Abstract
Undernutrition during pregnancy is implicated in the programming of offspring for the development of obesity and diabetes. We hypothesized that maternal programming causes epigenetic changes in fetal hypothalamic pathways regulating metabolism. This study used sheep to examine the effect of moderate maternal undernutrition (60 d before to 30 d after mating) and twinning to investigate changes in the key metabolic regulators proopiomelanocortin (POMC) and the glucocorticoid receptor (GR) in fetal hypothalami. Methylation of the fetal hypothalamic POMC promoter was reduced in underfed singleton, fed twin, and underfed twin groups (60, 73, and 63% decrease, respectively). This was associated with reduced DNA methyltransferase activity and altered histone methylation and acetylation. Methylation of the hypothalamic GR promoter was decreased in both twin groups and in maternally underfed singleton fetuses (52, 65, and 55% decrease, respectively). This correlated with changes in histone methylation and acetylation and increased GR mRNA expression in the maternally underfed singleton group. Alterations in GR were hypothalamic specific, with no changes in hippocampi. Unaltered levels of OCT4 promoter methylation indicated gene-specific effects. In conclusion, twinning and periconceptional undernutrition are associated with epigenetic changes in fetal hypothalamic POMC and GR genes, potentially resulting in altered energy balance regulation in the offspring.—Begum, G., Stevens, A., Smith, E. B., Connor, K., Challis, J. R. G., Bloomfield, F., White, A. Epigenetic changes in fetal hypothalamic energy regulating pathways are associated with maternal undernutrition and twinning.
Keywords: proopiomelanocortin, glucocorticoid receptor, methylation analysis
There is emerging evidence that epigenetic modifications of genes are implicated in fetal programming. Maternal events that result in programming of the fetus are clearly important factors in increasing the risk of the offspring developing type 2 diabetes (1) and cardiovascular disease as adults (2). The developmental origins of health and disease (DOHaD) hypothesis predicts that fetal recognition of maternal signals relating to the intrauterine environment, such as decreased nutrition, results in adaptations in fetal development (3). These adaptations may be beneficial for the intrauterine environment; however, if the postnatal environment has plentiful nutrition, the adaptations may cause the offspring to overcompensate (4). This has been demonstrated in several human and animal studies with the development of obesity and impaired glucose tolerance or type 2 diabetes in the adult offspring (5–7).
Twinning presents an ideal model to study an alternative effect of nutritional programming. Twinning is known to result in reduced birth weight, which is a common trait associated with fetal changes due to maternal programming (8, 9). Furthermore, twins have an increased propensity to develop abdominal obesity and type 2 diabetes in their adult life (5, 10). This suggests that twinning is an alternative impact that would cause epigenetic changes in the hypothalamus. Twin studies have also been vital for demonstrating that the occurrence of type 2 diabetes is strongly influenced by the fetal environment. This is exemplified by studies demonstrating strong relationships between coefficients of birth weight within-twin pairs and adult insulin sensitivity. There are also studies showing that the twin with the lower birth weight developed type 2 diabetes (11). If twinning is a nutritional paradigm, then any changes that we observe in the fetal hypothalami from mothers subjected to periconceptional undernutrition will be mirrored in twin fetal hypothalami. Therefore, we determined the epigenetic effects of twinning and of maternal undernutrition during the periconceptional period in sheep.
Studies investigating the effects of twinning and maternal nutritional insults on the development of obesity and type 2 diabetes have focused on peripheral metabolic pathways. While this is important, there is the potential for these insults to act at the arcuate nucleus in the hypothalamus, which is the central regulator of energy balance. A likely candidate for epigenetic changes is the hypothalamic neuropeptide proopiomelanocortin (POMC), which acts to inhibit food intake and modify glucose handling (12). Furthermore, the glucocorticoid receptor (GR) is known to be a regulator of this gene (13–15). Both these genes have been implicated as targets for programming particularly in rodent studies (16–19). We have chosen to use sheep because their hypothalami develop prenatally (20), as in humans, and the model has been fully validated by our previous studies (21). Also, POMC expression within the sheep hypothalamus has previously been shown as early as 110 d during gestation (22).
The POMC region selected is a highly conserved enhancer region that binds to RNA polymerase II and alters POMC expression in the pituitary (21, 23). The region is also closely associated with the neuronal promoter enhancer regions 1 and 2 (nPE1 and nPE2), known to regulate the expression of hypothalamic POMC (24). The GR region is also highly conserved and is associated with a CpG island linked to the transcriptional start sites (25). The interacting histone modifications play a vital role in the overall state of gene activation or repression (26). Key chromatin modifications of gene promoters include histone 3 lysine 9 acetylation (H3K9AC) and histone 3 lysine 4 trimethylation (H3K4me3). These histone states are associated with chromatin opening and gene activation (27, 28). In addition, histone 3 lysine 27 trimethylation (H3K27me3) has been extensively reported as being associated with gene inactivation and compact chromatin (27).
The findings from this investigation show that twinning and undernutrition in the mother around the time of conception alter the epigenetic status of POMC and GR in the hypothalamus. The changes have the potential to result in altered regulation of food intake in offspring when they reach adulthood, leading to the development of obesity. The outcome of this study presents twinning as a nutritional paradigm, leading to specific epigenetic changes in hypothalamic genes centrally involved in the regulation of energy balance.
MATERIALS AND METHODS
Animal management
Authorization for the study was provided by the Animal Ethics Committee at the University of Auckland. Multiparous Romney ewes were fed a concentrate feed of 65% lucerne, 30% barley, and limestone, molasses, and trace elements (CamTech, Cambridge, New Zealand). The ewes were then randomly separated into 2 groups: controls [fed ad libitum at 3–4% of body weight per day (bw/d)] and undernourished from 60 d before to 30 d after mating (−60 to +30). Undernourishment of the ewes was achieved by withholding food for 2 d, followed by individually determined concentrate feeds to induce and sustain a 10–15% reduction in maternal body weight. Initial food intake was at 1–2% bw/d, rising to 80% of that of the controls. After the period of undernourishment, the ewes were fed ad libitum.
