Abstract
The term “proteomics” encompasses the large-scale detection and analysis of proteins and their post-translational modifications. Driven by major improvements in mass spectrometric instrumentation, methodology, and data analysis, the proteomics field has burgeoned in recent years. It now provides a range of sensitive and quantitative approaches for measuring protein structures and dynamics that promise to revolutionize our understanding of cell biology and molecular mechanisms in both human cells and model organisms. The Proteomics Specification in Time and Space (PROSPECTS) Network is a unique EU-funded project that brings together leading European research groups, spanning from instrumentation to biomedicine, in a collaborative five year initiative to develop new methods and applications for the functional analysis of cellular proteins. This special issue of Molecular and Cellular Proteomics presents 16 research papers reporting major recent progress by the PROSPECTS groups, including improvements to the resolution and sensitivity of the Orbitrap family of mass spectrometers, systematic detection of proteins using highly characterized antibody collections, and new methods for absolute as well as relative quantification of protein levels. Manuscripts in this issue exemplify approaches for performing quantitative measurements of cell proteomes and for studying their dynamic responses to perturbation, both during normal cellular responses and in disease mechanisms. Here we present a perspective on how the proteomics field is moving beyond simply identifying proteins with high sensitivity toward providing a powerful and versatile set of assay systems for characterizing proteome dynamics and thereby creating a new “third generation” proteomics strategy that offers an indispensible tool for cell biology and molecular medicine.
Within the postgenomics fields, proteomics has a privileged role because it deals directly with the analysis of proteins, which are the key functional units of the cell. Proteomics has developed from diverse roots, which have now coalesced into an “omics” field of study, characterized more by its diversity than a common methodological or subject orientation. Nevertheless, the current, general definition of proteomics views it as the large-scale study of proteins and their modifications. More ambitious definitions of proteomics also include the goal of analyzing the entire spectrum of protein functions, but this essentially overlaps with the goal of biological sciences in general. We suggest that the large-scale study of endogenous proteins, their post-translational modifications, interactions and dynamic behavior in space and time, are indeed the core subject area of proteomics.
Proteomics Specification in Time and Space (PROSPECTS)1 is a five year collaborative project that commenced early in 2008, funded by the Research Directorate of the European Commission under the 7th Research Framework Program. PROSPECTS is coordinated by Matthias Mann and brings together ten top proteomics research groups from around Europe, as well as a leading mass spectrometry instrument manufacturer and chromatography company. The PROSPECTS project arose in part from the recognition that the proteomics field was rapidly developing toward a “second generation” state, where it was possible not only to identify proteins with high sensitivity, but increasingly to use proteomics technologies to assay the dynamic properties of proteins in high throughput and to characterize the structure and composition of large multiprotein complexes. PROSPECTS seeks to develop and optimize new technology and methodology in the proteomics field with a strong focus on how these can be deployed to maximum effect to advance our understanding of fundamental aspects of cell regulation and disease mechanisms, including stress responses and neurodegenerative syndromes.
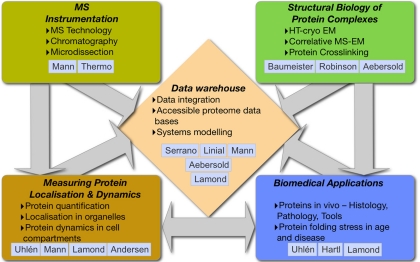
The PROSPECTS team brings together leading European proteomics and biology laboratories with complementary expertise and a strong track record of working together effectively (Fig. 1). The coordinating Mann group (MPIB, Martinsried) and the industrial partner Thermo Fisher Scientific, Bremen, have a longstanding collaboration concerning the improvement of instrumentation in mass spectrometry (MS). The Lamond (University of Dundee) and Andersen (University of Southern Denmark) groups have collaborated closely with the Mann group for over 15 years to apply cutting edge MS-based proteomics technology to important biological areas, producing several landmark publications that are now citation classics. Proxeon (now Thermo Fisher Scientific, Odense), a company collaborating with the Mann group for over ten years, are specialists in the design of hardware and software for proteome analysis, bringing expertise with new instrumentation for separation of peptides and proteins by liquid chromatography at the nano scale.
Fig. 1.
Structure and expertise in the PROSPECTS network.
The structural biology core is formed by the Baumeister group (MPIB, Martinsried), who, together with industrial partners, develops advanced instrumentation and methodology for structural studies on large protein complexes, working toward high-throughput analyses in electron microscopy. The Aebersold laboratory (ETH, Zurich) is a leader in mass spectrometry based proteomics and systems biology, and contribute their expertise in using MS technology to solve problems in structural biology. The Aebersold group has had close interactions with both the Mann and Baumeister groups over the last 15 years. The Robinson group (University of Oxford) are expert in using mass spectrometry to determine overall topology of protein complexes and thus bridges the structural and mass spectrometric efforts.
Biomedical expertise in the partnership is specifically contributed by the Uhlén group (KTH, Stockholm) working on the Human Protein Atlas project (www.proteinatlas.org), and systematically mapping the human proteome by characterizing antibodies specific to each open reading frame. The Protein Atlas project provides PROSPECTS with extensive pathology and histology expertise and access to validated antibodies specific for relevant protein targets studied by groups in the consortium. This adds the unique opportunity to cross-validate high-throughput data obtained by MS-based proteomics and immuno-histological findings with cognate antibodies against thousands of human proteins, using both tissue array data and confocal microscopy.
The Hartl group (MPIB, Martinsried) brings expertise in the biochemical analysis of protein folding, aggregation and molecular chaperones. They recently elucidated mechanisms underlying misfolding of proteins inducing aggregates that are causal in neurodegenerative diseases, including Alzheimer's, Parkinson's, and Huntington's disease.
The integration of large data sets into databases, in easily accessible and retrievable form, is aided by the bioinformatics expertise of the Linial group (Hebrew University of Jerusalem). The Serrano group (CRG, Barcelona) provides their know-how with systems modeling and integrating the experimental data generated within the PROSPECTS project in their modeling platform.
