Abstract
There is controversy as to the recommended daily intake of selenium (Se), and whether current New Zealand diets are adequate in this nutrient. Various functional single-nucleotide polymorphisms (SNPs) polymorphisms may affect the efficacy of Se utilisation. These include the glutathione peroxidases GPx1 rs1050450, GPx4 rs713041, as well as selenoproteins SEPP1 rs3877899, SEL15 rs5845, SELS rs28665122 and SELS rs4965373. This cross-sectional study measured serum Se levels of 503 healthy Caucasian men in Auckland, New Zealand, between ages 20–81. The Se distribution was compared with activities of the antioxidant enzymes glutathione peroxidase and thioredoxin reductase, and DNA damage as measured by the single cell gel electrophoresis assay, both without and with a peroxide-induced oxidative challenge. Serum Se was measured using inductively coupled plasma-dynamic reaction cell-mass spectrometry, while selenoprotein SNPs were estimated using TaqMan® SNP genotyping assays. While antioxidant enzyme activities and DNA damage recorded after a peroxide challenge increased with increasing serum selenium, the inherent DNA damage levels in leukocytes showed no statistically significant relationship with serum selenium. However, these relationships and dietary Se requirements at the individual level were modified by several different SNPs in genes for selenoproteins. The GPx1 rs1050450 C allele was significantly associated with GPx activity. Significant correlations between serum Se level and GPX activity were seen with all genotypes except for homozygous minor allele carriers, while the GPx1 rs1050450 CT genotype showed the highest correlation. Several genotypes showed significant correlations between serum Se and TR activity with SEPP1 rs3877899 GG genotype showing the highest correlation. A significant decreasing trend in DNA damage with increasing serum Se was seen among GPx1 rs1050450 CC and GPx4 rs713041 TT genotype carriers up to a serum Se level of 116 and 149 ng/ml, respectively. In the absence of this genetic information, we would recommend a serum Se concentration in the region of 100–150 ng/ml as providing a useful compromise.
Keywords: Selenium (Se), Glutathione peroxidase (GPx), Thioredoxin reductase (TR), Single nucleotide polymorphisms (SNPs)
Introduction
There is controversy over the potential role of the essential human micronutrient, selenium (Se), in the prevention of cancers such as prostate cancer. Se status varies substantially across different populations and different ethnic groups (Bleys et al. 2008; Cook et al. 2005; Johnson et al. 2010; Kant and Graubard 2007). A trace element, Se enters the food chain through plants, and the amount and bioavailability of Se in the soil typically reflects the plant level. It has been generally considered to be low in New Zealand soils and probably diets (Thomson et al. 2007). Although it is the South Island of New Zealand that is generally considered Se deficient, our own work has suggested that this may also be true for North Island populations (Karunasinghe et al. 2004, 2006).
There is reason to believe that individual requirements for Se will differ because of polymorphisms in selenoprotein genes (Ferguson and Karunasinghe 2011; Hesketh 2008; Hesketh and Meplan 2011). Se effects on prostate cancer development may act through antioxidant pathways, whose efficacy is related to single-nucleotide polymorphisms in certain genes for selenoproteins (Cooper et al. 2008; Li et al. 2005). Among the ~25 selenoproteins known to date, various functions are well characterised, of which several examples follow. These include Se transport (selenoprotein P), antioxidant/redox properties (glutathione peroxidases (GPxs), thioredoxin reductases and selenoprotein P) and anti-inflammatory properties (selenoprotein S and GPx4). Glutathione peroxidase 1 (GPx1) is the most abundant selenoperoxidase and is ubiquitously expressed in almost all tissues (Cheng et al. 1997, 1998). GPx1 can reduce hydrogen peroxide or fatty acid hydroperoxides rapidly from tissues (Brigelius-Flohé 1999). Unlike GPx1, Glutathione peroxidase 4 (GPx4), or phospholipid hydroperoxidase is a monomer, and is the only GPx enzyme that reduces phospholipid hydroperoxides (Bellinger et al. 2009) which are fatty acid hydroperoxides esterified to phospholipids commonly occurring in cell membranes undergoing oxidative stress (Thomas et al. 1990). GPx-4 controls the NF-kB-dependent interleukin 1 (IL-1) signalling and thereby inhibits the biosynthesis of leukotrienes and prostanoids. This is done by interfering with the activity of cyclooxygenases and lipoxygenases mediating the inflammatory process (Brigelius-Flohe 2006), producing lipoxygenase metabolites (Villette et al. 2002). Thioredoxin reductase (TR) is part of a major thiol-based redox pathway working alongside thioredoxin and NADPH. TR reduces selenates and selenites to selenides which are precursors for selenoprotein synthesis (Selenius et al. 2010). The Selenoprotein P (SEPP1) gene promoter interacts with cytokine and growth factor pathways, suggesting that inflammation can alter its’ activation (Al-Taie et al. 2002; Dreher et al. 1997; Mostert et al. 2001). Selenoprotein S is an endoplasmic reticulum protein involved in removing unfolded proteins (Kelly et al. 2009). The promoter region of the SELS gene has two sites for the binding of transcription factor NFκB, and is activated by pro inflammatory cytokines (Gao et al. 2006). The 15-kDa selenoprotein (SEL15 gene) is also considered as an endoplasmic reticulum protein involved in the unfolded protein response (Shchedrina et al. 2010).
The current analysis investigates 503 healthy men in Auckland, New Zealand, and relates serum Se level and certain selenoprotein genotypes to biomarkers of Se status and genomic stability (Ferguson et al. 2006). These biomarkers include activity of seleno-enzymes GPx and TR and leukocyte DNA damage levels. The single-nucleotide polymorphisms (SNPs) considered here are functional SNPs: GPx1 rs1050450, GPx4 rs713041, SEPP1 rs3877899, SEL15 rs5845, SELS rs28665122 and SELS rs4965373 (Martinez et al. 2008). The GPx1 rs1050450 SNP occurs within exon 1 of the GPx1 gene and forms structurally different subunits containing either prolein or leucine at codon 198 (Moscow et al. 1994). Located in the 3′UTR near the selenocysteine (Sec) insertion sequence element, the GPx4 rs713041 C > T polymorphism can modulate GPx4 activity by altering Sec insertion and protein binding to the 3′UTR (Meplan et al. 2008). The major allele G of SEPP1 rs3877899 favours the production of a Se-rich 60 kDa isoform, while allele A favours the 50-kDa low Se isoform (Meplan et al. 2009; Mostert et al. 1998), and may be an evolutionary adaptation for survival in low Se environments. Differential Se incorporation among SEL-15 rs5845 C and T alleles are also reported (Kumaraswamy et al. 2000). According to Curran et al. (2005), the G allele of SEL-S rs28665122 SNP was required to produce sufficient promoter activity in the presence of a stress stimulator, whereas the A allele has shown significantly lower activities.
Considering the above functional differences of gene polymorphisms, the current analysis was undertaken to investigate whether the biomarkers assessed have variations depending on the above gene polymorphisms and/or serum Se status.
Methods
Study population
A total of 503 male subjects from Auckland, New Zealand, self-reported as having European ancestry, took part in this study. They were of ages ranging from 20 to 81 years, have self-reported as having no history of cancers or any urological problem, and are not taking more than 50 μg Se/day as supplements. They are part of a group of volunteers recruited with informed consent for the Se supplementation trial carried out by the Discipline of Nutrition, University of Auckland (Ethics Ref: NTY/06/07/060). At entry to the study, the height and weight of the volunteers were noted, and they submitted answers to a health and lifestyle questionnaire. A summary of their demographic characteristics is given in Table 1.
