 5A visualizes the BSA restraints on a homology model obtained from ModBase (28). As shown in Fig. 5B, the majority of the observed distances conform to the expected span of the cross-linking reagent (Cα-Cα distance of 23 Å excluding any structural flexibility), and with the exception of two contacts, the bridged distances lie within 28 Å. The calculated span between residues 117 and 489 is 34.8 Å and could result from a deviation of the homology model to the actual structure, whereas the second outlier corresponds to a theoretical distance of 57.1 Å. Even if both cases are classified as false positives, the error rate would still be at an acceptable level of 3.6% (2 of 55).
Fig. 5.
A, Lys-Lys contacts identified by the multiprotease approach mapped onto a bovine serum albumin homology structure obtained from ModBase. Distances (Cα-Cα) of less than 30 Å, between 30 and 35 Å and above 35 Å are colored in black, orange and red, respectively. Visualization was performed using PyMOL 1.3 (Schrödinger LLC). B, Histogram showing the distribution of the BSA distance restraints shown in (A).
As summarized in Table II, 34 of the 55 Lys-Lys contacts were obtained from the trypsin sample. However, not all regions of the protein are covered equally well by this enzyme. We highlight two exemplary regions where additional proteases provide substantial new structural information. First, few cross-links in the N-terminal region were identified from the tryptic digest. Cross-links containing Lys28 and Lys36 were identified in a total of six restraints, but the next position included in a cross-link is Lys140 (excluding the ambiguous cross-link 117 × 489 mentioned above). Within this span of ∼100 residues, Asp-N and Glu-C derived contacts provide seven additional restraints (visualized in supplemental Fig. S3). A closer inspection of the amino acid composition reveals that the N-terminal region is relatively poor in arginine residues, but rich in lysines. Therefore, if exposed lysines are blocked following the reaction with the cross-linking reagent (therefore precluding cleavage by a protease) the resulting peptides would become excessively long. On the contrary, if lysines are not accessible in the native structure, these Lys-rich regions will present many cleavage sites upon denaturation, resulting in relatively short peptides that are challenging to identify and are underrepresented as a consequence of the SEC fractionation. The more homogeneous distribution of acidic residues offers better coverage of this region.
As a second example, contacts only identified with Asp-N and Lys-C also covered a region near the C terminus more extensively. Lysines 561 and 568 were connected in peptides distant in the primary sequence, defining the orientation of this region toward the rest of the structure with the contacts 495 × 561 and 520 × 568 (supplemental Fig. S4).
Even with the expanded coverage achieved by the use of five proteases, the number of experimentally observed contacts was still roughly an order of magnitude below the theoretically possible. A simulation by the tool Xwalk (29) revealed that in excess of 500 contacts are possible within a reasonable distance restraint of up to 30 Å. Eventually, it is crucial for a cross-linking workflow to provide input for refining unknown structures of proteins and protein complexes. To achieve this, it has to be considered that part of the cross-links that are identified yield little relevant structural information because residues adjacent in the primary amino acid sequence are connected. For example, in our case about 20% of the nonredundant restraints connect residues less than 20 amino acids apart. In this context, the expanded coverage that is obtainable by multiple proteases becomes even more relevant.
Biological Application of the Optimized Workflow
To demonstrate the benefit of both SEC fractionation and multiprotease digestion, we applied the optimized workflow to a multisubunit protein complex of high biological interest, the proteasome. The proteasome is the end point of the ubiquitin-proteasome pathway for protein degradation (30, 31). It consists of two main compartments, the 19S regulatory particle (RP), responsible for the recognition of polyubiquitinated substrates, and a barrel-shaped 20S core particle (CP). Chemical cross-linking has recently contributed important information for deriving the structure of the CP and the AAA-ATPases that are part of the RP (25). Here, to demonstrate the value of the refined workflow, we focused on the CP that consists of 14 different subunits (α1 - α7 and β1 - β7), each present in two copies, which assemble to four stacked heteroheptameric rings (αββα). High-resolution structures of CPs from several organisms are available (32, 33).
Samples From Two Species Were Evaluated. A rabbit 20S preparation was cross-linked with DSS at concentrations comparable to the standard proteins (using a starting amount of 50 μg at ∼0.8 mg ml−1). Additionally, S. pombe proteasome was reacted with DSS, although at much lower concentration, reflecting a typical sample-limited scenario. In this case, even after preconcentration, only around 10 μg of protein at ∼0.2 mg ml−1 was recovered from the preparation. It is well known that cross-linking kinetics is not very favorable in this concentration range; we commonly observe that reaction yields drop substantially below 0.5 mg ml−1. In addition, conditions for enzymatic digestion of such small sample amounts may also be suboptimal. In both cases, the cross-linked sample was split in half and digested with either trypsin or Asp-N. Asp-N was chosen because it provided the highest degree of complementary information to trypsin for the 8-protein mix. The digested samples were then fractionated by SEC and the two main fractions (0 and +1) were collected for LC-MS analysis.