Together, the groups working in this truly multidisciplinary consortium drive development of new proteomics strategies, i.e. pioneering generic, quantitative approaches for identification and functional characterization of the human proteome and its dynamics. Four years into the PROSPECTS project, it is possible to measure its achievements against the promises made at project start. Already more than 80 manuscripts have been published by the consortium, with many of these appearing in leading international journals. The publications in this special issue present some of the latest advances and results arising from the PROSPECTS project, illustrate the variety of approaches taken and point to some important future directions for the proteomics field.
Instrumentation
Much of modern proteomics is in fact an application of mass spectrometry, which has become the method of choice for high sensitivity protein detection and identification (1–3). MS-based proteomics builds upon the continued development of increasingly more sensitive and sophisticated instruments, which are a basis for an ever more comprehensive characterization of cell proteomes. Thermo Fisher Scientific, which is a partner in PROSPECTS, has worked with laboratories in the consortium to develop improved hybrid mass spectrometers that enhance the performance of the modern proteomics workflow. Building on previous joint efforts leading to the development of hybrid linear ion trap Orbitrap™ instruments (4, 5), in this special issue, Thermo and the Mann group describe the latest linear ion trap Orbitrap instrument, termed the Orbitrap ELITE (Michalski et al., this issue) (6).
The Orbitrap ELITE instrument employs an Orbitrap analyzer of roughly half the size, and hence almost twice the frequency, of the previous model. Together with an enhanced Fourier transform algorithm, the Orbitrap ELITE achieves fourfold higher resolution at the same transient length. This enables efficient top-down operation in a chromatographic time scale, as well as ultra-high resolution scans for routine liquid chromatography tandem MS (LC MS/MS) operation. Its increased speed enables acquisition of high resolution collision-induced dissociation (CID) and HCD scans in a time frame that was previously needed for a single, low resolution CID scan. This should dramatically increase the certainty and information content of MS/MS assignments, especially for modified peptides.
In the PROSPECTS consortium Thermo recently introduced a novel combination of a quadrupole mass filter with the Orbitrap analyzer, in a benchtop format, termed the Q Exactive. Unlike the LTQ Orbitrap instruments, the Q Exactive combines very fast mass selection “in space,” with high resolution measurements of both precursor and fragment spectra in the Orbitrap analyzer (7). Important features of the Q Exactive include its ability to multiplex narrow mass range analysis (SIM scans) with higher sensitivity than in the full mass range, as well as to multiplex different fragmentation spectra. With its smaller footprint and lower cost, the Q Exactive should make many of the strategies developed in PROSPECTS available to a broad biological user community.
In this issue, Nagaraj et al., describe the combination of high performance “single-shot” proteomics with MS analyses using the Q Exactive (8). The idea in single-shot proteomics is to rely exclusively on LC MS/MS as the analysis method, rather than performing prior up-front fractionation (9). Advantages are drastically reduced measurement time, sample consumption, and experimental simplicity, but conversely the demands on the performance of the chromatographic systems and the mass spectrometer are increased correspondingly. Coupling a novel ultrahigh pressure system (Easy nanoLC 1000, partner Proxeon, now Thermo Fisher), to high resolution columns, the majority (about 4000 of an estimated 4400 expressed proteins) of the yeast proteome could be quantified in single runs (8). This study also describes a “spike-in SILAC” mix, corresponding to a heavy isotope labeled version of the yeast proteome, prepared under different conditions. It provides a reference standard that can be spiked into other samples and was used to investigate the response of the yeast proteome to heat shock. Significantly, this prototypical proteome experiment only required one to two days of total MS measurement time, while generating sufficient data to reveal the system-wide response of the proteome to heat shock with exquisite accuracy.
In another contribution describing improvements to MS instrumentation, Graumann et al. (this issue) introduce so called “intelligent agents” into proteomics (10). These intelligent agents are autonomous software entities that are ubiquitous in computer science and are, for instance, applied in search engines that trawl the net for information. Here an intelligent agent is constructed that directs the mass spectrometer toward narrow mass range (SIM) acquisition for better quantification, as well as to the sequencing of specific peptides of interest. This study also introduces the first real time search engine, which scans the obtained MS/MS spectra against a sequence database on the fly within a few milliseconds, allowing the intelligent agent to base future decisions implemented by the instrument on this real time information. This capability is showcased on the directed sequencing of peptides that are offset from each other by a mass difference corresponding to a potential deamidation. Future applications of intelligent agents in proteomics will only be bounded by the imagination, especially when current constraints due to software architecture are removed in new generations of instruments.
Methods for Absolute Quantification
The emergence of what we can term “2nd generation” proteomics was characterized by the development of quantitative MS-based methods that moved the field on from primarily identifying proteins, to also providing measurements of relative changes in protein levels between different cell states and/or experimental conditions. However, in recent years the usefulness of absolute, rather than relative, quantification of protein levels has increasingly become appreciated. This issue introduces two new tools for absolute protein quantification developed within PROSPECTS.
Ludwig et al. (this issue) build on the Selected Reaction Monitoring technique to estimate absolute protein abundance in unlabeled and nonfractionated cell lysates (11). Their approach is based on the “best-flyer” hypothesis, which assumes that the specific MS signal intensity of the most intense tryptic peptides per protein is approximately constant throughout a whole proteome.
From a linear calibration curve, generated using a small number of anchor point proteins with known absolute concentrations, absolute abundances are inferred for any selected reaction monitoring-targeted protein of interest. The described method exploits inherent performance advantages of selected reaction monitoring, such as high sensitivity, selectivity, reproducibility, and dynamic range. Except for the generation of the calibration curve, the approach does not require synthesis and usage of cost- and labor-intensive isotopically labeled reference peptides or proteins, and can be applied to any nonlabeled protein sample of interest. However, the label-free workflow comes at the cost of measurement accuracy, i.e. it allows inference of protein abundances with a mean-fold error rate of twofold, as determined by Monte Carlo cross-validation.