Table 1.
Demographic characteristics of the study group
| N (%) | Mean (SD) | Range | |
|---|---|---|---|
| Age at joining study in years | 52.3 (13.9) | 20–81 | |
| <40 | 96 (19.1) | ||
| 40–60 | 242 (48.1) | ||
| >60 | 165 (32.8) | ||
| Height (cm) | 177.6 (6.4) | 161–201 | |
| Weight (kg) | 84.2 (12.5) | 55–145 | |
| BMI | 26.7 (3.7) | 17.2–44.8 | |
| <25 | 168 (34.8) | ||
| 25–30 | 240 (49.7) | ||
| >30 | 75 (15.5) | ||
| Serum Se level (ng/ml) | 111.6 (1.21)a | 63–305a | |
| Smoking status | |||
| Current | 27 (5.4) | ||
| Former | 148 (29.4) | ||
| Never | 328 (65.2) | ||
| Alcohol consumption | |||
| Yes | 438 (87.1) | ||
| No | 65 (12.9) | ||
| Family history of cancerb | |||
| With no family history of cancer | 165 (32.8) | ||
| With family history of cancer | 338 (67.2) | ||
| Dietary supplements | |||
| No dietary supplements taken | 275 (55.3) | ||
| Dietary supplements taken | 222 (44.7) | ||
aTo convert serum Se level in ng/ml to μmol/l multiply by 0.0127
bA positive family history of cancer was considered if one or more close relatives have recorded cancer (including grandparents, parents, siblings, children, paternal and maternal uncles and aunts)
Blood collection and processing
At the entry to the study, blood samples from each volunteer were collected in each of an EDTA, heparin, and plain vaccutainer tube. An aliquot of the EDTA sample was subsequently used for DNA extraction. Total genomic DNA was extracted from blood with the QIAamp DNA Blood Mini Kit (Qiagen) according to the manufacturer’s instructions, using a fully automated procedure on the QIAcube.
SNP genotyping
The TaqMan® SNP Genotyping Assay from Applied Biosystems was used in the SNP genotyping of the panel of genes selected in this study. The assays were obtained either as pre-designed from Applied Biosystems or custom-made through Assay-by-Design service by ABI. Samples were assayed along with no-template and HapMap controls, and run on the AB 7900HT Fast Real-Time PCR System using the conditions: 10 min 95°C enzyme activation followed by 40 cycles at 92°C for 15 s and 60°C for 1 min (annealing/extension). The allelic discrimination results were determined after the amplification by performing an endpoint read.
Enzyme activities
Erythrocyte lysates were prepared from an aliquot of EDTA bloods and were assayed for GPx activities using a modified protocol of Wendel (1981) as described by Karunasinghe et al. (2006). One unit of GPx activity is defined as 1 mmol NADPH oxidized/min at 37°C. TR activities on erythrocyte lysates were assayed using the method of Smith and Levander (2002) with minor modifications, as reported in Karunasinghe et al. (2006). One unit of TR activity is defined as 1 mmol 5-thio-2-nitrobenzoic acid formed/min at 37°C.
DNA damage
Heparinised blood was used to quantify DNA damage levels in leukocytes from fresh blood or peroxide challenged (200 μM) blood using the comet assay, as described in Karunasinghe et al. (2004) and Ferguson et al. (2010). DNA damage was quantified as percentage DNA in the comet tail.
Serum selenium
Plain tube blood was spun at 2,000 g for 10 min at 4°C to separate the serum. Serum Se level was assayed at Gribbles Veterinary Pathology, Hamilton, using a modified semi automated fluorometric assay based on Watkinson (1979), Watkinson and Brown (1979) and Rongpu et al. (1986). The fluorescence of the final benzopiazselenol extracted into cyclohexane was measured with an excitation wave length of 360 nm and emission wave length of 518 nm.
Statistical analysis
Since the serum Se concentrations from analysis were the log-transformed values, the estimated actual Se concentrations were determined by utilizing the exponential (anti-log) function (and will be hereafter referred to as serum Se level). Smoking status, dietary supplementation and alcohol consumption were investigated, in order to check whether there were significant effects of these on serum Se level and the biomarkers. Relationships between serum Se and biomarkers within genotypes were examined using Pearson's correlation statistics for simple correlations. Multiple regression analyses were carried out using generalised linear models (GLM), using the GLM procedure in SAS. Whether the relationship between the outcome and the predictor of serum Se concentration was linear was tested by fitting a quadratic polynomial. The GPx1 rs1050450 C/C and GPx4 rs713041 T/T genotypes show significant nonlinearity, so a ‘broken stick’ regression (Seber 1938) was fitted, with different linear regression models in two different regions constrained so that the fitted curve was continuous at the break point concentration. The optimal break point was chosen by taking the concentration which gave rise to the lowest RSS (Residual Sum of Squares). This occurred at 116.07 and 149.23 ng/ml serum Se (P = 0.044 and P = 0.042, respectively). None of the other genotypes showed a nonlinear distribution with this or any other marker. All analyses including broken stick regression were carried out using R (Ihaka and Gentleman 1996) and SAS (V9.1 SAS Institute, Cary, NC, USA).
Results
Demographic data
A summary of the demographic data for the study population is given in Table 1. The mean age of the study group was 52.3 ± 13.9y with a mean body mass index (BMI) of 26.7 ± 3.7. While 65% of the study population had never smoked, 5.4% were current smokers and 29.4% were past smokers (Table 1). The study population was generally a health conscious group, with 67% recording a family history of cancer and 44.7% of the subjects taking dietary supplements including single or multi vitamins and minerals, occasionally or regularly (Table 2). Among the supplement users, 39% have used Se supplements or multivitamin and mineral supplements (Table 2). According to the sales figures from Pharmacybrands Limited New Zealand (personal communication), 3.7% of multivitamins sold in New Zealand contain Se. Therefore, Se supplements and the multivitamin supplements were considered together as possible sources of Se and were considered as supplements with Se in the current analysis.
Table 2.
