Abstract
The signaling pathways that commit cells to migration are incompletely understood. We employed human mammary cells and two stimuli: epidermal growth factor (EGF), which induced cellular migration, and serum factors, which stimulated cell growth. In addition to strong activation of ERK by EGF, and AKT by serum, early transcription remarkably differed: while EGF induced early growth response-1 (EGR1), and this was required for migration, serum induced c-Fos and FosB to enhance proliferation. We demonstrate that induction of EGR1 involves ERK-mediated down-regulation of microRNA-191 and phosphorylation of the ETS2 repressor factor (ERF) repressor, which subsequently leaves the nucleus. Unexpectedly, knockdown of ERF inhibited migration, which implies migratory roles for exported ERF molecules. On the other hand, chromatin immunoprecipitation identified a subset of direct EGR1 targets, including EGR1 autostimulation and SERPINB2, whose transcription is essential for EGF-induced cell migration. In summary, EGR1 and the EGF-ERK-ERF axis emerge from our study as major drivers of growth factor-induced mammary cell migration.—Tarcic, G., Avraham, R., Pines, G., Amit, I., Shay, T., Lu, Y., Zwang, Y., Katz, M., Ben-Chetrit, N., Jacob-Hirsch, J., Virgilio, L., Rechavi, G., Mavrothalassitis, G., Mills, G. B., Domany, E., Yarden, Y. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF.
Keywords: growth factor, phosphorylation, transcription, negative feedback
Cells are continuously exposed to multiple extracellular signals and stressors, but only a fraction of this input elicits fate-determining decisions. Mechanisms that integrate all incoming signals and determine response specificity remain poorly understood (1, 2). To sense environmental changes, eukaryotic cells evolved various strategies, including relay systems based on secreted growth factors (GFs) and their cognate receptor tyrosine kinases (RTKs). A prototype of the RTK family is the ErbB subgroup, which comprises 4 members, including the epidermal growth factor receptor (EGFR; refs. 3, 4). The ErbB family simultaneously activates multiple signaling pathways. Among others, these include the phosphoinositol 3′-kinase (PI3K) pathway, which activates the apoptosis-inhibitory AKT/PKB signaling pathway (5) and several mitogen-activated protein kinase (MAPK) cascades (6), out of which the ERK pathway is the best understood. Once activated, a major shift in the ERK interactome occurs, thereby permitting differential phosphorylation of ERK substrates (7). Of the many substrates, ETS2 repressor factor (ERF), a member of the E twenty-six (ETS) group of transcription factors (TFs), appears to act as an important integrator, which is exported from the nucleus (8) to initiate a process critical for pathogenic EGFR signaling (9).
For simplicity, the transcriptional response to GFs may be divided into 3 temporal phases: the first phase comprises a group of rapidly induced TFs and other regulators encoded by the immediate early genes [IEGs; e.g., early growth response-1 (EGR1); ref. 10]. The flanking group of delayed early genes (45–150 min) comprises both positively and negatively acting components (11). The third group comprises secondary response genes (>120 min), which confer stable phenotypes. One example relates to the acquisition of a motile phenotype, which is mediated by a large set of TFs (12). Collectively, these regulators enable GFs to instigate several molecular switches, such as the loss of the epithelial cadherin, and gain of the mesenchymal N-cadherin (13). Another transcriptional switch replaces tensins, linkers of the actin cytoskeleton and the extracellular matrix, with Cten (14).
The initial response to a migration-promoting agent involves cytoskeleton rearrangements, followed by de novo synthesis of proteins that enable a sustained migratory phenotype (15). For example, hepatocyte growth factor (HGF) stimulates the ERK cascade to induce EGR1 expression, which, in turn, up-regulates Snail. The latter forms a negative feedback loop by repressing EGR1 expression (16). Similarly, EGF has been shown to promote cell migration under various physiological conditions, such as wound healing (17), trophoblast invasion (18), and morphogenesis (19). Furthermore, EGFR overexpression has been associated with the invasive phenotype of both glioblastoma (20) and breast cancer (21). The present study addressed gene expression programs and signaling pathways underlying EGF-induced cell migration. To this end, we employed the nontransformed MCF10A mammary epithelial cells, for which EGF acts as a promoter of migration, whereas animal serum induces their proliferation. Differential proteomic and transcriptomic analyses identified ERF, as well as EGR1, as linearly connected components of a pathway regulating mammary cell migration in response to EGF. This axis controls a subset of migration-promoting, as well as migration inhibitory genes, which are assembled into a rich network of regulatory loops.
MATERIALS AND METHODS
Cell lines and transfection
MCF10A cells were grown in DME:F12 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10 μg/ml insulin, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, 5% heat-inactivated horse serum (Gibco BRL) and 10 ng/ml EGF. Cells were seeded in 6-well plates at a density of 1 × 105 cells/well for siRNA transfection, using the Oligofectamine reagent (Invitrogen, Carlsbad, CA, USA).