A cooperation between the PROSPECTS partners Uhlén and Mann, which originated from informal discussions at one of the PROSPECTS annual retreats, makes use of the large resource of tens of thousands of short recombinant protein fragments that had been expressed in E. coli for the purpose of antibody generation in the human proteome atlas project (www.proteinatlas.org) (12). These so-called Protein Epitope Signature Tags (PrESTs) are selected as 100–150 amino-acid-long parts of the protein of interest, fused to a solubilization tag. As shown by Zeiler et al. in this issue, these protein fragments can be exploited for copy number determination of proteins with the robustness and accuracy associated with stable isotope labeling with amino acid in cell culture (SILAC)-based experiments (13). The solubilization tag functions as the basis of absolute quantification in a first SILAC experiment, which determines the amount of the SILAC-PrEST standard. Appropriate amounts of each of these proteins are then combined to create a master mix, which is produced in large amounts. Zeiler et al. (13) use such a standard mix to determine the copy numbers of 40 proteins in HeLa cells, ranging from the transcription factor FOS (6000 copies per cell), to the high abundance cytoskeletal protein vimentin (20 million copies per cell). Clearly, the ease of use and robustness of this technique makes it a potential replacement for the Western blot in many situations, as anticipated several years ago (14).
Proteomics and Protein Complexes/Structural Biology
The PROSPECTS consortium has taken a number of approaches to use proteomics technologies in novel ways that can provide important structural information regarding the conformation of both specific protein-protein interactions and the topology of larger, multiprotein complexes and macromolecular machines. For example, the combination of chemical cross-linking and mass spectrometry to study protein-protein interactions has gained more widespread interest recently (15, 16). Previously reported methods either only allowed for a limited coverage, even for samples of moderate complexity, or were labor-intensive. Leitner et al. (this issue) improve the methodology by introducing peptide-level size exclusion chromatography as a convenient fractionation technique to enrich for cross-linked peptides (17). This way, the number of distance constraints in the form of lysine-lysine contacts increased by more than twofold for a set of standard proteins, compared with the same sample directly analyzed by LC MS/MS. The yield was further increased by using proteases with complementary cleavage specificity, such as Asp-N and Glu-C, in addition to trypsin. This provided 240 lysine-lysine contacts in model proteins, averaging 30 per protein. The novel SEC fractionation approach was subsequently applied to analyze the 28-subunit 20S core particle from the proteasome, a >2 MDa complex responsible for protein degradation, whose structure has recently been elucidated by various structural probing techniques, including cross-linking (18, 19). One of these reports (18) arose from a collaboration between the Baumeister and Aebersold laboratories. Using only 50 pmol of starting material, 50 cross-linked peptides were identified from the 20S core particle of the proteasome. The new enhanced protocol forms part of an integrated pipeline for the high-throughput generation of cross-linking data established in the Aebersold laboratory.
The role of the Robinson group in the PROSPECTS consortium has been to integrate their work using MS-based techniques for characterizing intact multiprotein complexes with other approaches involving proteomics, electron microscopy, and protein cross-linking. This is typified by a recent collaboration between the Robinson and Baumeister groups, applying quantitative proteomics to study the assembly of the 19S regulatory particle of the proteasome (20). In this study the collaborators were able to define the role of chaperones in proteasome assembly and to define a new role for Ubp6.
The Robinson group has also demonstrated “proof of principle” experiments for the novel pairing of electron microscopy with MS studies of intact complexes. By depositing intact GroEL 14-mer onto EM grids they have visualized the complex after its passage in the gas phase by negative staining. The EM grid was placed to intersect the ion beam and this involved construction of new ion optics and steering lenses within the mass spectrometer (21). The results showed that the characteristic “double doughnut” architecture was preserved but further work was necessary to improve the quality of the EM images. To support the work involved in unraveling the complexity of transient and heterogeneous cellular complexes the Robinson group are also developing a new software package (Massign) for assigning the composition of complexes.
In the review “Joining Forces: Integrating proteomics and crosslinking with the mass spectrometry of intact complexes” (this issue) (22), Stengel, Aebersold, and Robinson outline complementary MS based techniques used in structural biology and provide an overview of how MS is facilitating the understanding of the dynamics and heterogeneity of protein assemblies. The authors highlight in particular the benefits that stem from integrating MS of intact complexes with proteomics, crosslinking and ion mobility MS. Recent studies are described where multiple MS approaches have unraveled the composition, architecture, and dynamics of a heterogeneous protein assembly. In addition, the paper demonstrates how integrating modern MS based technologies with advanced homology and coarse-grained modeling approaches holds great promise to fill the gap between high resolution x-ray analysis of recombinant complexes and low resolution EM structures of cellular assemblies. Such integrated MS based approaches raise expectations for satisfying the growing demand for hybrid technologies that are able to complement classical structural biology methods and thereby enable study of transient, dynamic, or polydisperse assemblies.
Deep Coverage of the Proteome
One of the perceived limitations of conventional proteomics, especially in comparison with genome-wide next generation sequencing technologies, has been the restricted depth of coverage when attempting to analyze complete cellular systems. Although this has not been an obstacle for determining either specific protein-protein interactions, or for identifying the components of purified protein complexes or organelles, proteomics approaches typically could not match the performance of transcriptomics based methods for cataloging complete gene expression landscapes. Recently, however, because of spectacular progress in MS-based proteomics technology, this situation has finally changed and very recently two PROSPECTS groups have published the identification of more than 10,000 different proteins in single-cell-line systems (23, 24). Indirect evidence indicated that this is already close to describing almost the entire proteome of a single cell line. Geiger et al. now report the identification of the proteome of each of 11 commonly studied cell lines (25). Making use of the high sequencing speed of a prototype of the Orbitrap ELITE mass spectrometer, which is described elsewhere in this issue (6), they cut down the MS measurement time required to 1 day per proteome for each cell line, while still achieving an equivalent depth of proteome coverage to the two previous tour de force projects described above. Remarkably, this project involved only about one month of total measurement time for the triplicate analysis of the 11 cell lines. This is now comparable to the time and resources currently required for an equivalent transcriptome measurement by RNA-seq (24).