The variation of biomarker levels with smoking status and dietary supplementation among the participants
| N | Mean (SD) | Range | P | Estimate (95% CI) | |
|---|---|---|---|---|---|
| Serum selenium | |||||
| Supplements | |||||
| No | 275 | 109.93 (1.2) | 63.96–195.03 | 0.052 | 0.95 (0.91–0.99) |
| Yes | |||||
| (without Se) | 135 | 111.35 (1.19) | 63.17–181.61 | 0.96 (0.91–1.01) | |
| (with Se) | 87 | 116.17 (1.22) | 78.96–222.67 | 1.00 | |
| Smoking status | |||||
| Current | 27 | 102.3 (1.2) | 65.5–150.0 | 0.020 | 0.92 (0.85–0.99) |
| Ex-smoker | 148 | 112.9 (1.2) | 63.1–195.0 | 0.506 | 1.01 (0.98–1.05) |
| Non smoker | 322 | 111.5 (1.2) | 72.6–222.7 | 1.00 | |
| TR activity | |||||
| Supplements | |||||
| No | 275 | 1.11 (0.76) | 0–3.78 | 0.912 | |
| Yes | |||||
| (without Se) | 135 | 0.99 (0.75) | 0–3.26 | ||
| (with Se) | 87 | 1.15 (0.76) | 0–3.26 | ||
| Smoking status | |||||
| Current | 27 | 1.08 (0.63) | 0–2.84 | 0.218 | |
| Ex-smoker | 148 | 1.10 (0.79) | 0–3.68 | ||
| Non smoker | 322 | 1.07 (0.76) | 0–3.78 | ||
| GPx activity | |||||
| Supplements | |||||
| No | 275 | 14.18 (6.97) | 0.83–37.57 | 0.616 | |
| Yes | |||||
| (without Se) | 135 | 14.93 (7.78) | 0.83–38.81 | ||
| (with Se) | 87 | 14.4 (7.18) | 2.06–33.86 | ||
| Smoking status | |||||
| Current | 27 | 13.58 (7.15) | 2.48–33.86 | 0.199 | |
| Ex-smoker | 148 | 13.64 (6.87) | 2.89–37.57 | ||
| Non smoker | 322 | 14.85 (7.38) | 0.83–38.81 | ||
| Fresh blood DNA damage | |||||
| Supplements | |||||
| No | 275 | 5.96 (1.41) | 3.12–12.37 | 0.431 | |
| Yes | |||||
| (without Se) | 135 | 6.01 (1.37) | 3.14–11.03 | ||
| (with Se) | 87 | 6.19 (1.34) | 3.72–11.57 | ||
| Smoking status | |||||
| Current | 27 | 5.96 (1.38) | 4.41–9.57 | 0.567 | |
| Ex-smoker | 148 | 5.92 (1.38) | 3.22–11.03 | ||
| Non smoker | 322 | 6.06 (1.4) | 3.12–12.37 | ||
| Peroxide challenged DNA damage | |||||
| Supplements | |||||
| No | 275 | 7.37 (1.6) | 3.83–12.19 | 0.408 | |
| Yes | |||||
| (without Se) | 135 | 7.57 (1.81) | 2.97–14.78 | ||
| (with Se) | 87 | 7.67 (1.82) | 4.53–13.22 | ||
| Smoking status | |||||
| Current | 27 | 7.52 (1.7) | 4.5–14.69 | 0.859 | |
| Ex-smoker | 148 | 7.36 (1.85) | 3.83–13.22 | ||
| Non smoker | 322 | 7.55 (1.78) | 2.97–14.78 | ||
Estimate is the relative mean difference between two groups
Bold values are statistically significant
Biomarker variation with lifestyle habits
The mean serum Se level among the participants was 111.6 ± 1.21 ng/ml (Table 1). Serum Se concentrations were significantly different among participants according to smoking status. The current smokers had lower levels of serum Se concentrations compared with participants who have never smoked (OR = 0.92, 95% CI = 0.85–0.99, P = 0.020), although no significant differences between former smokers and never smokers were observed (Table 2). There was no significant difference in serum Se level among those consuming alcoholic drinks compared with the others. Se-containing supplements have nonsignificantly increased the serum Se level by 5.7% compared with those who do not use any supplements or 4.3% compared with other supplement users (Table 2). GPx and TR activities and % tail DNA levels, with and without a peroxide challenge, were not significantly different according to smoking status and dietary supplementation status (Table 2).
Correlations among selenium concentrations, seleno enzyme activities and % tail DNA
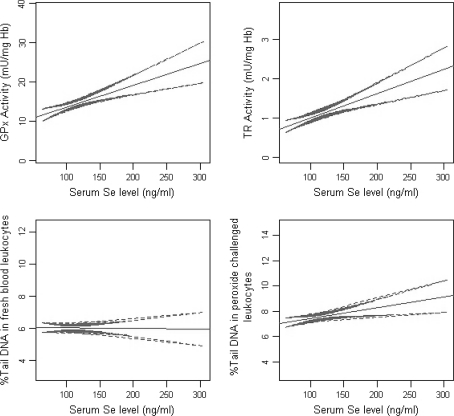
The variation of biomarker levels among subjects with increasing serum Se level is shown in Fig. 1. An increasing trend for the GPx and TR activity levels and peroxide challenged DNA damage is observed with increasing serum Se level. GPx and TR activities have not shown a plateau at this level. There were weak but positive and significant correlations, between overall serum Se concentration and GPx and TR activities and peroxide challenged DNA damage (Table 3). Overall % tail DNA in peroxide challenged leukocytes was significantly and positively associated with GPx and TR activities and % tail DNA in fresh blood leukocytes. The overall % tail DNA in fresh blood leukocytes was not directly related to serum Se level, GPx or TR activities.
Fig. 1.
Variation of biomarker levels with serum Se level among all subjects (line fitted with 95% CI)
Table 3.
Correlations among serum Se, biomarkers and DNA damage
| serum Se (ng/ml) | GPx activity | TR activity | Peroxide DNA | |
|---|---|---|---|---|
| GPx activity | r = 0.187 | |||
| P < 0.0001 | ||||
| TR activity | r = 0.169 | r = 0.159 | ||
| P = 0.0001 | P = 0.0004 | |||
| Peroxide DNA | r = 0.096 | r = 0.111 | r = 0.101 | |
| P = 0.033 | P = 0.014 | P = 0.021 | ||
| Fresh blood DNA | r = −0.018 | r = −0.002 | r = −0.032 | r = 0.168 |
| P = 0.697 | P = 0.963 | P = 0.478 | P = 0.0002 |
Peroxide DNA–DNA damage in peroxide challenged leukocytes
Fresh blood DNA–DNA damage in fresh blood leukocytes
Biomarker correlation with SNP genotypes
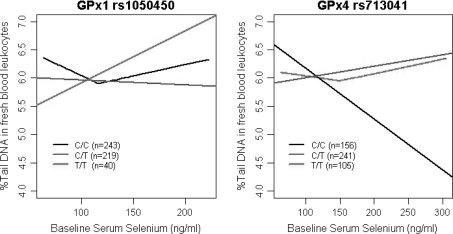
The data were stratified based on the genetic polymorphisms and analysed further. All except the homozygous minor alleles of the SNPs studied showed significant positive correlations between serum Se and GPx activity (P = 0.0003–0.049) (Table 4). Strong correlations were shown by GPx1 rs1050450 CT, SEP15 rs5845 CC, GPx4 rs713041 CT and SEPP1 rs3877890 GG and AG genotypes (P = 0.0003–0.003) (Table 4). TR activity was also significantly correlated with at least one genotype of each gene and was most strongly associated with SEPP1 rs3877899 GG genotype (P = 0.0001) (Table 5). Fresh blood DNA damage showed no significant correlations with serum Se when stratified according to genotype (Table 6). However, GPx1 rs1050450 CC and GPx4 rs713041 TT showed a significant quadratic function, and were fitted with a broken stick regression, with the lowest RSS at 116.07 and 149.23 ng/ml serum Se respectively (P = 0.044 and P = 0.042, respectively) (Fig. 2). Serum Se and DNA damage after a peroxide challenge was associated only with the SELS rs4965373 AG genotype (P = 0.003) (Table 7).
Table 4.