For the rabbit sample, the trypsinized aliquot yielded 42 nonredundant cross-linked peptides, corresponding to 36 nonredundant contact sites. Among these, 18 restraints were within the same subunit, with one exception exclusively within α-subunits. 18 other restraints were located between distinct subunits. Also for this subset, a majority of the restraints was located within α-subunits, three were located between an α- and a β-subunit and one between two β-subunits. The aliquot digested with Asp-N yielded a much smaller set of identifications: Six intra-subunit cross-links and three inter-subunit cross-links, all within or between α-subunits. The overlap between the two data sets was however minimal, as only one Lys-Lys contact was shared by both enzymes. The preference for α-subunits can be readily explained by the fact that the β-subunits are located in the two central rings and have significantly less exposed surface than the α-rings and, consequently, less exposed cross-linkable sites. Some additional cross-links were possibly missed because the data was searched against a database containing the human proteasome subunits, which are likely not completely identical to the rabbit sequences. All the identifications are summarized in supplemental Table S3, and annotated spectra are provided in supplemental Fig. S5.
From the S. pombe sample, seven contacts from the trypsin sample and four from the Asp-N sample were identified, reflecting the very low sample amounts. The trypsin data set yielded five intra-subunit cross-links on α-subunits (three on α2 plus additional ones on α6 and α7) plus one on the β1-subunit. An intersubunit cross-link between subunits α3 and α6 was also observed. The Asp-N cross-links, interestingly, preferentially covered the β-subunits, including an intrasubunit link on β7 and two inter-subunit links between β3 and β7. The remaining contact was observed between α4 and β7. In this case, no overlap between the trypsin and Asp-N restraints was found, and, importantly, all contacts were validated on a 20S homology model with Cα-Cα distances between 4.9 and 23.4 Å. Detailed information about the identifications is provided in Table III and supplemental Fig. S6.
Table III. Identified cross-linking sites in the S. pombe 20S proteasome. Shown are the amino acid sequences and cross-linking positions within the peptide (a = α chain, b = ß chain); the corresponding proteins and the absolute position of the cross-linking sites; experimentally observed mass (Mr), mass-to-charge ratio( m/z), and charge state (z) of the highest scoring identification; the deviation of experimental to theoretical mass in ppm; fraction of total ion current (TIC) explained; xQuest score; and Euclidean Cα-Cα distance calculated from the homology model.
| Sequences | Protein 1 | Pos. 1 | Protein 2 | Pos. 2 | Mr (exp.) | m/z (exp.) | z | Error (ppm) | %TIC | Score | Distance (Å) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypsin data | |||||||||||
| KIYNEYPPTK-KVAQTTYK-a1-b1 | PSA2_SCHPO | 99 | PSA2_SCHPO | 91 | 2327.237 | 582.817 | 4 | −1.7 | 0.43 | 28.50 | 10.5 |
| IITKEGVETR-RLLKLEEAMK-a4-b4 | PSB1_SCHPO | 212 | PSB1_SCHPO | 179 | 2512.421 | 503.492 | 5 | −3.4 | 0.36 | 26.65 | 14.6 |
| EYLEKNWKEGLSR-ASKAAR-a5-b3 | PSA7_SCHPO | 176 | PSA7_SCHPO | 168 | 2391.256 | 598.822 | 4 | 0.7 | 0.22 | 26.04 | 11.9 |
| KPTSELAIGASLEK-ATAIGKSSTAAK-a1-b6 | PSA2_SCHPO | 50 | PSA2_SCHPO | 165 | 2685.48 | 672.378 | 4 | −0.1 | 0.44 | 25.61 | 12.6 |
| KVAQTTYK-VLVDKSR-a1-b5 | PSA2_SCHPO | 91 | PSA2_SCHPO | 88 | 1891.072 | 473.776 | 4 | −2.9 | 0.15 | 25.51 | 5.1 |
| IITKEGVETR-LLKLEEAMK-a4-b3a | PSB1_SCHPO | 212 | PSB1_SCHPO | 179 | 2356.328 | 590.090 | 4 | −0.3 | 0.24 | 21.48 | 14.6 |
| ATSAGPKQTETINWLEK-KVPDKLIDASTVK-a7-b1 | PSA6_SCHPO | 169 | PSA6_SCHPO | 53 | 3423.852 | 856.971 | 4 | 0.7 | 0.28 | 19.88 | 14.5 |
| KVPDKLIDASTVK-NELEKLNFSSLK-a5-b5 | PSA6_SCHPO | 57 | PSA3_SCHPO | 177 | 2971.649 | 743.920 | 4 | 0.4 | 0.22 | 18.67 | 7.1 |
| Asp-N data | |||||||||||
| DKCIKRLVKGRQD-DRGTTAVLKE-a9-b9 | PSB3_SCHPO | 200 | PSB7_SCHPO | 248 | 2841.552 | 474.600 | 6 | 1.1 | 0.29 | 20.38 | 4.9 |
| DTTKNKMVCKIWKS-DIYKFVTVQ-a10-b4 | PSA4_SCHPO | 227 | PSB7_SCHPO | 253 | 2987.561 | 747.898 | 4 | 3.7 | 0.34 | 19.67 | 23.4 |