Antibodies, lysate preparation, and immunoblot analysis
Cell lysates were cleared by centrifugation and resolved by electrophoresis, followed by electrophoretic transfer to a nitrocellulose membrane. Membranes were blocked with TBS-T (Tris-buffered saline containing Tween-20) containing 1% low-fat milk, blotted with a primary antibody for 1 h, washed 3 times with TBS-T, incubated for 30 min with a secondary antibody linked to horseradish peroxidase (HRP), and washed with TBS-T. Immunoreactive bands were detected using the ECL reagent (Amersham Pharmacia Biotech, Little Chalfont, UK). Monoclonal antibodies to EGFR were from Alexis Biotech (London, UK). Polyclonal antibodies to phosphorylated EGFR and phosphorylated AKT were from Cell Signaling (Beverly, MA, USA). Phosphorylated ERK1/2 antibody was from Sigma-Aldrich (St. Louis, MO, USA) and antibodies to ERK2, ERF, EGR1, c-Fos, and AKT were from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Reverse-phase protein array (RPPA) analysis
Cell pellets were lysed in RPPA buffer (1% Triton X-100; 50 mM HEPES, pH 7.4; 150 mM NaCl; 1.5 mM MgCl2; 1 mM EGTA; 100 mM NaF; 10 mM Na-pyrophosphate; 1 mM Na3VO4; 10% glycerol; and protease and phosphatase inhibitors; Roche Diagnostics, Mannheim, Germany). Following centrifugation, protein concentration was assayed using the BCA reagent (Pierce, Rockford, IL, USA), and then 4XPSB (40% glycerol; 8% SDS; 0.25 M Tris-HCl, pH 6.8; and 10% 2-mercaptoethanol) was added to the cleared lysates, followed by boiling. Samples were analyzed and signals were quantified as described previously (11).
Real-time quantitative PCR and oligonucleotide microarray hybridization
cDNA was generated by using Invitrogen SuperScript II first-strand synthesis kit. Real-time PCR analysis was performed using SYBR Green I as a fluorescent dye. MicroRNA extraction and real-time PCR specific for microRNAs were performed as described previously (22). All experiments were carried out in triplicate and normalized to β-2 microglobulin RNA levels. Oligonucleotide microarray hybridization was performed as described previously (11). All values <50 were replaced by 50. Only probe sets that were up-regulated by ≥2-fold compared to time 0 were further analyzed. Expression values were mean-centered and divided by the standard deviation.
Transwell cell migration assays
MCF10A cells (6×104 cells/well) were plated in the upper compartment of a 24-well Transwell tray and allowed to adhere for 16 h at 37°C in full medium lacking EGF. Thereafter, the medium was either refreshed or replaced with EGF-containing medium, and cells were allowed to migrate for 16 h at 37°C through the intervening nitrocellulose membrane. The filter was removed, fixed in paraformaldehyde (3%), followed by cell permeabilization in Triton X-100 (0.05%, in saline) and staining with methyl violet (0.3%). Cells growing on the upper side of the filter were removed using a cotton swab, and cells on the bottom side of the filter were photographed and counted.
Cell proliferation assays
MCF10A cells grown in serum-containing medium were plated in 6-well plates (0.1×105 cells/well), and the medium was changed to either a starvation medium, starvation medium supplemented with EGF, or full medium that contained no EGF. Cell growth was determined by staining the cells with 0.3% methyl violet and quantification of color intensity.
Immunofluorescence and real-time immunofluorescence microscopy
Cells grown on coverslips were stained as described previously (14). All figures present middle sections of cells. Live cell fluorescence microscopy was carried out using the DeltaVision system (Applied Precision, Issaqua, WA, USA) and a ×63/1.3 objective.
Chromatin immunoprecipitation (ChIP)
Cells (3×106) were fixed in formaldehyde (1%) for 10 min, quenched with glycine (125 mM), and harvested. After washing, cells were lysed and sonicated until average DNA fragment size was <500 bp. The lysate was incubated overnight at 4°C with agarose beads prebound to an anti-EGR1 polyclonal antibody (Santa Cruz Biotechnology). After extensive washing, the lysate was incubated for 15 min at 65°C and cleared, and the supernatant was transferred to a new tube and incubated for 15 h at 65°C. Thereafter, proteinase K was added, and incubation proceeded for 2 h at 55°C. DNA was purified using the Wizard SV gel and PCR clean-up system (Promega, Fitchburg, WI, USA).
Luciferase reporter assay
Cells were cotransfected with a wild-type EGR1 promoter-reporter construct, along with the indicated combinations of expression plasmids and a pRL-Renilla plasmid, containing Renilla luciferase (Promega). Cells were harvested, and firefly and Renilla luciferase activities were assayed with the Promega dual-luciferase assay system.
Yeast 2-hybrid screens
The ERF cDNA fragment was cloned into the PGBKT7 vector (Clontech, Mountain View, CA, USA) within the Gal4 DNA-binding domain and introduced into the yeast strain PJ694-α (a gift from Dr. D. Alexandraki, University of Crete, Heraklion, Greece). A mouse cDNA-VP16 fusion library from 10.5 d embryo RNA was transformed in PJ694-α yeast cells and then inoculated in synthetic medium lacking leucine (Bio101). The library was screened by mating PJ694-α cells expressing Gal4-DNA-BD-ERF and the PJ694-α cells expressing the embryo mouse cDNA library. Cotransformants were plated onto Trp-Leu-His selective medium supplemented with 3-aminotriazole (4.5 mM). His-positive colonies were then assayed for β-galactosidase using a filter-lift assay. Positive clones were rescued into bacteria and retransformed into the PJ694 yeast strain to confirm interactions.