Correlation of serum Se and GPx activity by genotype of six selenoprotein genes
| Gene | SNP | Genotype | N | serum Se (ng/ml) | GPx_activity1 | Correlation | ||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | |||||
| GPx1 | rs1050450 | C/C | 243 | 112.1 (1.2) | 72.6–181.6 | 15.49 (7.24) | 0.83–38.81 | r = 0.151, P = 0.019 |
| C/T | 219 | 110.7 (1.2) | 63.2–195.0 | 13.40 (7.10) | 0.83–37.57 | r = 0.244, P = 0.0003 | ||
| T/T | 40 | 111.2 (1.2) | 64.0–222.7 | 13.56 (7.07) | 2.07–34.27 | r = 0.094, P = 0.564 | ||
| SELS | rs28665122 | A/A | 7 | 92.6 (1.1) | 75.8–102.6 | 10.44 (6.64) | 3.72–21.88 | r = 0.367, P = 0.418 |
| A/G | 87 | 108.2 (1.2) | 75.8–157.9 | 13.64 (7.93) | 0.83–37.16 | r = 0.215, P = 0.047 | ||
| G/G | 275 | 108.5 (1.2) | 65.5–157.9 | 13.72 (6.99) | 0.83–37.48 | r = 0.120, P = 0.049 | ||
| SEPP1 | rs3877899 | A/A | 24 | 107.1 (1.3) | 75.8–195.0 | 12.97 (4.63) | 4.96–26.84 | r = 0.132, P = 0.539 |
| A/G | 189 | 109.7 (1.2) | 63.2–170.5 | 14.74 (7.24) | 0.83–37.57 | r = 0.222, P = 0.002 | ||
| G/G | 285 | 113.0 (1.2) | 65.5–222.7 | 14.39 (7.39) | 0.83–38.81 | r = 0.175, P = 0.003 | ||
| SELS | rs4965373 | A/A | 57 | 112.2 (1.2) | 79.0–195.0 | 14.39 (7.26) | 2.07–38.81 | r = 0.208, P = 0.124 |
| A/G | 213 | 113.0 (1.2) | 64.0–222.7 | 14.55 (6.97) | 1.24–37.57 | r = 0.217, P = 0.002 | ||
| G/G | 232 | 109.8 (1.2) | 63.2–195.0 | 14.32 (7.48) | 0.83–37.48 | r = 0.161, P = 0.015 | ||
| SEP15 | rs5845 | C/C | 315 | 111.1 (1.2) | 63.2–222.7 | 14.20 (7.48) | 0.83–38.81 | r = 0.196, P = 0.0005 |
| C/T | 172 | 111.9 (1.2) | 72.6–195.0 | 14.67 (6.90) | 0.83–35.92 | r = 0.161, P = 0.036 | ||
| T/T | 15 | 114.0 (1.2) | 75.8–157.9 | 16.61 (4.76) | 9.08–22.30 | r = 0.416, P = 0.139 | ||
| GPx4 | rs713041 | C/C | 156 | 111.9 (1.2) | 72.6–157.9 | 14.48 (8.03) | 0.83–37.57 | r = 0.206, P = 0.010 |
| C/T | 241 | 110.4 (1.2) | 63.2–222.7 | 14.22 (6.53) | 0.83–37.16 | r = 0.223, P = 0.0005 | ||
| T/T | 105 | 113.3 (1.2) | 79.0–195.0 | 14.83 (7.55) | 2.89–38.81 | r = 0.088, P = 0.374 | ||
Correlations are based on linear regression line fitting (Selenium = serum Se level, SD = standard deviation)
Bold values is statistically significant
Table 5.
Correlation of serum Se and TR activity by genotype of six selenoprotein genes
| Gene | SNP | Genotype | N | serum Se (ng/ml) | TR Activity | Correlation | ||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | |||||
| GPx1 | rs1050450 | C/C | 243 | 112.1 (1.2) | 72.6–181.6 | 1.10 (0.79) | 0–3.15 | r = 0.189, P = 0.003 |
| C/T | 219 | 110.7 (1.2) | 63.2–195.0 | 1.08 (0.74) | 0–3.78 | r = 0.110, P = 0.105 | ||
| T/T | 40 | 111.2 (1.2) | 64.0–222.7 | 0.88 (0.75) | 0–2.57 | r = 0.258, P = 0.109 | ||
| SELS | rs28665122 | A/A | 7 | 92.6 (1.1) | 75.8–102.6 | 0.76 (0.53) | 0–1.37 | r = 0.258, P = 0.577 |
| A/G | 87 | 108.2 (1.2) | 75.8–157.9 | 1.01 (0.67) | 0–3.15 | r = 0.004, P = 0.970 | ||
| G/G | 275 | 108.5 (1.2) | 65.5–157.9 | 1.00 (0.77) | 0–3.78 | r = 0.137, P = 0.023 | ||
| SEPP1 | rs3877899 | A/A | 24 | 107.1 (1.3) | 75.8–195.0 | 1.10 (0.70) | 0–2.36 | r = 0.087, P = 0.688 |
| A/G | 189 | 109.7 (1.2) | 63.2–170.5 | 1.10 (0.76) | 0–3.78 | r = 0.063, P = 0.384 | ||
| G/G | 285 | 113.0 (1.2) | 65.5–222.7 | 1.06 (0.78) | 0–3.68 | r = 0.224, P = 0.0001 | ||
| SELS | rs4965373 | A/A | 57 | 112.2 (1.2) | 79.0–195.0 | 1.30 (0.81) | 0–2.68 | r = 0.201, P = 0.137 |
| A/G | 213 | 113.0 (1.2) | 64.0–222.7 | 1.01 (0.75) | 0–3.68 | r = 0.199, P = 0.004 | ||
| G/G | 232 | 109.8 (1.2) | 63.2–195.0 | 1.08 (0.76) | 0–3.78 | r = 0.108, P = 0.104 | ||
| SEP15 | rs5845 | C/C | 315 | 111.1 (1.2) | 63.2–22.7 | 1.06 (0.76) | 0–3.78 | r = 0.120, P = 0.068 |
| C/T | 172 | 111.9 (1.2) | 72.6–195.0 | 1.08 (0.76) | 0–3.26 | r = 0.148, P = 0.053 | ||
| T/T | 15 | 114.0 (1.2) | 75.8–157.9 | 1.42 (0.82) | 0–3.05 | r = 0.167,P = 0.029 | ||
| GPx4 | rs713041 | C/C | 156 | 111.9 (1.2) | 72.6–157.9 | 1.12 (0.80) | 0–3.68 | r = −0.020, P = 0.944 |
| C/T | 241 | 110.4 (1.2) | 63.2–222.7 | 1.10 (0.72) | 0–3.26 | r = 0.184, P = 0.004 | ||
| T/T | 105 | 113.3 (1.2) | 79.0–195.0 | 0.95 (0.81) | 0–3.78 | r = 0.204, P = 0.037 | ||
Correlations are based on linear regression line fitting (Selenium = serum Se level, SD = standard deviation)
Bold values are statistically significant
Table 6.