| DKCIKRLVKGRQD-DRGTTAVLKE-a5-b9 | PSB3_SCHPO | 196 | PSB7_SCHPO | 248 | 2841.559 | 569.320 | 5 | 3.5 | 0.19 | 19.21 | 13.6 |
| DEEKATPYRGYSKPN-ERATKQSKYTY-a13-b5 | PSB7_SCHPO | 226 | PSB7_SCHPO | 233 | 3265.594 | 654.127 | 5 | 1.5 | 0.13 | 18.34 | 19.1 |
a Redundant restraint.
Thus, although the increase in identifications from the use of a second enzyme was partially lower than for model proteins, it is apparent that additional contacts can be recovered using the same, well established cross-linking protocol. Practically, the starting amounts available for the S. pombe sample also seem to be the lower limit for providing any benefit by performing the fractionation step. However, the results from the rabbit proteasome compare favorably with a recent study by Kao et al. using the gas-phase cleavable cross-linking reagent, disuccinimidyl sulfoxide (21). In this work, similar starting amounts and concentrations (50 μl at a concentration of < 0.9 mg ml−1) were used, and identical instrumentation was employed. A total of 13 nonredundant cross-linked peptides were identified, which should be at least partly attributable to the fact that no enrichment step was included in the protocol.
Concluding Remarks
Although cross-linking methodology has improved considerably in recent years, it still can be expected that only a small fraction of potential cross-linking sites are currently detected in typical assays. Despite the expectation that the introduction of more sensitive mass spectrometers will drive up the numbers in the future and improved software should increase the fraction of spectra that can be assigned to a particular arrangement of two connected peptides, the adoption of relatively straightforward experimental protocols immediately provides a convenient approach to increase the depth of cross-linking studies. Here, we demonstrated that the introduction of SEC fractionation and the use of multiple proteases led to a more comprehensive coverage of cross-links. On model proteins, the number of cross-links identified increased more than 3-fold upon adoption of SEC fractionation using only trypsin as the protease. Because two SEC fractions contain a large majority of the cross-link fractions, the increased demand for instrument time is reasonable. In contrast to SCX fractionation, the procedure is highly amenable to automation and directly compatible with downstream MS analysis. The use of several proteases resulted in a further increase of more than 70% on the level of nonredundant cross-linking sites by using four other proteases, and more than 45% using Asp-N alone, for model proteins. Similar increases are achievable even for small sample amounts, as demonstrated by the application to the proteasome. Eventually, such improvements may depend on the particular properties of the protein(s) under study, mainly on the frequency and distribution of cross-linking and enzymatic cleavage sites.
The SEC fractionation described in this work has already been successfully applied in a previous study on the proteasome (25), and here we could show that the combined use of complementary proteases further increases the yield of cross-links. This is even the case for very low sample amounts, as could be demonstrated by the S. pombe sample, where the total number of Lys-Lys contacts increased from seven to eleven.
Acknowledgments
We thank Albert J. R. Heck (University of Utrecht) for providing a sample of Lys-N protease.
Footnotes
* This work was supported by the European Union 7th Framework project PROSPECTS (Proteomics Specification in Time and Space, grant HEALTH-F4-2008-201648). AL was partially supported by a Schrödinger fellowship of the Austrian Science Fund. The stay of RR was financially supported through a fellowship from the University of Vienna and the Österreichische Forschungsgemeinschaft (ÖFG). RA was supported by the ERC advanced grant “Proteomics v.3.0” (grant 233226).
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
1 The abbreviations used are:
- DSS
- disuccinimidyl suberate
- CP
- (proteasome) core particle
- RP
- (proteasome) regulatory particle
- SEC
- size exclusion chromatography.
