Abstract
Protein assemblies are critical for cellular function and understanding their physical organization is the key aim of structural biology. However, applying conventional structural biology approaches is challenging for transient, dynamic, or polydisperse assemblies. There is therefore a growing demand for hybrid technologies that are able to complement classical structural biology methods and thereby broaden our arsenal for the study of these important complexes. Exciting new developments in the field of mass spectrometry and proteomics have added a new dimension to the study of protein-protein interactions and protein complex architecture. In this review, we focus on how complementary mass spectrometry-based techniques can greatly facilitate structural understanding of protein assemblies.
Mass Spectrometry of Intact Assemblies
MS is an analytical technique that allows the measurement of the mass-to-charge (m/z) ratio of ions. In its simplest form, it comprises a source to produce ions, an analyzer to separate the various ions according to their m/z ratio, and a detector to count the ions emerging from the analyzer. This simple technique has over the last decades become the foundation for the study of proteins and their modifications. As such biological MS has evolved from being a purely auxiliary technique, used for the analysis of single peptide fragments for protein identification, to a major driving force in large scale proteomics and modern systems biology (1–4).
In parallel with these developments, MS is also emerging as a powerful approach for determining protein-protein interactions, subunit composition (2, 5–9), and architectural organization of intact protein complexes (5, 10–13). Protein-protein interactions have traditionally been investigated by affinity purification coupled with MS (AP-MS).1 AP-MS uses affinity tags to the proteins of interest or antibodies against to pull down the protein of interest and associated interactors, which are then analyzed and identified by proteomics-based MS (2, 5, 14). Using AP-MS, it is therefore possible to investigate the connectivity within a complex or even establish whole protein interaction networks.
By contrast, MS of intact assemblies usually focuses on a single protein complex or at least a relatively well defined assembly. However, for large heterogeneous complexes, MS of intact complexes is then capable of elucidating not only connectivity and mass but to also provide key information on absolute subunit stoichiometry, heterogeneity, and dynamical changes. The discovery in the early 1990s that noncovalent interactions could be maintained intact in the gas phase by the use of ESI (15) and the introduction of specialized instrumentation (16) and reliable protocols (17) enabled the use of MS in the study of intact complexes. These advancements paved the way to an area of research known as native MS or MS of intact protein assemblies (for general reviews, see Refs. 10, 11, 13, and 18).
The initial challenges in the analysis of macromolecular protein complexes by MS involved finding conditions for maintaining interactions. The protein complex, made up of multiple protein subunits and kept together by weak, noncovalent interactions, must survive the desolvation and ionization processes. Following these steps, the complex must be transferred intact into the gas phase. Finally, it has to pass through the instrument without dissociation and impinge upon the detector with sufficient impact for detection. Additionally, controlled dissociation is often required to elucidate fully the overall composition of the intact complex.
Many of these initial challenges have now been overcome. Briefly, gentle ionization takes place at atmospheric pressure, typically via nanoflow electrospray from buffered aqueous solutions to preserve the noncovalent interactions of the intact assembly (17). Ionized protein complexes, encapsulated in small droplets, then experience a free jet expansion during their traversal to the rough vacuum of the instrument. Here, optimization of instrument conditions, namely pressure and accelerating voltages, is required to transfer the protein complex of interest intact into the gas phase. This allows for its traversal through the instrument without dissociation. Because of their high impact, very large ions can reach kinetic energies of tens to hundreds of electron volts and consequently become defocused (19). Moreover, because of their very high kinetic energies, these ions cannot be focused by typical ion optics. The passage of protein complexes through the mass spectrometer therefore requires higher pressures in the instrument than one would normally use, in a process known as collisional cooling or focusing (20, 21). This results in collisions between large ions and small neutral gas molecules that now convert some of the kinetic energy into internal energy and help to focus the ion beam (19).
Having maintained all the interactions within the complex, an initial spectrum of the intact assembly is then recorded. This provides information on the overall subunit stoichiometry and homogeneity. Controlled activation and dissociation of the intact complex is then carried out. This permits interrogation of subcomplexes and protein-protein interactions, either through gas phase dissociation or solution-based disruption. Numerous techniques for controlled gas phase dissociation of protein complexes have been developed (22, 23). In collision-induced dissociation, the most commonly applied, dissociation is achieved by subjecting the protein complex to collisions with an inert gas within a collision cell. The exact mechanism of dissociation is the subject of much ongoing research (24–27) and is only discussed briefly here. From the study of a large variety of protein complexes, a predominant dissociation pathway has started to emerge, although exceptions have also been found. An increase in internal energy caused by collision events enables local unfolding, which increases the surface area. Mobile charges redistribute to this area to minimize coulombic repulsion, thus destabilizing further the regions close to the unfolded section. This leads to further disruption of tertiary and secondary structure until the subunit is essentially unfolded. The complex then dissociates in a sequential fashion by the expulsion of successive monomers, yielding a highly charged monomer and a relatively lowly charged “stripped” oligomer. Consequently, the overall charge of the parent ion of the complex limits the extent of asymmetric dissociation that can occur. Moreover, because mass is intrinsically coupled to the charge of the complex (28), this effectively limits the number of successive dissociation events. Consequently the amount of information that can be gained by gas phase dissociation alone is limited by the maximum charge that can be acquired on the surface of an intact complex.
Solution-based techniques take advantage of the sensitivity of subunit interfaces to changes in ionic strength, hydrophobicity, and pH (29). This parameter is often difficult to predict, and as a consequence optimal solution conditions, which effect subcomplex formation without precipitation, are usually found by trial and error. Once generated, these subcomplexes can be transferred intact into the mass spectrometer. Because these subcomplexes are not generated by gas phase dissociation, they will carry more charge than their collision-induced dissociation-derived siblings, allowing us to establish their composition by collision-induced dissociation. Subsequently if gas phase and solution-based information is combined, a complete picture of protein-protein interactions within a protein complex can be attained (29–32).
Because of its ability to provide key information on a wide range of intact high molecular mass complexes, including those complicated by polydispersity and dynamics (33), this approach is being applied increasingly in many areas of structural biology. Recently this has included complexes as large as intact viruses (34), as challenging as membrane bound assemblies (35, 36), and as complex as the eukaryotic translation factor Elf3 (31), clamp loader (37), ribosome (38, 39), exosome (29) spliceosome (40), RNA polymerase III (41), the anaphase-promoting complex (42), and the proteasome (43).
Although arguably a very powerful technique, MS of intact assemblies alone is often challenged when it comes to identifying component subunits unambiguously. This becomes increasingly apparent for complexes isolated directly from cells, without overexpression. Here often chaperones or other transient co-factors that are involved in their assembly or regulation are present. The main requirement for discounting many possible subunit combinations is a highly accurate mass measurement of the intact species such that unique assignments can be made for the subunit stoichiometry (44). Because conditions in the mass spectrometer need to be sufficiently gentle to maintain subunit interactions, buffer, water molecules, lipids, etc., remain trapped in subunit interfaces, in some cases introducing uncertainty in the mass measurement.