Correlation of serum Se and fresh blood DNA damage by genotype of six selenoprotein genes
| Gene | SNP | Genotype | N | serum Se (ng/ml) | Fresh blood DNA damage | Correlation | ||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | |||||
| GPx1 | rs1050450 | C/C | 243 | 112.1 (1.2) | 72.6–181.6 | 6.06 (1.48) | 3.12–12.37 | r = −0.079, P = 0.222* |
| C/T | 219 | 110.7 (1.2) | 63.2–195.0 | 5.97 (1.30) | 3.38–10.0 | r = −0.007, P = 0.919 | ||
| T/T | 40 | 111.2 (1.2) | 64.0–222.7 | 6.02 (1.31) | 3.70–8.74 | r = 0.133, P = 0.421 | ||
| SELS | rs28665122 | A/A | 7 | 92.6 (1.1) | 75.8–102.6 | 6.66 (1.28) | 4.31–8.27 | r = 0.034, P = 0.943 |
| A/G | 87 | 108.2 (1.2) | 75.8–157.9 | 5.92 (1.47) | 3.14–10.0 | r = −0.012, P = 0.910 | ||
| G/G | 275 | 108.5 (1.2) | 65.5–157.9 | 6.14 (1.48) | 3.12–12.37 | r = −0.062, P = 0.304 | ||
| SEPP1 | rs3877899 | A/A | 24 | 107.1 (1.3) | 75.8–195.0 | 6.38 (2.18) | 4.03–12.37 | r = −0.279, P = 0.197 |
| A/G | 189 | 109.7 (1.2) | 63.2–170.5 | 6.05 (1.39) | 3.14–10.85 | r = −0.063, P = 0.390 | ||
| G/G | 285 | 113.0 (1.2) | 65.5–222.7 | 5.97 (1.30) | 3.12–11.03 | r = 0.032, P = 0.593 | ||
| SELS | rs4965373 | A/A | 57 | 112.2 (1.2) | 79.0–195.0 | 6.14 (1.25) | 3.51–8.98 | r = −0.068, P = 0.618 |
| A/G | 213 | 113.0 (1.2) | 64.0–222.7 | 5.99 (1.41) | 3.22–12.37 | r = 0.036, P = 0.608 | ||
| G/G | 232 | 109.8 (1.2) | 63.2–195.0 | 6.01 (1.41) | 3.12–11.03 | r = −0.082, P = 0.219 | ||
| SEP15 | rs5845 | C/C | 315 | 111.1 (1.2) | 63.2–222.7 | 6.00 (1.41) | 3.12–12.37 | r = −0.008, P = 0.887 |
| C/T | 172 | 111.9 (1.2) | 72.6–195.0 | 6.06 (1.37) | 3.14–11.57 | r = −0.088, P = 0.256 | ||
| T/T | 15 | 114.0 (1.2) | 75.8–157.9 | 5.84 (1.36) | 3.89–9.18 | r = 0.085, P = 0.772 | ||
| GPx4 | rs713041 | C/C | 156 | 111.9 (1.2) | 72.6–157.9 | 6.03 (1.44) | 3.22–12.37 | r = −0.132, P = 0.100 |
| C/T | 241 | 110.4 (1.2) | 63.2–222.7 | 6.05 (1.41) | 3.14–11.57 | r = 0.013, P = 0.837 | ||
| T/T | 105 | 113.3 (1.2) | 79.0–195.0 | 5.93 (1.27) | 3.12–10.85 | r = 0.022, P = 0.828* | ||
Correlations are based on linear regression line fitting (Selenium = serum Se level, SD = standard deviation)
* showed the quadratic distribution therefore subsequently fitted with broken stick regression
Fig. 2.
Variation of biomarker levels with serum Se level among all subjects, according to genotype (line fitted with 95% CI), for those data sets best fitted by a broken stick regression. GPx1 rs1050450 C/C and GPx4 rs713041 T/T genotypes were better fitted using the broken stick regression with the lowest RSS at 116.07 and 149.23 ng/ml serum Se respectively (P = 0.044 and P = 0.042, respectively)
Table 7.
Correlation of serum Se and peroxide-challenged DNA damage, by genotype of six selenoprotein genes
| Gene | SNP | Genotype | N | serum Se (ng/ml) | DNA damage with peroxide challenge | Correlation | ||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Range | Mean (SD) | Range | |||||
| GPx1 | rs1050450 | C/C | 243 | 112.1 (1.2) | 72.6–181.6 | 7.59 (1.86) | 3.28–14.78 | r = 0.089, P = 0.169 |
| C/T | 219 | 110.7 (1.2) | 63.2–195.0 | 7.47 (1.65) | 4.35–13.19 | r = 0.024, P = 0.731 | ||
| T/T | 40 | 111.2 (1.2) | 64.0–222.7 | 7.63 (1.86) | 2.97–11.74 | r = 0.184, P = 0.255 | ||
| SELS | rs28665122 | A/A | 7 | 92.6 (1.1) | 75.8–102.6 | 7.49 (0.73) | 6.52–8.44 | r = −0.553, P = 0.198 |
| A/G | 87 | 108.2 (1.2) | 75.8–157.9 | 7.66 (1.62) | 4.74–13.22 | r = 0.043, P = 0.692 | ||
| G/G | 275 | 108.5 (1.2) | 65.5–157.9 | 7.32 (1.73) | 2.97–13.27 | r = −0.050, P = 0.412 | ||
| SEPP1 | rs3877899 | A/A | 24 | 107.1 (1.3) | 75.8–195.0 | 7.47 (2.14) | 2.97–11.29 | r = 0.173, P = 0.429 |
| A/G | 189 | 109.7 (1.2) | 63.2–170.5 | 7.42 (1.89) | 4.46–14.69 | r = 0.061, P = 0.406 | ||
| G/G | 285 | 113.0 (1.2) | 65.5–222.7 | 7.64 (1.66) | 3.28–14.78 | r = 0.065, P = 0.275 | ||
| SELS | rs4965373 | A/A | 57 | 112.2 (1.2) | 79.0–195.0 | 7.52 (1.80) | 4.35–13.19 | r = −0.061, P = 0.656 |
| A/G | 213 | 113.0 (1.2) | 64.0–222.7 | 7.55 (1.66) | 4.46–14.78 | r = 0.201, P = 0.003 | ||
| G/G | 232 | 109.8 (1.2) | 63.2–195.0 | 7.53 (1.87) | 2.97–14.69 | r = −0.003, P = 0.969 | ||
| SEP15 | rs5845 | C/C | 315 | 111.1 (1.2) | 63.2–222.7 | 7.51 (1.70) | 2.97–14.78 | r = 0.061, P = 0.281 |
| C/T | 172 | 111.9 (1.2) | 72.6–195.0 | 7.63 (1.88) | 4.35–14.69 | r = 0.102, P = 0.188 | ||
| T/T | 15 | 114.0 (1.2) | 75.8–157.9 | 7.08 (1.91) | 5.01–10.81 | r = −0.028, P = 0.924 | ||
| GPx4 | rs713041 | C/C | 156 | 111.9 (1.2) | 72.6–157.9 | 7.48 (1.80) | 3.84–14.78 | r = 0.044, P = 0.591 |
| C/T | 241 | 110.4 (1.2) | 63.2–222.7 | 7.58 (1.74) | 4.35–13.23 | r = 0.051, P = 0.433 | ||
| T/T | 105 | 113.3 (1.2) | 79.0–195.0 | 7.54 (1.81) | 2.97–13.22 | r = 0.177, P = 0.077 | ||
Correlations are based on linear regression line fitting (Selenium = serum Se level, SD = standard deviation)
Bold value are statistically significant
Discussion
A previous group of men from Auckland, New Zealand, that were considered high risk for prostate cancer, had a mean serum Se level of 96.3 ± 1.2 ng/ml (Karunasinghe et al. 2006). In comparison, the current study group, considered as having normal cancer risk, had a relatively higher serum Se level, of 111.6 ± 1.2 ng/ml. Given the time disparity, we cannot be certain whether these differences are associated with prostate cancer risk, or possibly imply a changing Auckland Se status. The current study also shows the possibility of smokers utilising their serum Se faster than nonsmokers and therefore ending up with significantly lower serum Se levels. Reduced levels of Se among smokers are also reported by Lloyd et al. (1983). This study has shown positive correlations of serum Se with GPx1 and TR activities in the study population, as previously described (Karunasinghe et al. 2006; Thomson et al. 2009). Se is known to increase DNA repair activities (Fischer et al. 2007; Kotsopoulos et al. 2010; Kumar et al. 2010), including base excision repair enzyme activities (Dziaman et al. 2009). The increase of DNA damage, quantitated in peroxide challenged leukocytes with increasing Se levels and subsequent TR and GPX activities, could be a consequence of increased DNA repair mechanisms taking place. As previously suggested (Hesketh 2008; Rayman 2009), selenoprotein activity, expression and functions could vary depending on selenoprotein SNPs. Therefore, we further analysed our data to see possible trends with a few relevant gene polymorphisms.