REFERENCES
- 1. Robinson C. V., Sali A., Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982 [DOI] [PubMed] [Google Scholar]
- 2. Konermann L., Pan J., Liu Y. H. (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 [DOI] [PubMed] [Google Scholar]
- 3. Konermann L., Stocks B. B., Pan Y., Tong X. (2010) Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 29, 651–667 [DOI] [PubMed] [Google Scholar]
- 4. Benesch J. L., Ruotolo B. T., Simmons D. A., Robinson C. V. (2007) Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem. Rev. 107, 3544–3567 [DOI] [PubMed] [Google Scholar]
- 5. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing Native Protein Structures by Chemical Cross-linking, Mass Spectrometry, and Bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petrotchenko E. V., Borchers C. H. (2010) Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 29, 862–876 [DOI] [PubMed] [Google Scholar]
- 7. Rappsilber J. (2011) The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mayne S. L., Patteron H. G. (2011) Bioinformatics tools for the structural elucidation of multi-subunit protein complexes by mass spectrometric analysis of protein–protein cross-links. Brief. Bioinform. 12, 660–671 [DOI] [PubMed] [Google Scholar]
- 9. Trester-Zedlitz M., Kamada K., Burley S. K., Fenyö D., Chait B. T., Muir T. W. (2003) A modular cross-linking approach for exploring protein interactions. J. Am. Chem. Soc. 125, 2416–2425 [DOI] [PubMed] [Google Scholar]
- 10. Kang S., Mou L., Lanman J., Velu S., Brouillette W. J., Prevelige P. E., Jr. (2009) Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 23, 1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nessen M. A., Kramer G., Back J., Baskin J. M., Smeenk L. E., de Koning L. J., van Maarseveen J. H., de Jong L., Bertozzi C. R., Hiemstra H., de Koster C. G. (2009) Selective enrichment of azide-containing peptides from complex mixtures. J. Proteome Res. 8, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Willis J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II–TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lauber M. A., Reilly J. P. (2010) Novel Amidinating Cross-Linker for Facilitating Analyses of Protein Structures and Interactions. Anal. Chem. 82, 7736–7743 [DOI] [PubMed] [Google Scholar]
- 15. Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., Bruce J. E. (2009) Identification of Protein-Protein Interactions and Topologies in Living Cells with Chemical Cross-linking and Mass Spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang X., Bruce J. E. (2010) A new cross-linking strategy: protein interaction reporter (PIR) technology for protein–protein interaction studies. Mol. BioSyst. 6, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng C., Yang L., Hoopmann M. R., Eng J. K., Tang X., Weisbrod C. R., Bruce J. E. (2011) Cross-linking measurements of in vivo protein complex topologies. Mol. Cell. Proteomics 10, M110.006841 (article number) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lauber M. A., Reilly J. P. (2011) Structural analysis of a prokaryotic ribosome using a novel amidinating cross-linker and mass spectrometry. J. Proteome Res. 10, 3604–3616 [DOI] [PubMed] [Google Scholar]
- 19. Santos L. F., Iglesias A. H., Gozzo F. C. (2011) Fragmentation features of intermolecular cross-linked peptides using N-hydroxy- succinimide esters by MALDI- and ESI-MS/MS for use in structural proteomics. J. Mass Spectrom. 46, 742–750 [DOI] [PubMed] [Google Scholar]
- 20. Trnka M. J., Burlingame A. L. (2010) Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene. Mol. Cell. Proteomics 9, 2306–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kao A., Chiu C., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., Huang L. (2011) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212 (article number) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tran B. Q., Hernandez C., Waridel P., Potts A., Barblan J., Lisacek F., Quadroni M. (2011) Addressing trypsin bias in large scale (phospho)proteome analysis by size exclusion chromatography and secondary digestion of large post-trypsin peptides. J. Proteome Res. 10, 800–811 [DOI] [PubMed] [Google Scholar]
- 24. Saeki Y., Isono E., Toh-E A. (2005) Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399, 215–227 [DOI] [PubMed] [Google Scholar]
- 25. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010) Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pedrioli P. G. (2010) Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol. Biol. 604, 213–238 [DOI] [PubMed] [Google Scholar]
- 27. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28. Pieper U., Webb B. M., Barkan D. T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E.C., Pettersen E. F., Huang C. C., Datta R. S., Sampathkumar P., Madhusudhan M. S., Sjölander K., Ferrin T. E., Burley S. K., Sali A. (2011) MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 39, D465–D474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kahraman A., Malmström L., Aebersold R. (2011) Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27, 2163–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finley D. (2009) Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weissman A. M., Shabek N., Ciechanover A. (2011) The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 33. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20S proteasome from yeast at 2.4Å resolution. Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]