RESULTS
EGF and serum stimulate migration and proliferation, respectively, of human mammary epithelial cells
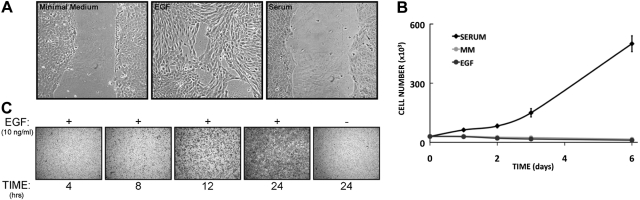
We previously characterized sets of mRNAs (11) and microRNAs (22) that mediate the short- and long-term biological effects of EGF on the well-established, nontumorigenic MCF10A human mammary epithelial cell line (23). Interestingly, following stimulation with EGF or other family members [e.g., β-cellulin and transforming growth factor α (TGFα), but not neuregulin-β1], these cells rearrange their actin cytoskeleton, scatter small colonies (Supplemental Fig. S1A, B), and close wounds introduced into monolayers (Fig. 1A). In contrast, we observed none of these effects following the addition of a mixture of serum factors (hereafter referred to as serum) or the platelet-derived growth factor. Instead, serum factors robustly augmented cell proliferation, unlike EGF-stimulated cells, whose multiplication was largely arrested under the conditions that we employed (Fig. 1B). These observations are consistent with previous reports (24, 25), and they offer a cellular system in which a migratory response could be contrasted with a proliferation outcome. Importantly, the migratory phenotype, as determined by the ability of EGF-treated cells to migrate through a porous filter, became apparent 8 h after the addition of EGF, and persisted for at least to 24 h (Fig. 1C), in line with involvement of newly synthesized proteins.
Figure 1.
EGF and serum stimulate migration and proliferation, respectively, of MCF10A mammary cells. A) Confluent monolayers of MCF10A cells were incubated for 24 h with minimal medium, and then wounded using a sterile tip, washed with saline, and incubated in the indicated media for another 24 h. Photos were taken using a light microscope. B) MCF10A cells (3×103) were seeded in a 6-well plate. After 12 h, the medium was replaced with a minimal medium (MM) and incubated for 24 h. This was followed by incubation of the cells with the indicated media. Cells were then counted at the indicated time intervals. C) MCF10A cells (0.6×105) were plated in the absence of EGF in a Transwell chamber. After 12 h, the medium was replaced with an EGF-containing medium, and cells were fixed and stained at the indicated intervals.
Proteomic analysis reveals differential EGF-induced activation of the ERK and AKT pathways
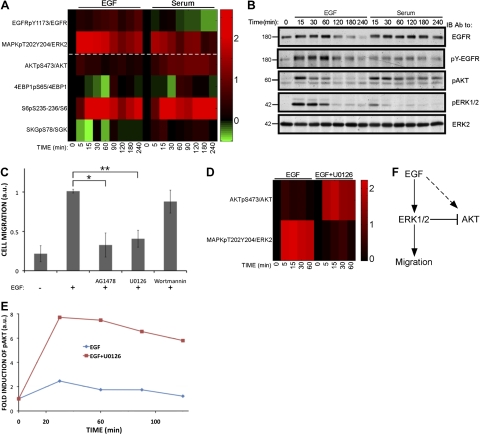
To comparatively identify the kinase cascades involved in EGF-induced migration of MCF10A cells, we used quantitative RPPAs (26). The experimental setup (see Supplemental Fig. S1C) comprised separate time courses of EGF and serum stimulation, followed by extraction of cells and RPPA analysis. The results presented in Fig. 2A imply that ERK1/2 serves as a major phosphorylation pathway activated by EGF, whereas serum more strongly activated the AKT pathway, and both stimuli induced phopshorylation of the ribosomal S6 protein. Direct, side-by-side comparison of these pathways using immunoblotting confirmed that EGF strongly induced persistent phosphorylation of ERK1/2, while serum caused only mild and transient ERK phosphorylation (Fig. 2B). The profile of AKT activation was reciprocal, with EGF being a transient stimulant and serum a persistent inducer of AKT. To determine whether these phosphorylation events were necessary for the migratory outcome, we employed inhibitors specific to EGFR, MEK, and AKT (AG1478, U0126, and wortmannin, respectively). In line with the dichotomy, treatment with EGFR or MEK inhibitors significantly reduced inducible colony scattering and cell migration, while the effect of AKT inhibition did not achieve statistical significance (Fig. 2C and Supplemental Fig. S1D).
Figure 2.
Differential activation of kinase cascades by EGF and by serum. A) MCF10A cells were serum starved for 24 h and then incubated with EGF (10 ng/ml) or serum (5%) for the indicated time intervals. Cells were then lysed and analyzed using RPPA and the indicated antibodies. Signals corresponding to phospho-specific antibodies were normalized to the signals obtained with the respective general antibodies. Log2 values of the respective fold induction are presented, relative to time 0. B) MCF10A cells were serum starved for 24 h, and then stimulated as indicated for different time intervals. Cell extracts were subjected to immunoblotting with the indicated antibodies. C) MCF10A cells (0.6×105) were plated in a Transwell chamber containing EGF and the inhibitors AG1478 (10 μM), U0126 (5 μM), or wortmannin (200 nM). After 24 h, cells were stained with crystal violet, photos were taken using a light microscope, and the migrated cells were counted. Normalized signals (arbitrary units) are presented. *P = 9.4 × 10−7, **P = 8.6 × 10−8; Student's t test. D) MCF10A cells were serum starved for 24 h and then incubated for the specified time intervals with EGF (10 ng/ml), in the presence or absence of U0126 (5 μM). Cell extracts were analyzed using RPPA and the indicated antibodies. Log2 values of the respective fold induction signals are presented, relative to time 0. E) MCF10A cells were treated as in D and subjected to immunoblotting with anti-AKT and anti-activated-AKT antibodies. Quantification of band intensities is shown. F) Scheme representing the proposed lateral inhibition of AKT by the EGF-stimulated ERK cascade.