A comparison of the label-free quantification data from the 11 cell lines revealed that qualitatively their respective proteomes are rather similar. This supports earlier findings of the Uhlén group, which also indicated that gene expression differences between cell lines are more a matter of degree, rather than on-or-off changes (26). Nevertheless, analysis of the results clearly revealed drastic quantitative changes even in household proteins and remnants of the functions of the original cell types from which these cell lines were derived. For example, there was relative overexpression of proteins with immune functions in Jurkat cells and metabolic and complement proteins in a hepatocyte cell line. The data also allowed detailed investigation of the notion of a “household proteome” for the first time. Even proteins involved in functions needed by all cells varied considerably in expression levels (25). Indeed this was true of an estimated 60–70% of the proteome overall. It will be interesting to determine whether similar proportions will be found also for the proteomes of different cell types in vivo.
Proteomic data such as these are of general interest for the cell biology community. In principle they enable researchers to check the expression level of each protein of interest in the cellular model that they employ. However, to achieve this utility, proteomics results must not be confined to large spreadsheets, where they would be difficult for biologists to access. With this in mind, this special issue introduces the MaxQB database (27), an online, searchable resource that contains the above mentioned 11 cell line proteome data. MaxQB features a modern and scalable database architecture and was, from the start, designed for high resolution, high confidence, and quantitative proteomics data. For each protein, or collection of proteins, MaxQB visualizes estimated absolute expression levels, as well as expression across the 11 cell lines. Results of a recent quantification of proteome levels of 26 mouse organs obtained from the SILAC mouse (28) are also provided and expression data of human-mouse homologs can be queried directly. For the proteomics community the peptide information can be useful as well, as it contains reference high resolution HCD spectra of peptides derived from proteins covering more than half of the genes in the human genome. Analysis of these peptide data in MaxQB revealed that the MS signals of the peptides identifying a given protein are highly consistent, with rank order coefficients generally in the range of 0.8. When this is not the case, for instance if the correlation is negative, then the protein is likely misidentified.
Parts of the proteomics community have recently advocated a chromosome by chromosome approach to characterizing the human proteome, where each chromosome is allocated for study by laboratories working in a specific nation, or group of nations (29). Although there is active discussion whether or not this is a good idea for shotgun MS-based approaches (which cannot easily be directed to specific chromosomes), it is clearly a good match for the antibody-based proteome approach mentioned above. In the PROSPECTS consortium, Uhlén and coworkers report the first largely complete chromosome project, the antibody based characterization of genes encoded on chromosome 21 (Uhlen et al., this issue) (30). PrESTs were used to generate antibodies against as many of the 240 ENSEMBL annotated genes on this chromosome as possible. Confocal microscopy was combined with immunohistochemistry and with transcript profiling using next generation sequencing data. A new method to independently detect isoforms, using a combination of antibody-based probing and isoelectric focusing, is also described. Interestingly, this major effort identified several new genes for which there had not been any previous evidence at the protein level. The project led by Uhlén and colleagues thus serves as a model for future efforts on larger chromosomes.
Proteins in Space and Time
The respective expression levels of proteins and their subcellular location are major determinants of cell function that can vary rapidly in response to a wide range of regulatory events and signaling mechanisms. Cells can often regulate protein abundance by post-transcriptional mechanisms affecting rates of protein synthesis or degradation, rather than by changing the rate of transcription of the cognate genes (31). An important goal of the PROSPECTS network has thus been to develop and apply new methods for the quantitative analysis of key properties of proteins, such as their subcellular localization, abundance and turnover rates, at a proteome-wide level.
One approach to studying the subcellular distribution of proteins is to purify specific organelles and characterize their protein composition in detail. This is illustrated, for example, by previous work from the Lamond, Andersen, and Mann PROSPECTS groups on studying the protein composition of human nucleoli, which introduced the concept of “time-lapse” proteomics where the organelle proteome was repeatedly evaluated at a series of time points following perturbation of the cells with drug treatments (32–34). An inventory of organelle components can uncover new functional interactions, which can in turn help to explain the molecular mechanisms of disease genes and can be used to identify causative genes mutated in affected families by targeted exome sequencing. This is illustrated within PROSPECTS by the proteomic characterization of the human centrosome by the Andersen and Mann groups (35), which has subsequently revealed regulators of centriole and cilia biogenesis and pinpointed genes associated with severe human diseases and developmental defects, known as ciliopathies (36). An important factor was the identification of protein candidates with high confidence and that their subcellular localization was correctly assigned. This was achieved for the centrosome studies using sucrose gradient centrifugation and protein correlation profiling (PCP) of collected fractions, combined with label-free MS-based protein quantitation. Recently, the Andersen and Uhlén groups in PROSPECTS have shown that even higher spatial resolution can be achieved with SILAC-based protein correlation profiling (PCP-SILAC), as validated by subcellular localization using complementary strategies based on antibodies from the Human Protein Atlas (37).
In this issue, Dengjel et al. (38) have applied a similar, cell fractionation based PCP-SILAC strategy to identify proteins associated with autophagosomes, which are organelles important for the execution of autophagy. Autophagy takes part in a wide variety of normal physiological processes, including energy metabolism, organelle turnover, and growth regulation. The realization that impaired autophagy contributes to diseases such as cardiomyopathy, neurodegeneration, microbial infection, and cancer has recently spurred an increase in autophagy related research. While these efforts have greatly improved our understanding of basic autophagy biology much still remains to be learned about this physiologically important process. For example, although functional and genetic screens, protein-protein interaction analysis, and biochemical studies have revealed important regulators of autophagy, we still do not know how many of these factors function at the molecular and cellular level and how they contribute to the formation of autophagosomes. Dengjel et al. address these questions using spatiotemporal proteomics in combination with genetic approaches to study the dynamic protein composition of autophagosomes induced by different stimuli. These studies were particularly challenging, both because of the exceptionally dynamic nature of autophagosomes, which are continually formed and rapidly degraded, and because of the morphological resemblance between autophagosomes and other cellular vesicles.