The significant association of GPx activity with serum Se level seen among GPx1 rs1050450 CC genotype individuals is similar to observations made by Xiong et al. (2010). There are other significant correlations between selenium and GPx activity, but these differ in exact nature according to genotype, as shown in Table 4. It is possible that the increases shown by the minor alleles were nonsignificant due to the low numbers of subjects carrying those alleles. Genotypic variation in correlations between Se and GPx activity have been reported before by Jablonska et al. 2009. Their study has shown correlation strengths of GPx rs1050450 genotypes in the order of the genotypes CC > CT > TT. However, in our study, the CT genotype has the highest correlation between GPx activity and serum Se level. Various GPx polymorphisms can alter the structure and the stability of the respective mRNAs and protein products (Zhuo and Diamond 2009). Using Caco-2 cell variants carrying either C or T alleles of GPx4 rs713041 SNP, Gautrey et al. (2011) have shown the variability of GPx4 expression among variants in relation to increasing Se. The relevance of the seleno SNPs analysed here is further emphasised by the fact that the minor alleles of some of them are associated with cancer etiologies (Ichimura et al. 2004, Zhuo and Diamond 2009, Ratnasinghe et al. 2000, Bermano et al. 2007, Méplan et al. 2010, Udler et al. 2007). The lower response in producing GPx activity in proportion to the available serum Se among homozygous minor allele carriers of GPx1 rs1050450 and GPx4 rs713041 SNPs could be at least partially responsible for the cancer etiologies mentioned above.
The current analysis also reports significant variability of TR activity levels among some genotypes, with the strongest correlation shown by SEPP1 rs3877899 GG genotype (P = 0.0001). Similar to our results, Meplan et al. (2007) has seen no significant difference in erythrocyte TR activity level between SEPP1 rs3877899 genotypes among men. However, their study showed that females with the SEPP1 rs3877899 GG genotype have a significantly higher erythrocyte TR activity compared with those with the GA genotype. SEPP1 rs3877899 SNP is located in the N-terminal domain of this gene and results in conversion of the alanine at position 234 to threonine (A234T, G > A), affecting the selenoprotein P levels in vivo (Al-Taie et al. 2004; Meplan et al. 2007). According to Meplan et al. (2009), the homozygous GG genotype produces more of 60 kDa SEPP1, while the GA genotype produces more of the 50-kDa isoform. The levels of TR could therefore be related to the 60-kDa selenoprotein P produced by the G allele.
Kumaraswamy et al. (2000) and Hu et al. (2001) have located rs5839 to be within the site proven as a functional selenocysteine insertion sequence element, and shown that the A allele has a better Se incorporation than the G allele in Se-depleted medium. Although a significant correlation between serum Se and TR activity is seen among carriers of SEL15 rs5845 T alleles, it was those with one or two C alleles that showed a significant correlation with GPx activity.
Those subjects with the SEL-S rs28665122 GG genotype showed a significant correlation of TR activity with serum Se, while those carrying one or two G alleles showed significant correlations with GPx activity. According to Curran et al. (2005), the G allele of this SNP was required to produce sufficient promoter activity in the presence of a stress stimulator, whereas the A allele leads to significantly lower activities. Typical with its location in a region on chromosome 15q26.3, described as carrying loci influencing inflammatory disorders, the promoter variants in the SELS gene are also associated with plasma levels of inflammatory cytokines IL-1β and TNF-α (Curran et al. 2005). Therefore, the G allele that produces sufficient promoter activity could be responsible for signalling the production of sufficient GPx and TR activities, which could be directly or indirectly related to keeping the cytokine production under control.
SNP genotype stratification has not shown any significant correlations between serum Se and DNA damage, except for increased damage shown by the SELS rs4965373 AG genotype when challenged with peroxide. This is the first reference to functionality of the SELS rs4965373 SNP. As the SELS gene is related to protein folding functions, it could be hypothesised that heterozygotes have an impaired function with increasing serum Se and causing stress in the cellular environment.
The broken stick regression shows that GPx1 rs1050450 CC and GPx4 rs713041 TT genotype carriers have a significant advantage against inherent DNA damage up to 116 and 149 ng/ml serum Se, respectively. These data could be used as a tool to optimize the Se requirements in the present study cohort, if they have either GPx1 rs1050450 CC or GPx4 rs713041 TT genotype. What is apparent from the other genotypes is that, while several respond positively to increasing Serum Se, there may be some that are significantly disadvantaged, as judged by the recorded seleno enzyme activity data and DNA stability levels.
In conclusion, this study has shown the relationship of the activity levels of GPx and TR to serum Se levels in a group of men without any cancers or urological problems, and the subsequent effects of the Se on stability of DNA. The present data, together with other studies on the effects of SNPs on selenoprotein metabolism (Hesketh 2008; Rayman 2009), suggest that the basic Se requirement in humans may vary with genotype for a number of variations in selenoprotein genes. However, in the absence of this information, we would recommend a serum Se concentration in the region of 100–150 ng/ml as providing a useful compromise.
Acknowledgments
The authors are grateful to the Cancer Society of New Zealand for the financial support provided for this study between June 2006 and July 2009. The incidental grants during 2006–2009 from the Auckland Medical Research Foundation are also appreciated. The authors also acknowledge the contributions made by Dr. Lotte Risom, and are grateful to the volunteers who took part in the study and appreciate their time and involvement in the study.