Early studies implicated the ERK cascade in the regulation of cell migration (27, 28), and although Ras-mediated activation of both ERK and PI3K are required for growth factor-induced disassembly of adherens junctions (29), a delicate balance between these pathways seems critical (reviewed in ref. 6). To uncover potential crosstalk between ERK and other signaling pathways, we performed an additional RPPA set of assays in the presence of a MEK1/2 inhibitor (U0126; Supplemental Fig S1C). The results obtained with this set identified an ERK-to-AKT crosstalk (Fig. 2D): U0126 not only caused a decrease in ERK1/2 activation, but also dramatically enhanced EGF-induced phosphorylation of AKT (Fig. 2E). In summary, the two pathways are functionally interconnected (Fig. 2F), in agreement with the observation that down-regulation of AKT1 can enhance ERK activation (25), and in analogy to the reverse crosstalk enabling AKT to inhibit RAF and the ERK pathway (30).
The ETS family member ERF regulates mammary cell migration
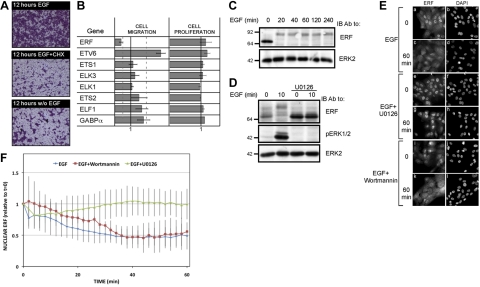
The marked differences between the cellular responses to EGF and to serum, along with the distinct downstream pathways, motivated us to delineate the potentially disparate transcriptional programs. We first confirmed that EGF-induced migration of MCF10A cells depends on newly synthesized proteins (Fig. 3A): inhibition of protein translation using cycloheximide almost fully abolished cell migration in a Transwell chamber. Previous genome-wide analysis of mRNA expression in response to EGF or serum uncovered links between immediate early genes and a later phase of transcription (11), and reanalysis of the data confirmed distinct wave-like patterns of new transcripts (Supplemental Fig. S2A). Conceivably, different sets of TFs are newly induced by each stimulant, in addition to preexisting TFs, which undergo activating phosphorylation (31). Because ERK emerged from our experiments as a major regulator of mammary cell migration, we addressed ERK-regulated preexisting TFs. The major candidate group of such TFs comprises the 26 members of the ETS family, which undergo rapid phosphorylation in response to activation of various MAPKs (32). Of the ETS family members, only 8 displayed detectable expression in MCF10A cells. Hence, each of the 8 was knocked down (Fig. 3B). Whereas none of the 8 ETS transcripts affected cell proliferation, 2 genes, namely ERF and ETV6, showed significant and reproducible modulation of migration on knockdown: ERF knockdown inhibited migration, whereas disabling ETV6 promoted migration of MCF10A cells in preliminary experiments, even in the absence of EGF.
Figure 3.
ERF is necessary for EGF-induced migration of mammary cells. A) MCF10A cells were incubated for 12 h in a Transwell chamber in the absence of EGF. Thereafter, the cells were treated for 12 h with EGF (10 ng/ml) and/or cycloheximide (CHX; 100 ng/ml) for 12 h and stained with crystal violet prior to photography. B) ETS family genes expressed in MCF10A cells were individually knocked down using specific siRNA oligonucleotides. For cell migration (middle column), 0.6 × 106 cells were incubated for 24 h in Transwell chambers, and cell migration through the filter was quantified after staining with crystal violet. Signals were normalized to the control (solid line). Dashed lines represent 0.5 or 2-fold change. For assessment of cell proliferation (right column), cell numbers were evaluated 4 d after siRNA transfection by staining with 0.3% methyl violet and quantifying color intensity. C) MCF10A cells were serum starved for 12 h and then stimulated with EGF (10 ng/ml) for the indicated time intervals. Whole-cell lysates were then subjected to immunoblotting with the indicated antibodies. D) MCF10A cells were serum starved for 12 h, preincubated for 30 min in the presence or absence of a MEK1/2 inhibitor (U0126, 5 μM), and then stimulated for 10 min with EGF (10 ng/ml) prior to immunoblotting. E) MCF10A cells were grown on coverslips, serum starved for 24 h, and stimulated with EGF (10 ng/ml) in the absence or presence of U0126 (5 μM) or wortmannin (200 nM), as indicated. Cells were then fixed, permeabilized, and stained with an anti-ERF antibody and DAPI prior to photography. F) MCF10A cells were grown on coverslips and transfected with a plasmid encoding a GFP-tagged ERF. At 24 h after transfection, cells were serum starved for 24 h and then stimulated as indicated. GFP intensity and localization were monitored using a Delta-Vision microscope. Nuclear localization of GFP-ERF was quantified and presented as a function of time after stimulation with EGF. Values are averages ± sd for 5 cells.
On stimulation, ERK phosphorylates ERF on multiple sites, which drives transport out of the nucleus (8). Direct analysis of ERF in EGF-treated MCF10A cells confirmed rapid and persistent phosphorylation, which was evident due to a large gel mobility shift (Fig. 3C). As expected, this modification strictly depended on ERK activity, as inhibition of MEK abolished ERF phosphorylation (Fig. 3D). Immunolocalization of the endogenous ERF protein confirmed coupling of ERF phosphorylation to nuclear export: both phosphorylation and export were induced by EGF and inhibited by U0126, but they were not influenced by wortmannin (Fig. 3E). To gain a dynamic view of ERF localization, we followed an ectopic GFP-tagged ERF in living cells (Fig. 3F). The results showed that ERF is rapidly exported out of the nucleus following EGF stimulation, reaching maximal export levels after 30 min.