In its classic implementation, the MS of intact assemblies is also not able to define the three-dimensional packing of subunits within a protein complex. More recently, however, it has been demonstrated that ion mobility, a technique originally developed for the separation of small molecules, can also be applied to protein complexes (45, 46). This can help to distinguish various packing arrangements of subunits including ring-shaped or collapsed (47), linear, or close packed (48), or changes caused by the binding of substrate (49). This information can delineate various possibilities and is particularly powerful when used in combination with electron microscopy.
With these limitations in mind, MS of intact assemblies is increasingly being combined with other approaches to generate comprehensive lists of interactors, as well as three-dimensional models of their architecture (5). In this review, we focus on the benefits that stem from integrating the MS of intact complexes with proteomics, cross-linking, and ion mobility MS. Specifically, we highlight how their integration compliments our understanding of the composition, architecture, and dynamics of heterogeneous protein assemblies.
The Synergy between MS of Intact Assemblies and Proteomics
MS-based proteomics is a vast and highly productive field that makes seminal contributions to all fields of biology. As such, it is one of the main drivers of modern systems-based biology. Recent advances and challenges exceed the scope of this article and are reviewed elsewhere (2–4, 14). Here we focus on the interface between MS of intact assemblies and other MS-based methods for the analysis of protein complexes (5, 50).
The first of these integrative studies presented here applied the MS of intact complexes (31) with proteomics (51) to the eukaryotic initiation factor (eIF) 3. This 800-kDa complex is involved in the initiation of protein synthesis through its interaction with the ribosome (52). After a comprehensive proteomics study of this complex (51), MS of intact complexes was used to assess their integration into the intact particle. Each subunit in this 13-subunit complex was found to be present as a single copy. A subunit interaction map was generated by manipulating the ionic strength of the complex-containing solution. This generated 27 subcomplexes enabling the subunit interactions of the intact eIF3 to be established with high confidence. To assign subcomplexes and to generate an interaction network, masses of subunits together with their stoichiometry were submitted to SUMMIT (44), an iterative search algorithm that computes all permutations of a given set of input proteins that sum to the target mass. The subunits are then joined by the shortest network pathway that connects all subunits with their interaction partners. In this way, the 27 eIF3 subcomplexes could be assigned, and the final model was not only consistent with previous studies on the core complex (53) but also revealed the modular assembly of additional complexes and their association with the core.
This “top-down” MS of intact assemblies was combined with a “bottom-up” proteomics strategy, in which the eIF3 was digested with trypsin and phosphopeptides were enriched prior to LC-MS. Here 29 phosphorylation sites were identified and assigned to specific residues within the eIF3 subunits (51). Considering the subunit interaction model from the MS of intact assemblies approach in the light of the proteomics analysis of eIF3 allowed mapping of phosphorylation events to subunit interfaces. Combining both data sets provided knowledge of the subunit interfaces that could be controlled by phosphorylation. Interestingly, all of the phosphorylation sites were located on subunits within the “core” of the complex, implying that these subunits respond to signaling events that promote assembly/disassembly reactions of the eIF3 core.
A major challenge when studying large protein assemblies arises from assigning masses to unique subunit stoichiometries (44). MS of intact complexes is optimal if all of the possible subunits are known, as in the case of a controlled in vitro environment. However, the real biological challenge lies in elucidating the architecture of protein assemblies in their native environment, where additional transient interactors may be present at substoichiometric levels. The problem is further aggravated, if many of the possible interacting subunits have similar masses, as is commonly the case for large protein complexes.
These ambiguities came to the fore in a recent MS study of the proteasome, a 2.5-MDa macromolecular machine responsible for protein degradation in cells (54). The intact 26 S proteasome is known to consist of at least 33 different canonical subunits in yeast contained within two modules: a highly conserved 20 S core particle containing a central catalytic cavity and two 19 S regulatory particles, together with many associated proteins (55). Cryo-electron microscopy (cryo-EM) images have vastly expanded our understanding of the intact 26 S proteasome (56–58, 122). An atomic structure of the full 26 S, however, has yet to be reported, presumably because of its heterogeneity and inherent dynamics. Recent studies have shed light on the assembly process of the 19 S regulatory particle (55, 59, 60) highlighting the role of chaperones in its formation.
To investigate the interaction of these chaperones with the different subunits of the 19 S, a hybrid approach was employed. MS of intact assemblies was used in combination with quantitative proteomics to probe the composition of the intact proteasome and its assembly intermediates (43). The idea behind this study was to improve the subunit assignments of the various intermediates, as well as to exemplify a more general method in which complexes could be defined from heterogeneous environments.
The procedure adopted is summarized briefly here. Following affinity purification of proteasomes, the various assembly intermediates were separated by a sucrose gradient to reduce complexity and to render the complex-containing solution amenable to interrogation by MS. A label-free proteomics approach was then used (56) to identify all proteins present within a particular fraction. This knowledge was subsequently used as an input to assign the stoichiometry of subunits within intact subcomplexes detected by MS of intact protein assemblies (Fig. 1). The results of the proteomics suggested the presence of Ubp6 within an intact base precursor and its role in the assembly of the base and the lid. Only by using this hybrid approach was it possible to solve the stoichiometry of an intact base precursor and assign its stoichiometry unambiguously. Following up on these indications with “classical” genetic and biochemical experiments, the authors could conclude that Ubp6 assists in the assembly of the regulatory particle and its incorporation into the intact proteasome via its catalytic activity in vivo and in vitro.
Fig. 1.
Hybrid approach combining MS of intact assemblies with quantitative proteomics used for the structural characterization of intermediates en route to the assembly of the proteasome. Here the proteasome is used as an example, although the approach is generic. Different aliquots of the same complex-containing purification are assessed by MS of intact complexes and quantitative proteomics. These data, in combination with information gained from in solution disruption experiments, are then used to generate a subunit interactome map. Information from the proteomics experiment is also used to aid the assignment of the spectra of intact complexes. Additional biochemical and molecular biology experiments can add functional information. The combined data shed light on the assembly pathway of the proteasome. This figure is reproduced from Ref. 43 with permission.
This study therefore highlights not only the combination of quantitative proteomics with MS of intact complexes but also its integration with established biochemical or molecular biology methods. In combination, these approaches can provide detailed insights into the assembly pathway of the proteasome and enable real advances of our understanding of an important molecular machine. In summary, combining quantitative proteomics with MS of intact complexes seems like a truly functional relationship; the reference library created by quantitative proteomics enables all potential candidates of a proteasomal assembly to be considered and consequently to anchor the various protein components within a functional module observed by the MS of intact assemblies.
Protein Complexes and Protein-Protein Interaction Networks
As discussed, MS of intact assemblies concentrates on the elucidation of the architectural map of a defined protein assembly. Other MS technologies are able to take a much broader view. In particular, AP-MS has established itself as a versatile player in modern systems biology and is nowadays probably the method of choice when looking at protein-protein interactions on a global scale (for selected reviews, see Refs. 2 and 5–9).