Ultrasound scanning was used to determine fetal number at 55 d. Fetal samples were collected by catheterization at 128 d for plasma analysis. Term in untreated ewes is ∼147 d, and so fetal tissue was collected at 131 d (twin fetuses) and 135 d (singleton fetuses) by giving a lethal dose of intravenous pentobarbitone to maternal ewes. Subsequently, the fetuses were weighed and dissected. As previously established, the hypothalamus was dissected and frozen to give an arcuate nucleus enriched ventral hypothalamic region, using the third ventricle as a guide (21, 22, 29). The hippocampus was dissected by taking the right lateral hippocampal region encompassing the dentate gyrus and CA1, CA2, and CA3 regions. The hypothalamic and hippocampal fetal samples from the singleton control and maternally undernourished samples used in this study were investigated previously (21), but for this comparison with twin samples, separate tissue sections were used.
Maternal ewes carrying singletons or twins and subjected to control or maternal undernutrition had similar weights at postmortem (Supplemental Table S1). As expected, there was a reduction in fetal weights in the twins compared with singletons.
Bioinformatic analysis
The POMC and GR marker regions used in this work were compared with genomic data from the Encyclopedia of DNA Elements (ENCODE) Consortium using the University of California, Santa Cruz (UCSC; Santa Cruz, CA, USA) genome browser. The ovine promoter sequence for POMC, GR, and OCT4 was located and determined as described previously (21).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was carried out with the Imprint ChIP kit (Sigma, St. Louis, MO, USA). To release the chromatin, 20 mg of hypothalamic or hippocampal tissue was homogenized and cross-linked with 1% formaldehyde. This was then digested with micrococcal nuclease (2 U/ml; Sigma) to provide ∼500 bp of fragmented genomic DNA. Fragmented chromatin was immunoprecipitated with either 1 μg of RNA polymerase 11 (R1530; Sigma) as the positive control, mouse IgG (M8695; Sigma) as the negative control, or rabbit polyclonal histone H3K27me3, H3K4me3, and H3K9AC (39917, 39155, and 39159, respectively; Active Motif, Carlsbad, CA, USA). Following hydrolyzation of the crosslinks, the DNA was collected and utilized for PCR reactions for POMC and GR. OCT4 was used as the control gene for the assay. The PCR reactions were set up as described previously (21), and the primers used were as follows: GR, 5′-TTTGGAGGGACTGTGGTCC-3′ and 5′-AGCAGGAGGTGGCAGGCC-3′, size = 230 bp; POMC, 5′-ACCCTCAGAGGTGAGAAGCT-3′ and 5′-GGAAGGAGACCGGAGCCG-3′, size = 160 bp; OCT4, 5′-CCTGGATGAGCTTCCAAGG-3′ and 5′-CCTCGGAGTTGCTCTCCCAC-3′, size = 223 bp. The PCR reactions were then resolved on a 2% agarose gel, which was maintained at 80 V for 1 h. ImageJ software (developed by Wayne Rasband, U.S. National Institutes of Health, Bethesda, MD, USA) was then used to quantify the intensity of the bands.
PCR-based methylation analysis
Genomic DNA was isolated from hypothalamic and hippocampal tissue using the AllPrep DNA/RNA kit (Qiagen, Valencia, CA, USA). The MethylCollector kit (Active Motif) was then used to separate methylated genomic DNA from total tissue-specific genomic DNA. The experiment involved digesting 50 ng/μl of total genomic DNA via an MSE1 digest (New England Biolabs, Beverly, MA, USA) to produce CpG methylated DNA fragments. These fragments, along with the positive and negative control DNA provided in the kit, were incubated with a His-tagged recombinant methyl-CpG binding domain 2b (MBD2b) protein. Nickel-coated magnetic beads were used to capture the His-tagged MBD2b-DNA complexes. The beads were then washed with a high-salt buffer to remove unmethylated fragments and incubated with proteinase K to allow the elution of the methylated DNA. To determine the amount of methylated DNA extracted from the total genomic DNA, PCR analysis was performed for POMC, GR, and OCT4, as described above. The initial MSE1-digested DNA was used as the input value and compared with the amount of tissue-specific methylation enriched DNA as the output value.
DNA methyltransferase (DNMT) activity/inhibition assay
Nuclear proteins were extracted from 20 mg hypothalamic tissue using the EpiQuik nuclear extraction kit (Epigentek Group, Farmingdale, NY, USA). The extracts were then utilized in the EpiQuik DNMT/inhibition assay kit (Epigentek). Briefly, the extracts were incubated in wells coated with a cytosine-rich DNA substrate and Adomet containing methyl groups. 5-Methyl cytosine antibody was added, followed by a capture antibody. A developing solution was then applied, allowing the absorbance to be measured colorometrically.
POMC ELISA
POMC levels in fetal samples were quantified by ELISA, as described previously (30, 31). The lower level of sensitivity of the POMC ELISA was determined as 10 pM.
mRNA expression analysis
RNA from hypothalamic and hippocampal tissue was isolated using the AllPrep DNA/RNA kit (Qiagen). RNA samples were reverse transcribed with the QuantiTect reverse transcription kit (Qiagen). The cDNA was used in a SYBR green mastermix (QuantiTect SYBR Green PCR; Qiagen). The primers used in the mastermix have been determined previously (32) and are as follows: POMC, forward 5′-GCTGCTGGTCTTGCTGCTTC-3′ and reverse 5′-CCTGACACTGGCTCGTCTCC-3′; GR, forward 5′-ACTGCCCCAAGTGAAAACAGA-3′ and reverse 5′-ATGAACAGAAATGGCAGACATTTTATT-3′; neuropeptide Y (NPY), forward 5′-TCATCACCAGGCAGAGATACGG-3′ and reverse 5′-GAGCAAGTTTCCCATCACC-3′; 18S, forward 5′-GATGCGGCGGCGTTATTCC-3′ and reverse 5′-CTCCTGGTGGTGCCCTTCC-3′. The reactions were then run on a StepOnePlus Real-Time PCR system thermal cycling block (Applied Biosystems, Foster City, CA, USA). mRNA expression was calculated according to the 2−ΔCt method (33).