To overcome these challenges, the PCP-SILAC method was applied to distinguish bona fide autophagosomal proteins from nonspecific, copurifying proteins that contaminate the biochemical preparations. These studies were complemented with the analysis of affinity-purified autophagosomes and the quantitative analysis of autophagosomes whose formation was induced by different stimuli and studied at different time points after induction. These experiments resulted in identification of a core set of autophagosomal protein candidates. The subcellular location in autophagosomes of candidate proteins was validated independently, which revealed cross-talk with the proteasome system. Regulatory proteins were further identified using complementary genetic screens in yeast and they were then further tested using assays for functional autophagy in human cells. The resulting information from these combined spatiotemporal proteomic and genetic data sets provide an important new resource for further characterization of autophagosome biogenesis and function.
Although organelle-based approaches provide valuable information about specific subcellular compartments in isolation, it is also important to study the protein composition of organelles in the context of the whole cell to obtain a system-wide view of proteome organization and dynamics. Recent work carried out in the PROSPECTS network has developed SILAC-based “spatial proteomics” approaches for quantifying the subcellular distribution of the proteome throughout the cell and for measuring rates of change in proteome localization in response to cellular responses such as DNA damage (39). This was achieved by SILAC-encoding specific subcellular compartments combined with cell fractionation. The ratio of SILAC isotope values for each peptide then reflects the relative levels of each protein in the respective compartments. By comparing parallel cell lines that differ only in the presence, or absence, of the p53 gene, it was possible to identify the subset of proteins whose relocation in response to DNA damage was p53-dependent (39). A previous collaboration involving the Andersen, Mann, and Lamond PROSPECTS groups showed that pulse-SILAC could be used to measure the turnover rates of human proteins in the nucleolar compartment (40). Now in this issue Boisvert et al. combine these pulse SILAC and spatial proteomics approaches to perform the first system-wide, quantitative study of protein half-lives that compares relative protein abundance with the rates of synthesis, degradation, and turnover of untagged, endogenous human proteins in specific subcellular compartments (41).
The study employed a pulse-SILAC and data analysis strategy whereby HeLa cells grown in “medium” label were pulsed with “heavy” label for varying times. At each of the time points analyzed, an equal number of HeLa cells from the heavy-medium pulse were mixed with unpulsed HeLa cells grown in normal “light” label. This provided an internal reference to establish measurement error limits and it allowed calculation of protein synthesis and degradation levels, as well as the net rate of turnover/replacement. Protein half-lives were also calculated from the intersection points of the separate protein synthesis and degradation rate curves and compared with the 50% turnover values obtained for 50% heavy/medium isotope replacement. For every time point analyzed, the mixture of 50% light isotope unpulsed cells and 50% heavy-medium pulsed cells were also fractionated into cytoplasmic, nucleoplasmic and nucleolar fractions and the SILAC values determined independently for proteins in each compartment, as well as for the whole cell. Using this approach, they quantified 8041 HeLa cell proteins.
The time taken for 50% protein turnover did not correlate with protein properties such as molecular weight or net charge, and also did not correlate with the specific amino acids present at either the amino or carboxyl termini of the proteins. However, turnover values did correlate with protein abundance, such that highly abundant proteins showed on average longer half-lives than low abundance proteins. Consequently, although the modal time for 50% protein turnover was about 20 h, it was calculated that roughly 24 h are required for 50% turnover of the entire HeLa proteome, because of the fact that the most abundant HeLa proteins had longer half-lives. This closely correlated with the 24.7 h cell division time measured empirically for these HeLa cells growing in the SILAC culture media. A comparison of protein turnover in the nucleus and cytoplasm showed that although most proteins found in both compartments have similar rates of turnover, consistent with extensive shuttling and nucleo-cytoplasmic transport, clear examples were identified of classes of proteins whose turnover rates varied according to location, including ribosomal proteins and RNA polymerase subunits. An important general feature emerged that the protein subunits of large multiprotein complexes that are assembled in a different compartment to where they function (such as ribosome subunits), are degraded much faster in the assembly compartment than in the functional compartment.
As already mentioned above, an important component of exploiting large scale proteomics data sets for biological knowledge discovery is the creation of software and user friendly graphical user interfaces to provide convenient tools for data visualization and analysis. The PepTracker software environment (www.peptracker.com) was developed as part of the PROSPECTS network to facilitate convenient management, analysis and correlation of information across large scale proteomics data sets and multiple experiments annotated with consistent metadata features. Boisvert et al. report the creation of a viewer within PepTracker to provide online access via a web-based interface to the HeLa turnover data, annotated at the peptide level (http://www.lamondlab.com/turnover/) (41). The application provides flexible search facilities and interactive charts and graphs allowing users to drill into the data from whole cell to subcellular compartment and protein fractionation level and from whole protein data to the measurements made on every individual peptide identified for each protein.
The protein turnover data illustrate that measurements of values made on a population of proteins (such as a whole cell extract), can mask the fact that distinct pools exist within the population that behave differently (as seen for the turnover rates of ribosomal proteins in the nucleus and cytoplasm). A collaborative study between the Lamond and Uhlén groups in the PROSPECTS network has therefore addressed the task of detecting distinct functional pools in protein populations, making use of the detailed metadata annotation of the above described large scale HeLa cell spatial proteomics turnover data (Ahmad et al., this issue) (42). They mined the turnover data for examples of either protein isoforms or distinct functional pools of the same protein that correlate with differences in measured protein properties, such as subcellular localization, post-translational modifications, or turnover values. Parallel approaches systematically compared SILAC isotope ratio values for subsets of peptides assigned to a protein with the mean ratio value for all peptides assigned to that protein, on the basis that isoforms will share some, but not all peptides. Therefore, where the SILAC isotope ratios can be related to protein properties such as localization (39), or turnover (41), subsets of peptides may have statistically different mean values compared with the mean value calculated using all of the peptides mapped to the same open reading frame, if more than one isoform is produced and the isoforms differ in the measured property, (such as localization). Ahmad et al. (42) showed multiple examples where isoforms were predicted by detailed analysis of the SILAC data at the peptide level and they used antibodies generated by the Uhlén group in the Human Protein Atlas Project to independently confirm the existence of such isoforms with different turnover rates and localization patterns. This provides a new approach for the functional characterization of the proteome based on direct protein detection and experimental measurements of protein properties, rather than relying on inferences based on high throughput RNA expression studies.