Ground-breaking studies pioneered global views of protein interaction networks (61–63), whereas very recently more than 3000 affinity purifications using different antibodies were used to define the human coregulator complexome (64). Contemporary large scale studies are designed often to address specific questions, such as the basic principles of proteome organization (65). Other focused studies have probed the dynamics of a ubiquitin ligase network (66) or provided a global view of protein kinases and phosphatases (67).
Many workflows have been established (7, 8, 68) based on the successful Tandem affinity purification (TAP) tag approach (69). Recent protocols allow relatively high throughput analysis of human and mammalian cell lines (64, 70, 71). Nowadays efforts often concentrate on getting high quality data sets (72) and quantitative information (73–75). However, when using AP-MS to study biologically relevant protein assemblies, additional biochemical fractionation or other purification techniques are usually required (5).
Combining the isolation of a complex with some quantification is a requirement to gain information on the stoichiometry of a given complex. If all the components of a complex are known, then synthetic peptides can be used for quantification (52, 76, 77) (for different strategies, see, for example, Ref. 5). However, quantitative interaction proteomics usually requires identifying, synthesizing, and validating isotope-labeled proteotypic peptides, which can be highly cost-intensive. Here, label-free approaches can pose attractive alternatives (78). Quantification also helps with a classical problem of AP-MS: that of distinguishing true interactors from the background. This shortcoming can either lead to a high rate of false positives or, if too stringent purification protocols are employed, a loss of weak and/or transient interactors. Quantitative interaction proteomics has been shown to discriminate efficiently between specific interactors and mere background without the drawback of requiring too stringent a purification procedure (74, 79, 80).
Two examples illustrate the power of quantitative interaction proteomics. One study combined AP-MS with multiplex absolute quantification technology in an elegant approach that served to elucidate the stoichiometry and architecture of the cullin-RING ubiquitin ligase network (66). Classic AP-MS provided the authors with a systematic proteomic analysis of the cullin-RING ubiquitin ligase network at steady state. The authors subsequently developed a multiplex absolute quantification platform with a library of 38 tryptic peptides, consisting of peptides that had been previously observed by LC-MS/MS for each of the cullins. These 38 peptides provided sufficient sequence coverage to generate a quantitative picture of cullin-RING ubiquitin ligase architecture upon deneddylation and to determine the occupancy of individual subunits within the network.
Other improvements toward a robust and affordable quantitative interactions workflow include an additional peptide, known as SH-quant. This is integrated into the affinity tag and is released upon tryptic digestion, and subsequently used for quantification (73). Isotope-labeled SH-quant peptides are added to calculate the amount of bait protein and to provide a correction factor for potential losses during sample handling. The attraction of this approach lies in the fact that once a protein has been used as a bait and its absolute quantity determined, its proteotypic peptides can serve as a calibration curve. The protein can then be quantified in any other affinity purification study by a process the authors term “correlational quantification.” Such an approach is extremely powerful when one major protein complex is studied with multiple pulldowns. The authors tested their correlational quantification approach by investigating the human protein phosphatase 2A (PP2A) interaction network (Fig. 2). Using this method, the interaction stoichiometries and complex composition of PP2A under different growth conditions were investigated. Very subtle changes in protein-protein interactions could be detected. For example, it was possible to detect that the regulatory subunit of PP2A not only interact with its catalytic counterpart, but also, in less than 2% of all cases, with the catalytic subunit of the protein phosphatase 4.
Fig. 2.
Quantitative interaction proteomics. A, quantitative representation of a PP2A protein interaction network. The thickness and color of the edges corresponds to the amount of prey protein normalized to the amount of the respective bait protein. Each arrow represents an average of two purifications. PPP2CA and PPP2CB cannot be distinguished in the quantification calculations because they have 97.7% identical amino acid sequence. B, average abundance of proteins interacting with PPP2R1A from two purifications normalized to the abundance of the bait in untreated cells or those treated for 3 h with 100 nm okadaic acid. C, abundance of canonical PPP2C-PPP2R1A/PPP2R2A complexes compared with hybrid PPP4C-PPP2R1A/PPP2R2A complexes. The estimation relied on the stoichiometry data from the PPP2R1A and PPP2R2A purifications. This figure is reproduced from Ref. 73 with permission.
Quantitative interaction proteomics at first glance would appear to be a congenial partner for the MS of intact assemblies. Both are able to assess stoichiometries of protein complexes. However, to date this combination has not been realized. One problem lies in the different concentrations that are needed. For the MS of intact complexes, 1–2 μl of submicromolar concentration solutions are in theory required. Handling such low quantities without significant losses, however, becomes problematic, often necessitating larger quantities of protein complexes. Lower quantities of protein are typically required for AP-MS. However, in combination quantitative interaction proteomics could enable placing the architectural map of an intact protein assembly into a wider context.
Merging MS of Intact Assemblies with X-linking
Cross-linking as a technique, where two or more reactive groups are linked covalently, has been a well established biochemical technique for many years (81). Moreover, coupling cross-linking with mass spectrometry (CXMS) has been demonstrated as a working concept for more than a decade (82). In the standard workflow, the protein complex is cross-linked and then enzymatically digested. This is usually followed by an enrichment step to improve the yield of cross-linked peptides. After MS analysis, the data need to be analyzed to identify cross-linked peptides. This final step is one of the biggest bottlenecks in modern CXMS.
In the course of this last decade, multiple improvement steps to this protocol have been introduced (83), and significant advances have been made in this field (see recent reviews in Refs. 84–90). This progress is set to build on ever more powerful mass spectrometers with increased sensitivity, mass resolution, and accuracy. Today it is possible to study multiprotein complexes that are difficult to study with conventional structural techniques. For example, CXMS was used to map two coiled-coil domain-containing complexes, which often pose problems for crystallography (91). Insights gained from this CXMS study helped with the construction of a recombinant “bonsai” crystallized version of a kinetochore complex, the 176-kDa, four-protein Ndc80 complex (92). Other very large protein complexes such as the GroEL-GroES system (93) or the RNA polymerase I (94) have already been successfully probed by CXMS. In this last benchmark study, the 15-subunit, 670-kDa complex of Pol II in complex with the initiation factor TFIIF was examined. The interaction contacts were identified at the level of individual peptides, clearly establishing CXMS as a highly valuable tool for studying multiprotein assemblies (94).
Despite these remarkable achievements, the number of possible cross-links invariably exceeds the number of experimentally observed cross-links by at least one order of magnitude (84). Moreover, despite ongoing efforts to improve the bioinformatics (91, 95–100), the complexity of processing and interpreting cross-linked data sets remains the critical point of the CMXS approach. For further reading and also for in-depth discussion on the related problem of how to evaluate the confidence of a cross-link and how to reduce the number of false positives, see recent reviews in Refs. 84, 89, and 101. Briefly, a database of possible cross-links has to consider every residue that is capable of being cross-linked in the actual experiment and then calculate all possible pair-wise combinations. This leads to (n2 + n)/2 possible cross-links for n peptides. This problem makes it very difficult to search a potential cross-link against a large unbiased database, because the risk of a random match increases, and computation becomes very expensive (84, 101). One strategy is to use cross-linkers that contain labile bonds that fragment during MS analysis to yield reporter ions. This has two advantages. On the one hand a reporter ion is released facilitating the identification of spectra containing cross-linked peptides. On the other hand, the released peptides, which were formerly cross-linked, fragment independently of each other and can be searched and identified using standard database search algorithms (102). Using such a strategy, it was possible to map protein interactions in Escherichia coli and to measure structures and topologies of proteins and complexes in vivo (103).