Statistical analysis
All statistical analysis was carried out using GraphPad Prism software (GraphPad, La Jolla, CA, USA). All data are shown as means ± se. Analysis was carried out via the 2-way ANOVA. This was followed by the Bonferroni post hoc test. Values of P < 0.05 were considered significantly different.
RESULTS
Selection of the POMC and GR gene promoter marker regions for epigenetic analysis
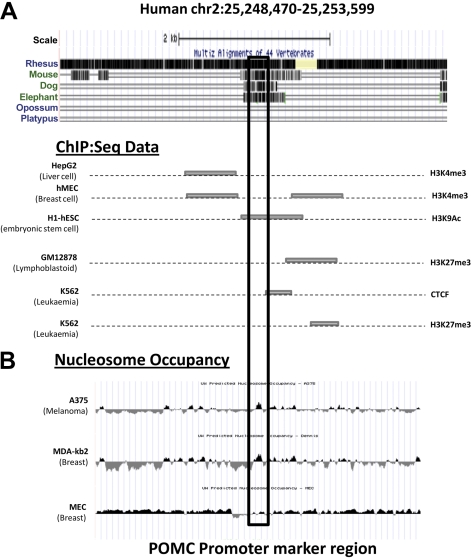
The availability of sequence data in the sheep is limited. Nevertheless, the POMC marker region selected shows high homology across all species analyzed (Fig. 1A). We examined the available data from ENCODE, and while there was no data on hypothalamic cells, ENCODE data were available on 3 cell lines (34–37).
Figure 1.
Summary of ENCODE data from different human cell lines. Data analysis from the UCSC genome browser depicts cell line-specific changes in chromatin over the POMC promoter marker region. A) ChiP:Seq from multiple ENCODE tracks. B) Nucleosome occupancy (28–30).
Multiple sets of ENCODE ChIP sequence (ChIP:seq) data demonstrated cell type-specific changes in histone modifications (Fig. 1A; refs. 37, 38). The presence of histone H3K4me3 and H3K3AC was identified in an embryonic stem cell line, a breast cell line, and a liver cell line, indicating a potentially active enhancer region (26). In contrast, other cell lines demonstrated an increase in H3K27me3 (26) or interaction with the transcriptional repressor CTCF, indicating transcriptionally inactive chromatin (39). The ChIP:seq data also correlated with ENCODE chromatin state segmentation (CSS) data (40, 41). Open heterochromatin was shown to be present across the POMC marker region in HepG2 cells, H1-hESC cells, and HMEC cells, whereas repressed chromatin was present in GM12878 cells and K562 cells (Fig. 1A). CSS also highlighted a putative weak enhancer site immediately 5′ of the POMC marker region.
Nucleosome positioning within promoter and enhancer regions is essential for control of transcriptional activity due to regulation of accessibility to the DNA by transcription factors (34). Nucleosome occupancy within the POMC marker region was found in A375 and MDA-kb2 cells but not in mammary epithelial cells. This suggests that there is some cell type-dependent variation in nucleosome positioning at this site (Fig. 1B).
The POMC gene marker region used in this study also colocalized with an enhancer region that has recently been shown to increase pituitary expression of POMC, with a preference for corticotrope cells (23).
The GR marker region showed evidence of distinct enhancer/promoter activity in all cell lines with available information within the ENCODE database (Supplemental Fig. S1).
Histone modifications of the fetal POMC enhancer region in response to twinning and undernutrition in the mother around conception
To establish a mechanistic insight into the chromatin modifications of the hypothalamic POMC gene, maternal ewes were fed ad libitum or periconceptionally undernourished from 60 d before until 30 d during pregnancy to maintain a 10–15% reduction in body weight. Fetal ventral hypothalami enriched for the arcuate nucleus from twins were compared with tissue samples taken from the ventral hypothalami of control and maternally undernourished singleton fetuses optimized previously (21). This allowed us to determine reproducibility of analysis of multiple hypothalamic biopsies.
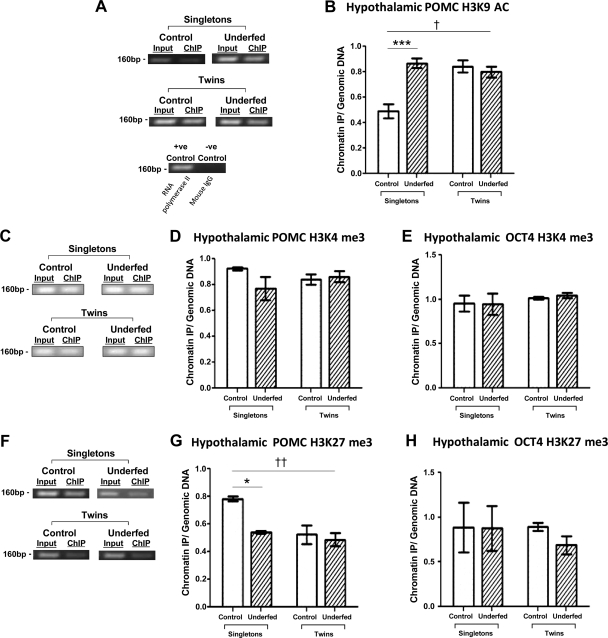
Initially, ChIP analysis of the selected POMC region was assessed for H3K9AC and H3K4me3 as markers of open chromatin in the hypothalami from singleton and twin fetuses. H3K9AC levels for the POMC amplicon were found to be significantly increased in the control twin (43%), underfed twin (40%), and underfed singleton (44%) fetal groups (Fig. 2A, B), compared with the control singletons. Analysis by 2-way ANOVA revealed a significant interaction of singleton-twin status (P<0.05), in that maternal undernutrition did not induce a further additive effect to twinning in the levels of POMC H3K9AC levels. Another marker for transcriptional activation, H3K4me3, demonstrated no alterations in POMC for any of the groups (Fig. 2C, D).