Translational and Modeling Applications
A specific aim of the PROSPECTS network is the development and application of proteomics methods that can address problems of clinical significance. This was already illustrated above by the work from the Andersen group characterizing centrosome proteins and thus helping to identify genes linked to ciliopathies.
Inhibition of heat shock proteins has been shown to be a clinically promising avenue in cancer treatment (43). However, there are many suggestions how selective toxicity of cancer cells is achieved, and in the absence of accurate, proteome-wide measurements, it was difficult to separate minor from major effects on a cellular scale. Here, the Hartl and Mann groups report a SILAC based screen for proteome and phosphoproteome changes resulting from application of a prototypical HSP90 inhibitor (Sharma et al., this issue) (44). Bioinformatic analysis of the results in the Perseus environment, which is a module of MaxQuant, clearly indicated a heat shock response, as expected, but also indicated activation of proteolytic responses, presumably to degrade irretrievably misfolded proteins. Interestingly, kinases together with proteins involved in DNA repair emerged as the protein classes and functions most vulnerable to HSP90 inhibition. Concordantly, changes at the phosphoproteome level were highly asymmetric, with many more sites down-regulated than up-regulated. This supports a rationale for combination therapy involving HSP90 inhibition, because it will be very difficult for cancer cells to acquire mutations that protect them from such broad down-regulation of cellular signaling events. This study also illustrates how proteomics can uniquely inform drug therapy: as shown here, it directly picks up effects on the proteome even though they may not be mediated through transcript level changes. Furthermore, it can provide a rational framework for understanding not only the nature of functional proteome changes at a systems-wide level, but also provide a list of the specific proteins involved in the process together with the quantitative magnitude of their changes.
Small molecule inhibitors of protein function are widely used as therapeutic agents but also have important uses as chemical tools for dissecting cellular mechanisms. Quantitative proteomics strategies provide an unbiased approach that can systematically evaluate both direct and indirect effects of drug treatments and thereby reveal unanticipated new aspects of cell regulation and molecular interactions. A collaboration between the Lamond and Uhlén groups in PROSPECTS has used an unbiased SILAC screen to characterize novel interactions revealed using MLN4924, a small molecule inhibitor of NEDD8 conjugation (Larance et al., this issue) (45).
NEDD8 is a ubiquitin-like small peptide modifier that can be post-translationally conjugated to lysine residues by specific E1 and E2 enzyme complexes. MLN4924 is a small molecule inhibitor of the NEDD8 E1 enzyme complex that is in clinical trials as an anticancer agent (46). It rapidly blocks NEDDylation of substrate proteins, including Cullin E3 ligases, which are the major known targets regulated by NEDD modification. A recent large-scale study used combined proteomic and transcriptomic methods to identify 38 proteins important for the cytotoxic effects of MLN4924 in human A375 melanoma cells (47).
Larance et al. (45) now report that the stability of both the MRFAP1 and MORF4L1 proteins is dramatically increased upon inhibiting NEDDylation with MLN4924 in a range of human cell lines. The MRFAP1 and MORF4L1 proteins are direct interaction partners and were shown to be among the most rapidly degraded cell proteins, both here and in the large scale HeLa turnover study (41). Using SILAC-based immunoprecipitation and data analysis strategies optimized in work previously carried out in the PROSPECTS project (48), Larance et al. (45) interrogated the protein-protein interaction networks for both proteins, which showed that the MORF4L1 protein binds in a mutually exclusive fashion to either MRFAP1, or to MRGBP and other protein components of the NuA4 histone acetyl transferase complex. The NuA4 complex is recruited to chromatin by MORF4L1, where it can acetylate histone H4 and alter chromatin compaction levels. This prompts a model in which MRFAP1 can contribute to regulation of chromatin structure and gene expression by competing for binding to MORF4L1 and hence preventing it recruiting the NuA4 complex to chromatin.
Support for the mutually exclusive binding model and its physiological relevance was provided by the human tissue expression pattern for MRFAP1, using histopathology data from the human protein atlas project. MRFAP1 is expressed in only a restricted set of human tissues and cell types, including spermatogonia. Detailed microscopy analyses of human testes showed that MORF4L1 was expressed throughout testes development, whereas the MRFAP1 and MRGBP proteins were expressed in a mutually exclusive fashion. The data suggest that recruitment of the NuA4 complex to chromatin may be restricted to late stages of spermatogenesis, correlating with data in rodents showing that hyperacetylation of histone H4 is important for the replacement of histones with protamines in sperm that results in the extensive chromatin compaction found in the small nuclei of sperm cells.
Another major focus of the PROSPECTS network is how quantitative proteomic data can best be used to enhance our understanding of biology through providing the accurate information needed to construct robust models that can predict the behavior of biological systems. The Serrano group is aiming to develop a quantitative understanding of biological systems where one is able to predict systemic features, with the ultimate aim to rationally design and modify their behavior. Signaling networks triggered by TNF-family receptors (TNF-R) are among the model systems studied. This involves either blocking cell proliferation signals or stimulating cell-suicide (apoptosis) signals with specifically designed proteins or peptides. The aim is to enhance our understanding of these networks and develop new therapeutic strategies to treat diseases caused by deregulation of TNF-R signaling networks, including cancer.