A powerful solution to the identification of cross-linked peptides is xQuest, a software suite originally developed in the Aebersold group (95). Here, isotopically labeled cross-linkers are used to guide the search for cross-linked peptides by their characteristic isotopic shift in their MS1 precursor ion spectra before they are subjected to separate MS2 sequencing. By comparison of the two MS2 spectra, common pre-cross-link and post-cross-link fragment ions can be identified. The power of this approach lies in the fact that it allows the combination of a low-stringency search for candidate peptides using those common ions followed by stringent spectrum matching using the full reconstructed MS2 spectrum against the compacted library of identified candidates. With this software, it was possible to identify cross-linked peptides in bacterial whole cell lysates using an unrestricted database search (95).
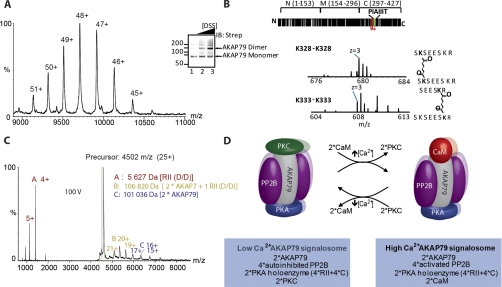
In cases where the identity of potential interactors is limited, and in principle known, as it is for many protein complexes, this problem is simplified. Only those proteins present in the complex need to be considered (101). This was the case in a recent study where the MS of intact assemblies was combined with CXMS to study the architecture and dynamics of a major signaling complex (104) (Fig. 3). Here, MS of the intact AKAP79 assemblies, expressed recombinantly, established the dimerization of AKAP79. It was also possible to demonstrate that one AKAP79 molecule binds to two fragments of its regulatory subunits. Using a CMXS strategy that employs the lysine linker disuccinimidylsuberate, dimeric AKAP79 cross-linking sites were identified with xQuest after selecting for homodimeric peptides. This procedure was used to verify the dimerization properties of AKAP79 identified in the MS of the intact complex. The detection of symmetrical cross-linked pairs by CXMS suggested that the AKAP79 dimer is aligned in parallel. Additional validation was carried out using in silico digestion of AKAP79 and consideration of all possible cross-linked peptides within a certain threshold. This confirmed the cross-linked peptides indentified by xQuest and provided further evidence for this unexpected dimerization of AKAP79. With the stoichiometry of this subcomplex firmly established, it was possible to assign the quaternary structure to the mass determined for the intact AKAP79 signaling complex. This study serves to highlight how combining MS of intact assemblies with CXMS not only adds an internal validation to protein-protein interactions at the subunit level but also helps to integrate the precise location of an interaction within the landscape of a multiprotein assembly.
Fig. 3.
Combining MS of protein assemblies with CXMS. A, mass spectrometry of intact complexes reveals the stoichiometry of the intact AKAP complex: 2 × [AKAP79 + PP2B (A+B) + 2RII D/D + CaM] and demonstrates the dimerization of AKAP 79. B and C, confirmation of the AKAP dimer by chemical cross-linking and MS also suggests that the dimer is aligned in parallel. D, a proposed model for the architecture and dynamics of the core AKAP79 signaling complex, in conditions of both high and low [Ca2+]. The second AKAP79 monomer and associated proteins are in the background. These figures are reproduced from Ref. 104 with permission.
Both CXMS and MS of intact complexes are on many levels complementary. It is easy to see how a combined use would be mutually beneficial. Both techniques are able to identify protein-protein interaction partners with high fidelity, one at peptide resolution (CXMS), whereas the other is able to solve the absolute stoichiometry of the intact complex (MS of intact assemblies). Moreover, both techniques can be applied to heterogeneous environments, caused by multiple conformations, additional protein impurities, or polydispersity of the complex itself. Even more importantly, both methods are able to provide data on large protein assemblies in their native and quaternary structure. This is immediately apparent for CXMS, where the cross-linking event takes place in solution itself (84). For MS of intact assemblies, it is also now well established that aspects of the native conformation are retained in the gas phase (11, 105), allowing the stoichiometry of protein subunits within intact assemblies to be defined. In the next section, we consider how this can be extended to include topological information.
Combining Topological Information with EM
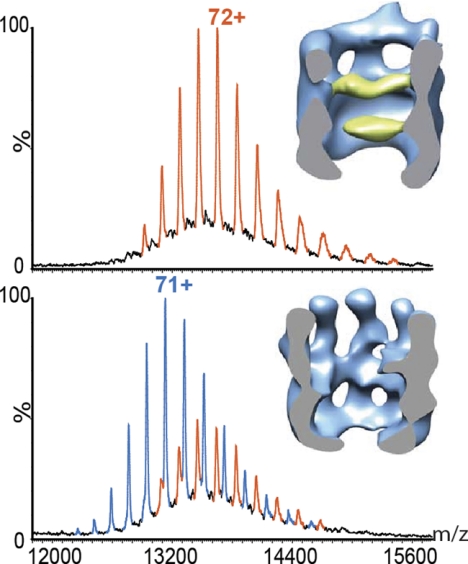
Traditionally, ion mobility has been applied to elucidate shape and conformation of small molecules and single proteins (106). Over the course of the last few years ion mobility (IM)-MS has been successfully applied to the study of macromolecular assemblies (for recent reviews, see Refs. 45 and 46). In a landmark paper it was shown that the quaternary structure of a large ring-shaped protein complex could be maintained in the gas phase of a mass spectrometer (47). This study investigated the 11-subunit TRAP (trpRNA-binding protein) and found that different topologies of the complex were induced by the binding of tryptophan and the addition of a specific RNA molecule. These different sized protein complexes were maintained in the absence of bulk solvent and could be separated by IM-MS. This study therefore established IM-MS as a tool with which the shape and topology of intact protein assemblies can be investigated.
In IM-MS, ions are separated not only according to their m/z ratio but also by their ability to traverse a region of neutral gas, where ions are separated under the influence of a weak electric field. Large ions will experience more collisions with the inert gas molecules than small ones. They therefore need a longer time period to drift through this region of the mass spectrometer. The drift time that is measured for ions of the same charge state is directly linked to an orientationally averaged collision cross-section, thus providing information on the overall shape of a protein or complex (107, 108). Experimental collision cross-sections from ion mobility measurements have been shown to be consistent with the crystal structure of proteins (47, 109–111). This makes IM-MS an attractive tool for probing the topology of complexes, particularly those for which there is only limited high resolution structural information.