Figure 2.
Histone modifications of the POMC promoter in response to twinning and periconceptional maternal undernutrition. A, C, F) POMC amplicon levels via ChIP analysis as input genomic DNA compared with the enriched output DNA from the singleton control, singleton underfed, twin control, and twin underfed fetal groups for H3K9AC (A), H3K4me3 (C), and H3K27me3 (F). RNA polymerase II was used as positive control; mouse IgG as negative control. Results are representative of n=3 experiments. B, D, G) ChIP analysis of hypothalamic POMC promoter H3K9AC (B), H3K4me3 (D), and H3K27me3 (G) in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs, i.e., 14), and twin underfed (n=8 pairs) groups. E, H) ChIP analysis of the OCT4 promoter H3K4me3 (E) and H3K27me3 (H) levels in the singleton and twin control and underfed groups. Groups were compared with control via 2-way ANOVA with Bonferroni post hoc test. *P < 0.05, ***P < 0.005 for nutritional effect; †P < 0.05, ††P < 0.01 for twin effect.
To further analyze the open or closed status of the POMC chromatin, we measured the H3K27me3 levels as a marker for transcriptional inactivation. In comparison with the control singleton group, H3K27me3 was found to be markedly decreased in the control twin and underfed twin fetal groups to a level similar to the singleton underfed group (34, 39, and 31% decrease, respectively; Fig. 2F, G). The interaction of singleton-twin status was also found to be significantly different (P<0.01). To provide evidence for the role of the selected POMC region in transcription, RNA polymerase II was used as a positive control in the ChIP assay. It was observed that RNA polymerase II consistently bound to the selected POMC region (Fig. 2A).
To establish whether the epigenetic changes we found were gene specific, OCT4, a transcription factor necessary for pluripotency, was used as a control gene for methylation analysis (42). OCT4 presented itself as an ideal candidate gene, as it has previously been shown to be hypermethylated in humans (43). The marker chosen to target this gene is situated within the OCT4 CpG island, with a high level of mammalian conservation (21). The H3K4me3 and H3K27me3 levels of this region remained unchanged across all fetal groups (Fig. 2E, H).
Twinning alters DNA methylation levels of the hypothalamic POMC region in the fetuses
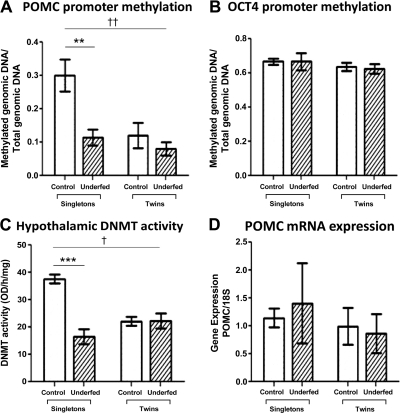
To further characterize the epigenetic status of the POMC gene as a result of twinning, PCR-associated methylation analysis was carried out. POMC enhancer methylation levels in the arcuate nucleus enriched hypothalamic region demonstrated a significant reduction in the control (60%) and twin underfed (73%) twin groups to a level similar to the singleton group from mothers underfed around conception (63%; Fig. 3A). There was also a significant interaction between nutritional status and singleton-twin status (P<0.01). A decrease in POMC promoter methylation levels in these groups was also associated with a reduction in the overall hypothalamic levels of DNMT activity (Fig. 3C).
Figure 3.
DNA methylation and expression levels of fetal hypothalamic neuropeptides. A) POMC promoter methylation levels, in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. B) OCT4 methylation levels in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. C) Overall DNMT activity in the fetal hypothalamus following twinning and periconceptional maternal undernutrition. Levels of DNMT activity were established in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. D) POMC mRNA expression levels determined using qRT-PCR. Groups were compared with control via 2-way ANOVA with Bonferroni post hoc test. **P < 0.01, ***P < 0.005 for nutritional effect; †P < 0.05, ††P < 0.01 for twin effect.
To determine whether the methylation changes associated with the POMC promoter were gene specific, we measured OCT4 amplicon methylation status. No changes were found in the levels of OCT4 marker methylation across any of the groups (Fig. 3B), suggesting that twinning and maternal undernutrition induce gene-specific alterations.
Transcriptional activity of the POMC locus is not activated at the fetal stage
We analyzed mRNA expression of the POMC gene in the ventral hypothalamus by quantitative RT-PCR at the fetal stage of 131 d to establish whether the epigenetic changes translated into changes in transcriptional activity of POMC at this early stage. No changes in POMC were found in the twins or in the groups from undernourished mothers (Fig. 3D). Similarly, no changes were found in NPY, analyzed as an alternative neuropeptide with effects on food intake (Supplemental Fig. S2).
Twinning and nutritional status are associated with histone modifications of the hypothalamic GR
Because of the well-established influence of GR on regulation of food intake, it is a key gene that may be influenced by twinning and maternal undernutrition in the periconceptional period. To study the epigenetic status of GR, a marker for the GR gene promoter situated 5 kb upstream of the exon 2 translational start site was used (Supplemental Fig. S1). Previously published data on the sheep GR promoter revealed this region to be highly conserved and CpG dense (21).