In light of the nuclear export and the observed inhibition of inducible migration on ERF knockdown (Fig. 3B), we assume that this repressor inhibits transcription of critical migration-inhibitory genes. Alternatively, ERF may function in the cytoplasm as an inducer of cell migration, in similarity with MYC-Nick, a cleaved form of MYC that exits the nucleus to interact with microtubules and drive muscle differentiation (33). To preliminarily test this scenario, we screened a yeast 2-hybrid library for cytoplasmic partners of ERF and found reproducible interactions with filamins (12 clones isolated independently), the tight junction protein ZO-1 (6 clones isolated independently) and additional candidates (such as HSPA8, vimentin, 14-3-3-E, fibronectin, cadherin, and fibulin). In summary, ERF stands out among the preexisting TFs, which are rapidly activated by EGF in migrating cells: activation of ERK leads to phosphorylation and translocation of ERF to the cytoplasm, but the mechanism by which this translates to initiation of cell migration remains to be elucidated.
Dissection of the transcriptional response identifies EGR1 as an early regulator of mammary cell migration
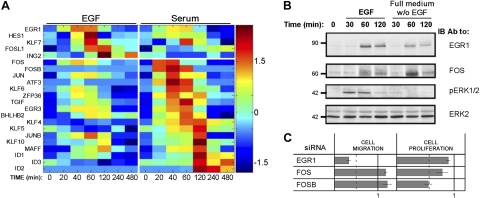
In addition to preexisting TFs, like ERF, the EGF-induced transcription program leading to cell migration likely involves newly synthesized TFs. Focusing on genes annotated as direct controllers of RNA abundance (namely, TFs and RNA-binding proteins included in Supplemental Fig. S2A), we compiled two groups of genes, one specifically induced by EGF and the other induced by serum (Fig. 4A). This analysis revealed that the earliest TF induced by EGF was EGR1, whereas serum induced 2 AP-1 subunits, c-Fos and FosB. Notably, both EGF and serum up-regulated all 3 genes to some extent, but RT-PCR confirmed that EGF more robustly induced EGR1 (Supplemental Fig. S2B), while c-Fos and FosB were more potently induced by serum (Supplemental Fig. S2C, D). In line with these observations, we detected relatively weak induction of the EGR1 protein by serum and similarly weak induction of the c-Fos protein by EGF (Fig. 4B). Consistently, immunoblot analysis confirmed early, MEK-dependent synthesis of the EGR1 protein (Supplemental Fig. S3A). Furthermore, treatment of MCF10A cells with an alternative inducer of the ERK pathway, namely phorbol 12-myristate 13-acetate (PMA), not only transiently induced ERK activation and increased EGR1 expression, but also enhanced cellular migration (Supplemental Fig. S3B, C), in line with coupling of EGR1 to the induction of cell migration. To firmly establish a linear ERK-to-EGR1 cascade leading to mammary cell migration, we knocked down EGR1 and observed >60% reduction in EGF-induced cell migration (Fig. 4C). In contrast, silencing c-Fos and FosB exerted no statistically significant effect on cell migration, but consistently reduced cell proliferation in the case of FosB. Taken together, the data presented in Fig. 4 and Supplemental Figs. S2 and S3 indicate that EGR1 is regulated by the ERK pathway, and it serves as a positive regulator of EGF-induced cell migration. In contrast, serum stimulation activates the IEGs c-Fos and FosB to regulate cell proliferation.
Figure 4.
Migratory response of MCF10A cells to EGF associates with early induction of EGR1. A) Heatmap of the expression of mRNAs encoding transcription factors and RNA-binding proteins, the expression of which was altered by ≥2-fold by either EGF or serum. Genes were organized according to the timing of their peak expression on stimulation with EGF or serum (above and below horizontal line, respectively). B) MCF10A cells were serum starved for 24 h, and then were incubated for different time intervals in the presence or absence of EGF. Cells were then lysed and subjected to immunoblotting. C) MCF10A cells were transfected with siRNA oligonucleotides specific to EGR1, c-Fos, or FosB. Both cell migration (middle column) and cell proliferation (at d 4; right column) were assayed in 3 different experiments, and the results were normalized to control oligonucleotides. Dotted lines represent 0.5-fold change. Values are averages ± sd.
EGR1 target genes critically regulate mammary cell migration
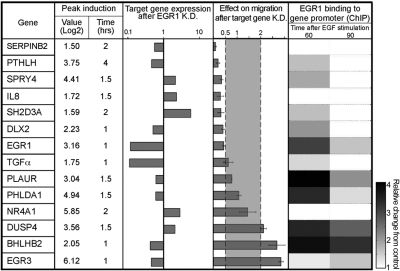
To identify EGR1-regulated genes involved in the inducible migratory phenotype, we knocked down EGR1 and analyzed effects on transcription using DNA arrays (Supplemental Fig. S3D, E). In the first step, we compared the transcripts in the control siRNA time series to those induced by EGF (Supplemental Fig. S2A), and assembled a list of 103 overlapping genes (validated EGF signature genes; Supplemental Fig. S3F). Next, expression of the 103 genes was analyzed in the EGR1 knocked-down cells. Genes that differed at any time point by ≥2-fold were grouped (denoted EGR1 signature genes). The resulting list comprised 18 of the 103 genes. Out of these, only 14 could be validated by RT-PCR (Fig. 5, left column). Consistent with early induction, 5 of the 14 validated genes (including EGR1) displayed peak expression within the first 60 min after stimulation (Fig. 5, second column). Another feature of this list was reciprocality: while the majority of the genes that peaked within 60 min displayed decreased expression on EGR1 knockdown, most of the rest peaked at 1.5–2 h and displayed an increase in mRNA expression under the same conditions. This is reminiscent of the dual effect of EGR1, which acts both as a transcription activator, as well as a repressor (16, 34).