For the protein complex of interest, a number of potential models are generated using molecular modeling. Candidate structures are then filtered according to distance and connectivity constraints imposed by IM-MS data (109). This approach is particularly powerful when used in conjunction with other structural information, for example an atomic structure of one of the subunits. Used either as a stand-alone method or in conjunction with other structural biology approaches, IM-MS is beginning to generate model structures of large protein complexes (31, 46, 48, 49, 110, 112, 113). An example of this hybrid structural approach was a study where classical proteomics, MS of intact complexes (31) and IM-MS were combined to place subcomplexes within a low-resolution EM density map of human eIF3 (114). Here, IM constrains were used as an input to distinguish between linear and close packed trimers and consequently to place these within EM density maps (31).
A recent study of recombinant anaphase-promoting complex or cyclosome (APC/C) (42) also benefitted from combining MS with EM. This multisubunit protein complex is a key regulator of the cell cycle (115). EM studies were able to improve our understanding of the three-dimensional structure of this important cellular machine (42, 116, 117). Because of the absence of a structure at atomic resolution, however, ambiguities in subunit localization remained. MS of intact assemblies and subcomplexes was used to obtain a quantitative estimate of the mass and subunit stoichiometry of a recombinant subcomplex of the APC. Combining these data with data derived from other sources, namely crystal structures on single subunits and dimeric complexes, enabled an estimate of the exact subunit stoichiometry for almost all components of the intact APC. Combining knowledge on stoichiometry with molecular docking of crystallographic and homology-derived subunits resulted in the generation of a pseudoatomic model for 70% of the APC/C (42).
Other examples where mass spectrometry has been able to complement EM include an interesting combination with CXMS applied to the 26 S proteasome from Schizosaccharomyces pombe (58). This study combined spatial information from cross-linking with a EM density map. This spatial information arises because to be cross-linked, residues must be within a certain distance of each other. This is determined by the cross-linking agent and the flexibility of the linked amino acid (84, 85, 101, 103). A cross-link between two known residues therefore provides a valuable experimental constraint for modeling. In particular a cross-link between Lys49 in the coiled-coil region of Rpt3 and Lys281 of Rpn11 was used to help clarify the spatial relationship between these subunits. As such, it was possible to constrain the topology of the AAA-ATPase module within the 19 S regulatory particle.
A further illustration of the power of CXMS and the MS of intact complexes in combination with EM is shown in Fig. 4, where these techniques were applied to study the binding of tubulin by bovine Tric complex (118). Comparisons of nondenaturing MS spectra of intact bovine Tric complex before and after ATP incubation were used to show that the Tric complex is able to bind one molecule of substrate. Additional details of this Tric-tubulin interaction were subsequently confirmed by CXMS, again using the xQuest algorithm (95). This information could be used to interpret densities in the cryo-EM three-dimensional reconstitution of the purified complex and helped to confirm substrate binding in the central cavity of the Tric complex.
Fig. 4.
Binding of tubulin inside the central cavity of bovine Tric complex revealed by mass spectrometry. Comparison of nondenaturing MS spectra of intact bovine Tric complex before (upper panel) and after (lower panel) ATP incubation and SEC purification shows that the complex has one molecule of substrate bound. The two series are measured as 947.5 (blue) and 997.7 kDa (orange), respectively. Insets across the cryo-EM three-dimensional reconstitution at 30A of the purified complex (blue) also show additional EM density (yellow), in line with the MS data. These figures are reproduced from Ref. 118 with permission.
Combining Multiple MS Technologies to Derive Structural Models
These examples demonstrate that modern MS-based technologies, especially when used in conjunction with modeling and EM, represent a powerful hybrid approach for generating structural information. A potential work flow is depicted in Fig. 5. After purification of the sample, involving multiple biochemical and affinity purification steps, subunit interaction maps can be generated following analysis by CXMS, quantitative AP-MS, and MS of intact complexes. Each of these technologies has different strengths and weaknesses. AP-MS is able to detect subtle changes in interaction partners and dynamics and is able to deal with lowly expressed complexes. MS of intact assemblies is able to assess the absolute stoichiometry of the intact complex and multiple subcomplexes. CXMS is capable of providing many interactions for a single protein-protein interaction and at peptide resolution. Combining the different data sets could generate complete interaction maps for subsequent merging with topological information from IM-MS and CXMS.
Fig. 5.
Combining multiple MS-based methods for the analysis of protein complexes. A, after purification of the complex/assembly, usually by affinity purification in combination with one or more biochemical fractionation techniques, the sample is analyzed using the following methods. B–H, quantitative proteomics (B) to assess the subunit content assists the process in C, the assignment of the absolute stoichiometry of the intact complex and subcomplexes by MS of intact assemblies. Additional protein-protein interaction data are generated by AP-MS (D) and CXMS (E). Combining the data will give a complete and validated map of the subunit interactome (F). Topological information from CXMS and IM-MS (G) can be used to provide additional local and global restraints that can be used to together with the protein-protein interaction data to guide molecular modeling of the complex (H).
Because both cross-linking and ion mobility provide topological information, on local and global levels, respectively, these restraints could be used in tandem with protein-protein interaction data to generate three-dimensional models for protein complexes. In summary, by integrating data from a number of low resolution experimental MS-based methods, to provide distance and interaction restraints, with homology modeling, architectural or even pseudoatomic models can be assembled (44, 109).
Outlook
There is growing interest in understanding the so-called functional modules of the cell. These modules can be either protein complexes or larger protein assemblies. Because of their ability to fine tune their composition or post-translational modification to respond to the cellular environment, these complexes are often heterogeneous and dynamic in nature. Their study therefore requires a combination of technologies that can not only deal with these dynamics and heterogeneity but can also provide higher throughput than conventional structural biology methods. A further advantage of applying multiple methods comes from cross-validation of results. This can decrease the number of false positive results and consequently increase confidence in resulting data sets. Consequently this helps fulfill the requirement for higher resolution and higher precision data sets (4).
A powerful combination is that of the MS of intact assemblies with proteomics, in which lists of all proteins present during a particular isolation can be generated and combined with knowledge of stoichiometry of the intact complex and its subcomplexes (43). A strength of proteomics includes the ability to identify and quantify all proteins present in an assembly or complex (119). Proteomic methods, however, cannot readily reveal the integration and stoichiometry of subunits within different subcomplexes. This latter capability is the hallmark of the MS of intact complexes. Their marriage therefore compliments the shortcomings in both approaches with proteomics providing excellent definition of all proteins present at a particular time and MS of intact complexes yielding definitive knowledge of their integration and stoichiometry within an assembly.
Over the course of the last few years, further contribution to the MS platform has come to the fore with the emergence of reliable CXMS (58, 83, 84, 91, 94, 104, 120, 121) and IM-MS (45, 46). CXMS gives more precise information concerning the interactions between subunits than MS of intact complexes and consequently enables improved modeling and docking of subunits. This is particularly powerful when atomic structures are known of subunits and can enable construction of architectural models with atomic detail in some cases. The application of IM-MS to the study of macromolecular assemblies provides a means of generating additional information on the overall topology of protein assemblies and their subcomplexes (47, 107). Together with knowledge of interactions between subunits and the overall topology of subcomplexes generated from larger assemblies, it is becoming possible to generate architectural models that can be aligned within EM density maps or other high resolution structures. Here, the computational integration of the diverse data sets often lags behind. However, designing and implementing novel computational suites that are able to generate high fidelity three-dimensional models of large, heterogeneous protein complexes by integrating data from these various MS based sources remains a huge challenge.