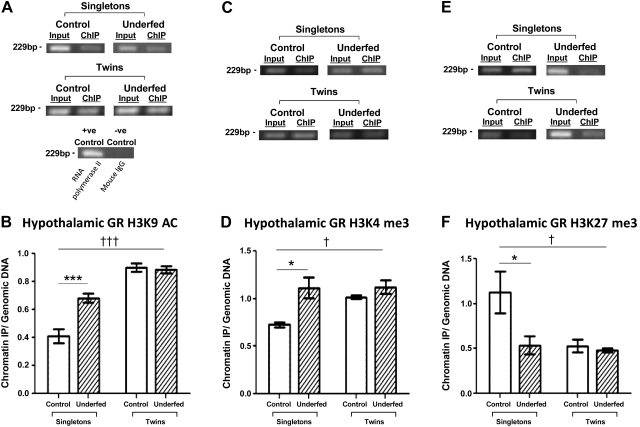
Epigenetic interactions with the GR gene in response to twinning were assessed by ChIP analysis of H3K9AC, H3K4me3, and H3K27me3. The selected GR region was found to have higher levels of H3K9AC in the hypothalami from the control and maternally underfed twin groups compared with the control and maternally underfed singleton groups (Fig. 4A, B). Singleton fetuses from maternal ewes subjected to periconceptional undernutrition also had higher levels of H3K9AC compared with singleton controls (singleton controls n=7 and singleton underfed n=9; P<0.005). The interaction was established as being significantly different (P<0.005). Hypothalamic GR H3K4me3 levels were significantly greater in the control twin (29%) and maternally underfed twin (36%) and underfed singleton (35%) groups (Fig. 4C, D). The levels of variation between being a singleton or twin was found to be significantly different (P<0.05). The changes in the markers for increased chromatin accessibility were correlated with a decrease in GR H3K27me3 levels in the singleton underfed and control twin groups and the underfed twin groups (53, 53, and 57% decrease, respectively; Fig. 4E, F).
Figure 4.
Changes in the histone patterns of the GR promoter as a result of twinning and maternal periconceptional undernutrition. A, C, E) PCR blots following ChIP enrichment for GR promoter H3K9AC (A), H3K4me3 (C), and H3K27me3 (E). RNA polymerase II was used as positive control; mouse IgG as negative control. B, D, F) Levels of fetal hypothalamic H3K9AC (B), H3K4me3 (D), and H3K27me3 (F) of the GR promoter in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups following ChIP analysis. Groups were compared with control via 2-way ANOVA with Bonferroni post hoc test. *P < 0.05, ***P < 0.005 for nutritional effect; †P < 0.05, †††P < 0.005 for twin effect.
Twinning and maternal undernutrition around conception influence DNA methylation and transcriptional activity of the GR locus
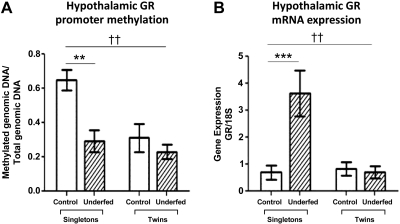
Ventral hypothalamic GR promoter methylation analysis showed that relative to the total genomic DNA, the levels of methylated GR marker DNA were significantly decreased in the control and underfed twin groups (52 and 65% decrease, respectively) and the singleton underfed group (55% decrease) compared with the control singleton group (Fig. 5A). This was associated with a significant interaction in singleton-twin status (P<0.01). Quantitative (q)RT-PCR analysis also demonstrated a 5-fold increase in the GR mRNA expression levels in the singleton underfed group (singleton control n=7 and singleton underfed n=9; P<0.005) but not the twin groups (Fig. 5B).
Figure 5.
Effects of twinning and periconceptional maternal undernutrition on fetal hypothalamic GR promoter methylation and GR mRNA expression levels. A) GR promoter methylation levels were determined for the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) fetal groups. B) Fetal hypothalamic GR mRNA analysis in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups established using qRT-PCR. Groups were compared with control via 2-way ANOVA with Bonferroni post hoc test. **P < 0.01, ***P < 0.005 for nutritional effect; ††P < 0.01 for twin effect.
Changes in the GR locus are tissue specific and do not affect the hypothalamic-pituitary-adrenal (HPA) axis
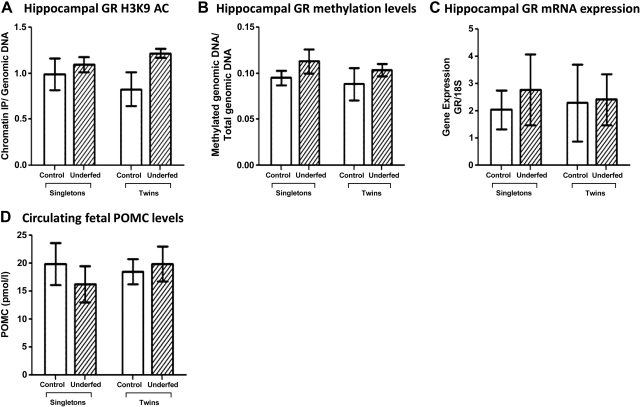
The epigenetic status of the GR marker region in the hippocampus was not affected by twinning or maternal periconceptional undernutrition (Fig. 6A, B). Following qRT-PCR, it was observed that the levels of hippocampal GR expression were similar across all groups (Fig. 6C). To determine whether the HPA axis activity was altered in response to twinning and maternal undernutrition, the plasma POMC levels were measured in the control and underfed fetal sheep groups. It was found that the levels of POMC were also similar across all groups (Fig. 6D). All the hypothalamic and hippocampal data were further analyzed according to gender. However, no apparent differences were found between male and female fetuses (data not shown).
Figure 6.
Fetal HPA axis dynamics following twinning and maternal periconceptional undernutrition. A) Fetal hippocampal GR H3K9AC levels were determined using ChIP analysis for the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. B) Fetal hippocampal GR methylation levels were established using PCR-based methylation enrichment analysis for the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. C) Hippocampal GR mRNA expression was quantitated using qRT-PCR in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups. D) Fetal circulating POMC plasma levels in the singleton control (n=7), singleton underfed (n=9), twin control (n=7 pairs), and twin underfed (n=8 pairs) groups was measured using an ELISA-based method. Groups were compared with control via 2-way ANOVA with Bonferroni post hoc test.
DISCUSSION
We provide the first evidence of epigenetic changes in fetal hypothalamic pathways regulating energy balance as a consequence of twinning and maternal undernutrition around conception. The importance of these findings lie in the potential long-term consequences for the fetus should it, in postnatal life, be exposed to a nutritional environment that is mismatched to that experienced in utero, when these changes in pathways regulating energy balance may lead to an increased propensity to become obese and/or develop type 2 diabetes.