Figure 5.
Functional analyses of the putative EGR1 signature genes. Candidate genes are listed in left column, along with features of their maximal induction. Effect of knocking down EGR1 expression on the level of expression of each gene is shown on a logarithmic scale (middle column). Involvement of each candidate gene in cell migration was tested by knocking down the respective gene using siRNA oligonucleotides. Bars represent mean ± sd values of duplicate determinations. To determine EGR1 binding to the respective promoter, MCF10A cells were serum starved and stimulated with EGF for 60 or 90 min. ChIP (right column) was performed using an anti-EGR1 antibody and qPCR with primers specific to the predicted EGR1 binding motifs, located up to 1 kb upstream to the transcription start site of the respective gene.
To test the prediction that EGR1 directly regulates the promoters of individual members of the 14-gene signature, we employed a ChIP assay (Fig. 5, right column). The results indicated that the majority of genes (11 of 14) were physically bound by EGR1. Interestingly, we found that EGR1 also binds to its own promoter, implying autoregulation. Next, we assessed the effect of each gene on cell migration by using RNA interference (Fig. 5, 4th column). Of the 14 genes, 10 demonstrated significant (>2-fold) and reproducible effects on cell migration, primarily a decrease when disabled, in line with the effect of EGR1 knockdown (Fig. 4C). To complete the functional characterization of the group of EGR1 target genes, we quantified the effect of their knockdown on cell proliferation (Supplemental Fig. S4A). With the exception of one gene, IL8, none of the genes exerted a significant effect on cell proliferation. Taken together, the results presented in Fig. 5 identify a group of EGR1 target genes specifically involved in the regulation of cell migration by EGF.
Regulation of EGR1 by microRNA-191 and feedback loops
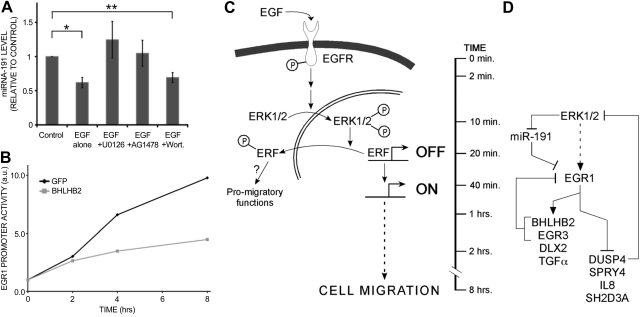
Because of the pivotal migratory roles that our analyses attributed to EGR1, we investigated how EGR1 itself is regulated. Our previous studies found that EGR1's transcript is directly controlled by microRNA-191 via multiple nucleotide sequences located at the 3′ untranslated region of the corresponding mRNA. Moreover, microRNA-191 undergoes rapid degradation in EGF-stimulated MCF10A cells prior to the initiation of migration (22). The results presented in Fig. 6A confirm this effect, and also indicate that the rapid turnover of microRNA-191 is mediated by the MEK-ERK pathway, but not by the PI3K-AKT pathway. Other feedback mechanisms regulate EGR1 by means of its downstream targets. Three of the 14 EGR1 signature genes showed an increase in cell migration on knockdown (Fig. 5), suggesting that they act as negative regulators. Of these, EGR3 has previously been shown to negatively regulate EGR1 transcription (35), and DUSP4 is a known attenuator of the ERK cascade (36). Our results indicate that the third, the transcriptional repressor BHLHB2 (37), directly regulates the EGR1 promoter: knockdown of BHLHB2 up-regulated EGR1 (Supplemental Fig. S4B). On the other hand, a previously described EGR1 reporter plasmid (38) showed that BHLHB2 can suppress EGR1 expression (Fig. 6B), in line with the existence of a single BHLHB2 binding element in the promoter of EGR1 (Supplemental Fig. S4C).
Figure 6.
Complex regulation of EGR1 and mammary cell migration. A) MCF10A cells were serum starved for 24 h; preincubated for 30 min with AG1478 (10 μM), U0126 (5 μM), or Wortmannin (200 nM); and then stimulated with EGF (10 ng/ml, 15 min). Total RNA was extracted and reverse transcribed; RT-PCR with microRNA-191-specific primers was performed. *P = 0.018, **P = 0.026; Student's t test. B) HeLa cells were cotransfected with plasmids encoding GFP or BHLHB2, along with a luciferase reporter containing the EGR1 promoter. At 24 h after transfection, cells were serum starved for 16 h and then stimulated with EGF (20 ng/ml) for the indicated intervals. C, D) Models depicting the complex regulation of mammary cell migration by the EGF-ERK-ERF to the EGR1 axis. C) Binding of EGF to the EGFR rapidly stimulates the RAS-RAF-MEK pathway, which culminates in dual phosphorylation of ERK. The latter translocates to the cell nucleus and phosphorylates ERF on multiple sites. As a result, the transcriptional inhibition imposed by ERF is relieved, and the repressor translocates to the cytoplasm, where it may enhance promigratory functions. D) Wiring diagram presents the complex internal regulation of the EGR1 signature genes by microRNA-191 and intrinsic control loops. Vertical line represents the approximate timing of events.