References
- Al-Taie OH, Seufert J, Mork H, Treis H, Mentrup B, Thalheimer A, Starostik P, Abel J, Scheurlen M, Köhrle J, Jakob F. A complex DNA-repeat structure within the Selenoprotein P promoter contains a functionally relevant polymorphism and is genetically unstable under conditions of mismatch repair deficiency. Eur J Hum Genet. 2002;10:499–504. doi: 10.1038/sj.ejhg.5200811. [DOI] [PubMed] [Google Scholar]
- Al-Taie OH, Uceyler N, Eubner U, Jakob F, Mörk H, Scheurlen M, Brigelius-Flohe R, Schöttker K, Abel J, Thalheimer A, Katzenberger T, Illert B, Melcher R, Köhrle J. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer. 2004;48:6–14. doi: 10.1207/s15327914nc4801_2. [DOI] [PubMed] [Google Scholar]
- Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422:11–22. doi: 10.1042/BJ20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermano G, Pagmanditis V, Holloway N, Kadri S, Mowat NA, Shiel RS, Arthur JR, Mathers JC, Daly AK, Broom J, Hesketh JE. Evidence that a polymorphism within the 3_UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutr. 2007;2:227–232. doi: 10.1007/s12263-007-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleys J, Navas-Acien A, Guallar E. Serum selenium levels and all-cause, cancer, and cardiovascular mortality among US adults. Arch Intern Med. 2008;168:404–410. doi: 10.1001/archinternmed.2007.74. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R. Glutathione peroxidases and redoxregulated transcription factors. Biol Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohé R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/S0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Ho YS, Ross DA, Valentine BA, Combs GF, Lei XG. Cellular glutathioneperoxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127:1445–1450. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Combs GF, Lei XG. Knockout of cellular glutathione peroxidase affects selenium-dependent parameters similarly in mice fed adequate and excessive dietary selenium. Biofactors. 1998;7:311–321. doi: 10.1002/biof.5520070403. [DOI] [PubMed] [Google Scholar]
- Cook ED, Moody-Thomas S, Anderson KB, Campbell R, Hamilton SJ, Harrington JM, Lippman SM, Minasian L, Paskett E, Craine S, Arnold KB, Probstfield JL. Minority recruitment to the Selenium and Vitamin E Cancer Prevention Trial (SELECT) Clinical trials (London, England) 2005;2:436–442. doi: 10.1191/1740774505cn111oa. [DOI] [PubMed] [Google Scholar]
- Cooper ML, Adami H-O, Gronberg H, Wiklund F, Green FR, Rayman MP. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 2008;68:10171–10177. doi: 10.1158/0008-5472.CAN-08-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JE, Jowett JB, Elliott KS, Gao Y, Gluschenko K, Wang J, Abel Azim DM, Cai G, Mahaney MC, Comuzzie AG, Dyer TD, Walder KR, Zimmet P, MacCluer JW, Collier GR, Kissebah AH, Blangero J. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37:1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- Dreher I, Jakobs TC, Köhrle J. Cloning and characterization of the human selenoprotein P promoter. J Biol Chem. 1997;272:29364–29371. doi: 10.1074/jbc.272.46.29364. [DOI] [PubMed] [Google Scholar]
- Dziaman T, Huzarski T, Gackowski D, Rozalski R, Siomek A, Szpila A, Guz J, Lubinski J, Wasowicz W, Roszkowski K, Olinski R. Selenium supplementation reduced oxidative DNA damage in adnexectomized BRCA1 mutations carriers. Cancer Epidemiol Biomarkers Prev. 2009;18:2923–2928. doi: 10.1158/1055-9965.EPI-09-0529. [DOI] [PubMed] [Google Scholar]
- Ferguson LR, Karunasinghe N (2011) Nutrigenetics, nutrigenomics, and selenium. Front Genet 2:1–11 [DOI] [PMC free article] [PubMed]
- Ferguson LR, Philpott M, Karunasinghe N. Oxidative DNA damage and repair: significance and biomarkers. J Nutr. 2006;136:2687S–2689S. doi: 10.1093/jn/136.10.2687S. [DOI] [PubMed] [Google Scholar]
- Ferguson LR, Han DY, Fraser AG, Huebner C, Lam WJ, Morgan AR, Duan H, Karunasinghe N. Genetic factors in chronic inflammation: single nucleotide polymorphisms in the STAT-JAK pathway, susceptibility to DNA damage and Crohn’s disease in a New Zealand population. Mutat Res Fundam Mol Mech Mutagen. 2010;690:108–115. doi: 10.1016/j.mrfmmm.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Fischer JL, Mihelc EM, Pollok KE, Smith ML. Chemotherapeutic selectivity conferred by selenium: a role for p53-dependent DNA repair. Mol Cancer Ther. 2007;6:355–361. doi: 10.1158/1535-7163.MCT-06-0472. [DOI] [PubMed] [Google Scholar]
- Gao Y, Hannah N, Wanyoni S, Konstantopolous N, Pagnon J, Feng HC, Jowett JBM, Walder K, Collier GR. Activation of the Selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells. Cytokine. 2006;33:246–251. doi: 10.1016/j.cyto.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Gautrey HNF, Sneddon AA, Hall J, Hesketh J. A T/C polymorphism in the GPX4 3′UTR affects the selenoprotein expression pattern and cell viability in transfected Caco-2 cells. Biochim Biophys Acta. 2011;10:284–291. doi: 10.1016/j.bbagen.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh J. Nutrigenomics and selenium: gene expression patterns, physiological targets, and genetics. Annu Rev Nutr. 2008;28:157–177. doi: 10.1146/annurev.nutr.28.061807.155446. [DOI] [PubMed] [Google Scholar]
- Hesketh J, Meplan C (2011) Transcriptomics and functional genetic polymorphisms as biomarkers of micronutrient function: focus on selenium as an exemplar. Proc Nutr Soc 3:1–9 [DOI] [PubMed]
- Hu YJ, Korotkov KV, Mehta R, Hatfield DL, Rotimi CN, Luke A, Prewitt TE, Cooper RS, Stock WV, Everett E, Dolan ME, Gladyshev VN, Diamond AM. Distribution and functional consequences of nucleotide polymorphisms in the 3′-untranslated region of the human Sep15 gene. Cancer Res. 2001;61:2307–2310. [PubMed] [Google Scholar]
- Ichimura Y, Habuchi T, Tsuchiya N, Wang L, Oyama C, Sato K, Nishiyama H, Ogawa O, Kato T. Increased risk of bladder cancer associated with a glutathione peroxidase 1 codon 198 variant. J Urol. 2004;172:728–732. doi: 10.1097/01.ju.0000130942.40597.9d. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman RR. A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. doi: 10.2307/1390807. [DOI] [Google Scholar]
- Jablonska E, Gromadzinska J, Reszka E, Wasowicz W, Sobala W, Szeszenia-Dabrowska N, Boffetta P. Association between GPx1 Pro198Leu polymorphism, GPx1 activity and plasma selenium concentration in humans. Eur J Nutr. 2009;48:383–386. doi: 10.1007/s00394-009-0023-0. [DOI] [PubMed] [Google Scholar]
- Johnson CC, Fordyce FM, Rayman MP. Symposium on Geographical and geological influences on nutrition: Factors controlling the distribution of selenium in the environment and their impact on health and nutrition. Proc Nutr Soc. 2010;69:119–132. doi: 10.1017/S0029665109991807. [DOI] [PubMed] [Google Scholar]
- Kant AK, Graubard BI. Ethnicity is an independent correlate of biomarkers of micronutrient intake and status in American adults. J Nutr. 2007;137:2456–2463. doi: 10.1093/jn/137.11.2456. [DOI] [PubMed] [Google Scholar]
- Karunasinghe N, Ryan J, Tuckey J, Masters J, Jamieson M, Clarke C, Marshall R, Ferguson LR. DNA Stability and Serum Selenium Levels in a High-Risk Group for Prostate Cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:391–397. [PubMed] [Google Scholar]
- Karunasinghe N, Ferguson LR, Tuckey J, Masters J. Hemolysate thioredoxin reductase and glutathione peroxidase activities correlate with serum selenium in a group of New Zealand men at high prostate cancer risk. J Nutr. 2006;136:2232–2235. doi: 10.1093/jn/136.8.2232. [DOI] [PubMed] [Google Scholar]