The association of twinning with abdominal obesity (5, 10) led us to hypothesize that twinning is a nutritional paradigm that may affect the hypothalamic pathways involved in energy balance. The overall net effect of all the epigenetic changes identified suggests that the POMC and GR chromatin is open, increasing the accessibility of transcriptional activators and repressors. It is difficult to speculate on the mechanism whereby twinning and alterations in maternal diet result in changes in the fetal epigenome. However, we found a significant reduction in the levels of ventral hypothalamic DNMT activity in twins and in fetuses of periconceptionally undernourished mothers. In addition, alterations in intermediate metabolites can influence the activity of histone-associated enzymes (44). Therefore, it is possible that twinning and maternal undernutrition cause changes in fetal metabolites that alter epigenetic mechanisms impacting on hypothalamic pathways. The greater implication of these findings is that twin conception results in programming of fetuses that result in epigenetic changes similar to that of maternal undernutrition. These data suggest that twins are more likely to develop obesity and consequential diseases later in life regardless of additional periconceptional maternal undernutrition effects.
In focusing on regulation of hypothalamic neuropeptides involved in energy balance, we chose POMC because it is a necessary component of the hypothalamic networks, acting to reduce food intake and increase energy expenditure (12). There is good evidence in rodents for the programming of POMC by maternal undernutrition (16), supporting the concept of epigenetic changes. Neonatal overfeeding in rodents has also identified hypermethylation of the POMC promoter (18), and maternal low-protein food restriction in rats is associated with reduced methylation in specific CpG sites in the POMC promoter in the offspring (17). We have also previously shown decreased methylation in the POMC promoter in fetal sheep hypothalami (21). Our current data further the limited evidence of the epigenetic alterations of POMC in the hypothalamus. The changes suggest that the selected POMC promoter chromatin is open, increasing the possibility of regulation of this region and transcription factor binding (45, 46).
Epigenetic methylation patterns during fetal development are understood to be maintained into adulthood (47). As a result, the observed epigenetic changes in the current study would have implications for the control of food intake and energy balance after birth, when the offspring feeds independently, and any abnormalities would be manifest later in life due to age-dependent or pathologically induced changes in transcription factor expression.
In our study, hypomethylation of the hypothalamic POMC region did not translate into changes in POMC mRNA expression in the fetuses, as expected. This has also been observed in a study looking at prenatal undernutrition in rats (48). Any resulting changes in POMC expression due to epigenetic effects may become apparent after birth when they could modulate transcription factor accessibility. For example, in rodents where the mothers were undernourished, the postnatal offspring had decreased POMC (16). These data also reinforce the suggestion that due to decreased methylation, the increased accessibility of the marker region in the POMC promoter could lead to a down-regulation of the POMC gene, causing an increase in food consumption.
POMC neurons in the arcuate nucleus also have direct projections to the dorsal vagal complex, and POMC is known to decrease hepatic glucose production and peripheral glucose uptake (49). Therefore, once the offspring are feeding independently, the observed changes in epigenetic status of POMC could induce alterations in the regulation of glucose homeostasis. Indeed, twins and singletons from mothers undernourished around conception, which were on the same regime as the sheep in this study, have impaired glucose tolerance in adult life (7).
The POMC promoter region chosen for this study is thought to be associated with the hypothalamic-specific regulatory region (24). Hypomethylation of our chosen promoter marker region could also reflect epigenetic changes present in this hypothalamic-specific promoter region, altering the control of food intake. It has been shown that GR binds proximally to our selected POMC promoter marker, which is an enhancer region in rats (23). GR is also predicted to bind within the marker region used in this work (glucocorticoid response elements with <15% dissimilarity; refs. 50, 51). Therefore, it is interesting to speculate that if a GR-regulated functional enhancer region is epigenetically modified in the arcuate nucleus, it may allow GR inhibition of POMC expression, with a concomitant increase in food intake. These changes would only become apparent after birth, when energy balance is regulated by the hypothalamus.
There is good evidence that glucocorticoids regulate POMC in the hypothalamus (13, 14). Therefore, changes in the epigenetic status of GR could lead to altered regulation of energy balance increasing the likelihood of the development of obesity. The epigenetic changes in GR found in this study indicate that the chromatin is open, thus allowing for changes in gene expression modulated by the change in transcription factor accessibility. The promoter region is closely associated with the GR 17 promoter region, which has been linked to altered hippocampal GR expression in response to changes in maternal care (19). Therefore, the promoter region in this investigation could be reflective of changes in the GR 17 promoter region.
In the current model, it could be hypothesized that in the adults subjected to maternal programming, GR in the hypothalamus modulates neuropeptides to cause an increase in food intake. However, the precise mechanism for GR regulation of POMC neurons is conflicting with different studies showing an increase or a decrease in POMC expression following adrenalectomy (which would remove the glucocorticoids; refs. 13, 14). It may be that GR regulation of NPY is more relevant, given that there is evidence that NPY requires the presence of glucocorticoids to increase food intake (15). Therefore, the epigenetic alterations in hypothalamic GR may influence neuropeptide regulation of energy balance by decreasing POMC and increasing NPY in offspring of mothers that were underfed.
Previous evidence has indicated that maternal undernutrition induces programming effects in the HPA axis (52). However, in this investigation, maternal undernutrition before and during early pregnancy did not produce any epigenetic changes in the GR marker in the hippocampus. This is in line with another study that used the same levels of maternal undernutrition over the same period and found similar levels of plasma ACTH and cortisol concentrations (53). This suggests that twinning and maternal undernutrition in the current model induce specific hypothalamic programming effects.
We hypothesize that the epigenetic changes found at the fetal stage in this study may persist into adulthood, influencing the development of metabolic disease. This is because sheep subjected to the same level of maternal undernutrition as those in this study have produced offspring that are overweight compared with control offspring at 10 mo of age (7). Furthermore, twins and singletons from undernourished mothers also have impaired glucose tolerance, which was found in late gestation fetuses and adult offspring (7, 54).
In summary, our work demonstrates that twinning and periconceptional maternal undernutrition induce specific epigenetic modifications in the fetal hypothalamic pathways regulating energy balance. This suggests that twins may undergo a nutritional programming event leading to altered physiology of the hypothalamic pathways. As a result, twins and maternally undernourished offspring may have an increased propensity to develop obesity and/or type 2 diabetes later in life.