In this issue, Szegezdi et al. describe how apoptosis induction by the Tumor necrosis factor family ligand TRAIL can be determined by the specificity properties of the cognate receptors (49). Apoptosis induction by TRAIL involves five receptors belonging to the TNF-R receptor family. Two of these receptors induce apoptosis (DR4 and DR5) and three receptors counteract the induction of apoptosis. Using the protein design software package FoldX that was developed in the Serrano group, receptor selective TRAIL variants were created that reduce the complexity of this signaling network and allowed the individual contribution of these receptors to be studied. Employing their biochemical network simulator (SmartCell) to simulate TRAIL signaling, they noticed that in a less complex network (for example using a receptor selective variant), apoptosis induction was several-fold faster than in a more complex network (using TRAIL WT), even when the receptor selective variant and WT show the same receptor binding kinetics toward the target receptor. With the TRAIL receptor-selective variant D269H/E195R, they demonstrated that this was indeed the case. This demonstrates in principle that more effective treatments could be achieved using protein therapeutics simply by altering specificity. It also presents a warning when simulating other promiscuous receptor ligand systems (or any other system comprising multiple binding targets): failure to include all the relevant binding partners will severely affect the outcome of the simulation.
Conclusion and Outlook
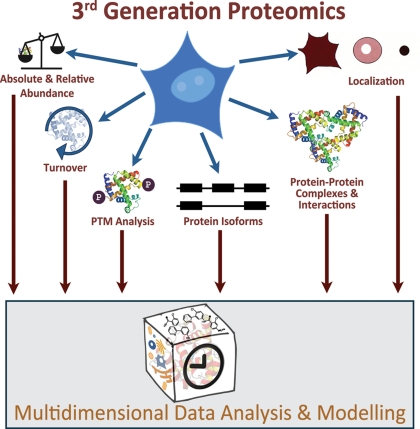
The PROSPECTS project demonstrates that proteomics technology has now become truly quantitative, multidimensional and amazingly versatile. The simple goal of protein identification has thus been superseded by more ambitious aims involving large-scale protein quantification in a panoply of applications (Fig. 2). Unlike transcriptomics, proteomics can directly address the actual levels of proteins expressed in a cell and how these change over time in response to cell signaling events and other perturbations. With further development, we anticipate that direct quantification of the myriad splice variants and post-translationally modified forms of proteins will also become more commonplace. Another conceptual difference from microarray and RNA-seq studies concerns the ability of proteomics to determine the subcellular localization of proteins, both on a large scale and under endogenous conditions. Thus, in our view, proteomics does not simply confirm the results of transcriptome studies, but rather provides unique and indispensible new quantitative information that promises to bring hitherto unprecedented insights into cell biology.
Fig. 2.
An overview of third generation proteomics applied to cell biology. Protein localization, turnover, modifications, isoforms, absolute and relative abundance, PTM, post-translational modifications can increasingly be studied in relation to each other.
Historically, developing the technology to enable efficient protein identification and quantification has dominated the proteomics field and will continue as an important goal for the foreseeable future. This is highlighted within PROSPECTS by the new developments of the Orbitrap family of mass spectrometers that are shown here to improve resolution, accuracy and analysis speed through a combination of hardware and software improvements. The advent of the Q Exactive instrument represents a particularly important advance, creating an extremely powerful and flexible mass spectrometer in a benchtop format. We anticipate that this will promote the incorporation of mass spectrometry instruments and applications directly in many cell biology laboratories, rather than just in specialized facilities and in technology oriented groups. The ability of biology groups to integrate MS-based proteomics technology in their research has also been facilitated through PROSPECTS by the development and free distribution of the powerful MaxQuant software (50).
A corollary of the improved mass spectrometry instrumentation and software that is exploited by the PROSPECTS groups is the ability to perform single-shot, deep proteome coverage with much reduced analysis time. We foresee that this will increasingly lead to the replacement of current methods for routine protein analysis, such as protein blotting procedures, by quantitative MS-based proteomics. Obvious advantages are unambiguous protein identification and the ability to detect large numbers of proteins reliably in a single experiment. As the cost decreases and analysis speed increases coupled to easier access to MS instruments and user friendly software, we anticipate that there will be a large expansion in the use of direct, MS-based protein analysis by biologists. This will have a major impact in cell biology for improving the accuracy and rigor with which evidence based experiments are performed, and hypotheses tested. PROSPECTS has developed and illustrated this potential by the conception of new MS-based methods for quantifying the subcellular distribution of the proteome and its changes in response to cellular responses and perturbations. We predict that quantitative, unbiased MS-based screens will increasingly be used to augment the more traditional genetic screens to identify new interactions and generate new hypotheses that can then be evaluated.
In summary, we argue here that the most exciting future direction of proteomics extends beyond the obvious benefits of continued improvements in high sensitivity protein detection. Recent advances from the PROSPECTS network, including methods for rapid and efficient protein quantification, as well as identification, open the door to a wide range of new applications for the systematic analysis of cell proteins. They promise to revolutionize our understanding of the functional interpretation of genomic information. This leads us into what we can define as the third generation phase of modern MS-based proteomics. The first generation was marked by the development of MS-based technologies for peptide analysis that enhanced the sensitivity of protein identification. The second generation arose from the development of SILAC and other quantitative methods that allowed the use of MS analysis for high throughput measurement of protein dynamics and interactions, rather than just identification. As we now enter the third generation phase, we anticipate that MS-based proteomics will be used routinely to measure absolute protein levels, to survey the dynamic responses of entire proteomes in both space and time and to evaluate the structures of multi-protein complexes and intricate patterns of reversible post-translational modifications. We propose that one of the largest challenges that will arise in this third generation phase will be how best to manage and integrate the large volumes of quantitative proteomics data that will emerge, such as the Super Experiment approach developed within PROSPECTS. This is a challenge best addressed at the community level and the success of the PROSPECTS network illustrates one model in which concerted and collaborative interactions between groups can have a major impact on shaping the future directions of the field.
Acknowledgments
AIL is a Wellcome Trust Principal Research Fellow.
Footnotes
* This work was funded in part by the EU FP7 PROSPECTS network (Grant#: HEALTH-F4-2008-201648).