In summary, our results highlight the complex wiring scheme underlying growth factor-induced migration of human mammary cells (Fig. 6C, D). We showed that the EGF-to-ERK cascade relays signals culminating in nuclear export of ERF and a multigene transcriptional program. Within this temporal program of gene expression, EGR1 appears to act as a homeostatic hub, which is richly regulated by intrinsic control circuitries.
DISCUSSION
Approximately 10–12 major signaling pathways relay the biochemical messages of several hundred growth factors and other cytokines, but mechanisms that encode signal specificity remain incompletely understood (1). One attractive mechanism, which was exemplified in pheochromocytoma PC-12 cells stimulated by either the nerve growth factor or EGF, entails feedback regulatory loops that determine signaling duration (39). Accordingly, a complex interplay of early covalent modifications (e.g., phosphorylation) and delayed transcriptional responses modulates the amplitude and frequency of the response, thereby achieving specificity (reviewed in ref. 40). Here, we employed EGF and MCF10A mammary cells to resolve the cascade of intracellular events leading to cell migration. When stimulated with EGF, MCF10A cells migrate and invade across tissue barriers, as opposed to cell proliferation initiated by serum factors (Fig. 1). This dichotomy enabled us to contrast the underlying biochemical pathways at the phosphoproteome (Fig. 2) and transcriptome (Fig. 4) levels. Accordingly, our screens identified a homeostatic hub, EGR1, which regulates both activators and inhibitors of cell migration (see summary models in Fig. 6).
Interestingly, while treatment with EGF primarily activated the ERK pathway, stimulation with serum resulted in AKT activation (Fig. 2B). Intriguingly, when ERK activation was inhibited in EGF-stimulated cells, AKT signals underwent an extensive enhancement (Fig. 2D, E), indicating a crosstalk mechanism that polarizes the responses to EGF and serum. Consistent with this notion, EGF-induced cellular migration depended on ERK activation, while the AKT pathway played no significant role in this process (Fig. 2C). Likewise, the transcriptional events emanating from each stimulus exhibited high functional specificity. Although both stimuli induced a similar set of IEGs, the amplitude of the induction profiles differed (Fig. 4A and Supplemental Fig. S2B–D). While EGR1 regulated primarily cell migration, the main serum-induced IEGs, c-Fos and FosB, did not affect this phenotype, and FosB regulated cell proliferation (Fig. 4C). Altogether, we demonstrate that signal-specific activation patterns are achieved by different stimuli; however, in analogy to the PC-12 cell system, the recognition of specificity may be fully appreciated when different signals are contrasted (41).
Screening for potential ERK-activated TFs (Fig. 3B) identified ERF, a ubiquitously expressed ETS domain transcriptional repressor, as a main regulator of cell migration. In analogy, ERF mediates neuronal differentiation of PC-12 cells through sustained activation of ERK (7). Likewise, in HeLa cells, EGF has been shown to induce ERF phosphorylation (42). In line with these observations, examination of ERF activation profiles in MCF10A cells revealed temporal and spatial regulation (Fig. 3D, F). Conceivably, ERF is able to translate ERK activation profiles to the induction of gene transcription. Because ERK is sensitive to fold-changes rather than to changes in absolute levels (43), ERF activation might share similar features. This issue and the possibility that nuclear and cytoplasmic pools of ERF fulfill different biological functions remain open.
To identify the downstream transcriptional events that initiate the EGF-induced migratory response, we focused on EGR1. Expression arrays of EGR1-depleted cells identified a group of genes, denoted EGR1 signature genes, whose knockdown displayed a significant effect on cell migration (Fig. 5), but not on cell proliferation (Supplemental Fig. S4A). Of this group, most of the respective promoters were bound by EGR1, as has previously been shown (44). These results, along with the previously reported roles played by EGR1 in motility induced by either HGF (45), or by viral SRC (46), provide compelling evidence that the gene signature that we identified is critical for mammary cell migration. Plotting the relationships between EGR1 and its target gene expression unveiled a dynamic function, which may be reminiscent of EGR1's duality: a tumor suppressor and an oncogene (47). Initially, EGR1 acts as an activator of transcription; however, from 90 to 120 min poststimulation, EGR1 may switch to function as a repressor of gene transcription. Accordingly, EGR1 controls both migration-promoting genes and negative regulators that initiate feedback at various levels to accelerate or dampen the response (see a scheme of feedback loops in Fig. 6D). Thus, TFs EGR3 (35) and BHLHB2 (Fig. 6B), both activated by EGR1, intiate feedback to down-regulate EGR1. Likewise, DUSP4, a MAPK-specific phosphatase, which is repressed by EGR1, dampens ERK activation (36). Yet another control layer comprises an ERK-regulated microRNA, miR-191 (Fig. 6A), which enhances degradation of the EGR1 mRNA and establishes a coherent feed-forward loop.