Supplementary Material
Acknowledgments
The study was supported by the UK National Institute of Health Research Manchester Biomedical Research Center, the Health Research Council of New Zealand, the New Zealand National Research Centre for Growth and Development, and the Canadian Institutes for Health Research (to J.R.G.C.). The authors also thank Ngapouri Research Station staff and Hui Hui Phua for technical assistance.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ChIP
- chromatin immunoprecipitation
- DNMT
- DNA methyltransferase
- DOHaD
- developmental origins of health and disease
- ENCODE
- Encyclopedia of DNA Elements
- GR
- glucocorticoid receptor
- H3K9AC
- histone 3 lysine 9 acetylation
- H3K4me3
- histone 3 lysine 4 trimethylation
- H3K27me3
- histone 3 lysine 27 trimethylation
- HPA
- hypothalamic-pituitary-adrenal
- nPE
- neuronal promoter enhancer region
- POMC
- proopiomelanocortin.
REFERENCES
- 1. Hales C. N., Barker D. J. (1992) Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35, 595–601 [DOI] [PubMed] [Google Scholar]
- 2. Gluckman P. D., Hanson M. A., Cooper C., Thornburg K. L. (2008) Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 359, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barker D. J. (2007) The origins of the developmental origins theory. J. Intern. Med. 261, 412–417 [DOI] [PubMed] [Google Scholar]
- 4. Hales C. N., Barker D. J. (2001) The thrifty phenotype hypothesis. Br. Med. Bull. 60, 5–20 [DOI] [PubMed] [Google Scholar]
- 5. Poulsen P., Grunnet L. G., Pilgaard K., Storgaard H., Alibegovic A., Sonne M. P., Carstensen B., Beck-Nielsen H., Vaag A. (2009) Increased risk of type 2 diabetes in elderly twins. Diabetes 58, 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The N. S., Adair L. S., Gordon-Larsen P. (2010) A study of the birth weight-obesity relation using a longitudinal cohort and sibling and twin pairs. Am. J. Epidemiol. 172, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Todd S. E., Oliver M. H., Jaquiery A. L., Bloomfield F. H., Harding J. E. (2009) Periconceptional undernutrition of ewes impairs glucose tolerance in their adult offspring. Pediatr. Res. 65, 409–413 [DOI] [PubMed] [Google Scholar]
- 8. Buckler J. M., Green M. (2004) A comparison of the early growth of twins and singletons. Ann. Hum. Biol. 31, 311–332 [DOI] [PubMed] [Google Scholar]
- 9. Van Dommelen P., de Gunst M., van der Vaart A., van Buuren S., Boomsma D. (2008) Growth references for height, weight and body mass index of twins aged 0–2.5 years. Acta Paediatr. 97, 1099–1104 [DOI] [PubMed] [Google Scholar]
- 10. Poulsen P., Vaag A. A., Kyvik K. O., Moller Jensen D., Beck-Nielsen H. (1997) Low birth weight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia 40, 439–446 [DOI] [PubMed] [Google Scholar]
- 11. Monrad R. N., Grunnet L. G., Rasmussen E. L., Malis C., Vaag A., Poulsen P. (2009) Age-dependent nongenetic influences of birth weight and adult body fat on insulin sensitivity in twins. J. Clin. Endocrinol. Metab. 94, 2394–2399 [DOI] [PubMed] [Google Scholar]
- 12. Brady L. S., Smith M. A., Gold P. W., Herkenham M. (1990) Altered expression of hypothalamic neuropeptide mRNAs in food-restricted and food-deprived rats. Neuroendocrinology 52, 441–447 [DOI] [PubMed] [Google Scholar]
- 13. Wardlaw S. L., McCarthy K. C., Conwell I. M. (1998) Glucocorticoid regulation of hypothalamic proopiomelanocortin. Neuroendocrinology 67, 51–57 [DOI] [PubMed] [Google Scholar]
- 14. Beaulieu S., Gagne B., Barden N. (1988) Glucocorticoid regulation of proopiomelanocortin messenger ribonucleic acid content of rat hypothalamus. Mol. Endocrinol. 2, 727–731 [DOI] [PubMed] [Google Scholar]
- 15. Zakrzewska K. E., Sainsbury A., Cusin I., Rouru J., Jeanrenaud B., Rohner-Jeanrenaud F. (1999) Selective dependence of intracerebroventricular neuropeptide Y-elicited effects on central glucocorticoids. Endocrinology 140, 3183–3187 [DOI] [PubMed] [Google Scholar]
- 16. Delahaye F., Breton C., Risold P. Y., Enache M., Dutriez-Casteloot I., Laborie C., Lesage J., Vieau D. (2008) Maternal perinatal undernutrition drastically reduces postnatal leptin surge and affects the development of arcuate nucleus proopiomelanocortin neurons in neonatal male rat pups. Endocrinology 149, 470–475 [DOI] [PubMed] [Google Scholar]
- 17. Coupe B., Amarger V., Grit I., Benani A., Parnet P. (2010) Nutritional programming affects hypothalamic organization and early response to leptin. Endocrinology 151, 702–713 [DOI] [PubMed] [Google Scholar]
- 18. Plagemann A., Harder T., Brunn M., Harder A., Roepke K., Wittrock-Staar M., Ziska T., Schellong K., Rodekamp E., Melchior K., Dudenhausen J. W. (2009) Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J. Physiol. 587, 4963–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weaver I. C., Cervoni N., Champagne F. A., D'Alessio A. C., Sharma S., Seckl J. R., Dymov S., Szyf M., Meaney M. J. (2004) Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 [DOI] [PubMed] [Google Scholar]
- 20. Symonds M. E., Budge H. (2009) Nutritional models of the developmental programming of adult health and disease. Proc. Nutr. Soc. 68, 173–178 [DOI] [PubMed] [Google Scholar]
- 21. Stevens A., Begum G., Cook A., Connor K., Rumball C., Oliver M., Challis J., Bloomfield F., White A. (2010) Epigenetic changes in the hypothalamic proopiomelanocortin and glucocorticoid receptor genes in the ovine fetus after periconceptional undernutrition. Endocrinology 151, 3652–3664 [DOI] [PubMed] [Google Scholar]
- 22. Muhlhausler B. S., McMillen I. C., Rouzaud G., Findlay P. A., Marrocco E. M., Rhind S. M., Adam C. L. (2004) Appetite regulatory neuropeptides are expressed in the sheep hypothalamus before birth. J. Neuroendocrinol. 16, 502–507 [DOI] [PubMed] [Google Scholar]