- Kelly E, Greene CM, Carroll TP, McElvaney NG, O’Neill SJ. Selenoprotein S/SEPS1 modifies endoplasmic reticulum stress in Z variant alpha1-antitrypsin deficiency. J Biol Chem. 2009;284:16891–16897. doi: 10.1074/jbc.M109.006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsopoulos J, Chen Z, Vallis KA, Poll A, Ghadirian P, Kennedy G, Ainsworth P, Narod SA. Toenail selenium status and DNA repair capacity among female BRCA1 mutation carriers. Cancer Causes Control. 2010;21:679–687. doi: 10.1007/s10552-009-9495-8. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pollok KE, Smith ML. Selenomethionine or methylseleninic acid inhibits mutagenesis of a reporter gene in mouse bone marrow. Anticancer Res. 2010;30:291–293. [PubMed] [Google Scholar]
- Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, Hatfield DL, Diamond AM, Gladyshev VN. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J Biol Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- Li H, Kantoff PW, Giovannucci E, Leitzmann MF, Gaziano JM, Stampfer MJ, Ma J. Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 2005;6:2498–2504. doi: 10.1158/0008-5472.CAN-04-3535. [DOI] [PubMed] [Google Scholar]
- Lloyd B, Lloyd RS, Clayton BE. Effect of smoking, alcohol and other factors on the selenium status of a healthy population. J Epidemiol Community Health. 1983;37:213–217. doi: 10.1136/jech.37.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Santiago JL, Varade J, Marquez A, Lamas JR, Mendoza JL, la Calle H, Diaz-Rubio M, Concha EG, Fernandez-Gutierrez B, Urcelay E. Polymorphisms in the selenoprotein S gene: lack of association with autoimmune inflammatory diseases. BMC Genomics. 2008;9:329. doi: 10.1186/1471-2164-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meplan C, Crosley LK, Nicol F, Beckett GJ, Howie AF, Hill KE, Horgan G, Mathers JC, Arthur JR, Hesketh JE. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study) FASEB J. 2007;21:3063–3074. doi: 10.1096/fj.07-8166com. [DOI] [PubMed] [Google Scholar]
- Meplan C, Crosley LK, Nicol F, Horgan GW, Mathers JC, Arthur JR, Hesketh JE. Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: interaction with sex. Am J Clin Nutr. 2008;87:1019–1027. doi: 10.1093/ajcn/87.4.1019. [DOI] [PubMed] [Google Scholar]
- Meplan C, Nicol F, Burtle BT, Crosley LK, Arthur JR, Mathers JC, Hesketh JE. Relative abundance of selenoprotein P isoforms in human plasma depends on genotype, Se intake and cancer status. Antioxid Redox Signal. 2009;11:2631–2640. doi: 10.1089/ars.2009.2533. [DOI] [PubMed] [Google Scholar]
- Méplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavatá I, Vrána D, Vodicka P, Hesketh JE. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis. 2010;31:1074–1079. doi: 10.1093/carcin/bgq076. [DOI] [PubMed] [Google Scholar]
- Moscow JA, Schmidt L, Ingram DT, Gnarra J, Johnson B, KH C. Loss of heterozygosity of the human cytosolic glutathione peroxidase I gene in lung cancer. Carcinogenesis. 1994;15:2769–2773. doi: 10.1093/carcin/15.12.2769. [DOI] [PubMed] [Google Scholar]
- Mostert V, Lombeck I, Abel J. A novel method for the purification of selenoprotein P from human plasma. Arch Biochem Biophys. 1998;357:326–330. doi: 10.1006/abbi.1998.0809. [DOI] [PubMed] [Google Scholar]
- Mostert V, Wolff S, Dreher I, Kohrle J, Abel J. Identification of an element within the promoter of human selenoprotein P responsive to transforming growth factor-beta. Eur J Biochem. 2001;268:6176–6181. doi: 10.1046/j.0014-2956.2001.02565.x. [DOI] [PubMed] [Google Scholar]
- Ratnasinghe D, Tangrea JA, Andersen MR, Barrett MJ, Virtamo J, Taylor PR, Albanes D. Glutathione peroxidase codon 198 polymorphism variant increases lung cancer risk. Cancer Res. 2000;60:6381–6383. [PubMed] [Google Scholar]
- Rayman MP. Selenoproteins and human health: insights from epidemiological data. Biochim Biophys Acta. 2009;1790:1533–1540. doi: 10.1016/j.bbagen.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Rongpu Y, Jiachen H, Gongkan F, Zhengen M. Fluorometric determination of micro-amounts of selenium in human blood, using 2, 3-diaminonaphthalene. Med Lab Sci. 1986;43:331–334. [PubMed] [Google Scholar]
- Seber GAF (1938) Linear regression analysis. In: Seber GAF, Lee AJ (eds), 2nd edn. pp 159–161. Wiley-Interscience, Hoboken, NJ: c2003
- Selenius M, Rundlöf AK, Olm E, Fernandes AP, Björnstedt M. Selenium and the selenoprotein thioredoxin reductase in the prevention, treatment and diagnostics of cancer. Antioxid Redox Signal. 2010;12:867–880. doi: 10.1089/ars.2009.2884. [DOI] [PubMed] [Google Scholar]
- Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, Gladyshev VN. Structure-function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid Redox Signal. 2010;12:839–849. doi: 10.1089/ars.2009.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Levander OA. High-throughput 96-well microplate assays for determining specific activities of glutathione peroxidase and thioredoxin reductase. Methods Enzymol. 2002;347:113–121. doi: 10.1016/S0076-6879(02)47012-7. [DOI] [PubMed] [Google Scholar]
- Thomas JP, Geiger PG, Maiorino M, Ursini F, Girotti AW. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim Biophys Acta. 1990;1045:252–260. doi: 10.1016/0005-2760(90)90128-k. [DOI] [PubMed] [Google Scholar]
- Thomson CD, McLachlan SK, Parnell WR, Wilson N, Wohlers M, Scragg R, Schaaf D, Fitzgerald ED. Serum selenium concentrations and dietary selenium intake of New Zealand children aged 5–14 years. Br J Nutr. 2007;97:357–364. doi: 10.1017/S0007114507336738. [DOI] [PubMed] [Google Scholar]
- Thomson CD, Campbell JM, Miller J, Skeaff SA, Livingstone V. Selenium and iodine supplementation: effect on thyroid function of older New Zealanders. Am J Clin Nutr. 2009;90:1038–1046. doi: 10.3945/ajcn.2009.28190. [DOI] [PubMed] [Google Scholar]
- Udler M, Maia AT, Cebrian A, Brown C, Greenberg D, Shah M, Caldas C, Dunning A, Easton D, Ponder B, Pharoah P. Common germline genetic variation in antioxidant defense genes and survival after diagnosis of breast cancer. J Clin Oncol. 2007;25:3015–3023. doi: 10.1200/JCO.2006.10.0099. [DOI] [PubMed] [Google Scholar]
- Villette S, Kyle JAM, Brown KM, Pickard K, Milne JS, Nicol F, Arthur JR, Hesketh JE. A novel single nucleotide polymorphism in the 3′ untranslated region of human glutathione peroxidase 4 influences lipoxygenase metabolism. Blood Cells Mol Dis. 2002;29:174–178. doi: 10.1006/bcmd.2002.0556. [DOI] [PubMed] [Google Scholar]
- Watkinson JH. Semi-automated fluorimetric determination of nanogram quantities of selenium in biological materials. Anal Chim Acta. 1979;105:319–325. doi: 10.1016/S0003-2670(01)83763-6. [DOI] [Google Scholar]
- Watkinson JH, Brown MW. New phase separating device and other improvements in the semi-automated fluorimetric determination of selenium. Anal Chim Acta. 1979;105:451–454. doi: 10.1016/S0003-2670(01)83782-X. [DOI] [Google Scholar]
- Wendel A. Glutathione peroxidase. Methods Enzymol. 1981;77:325–333. doi: 10.1016/S0076-6879(81)77046-0. [DOI] [PubMed] [Google Scholar]
- Xiong YM, Mo XY, Zou XZ, Song RX, Sun WY, Lu W, Chen Q, Yu YX WJZ. Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin-Beck disease. Osteoarthr Cartil. 2010;18:817–824. doi: 10.1016/j.joca.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Zhuo P, Diamond AM. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochimica et Biophysica Acta, General Subjects. 2009;1790:1546–1554. doi: 10.1016/j.bbagen.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]