Combining modern MS-based technologies with advanced modeling holds great promise to fill the gap between high resolution x-ray analysis of recombinant complexes and low resolution EM structures. As such, it raises expectations for the ultimate goal of high throughput, high resolution structural biology.
Footnotes
* This work was supported by funding from the European Union 7th Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648). This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
1 The abbreviations used are:
- AP-MS
- affinity purification coupled with mass spectrometry
- APC/C
- anaphase-promoting complex or cyclosome
- AKAP
- A-kinase anchoring protein
- CXMS
- cross-linking coupled to mass spectrometry
- eIF
- eukaryotic initiation factor
- IM-MS
- ion mobility-mass spectrometry
- MS/MS
- tandem mass spectrometry
- nESI
- nanoflow ESI
- PP2A
- human protein phosphatase 2A
- MS
- mass spectrometry
- ESI
- electrospray ionization.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Gstaiger M., Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 3. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 5. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 6. Seebacher J., Gavin A. C. (2011) SnapShot: Protein-protein interaction networks. Cell 144, 1000, 1000, e1001 [DOI] [PubMed] [Google Scholar]
- 7. Gavin A. C., Maeda K., Kühner S. (2011) Recent advances in charting protein-protein interaction: Mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 [DOI] [PubMed] [Google Scholar]
- 8. Kaake R. M., Wang X., Huang L. (2010) Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol. Cell. Proteomics 9, 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Köcher T., Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: From molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 10. Sharon M., Robinson C. V. (2007) The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 76, 167–193 [DOI] [PubMed] [Google Scholar]
- 11. Benesch J. L., Ruotolo B. T., Simmons D. A., Robinson C. V. (2007) Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem. Rev. 107, 3544–3567 [DOI] [PubMed] [Google Scholar]
- 12. Wyttenbach T., Bowers M. T. (2007) Intermolecular interactions in biomolecular systems examined by mass spectrometry. Annu. Rev. Phys. Chem. 58, 511–533 [DOI] [PubMed] [Google Scholar]
- 13. Heck A. J. (2008) Native mass spectrometry: A bridge between interactomics and structural biology. Nat. Methods 5, 927–933 [DOI] [PubMed] [Google Scholar]
- 14. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 15. Ganem B., Li Y. T., Henion J. D. (1991) Detection of noncovalent receptor ligand complexes by mass spectrometry. J. Am. Chem. Soc. 113, 6294–6296 [Google Scholar]
- 16. Sobott F., Hernández H., McCammon M. G., Tito M. A., Robinson C. V. (2002) A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 [DOI] [PubMed] [Google Scholar]
- 17. Hernández H., Robinson C. V. (2007) Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 [DOI] [PubMed] [Google Scholar]
- 18. Barrera N. P., Robinson C. V. (2011) Advances in the mass spectrometry of membrane proteins: From individual proteins to intact complexes. Annu. Rev. Biochem. 80, 247–271 [DOI] [PubMed] [Google Scholar]
- 19. Chernushevich I. V., Thomson B. A. (2004) Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 76, 1754–1760 [DOI] [PubMed] [Google Scholar]
- 20. Rostom A. A., Robinson C. V. (1999) Detection of the intact GroEL chaperonin assembly by mass spectrometry. J. Am. Chem. Soc. 121, 4718–4719 [Google Scholar]
- 21. Tahallah N., Pinkse M., Maier C. S., Heck A. J. (2001) The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 15, 596–601 [DOI] [PubMed] [Google Scholar]
- 22. van Duijn E., Simmons D. A., van den Heuvel R. H., Bakkes P. J., van Heerikhuizen H., Heeren R. M., Robinson C. V., van der Vies S. M., Heck A. J. (2006) Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J. Am. Chem. Soc. 128, 4694–4702 [DOI] [PubMed] [Google Scholar]
- 23. Benesch J. L. (2009) Collisional activation of protein complexes: Picking up the pieces. J. Am. Soc. Mass Spectrom. 20, 341–348 [DOI] [PubMed] [Google Scholar]
- 24. Benesch J. L., Ruotolo B. T., Sobott F., Wildgoose J., Gilbert A., Bateman R., Robinson C. V. (2009) Quadrupole-time-of-flight mass spectrometer modified for higher-energy dissociation reduces protein assemblies to peptide fragments. Anal. Chem. 81, 1270–1274 [DOI] [PubMed] [Google Scholar]
- 25. Jurchen J. C., Garcia D. E., Williams E. R. (2003) Gas-phase dissociation pathways of multiply charged peptide clusters. J. Am. Soc. Mass Spectrom. 14, 1373–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pagel K., Hyung S. J., Ruotolo B. T., Robinson C. V. (2010) Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal. Chem. 82, 5363–5372 [DOI] [PubMed] [Google Scholar]
- 27. Felitsyn N., Kitova E. N., Klassen J. S. (2002) Thermal dissociation of the protein homodimer ecotin in the gas phase. J. Am. Soc. Mass Spectrom. 13, 1432–1442 [DOI] [PubMed] [Google Scholar]
- 28. de la Mora J. F. (2000) Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism. Anal. Chim. Acta 406, 93–104 [Google Scholar]
- 29. Hernández H., Dziembowski A., Taverner T., Séraphin B., Robinson C. V. (2006) Subunit architecture of multimeric complexes isolated directly from cells. EMBO Reports 7, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharon M., Mao H., Boeri Erba E., Stephens E., Zheng N., Robinson C. V. (2009) Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17, 31–40 [DOI] [PubMed] [Google Scholar]
- 31. Zhou M., Sandercock A. M., Fraser C. S., Ridlova G., Stephens E., Schenauer M. R., Yokoi-Fong T., Barsky D., Leary J. A., Hershey J. W., Doudna J. A., Robinson C. V. (2008) Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 105, 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lane L. A., Fernández-Tornero C., Zhou M., Morgner N., Ptchelkine D., Steuerwald U., Politis A., Lindner D., Gvozdenovic J., Gavin A. C., Müller C. W., Robinson C. V. (2011) Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure 19, 90–100 [DOI] [PubMed] [Google Scholar]
- 33. Stengel F., Baldwin A. J., Painter A. J., Jaya N., Basha E., Kay L. E., Vierling E., Robinson C. V., Benesch J. L. (2010) Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. U.S.A. 107, 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uetrecht C., Versluis C., Watts N. R., Roos W. H., Wuite G. J., Wingfield P. T., Steven A. C., Heck A. J. (2008) High-resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. U.S.A. 105, 9216–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barrera N. P., Di Bartolo N., Booth P. J., Robinson C. V. (2008) Micelles protect membrane complexes from solution to vacuum. Science 321, 243–246 [DOI] [PubMed] [Google Scholar]