- 23. Langlais D., Couture C., Sylvain-Drolet G., Drouin J. (2011) A pituitary-specific enhancer of the POMC gene with preferential activity in corticotrope cells. Mol. Endocrinol. 25, 348–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Souza F. S., Santangelo A. M., Bumaschny V., Avale M. E., Smart J. L., Low M. J., Rubinstein M. (2005) Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol. Cell. Biol. 25, 3076–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geng C. D., Schwartz J. R., Vedeckis W. V. (2008) A conserved molecular mechanism is responsible for the auto-up-regulation of glucocorticoid receptor gene promoters. Mol. Endocrinol. 22, 2624–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Portela A., Esteller M. (2010) Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 27. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 28. Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 29. Stevens A., Begum G., White A. (2011) Epigenetic changes in the hypothalamic pro-opiomelanocortin gene: a mechanism linking maternal undernutrition to obesity in the offspring? Eur. J. Pharmacol. 660, 194–201 [DOI] [PubMed] [Google Scholar]
- 30. Schwartz J., Rose J. C. (1998) Development of the pituitary adrenal axis in fetal sheep twins. Am. J. Physiol. 274, R1–R8 [DOI] [PubMed] [Google Scholar]
- 31. Crosby S. R., Stewart M. F., Ratcliffe J. G., White A. (1988) Direct measurement of the precursors of adrenocorticotropin in human plasma by two-site immunoradiometric assay. J. Clin. Endocrinol. Metab. 67, 1272–1277 [DOI] [PubMed] [Google Scholar]
- 32. Sebert S. P., Hyatt M. A., Chan L. L., Patel N., Bell R. C., Keisler D., Stephenson T., Budge H., Symonds M. E., Gardner D. S. (2009) Maternal nutrient restriction between early and midgestation and its impact upon appetite regulation after juvenile obesity. Endocrinology 150, 634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34. Gupta S., Dennis J., Thurman R. E., Kingston R., Stamatoyannopoulos J. A., Noble W. S. (2008) Predicting human nucleosome occupancy from primary sequence. PLoS Comput. Biol. 4, e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dennis J. H., Fan H. Y., Reynolds S. M., Yuan G., Meldrim J. C., Richter D. J., Peterson D. G., Rando O. J., Noble W. S., Kingston R. E. (2007) Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome Res. 17, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ozsolak F., Song J. S., Liu X. S., Fisher D. E. (2007) High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 25, 244–248 [DOI] [PubMed] [Google Scholar]
- 37. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 38. Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., Huebert D. J., McMahon S., Karlsson E. K., Kulbokas E. J., 3rd, Gingeras T. R., Schreiber S. L., Lander E. S. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 [DOI] [PubMed] [Google Scholar]
- 39. Ohlsson R., Bartkuhn M., Renkawitz R. (2010) CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma 119, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ernst J., Kellis M. (2010) Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M., Ku M., Durham T., Kellis M., Bernstein B. E. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Surani M. A., Hayashi K., Hajkova P. (2007) Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 [DOI] [PubMed] [Google Scholar]
- 43. Schneider E., Pliushch G., El Hajj N., Galetzka D., Puhl A., Schorsch M., Frauenknecht K., Riepert T., Tresch A., Muller A. M., Coerdt W., Zechner U., Haaf T. (2010) Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 38, 3880–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Turner B. M. (2009) Epigenetic responses to environmental change and their evolutionary implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 3403–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Newell-Price J. (2003) Proopiomelanocortin gene expression and DNA methylation: implications for Cushing's syndrome and beyond. J. Endocrinol. 177, 365–372 [DOI] [PubMed] [Google Scholar]
- 46. Ho S. M., Tang W. Y. (2007) Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod. Toxicol. 23, 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Razin A., Cedar H. (1993) DNA methylation and embryogenesis. EXS 64, 343–357 [DOI] [PubMed] [Google Scholar]
- 48. Ikenasio-Thorpe B. A., Breier B. H., Vickers M. H., Fraser M. (2007) Prenatal influences on susceptibility to diet-induced obesity are mediated by altered neuroendocrine gene expression. J. Endocrinol. 193, 31–37 [DOI] [PubMed] [Google Scholar]
- 49. Marino J. S., Xu Y., Hill J. W. (2011) Central insulin and leptin-mediated autonomic control of glucose homeostasis. Trends Endocrinol. Metab. 22, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M. M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 51. Farre D., Roset R., Huerta M., Adsuara J. E., Rosello L., Alba M. M., Messeguer X. (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edwards L. J., McMillen I. C. (2002) Impact of maternal undernutrition during the periconceptional period, fetal number, and fetal sex on the development of the hypothalamo-pituitary adrenal axis in sheep during late gestation. Biol. Reprod. 66, 1562–1569 [DOI] [PubMed] [Google Scholar]
- 53. Rumball C. W., Bloomfield F. H., Oliver M. H., Harding J. E. (2009) Different periods of periconceptional undernutrition have different effects on growth, metabolic and endocrine status in fetal sheep. Pediatr. Res. 66, 605–613 [DOI] [PubMed] [Google Scholar]
- 54. Rumball C. W., Harding J. E., Oliver M. H., Bloomfield F. H. (2008) Effects of twin pregnancy and periconceptional undernutrition on maternal metabolism, fetal growth and glucose-insulin axis function in ovine pregnancy. J. Physiol. 586, 1399–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.