1 The abbreviations used are:
- PROSPECTS
- Proteomics Specification in Space and Time
- EM
- electron microscopy
- PCP-SILAC
- protein correlation profiling-SILAC
- PrEST
- protein epitope tags
- SIM
- selected ion monitoring.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 3. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 4. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 5. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michalski A., Damoc E., Lange O., Denisov E., Nolting D., Mueller M., Viner R., Schwartz J., Remes P., Belford M., Dunyach J. J., Cox J., Horning S., Mann M., Makarov A. (2011) Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics. doi: 10.1074/mcp.O111.013698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagaraj N., Kulak N. A., Cox J., Neuhaus N., Mayr K., Hoerning O., Vorm O., Mann M. (2011) Systems-wide perturbation analysis with near complete coverage of the yeast proteome by single-shot UHPLC runs on a bench-top Orbitrap. Mol. Cell. Proteomics. doi: 10.1074/mcp.M111.013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Fröhlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Graumann J., Scheltema R. A., Zhang Y., Cox J., Mann M. (2011) A framework for intelligent data acquisition and real-time database searching for shotgun proteomics. Mol. Cell. Proteomics. doi: 10.1074/mcp.M111.013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ludwig C., Claassen M., Schmidt A., Aebersold R. (2011) Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell. Proteomics. doi: 10.1074/mcp.M111.013987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 13. Zeiler M., Straube W. L., Lundberg E., Uhlen M., Mann M. (2011) A protein epitope signature Tag (PrEST) library allows SILAC-based absolute quantification and multiplexed determination of protein copy numbers in cell lines. Mol. Cell. Proteomics. doi: 10.1074/mcp.O111.009613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mann M. (2008) Can proteomics retire the western blot? J. Proteome Res. 7, 3065. [DOI] [PubMed] [Google Scholar]
- 15. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rappsilber J. (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Structural Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leitner A., Reischl R., Walzthoeni T., Herzog F., Bohn S., Foerster F., Aebersold R. (2012) Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics. doi: 10.1074/mcp.M111.014126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pathare G. R., Nagy I., Bohn S., Unverdorben P., Hubert A., Körner R., Nickell S., Lasker K., Sali A., Tamura T., Nishioka T., Förster F., Baumeister W., Bracher A. (2012) The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl. Acad. Sci. U. S. A. 109, 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nat. Advance doi:10.1038/nature10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakata E., Stengel F., Fukunaga K., Zhou M., Saeki Y., Förster F., Baumeister W., Tanaka K., Robinson C. V. (2011) The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol. Cell 42, 637–649 [DOI] [PubMed] [Google Scholar]
- 21. Benesch J. L., Ruotolo B. T., Simmons D. A., Barrera N. P., Morgner N., Wang L., Saibil H. R., Robinson C. V. (2010) Separating and visualising protein assemblies by means of preparative mass spectrometry and microscopy. J. Structural Biol. 172, 161–168 [DOI] [PubMed] [Google Scholar]
- 22. Stengel F., Aebersold R., Robinson C. V. (2011) Joining forces: Integrating proteomics and crosslinking with the mass spectrometry of intact complexes. Mol. Cell. Proteomics doi: 10.1074/mcp.R111.014027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schaab C., Geiger T., Stoehr G., Cox J., Mann M. (2012) Analysis of high-accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.014068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fassler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 29. Editorial (2010) The call of the human proteome. Nat. Methods 7, 661. [DOI] [PubMed] [Google Scholar]
- 30. Uhlen M., Oksvold P., Algenas C., Hamsten C., Fagerberg L., Klevebring D., Lundberg E., Odeberg J., Ponten F., Kondo T., Sivertsson A. (2011) Antibody-based protein profiling of the human chromosome 21. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.013458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 32. Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Current Biology 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 33. Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., Lamond A. I. (2002) Paraspeckles: a novel nuclear domain. Current Biol. 12, 13–25 [DOI] [PubMed] [Google Scholar]
- 34. Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 35. Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 36. Nigg E. A., Raff J. W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 [DOI] [PubMed] [Google Scholar]
- 37. Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L. G., Bennetzen M., Westendorf J., Nigg E. A., Uhlen M., Hyman A. A., Andersen J. S. (2011) Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dengjel J., Høyer-Hansen M., Nielsen M. O., Eisenberg T., Harder L. M., Schandorff S., Farkas T., Kirkegaard T., Becker A. C., Schroeder S., et al. (2012). Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.014035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Current Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boisvert F. M., Ahmad Y., Gierlinski M., Charriere F., Lamont D., Scott M., Barton G., Lamond A. I. (2011) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahmad Y., Boisvert F. M., Lundberg E., Uhlen M., Lamond A. I. (2011) Systematic analysis of protein pools, isoforms and modifications affecting turnover and subcellular localisation. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.013680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitesell L., Lindquist S. L. (2005) HSP90 and the chaperoning of cancer. Nature reviews. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 44. Sharma K., Vabulas R. M., Macek B., Pinkert S., Cox J., Mann M., Hartl F. U. (2011) Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.014654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Larance M., Kirkwood K. J., Xirodimas D. P., Lundberg E., Uhlen M., Lamond A. I. (2011) Characterization of MRFAP1 Turnover and Interactions Downstream of theNEDD8 Pathway. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.014407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 47. Liao H., Liu X. J., Blank J. L., Bouck D. C., Bernard H., Garcia K., Lightcap E. S. (2011) Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Mol. Cell. Proteomics 10, M111.009183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boulon S., Ahmad Y., Trinkle-Mulcahy L., Verheggen C., Cobley A., Gregor P., Bertrand E., Whitehorn M., Lamond A. I. (2010) Establishment of a protein frequency library and its application in the reliable identification of specific protein interaction partners. Mol. Cell. Proteomics 9, 861–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Szegezdi E., van der Sloot A. M., Mahalingam D., O'Leary L., Cool R. H., Munoz I. G., Montoya G., Quax W. J., de Jong S., Samali A., Serrano L. (2012) Kinetics in signal transduction pathways involving promiscuous oligomerizing receptors can be determined by receptor specificity: Apoptosis induction by TRAIL. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]