- 36. Barrera N. P., Isaacson S. C., Zhou M., Bavro V. N., Welch A., Schaedler T. A., Seeger M. A., Miguel R. N., Korkhov V. M., van Veen H. W., Venter H., Walmsley A. R., Tate C. G., Robinson C. V. (2009) Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat. Methods 6, 585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park A. Y., Jergic S., Politis A., Ruotolo B. T., Hirshberg D., Jessop L. L., Beck J. L., Barsky D., O'Donnell M., Dixon N. E., Robinson C. V. (2010) A single subunit directs the assembly of the Escherichia coli DNA sliding clamp loader. Structure 18, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rostom A. A., Fucini P., Benjamin D. R., Juenemann R., Nierhaus K. H., Hartl F. U., Dobson C. M., Robinson C. V. (2000) Detection and selective dissociation of intact ribosomes in a mass spectrometer. Proc. Natl. Acad. Sci. U. S. A. 97, 5185–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gordiyenko Y., Deroo S., Zhou M., Videler H., Robinson C. V. (2008) Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 380, 404–414 [DOI] [PubMed] [Google Scholar]
- 40. Hernández H., Makarova O. V., Makarov E. M., Morgner N., Muto Y., Krummel D. P., Robinson C. V. (2009) Isoforms of U1–70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS One 4, e7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lorenzen K., Vannini A., Cramer P., Heck A. J. (2007) Structural biology of RNA polymerase III: Mass spectrometry elucidates subcomplex architecture. Structure 15, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 42. Schreiber A., Stengel F., Zhang Z., Enchev R. I., Kong E. H., Morris E. P., Robinson C. V., da Fonseca P. C., Barford D. (2011) Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470, 227–232 [DOI] [PubMed] [Google Scholar]
- 43. Sakata E., Stengel F., Fukunaga K., Zhou M., Saeki Y., Förster F., Baumeister W., Tanaka K., Robinson C. V. (2011) The catalytic activity of ubp6 enhances maturation of the proteasomal regulatory particle. Mol. Cell 42, 637–649 [DOI] [PubMed] [Google Scholar]
- 44. Taverner T., Hernández H., Sharon M., Ruotolo B. T., Matak-Vinković D., Devos D., Russell R. B., Robinson C. V. (2008) Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 41, 617–627 [DOI] [PubMed] [Google Scholar]
- 45. Park A. Y., Robinson C. V. (2011) Protein-nucleic acid complexes and the role of mass spectrometry in their structure determination. Crit. Rev. Biochem. Mol. Biol. 46, 152–164 [DOI] [PubMed] [Google Scholar]
- 46. Uetrecht C., Rose R. J., van Duijn E., Lorenzen K., Heck A. J. (2010) Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 [DOI] [PubMed] [Google Scholar]
- 47. Ruotolo B. T., Giles K., Campuzano I., Sandercock A. M., Bateman R. H., Robinson C. V. (2005) Evidence for macromolecular protein rings in the absence of bulk water. Science 310, 1658–1661 [DOI] [PubMed] [Google Scholar]
- 48. Pukala T. L., Ruotolo B. T., Zhou M., Politis A., Stefanescu R., Leary J. A., Robinson C. V. (2009) Subunit architecture of multiprotein assemblies determined using restraints from gas-phase measurements. Structure 17, 1235–1243 [DOI] [PubMed] [Google Scholar]
- 49. van Duijn E., Barendregt A., Synowsky S., Versluis C., Heck A. J. (2009) Chaperonin complexes monitored by ion mobility mass spectrometry. J. Am. Chem. Soc. 131, 1452–1459 [DOI] [PubMed] [Google Scholar]
- 50. Zhou M., Robinson C. V. (2010) When proteomics meets structural biology. Trends Biochem. Sci. 35, 522–529 [DOI] [PubMed] [Google Scholar]
- 51. Damoc E., Fraser C. S., Zhou M., Videler H., Mayeur G. L., Hershey J. W., Doudna J. A., Robinson C. V., Leary J. A. (2007) Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics 6, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 52. Hinnebusch A. G. (2006) eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562 [DOI] [PubMed] [Google Scholar]
- 53. Masutani M., Sonenberg N., Yokoyama S., Imataka H. (2007) Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 26, 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 55. Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 56. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature January 11; 482(7384), 186–91 doi: 10.1038/nature10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. da Fonseca P. C., Morris E. P. (2008) Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 283, 23305–23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., Nickell S. (2010) Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saeki Y., Toh-E. A., Kudo T., Kawamura H., Tanaka K. (2009) Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 60. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 62. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 63. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 64. Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., Chan D. W., Ding C., Shi Y., Yucer N., Krenciute G., Kim B. J., Li C., Chen R., Li W., Wang Y., O'Malley B. W., Qin J. (2011) Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kühner S., van Noort V., Betts M. J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., Castaño-Diez D., Chen W. H., Devos D., Güell M., Norambuena T., Racke I., Rybin V., Schmidt A., Yus E., Aebersold R., Herrmann R., Böttcher B., Frangakis A. S., Russell R. B., Serrano L., Bork P., Gavin A. C. (2009) Proteome organization in a genome-reduced bacterium. Science 326, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 66. Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guerrero C., Tagwerker C., Kaiser P., Huang L. (2006) An integrated mass spectrometry-based proteomic approach: Quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 69. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 70. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) An integrated workflow for charting the human interaction proteome: Insights into the PP2A system. Mol. Syst Biol. 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Poser I., Sarov M., Hutchins J. R., Hériché J. K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A. W., Pelletier L., Kittler R., Hua S., Naumann R., Augsburg M., Sykora M. M., Hofemeister H., Zhang Y., Nasmyth K., White K. P., Dietzel S., Mechtler K., Durbin R., Stewart A. F., Peters J. M., Buchholz F., Hyman A. A. (2008) BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wepf A., Glatter T., Schmidt A., Aebersold R., Gstaiger M. (2009) Quantitative interaction proteomics using mass spectrometry. Nat. Methods 6, 203–205 [DOI] [PubMed] [Google Scholar]
- 74. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pflieger D., Jünger M. A., Müller M., Rinner O., Lee H., Gehrig P. M., Gstaiger M., Aebersold R. (2008) Quantitative proteomic analysis of protein complexes: Concurrent identification of interactors and their state of phosphorylation. Mol. Cell. Proteomics 7, 326–346 [DOI] [PubMed] [Google Scholar]
- 76. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Beynon R. J., Doherty M. K., Pratt J. M., Gaskell S. J. (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 [DOI] [PubMed] [Google Scholar]
- 78. Rinner O., Mueller L. N., Hubálek M., Müller M., Gstaiger M., Aebersold R. (2007) An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat. Biotechnol. 25, 345–352 [DOI] [PubMed] [Google Scholar]
- 79. Vermeulen M., Hubner N. C., Mann M. (2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr. Opin. Biotechnol. 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 80. Ranish J. A., Yi E. C., Leslie D. M., Purvine S. O., Goodlett D. R., Eng J., Aebersold R. (2003) The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33, 349–355 [DOI] [PubMed] [Google Scholar]
- 81. Clegg C., Hayes D. (1974) Identification of neighbouring proteins in the ribosomes of Escherichia coli: A topographical study with the cross-linking reagent dimethyl suberimidate. Eur. J. Biochem. 42, 21–28 [DOI] [PubMed] [Google Scholar]
- 82. Young M. M., Tang N., Hempel J. C., Oshiro C. M., Taylor E. W., Kuntz I. D., Gibson B. W., Dollinger G. (2000) High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 97, 5802–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Leitner A., Reischl R., Walzthoeni T., Herzog F., Bohn S., Förster F., Aebersold R. (2012) Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 86. Jin Lee Y. (2008) Mass spectrometric analysis of cross-linking sites for the structure of proteins and protein complexes. Mol. Biosyst. 4, 816–823 [DOI] [PubMed] [Google Scholar]
- 87. Tang X., Bruce J. E. (2009) Chemical cross-linking for protein-protein interaction studies. Methods Mol. Biol. 492, 283–293 [DOI] [PubMed] [Google Scholar]
- 88. Mouradov D., King G., Ross I. L., Forwood J. K., Hume D. A., Sinz A., Martin J. L., Kobe B., Huber T. (2008) Protein structure determination using a combination of cross-linking, mass spectrometry, and molecular modeling. Methods Mol. Biol. 426, 459–474 [DOI] [PubMed] [Google Scholar]
- 89. Singh P., Panchaud A., Goodlett D. R. (2010) Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal. Chem. 82, 2636–2642 [DOI] [PubMed] [Google Scholar]
- 90. Sinz A. (2007) Investigation of protein-ligand interactions by mass spectrometry. ChemMedChem 2, 425–431 [DOI] [PubMed] [Google Scholar]
- 91. Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 92. Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J. G., De Wulf P., Salek M., Rappsilber J., Moores C. A., Salmon E. D., Musacchio A. (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Trnka M. J., Burlingame A. L. (2010) Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene: The power of high resolution electron transfer dissociation for determination of both peptide sequences and their attachment sites. Mol. Cell. Proteomics 9, 2306–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu H., Hsu P. H., Zhang L., Tsai M. D., Freitas M. A. (2010) Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J. Proteome Res. 9, 3384–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Petrotchenko E. V., Borchers C. H. (2010) ICC-CLASS: Isotopically-coded cleavable cross-linking analysis software suite. BMC Bioinformatics 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. McIlwain S., Draghicescu P., Singh P., Goodlett D. R., Noble W. S. (2010) Detecting cross-linked peptides by searching against a database of cross-linked peptide pairs. J. Proteome Res. 9, 2488–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: A bioinformatics strategy for identification of cross-linked peptides. Mol. Cell. Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rappsilber J. (2011) The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tang X., Munske G. R., Siems W. F., Bruce J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 103. Zheng C., Yang L., Hoopmann M. R., Eng J. K., Tang X., Weisbrod C. R., Bruce J. E. (2011) Cross-linking measurements of in vivo protein complex topologies. Mol. Cell. Proteomics 10.1074/mcp.M110.006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gold M. G., Stengel F., Nygren P. J., Weisbrod C. R., Bruce J. E., Robinson C. V., Barford D., Scott J. D. (2011) Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc. Natl. Acad. Sci. U.S.A. 108, 6426–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ruotolo B. T., Robinson C. V. (2006) Aspects of native proteins are retained in vacuum. Curr. Opin. Chem. Biol. 10, 402–408 [DOI] [PubMed] [Google Scholar]
- 106. von Helden G., Wyttenbach T., Bowers M. T. (1995) Conformation of macromolecules in the gas phase: Use of matrix-assisted laser desorption methods in ion chromatography. Science 267, 1483–1485 [DOI] [PubMed] [Google Scholar]
- 107. Ruotolo B. T., Benesch J. L., Sandercock A. M., Hyung S. J., Robinson C. V. (2008) Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 [DOI] [PubMed] [Google Scholar]
- 108. Bush M. F., Hall Z., Giles K., Hoyes J., Robinson C. V., Ruotolo B. T. (2010) Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 [DOI] [PubMed] [Google Scholar]
- 109. Politis A., Park A. Y., Hyung S. J., Barsky D., Ruotolo B. T., Robinson C. V. (2010) Integrating ion mobility mass spectrometry with molecular modelling to determine the architecture of multiprotein complexes. PLoS One 5, e12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang S. C., Politis A., Di Bartolo N., Bavro V. N., Tucker S. J., Booth P. J., Barrera N. P., Robinson C. V. (2010) Ion mobility mass spectrometry of two tetrameric membrane protein complexes reveals compact structures and differences in stability and packing. J. Am. Chem. Soc. 132, 15468–15470 [DOI] [PubMed] [Google Scholar]
- 111. Loo J. A., Berhane B., Kaddis C. S., Wooding K. M., Xie Y., Kaufman S. L., Chernushevich I. V. (2005) Electrospray ionization mass spectrometry and ion mobility analysis of the 20S proteasome complex. J. Am. Soc. Mass Spectrom. 16, 998–1008 [DOI] [PubMed] [Google Scholar]
- 112. Ekeowa U. I., Freeke J., Miranda E., Gooptu B., Bush M. F., Pérez J., Teckman J., Robinson C. V., Lomas D. A. (2010) Defining the mechanism of polymerization in the serpinopathies. Proc. Natl. Acad. Sci. U.S.A. 107, 17146–17151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Koschubs T., Lorenzen K., Baumli S., Sandström S., Heck A. J., Cramer P. (2010) Preparation and topology of the mediator middle module. Nucleic Acids Res. 38, 3186–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Siridechadilok B., Fraser C. S., Hall R. J., Doudna J. A., Nogales E. (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515 [DOI] [PubMed] [Google Scholar]
- 115. Barford D. (2011) Structure, function and mechanism of the anaphase promoting complex (APC/C). Q Rev. Biophys. 44, 153–190 [DOI] [PubMed] [Google Scholar]
- 116. Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J. M. (2009) Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323, 1477–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. da Fonseca P. C., Kong E. H., Zhang Z., Schreiber A., Williams M. A., Morris E. P., Barford D. (2011) Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature 470, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Muñoz I. G., Yébenes H., Zhou M., Mesa P., Serna M., Park A. Y., Bragado-Nilsson E., Beloso A., de Cárcer G., Malumbres M., Robinson C. V., Valpuesta J. M., Montoya G. (2011) Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 18, 14–19 [DOI] [PubMed] [Google Scholar]
- 119. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 120. Kisko K., Brozzo M. S., Missimer J., Schleier T., Menzel A., Leppänen V. M., Alitalo K., Walzthoeni T., Aebersold R., Ballmer-Hofer K. (2011) Structural analysis of vascular endothelial growth factor receptor-2/ligand complexes by small-angle X-ray solution scattering. FASEB J. 25, 2980–2986 [DOI] [PubMed] [Google Scholar]
- 121. Buey R. M., Mohan R., Leslie K., Walzthoeni T., Missimer J. H., Menzel A., Bjelic S., Bargsten K., Grigoriev I., Smal I., Meijering E., Aebersold R., Akhmanova A., Steinmetz M. O. (2011) Insights into EB structure and the role of its C-terminal domain in discriminating microtubule tips from lattice. Mol. Biol. Cell 22, 2912–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. January 31; 109(5), 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]