Predictably, the migration-promoting pathway that we uncovered, namely, a cascade linking the ERK-to-ERF axis with EGR1 and culminating in a small group of target genes, plays critical roles in late-progression stages of mammary and other types of tumors. For example, it has been reported that EGR1 regulates expression of heparanase, an endoglycosidase that degrades heparan sulfate proteoglycans, a key component of extracellular matrix (48). Similarly, several EGR1-regulated genes listed in Fig. 5 are known as positive regulators of tumor progression. For example, PLAUR, the urokinase receptor (also called u-PAR) mediates tumor-associated proteolysis, invasion, and metastasis (49). Interestingly, EGR1 also up-regulates PLAUR in prostate cancer cells (50), and we report regulation of yet another component of the plasminogen system, namely SERPINB2 (also called PAI2), a protease inhibitor. Another EGR1-regulated gene is the parathyroid hormone-related protein (PTHLH), which is produced by mammary tumors to stimulate osteolytic bone resorption and metastasis (51). In addition to EGR1-controlled transcription of TGF-α in mammary cells (Fig. 5), this TF also regulates expression of several other growth factors involved in tumor growth and spread, namely TGF-β, PDGF-α, IGF2, and 3 additional ligands of EGFR (epiregulin, HB-EGF, and amphiregulin; ref. 52). In line with pivotal roles of EGR1 in tumor progression, earlier studies demonstrated that the corresponding transcript is elevated in Wilm's tumors (53), and diminished EGR1 delays progression in a prostate cancer model (54).
In summary, we propose an EGR1-centered, migration-dedicated module of the EGF signaling pathway (55). Although EGF simultaneously ignites several cytoplasmic signaling pathways, only one of these pathways, the RAF-MEK-ERK cascade, appears to be responsible for the mammary cell migratory phenotype. Through multiple-site phosphorylation and export from the nucleus, the ERF transcriptional repressor links the rapid membranal and cytoplasmic events to the long-term transcriptional program. Our results indicate that the EGF-to-EGR1 migratory pathway is distinct from the signaling route, leading to cell proliferation, in terms of both the kinases involved and the underlying transcriptional programs. In addition, the migration-encoding pathway is tuned by several feedback and feed-forward loops, which likely ensure robustness and specificity. Future work will address the prediction that major oncogenic processes, like angiogenesis and evasion from apoptosis, unleash the ERK-ERF pathway through engagement of tissue-specific coactivators of EGR1, as well as by deregulating specific oncogenes and tumor suppressors.
Supplementary Material
Acknowledgments
The authors acknowledge research funding by the U.S. National Cancer Institute (CA072981, CA121994-01, and CA120248-01); the European Commission; the German-Israeli Project Cooperation; the Israel Cancer Research Fund; the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; the Kekst Family Institute for Medical Genetics; the Kirk Center for Childhood Cancer and Immunological Disorders; the Women's Health Research Center, funded by the Bennett-Pritzker Endowment Fund; the Marvelle Koffler Program for Breast Cancer Research; the Leir Charitable Foundation; the M. D. Moross Institute for Cancer Research; and The Susan G. Komen Foundation. Y.Y. is the incumbent of the Harold and Zelda Goldenberg Professorial Chair. E.D. is the recipient of the Henry J. Leir Professorial Chair.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ChIP
- chromatin immunoprecipitation
- EGFR
- epidermal growth factor receptor
- EGR1
- early growth response-1
- ERF
- ETS2 repressor factor
- ETS
- E twenty-six
- GF
- growth factor
- HGF
- hepatocyte growth factor
- IEG
- immediate early gene
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphoinositol 3′-kinase
- RPPA
- reverse-phase protein array
- TF
- transcription factor
- TGF
- transforming growth factor.
REFERENCES
- 1. Kholodenko B. N., Hancock J. F., Kolch W. (2010) Signalling ballet in space and time. Nat. Rev. Mol. Cell. Biol. 11, 414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avraham R., Yarden Y. (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell. Biol. 12, 104–117 [DOI] [PubMed] [Google Scholar]
- 3. Hynes N. E., MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 4. Yarden Y., Sliwkowski M. X. (2001) Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 5. Yuan T. L., Cantley L. C. (2008) PI3K pathway alterations in cancer: variations on a theme. Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katz M., Amit I., Yarden Y. (2007) Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta 1773, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Von Kriegsheim A., Baiocchi D., Birtwistle M., Sumpton D., Bienvenut W., Morrice N., Yamada K., Lamond A., Kalna G., Orton R., Gilbert D., Kolch W. (2009) Cell fate decisions are specified by the dynamic ERK interactome. Nat. Cell Biol. 11, 1458–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Gallic L., Sgouras D., Beal G., Jr., Mavrothalassitis G. (1999) Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol. Cell. Biol. 19, 4121–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chou Y. T., Lin H. H., Lien Y. C., Wang Y. H., Hong C. F., Kao Y. R., Lin S. C., Chang Y. C., Lin S. Y., Chen S. J., Chen H. C., Yeh S. D., Wu C. W. (2010) EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 70, 8822–8831 [DOI] [PubMed] [Google Scholar]
- 10. Herschman H. R. (1991) Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 60, 281–319 [DOI] [PubMed] [Google Scholar]
- 11. Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., Amariglio N., Vaisman N., Segal E., Rechavi G., Alon U., Mills G. B., Domany E., Yarden Y. (2007) A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39, 503–512 [DOI] [PubMed] [Google Scholar]
- 12. Yilmaz M., Christofori G. (2009) EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis. Rev. 28, 15–33 [DOI] [PubMed] [Google Scholar]
- 13. Nieman M. T., Prudoff R. S., Johnson K. R., Wheelock M. J. (1999) N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 147, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., Ben-Chetrit N., Tarcic G., Lindzen M., Avraham R., Liao Y. C., Trusk P., Lyass A., Rechavi G., Spector N. L., Lo S. H., Schmitt F., Bacus S. S., Yarden Y. (2007) A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 9, 961–969 [DOI] [PubMed] [Google Scholar]
- 15. Bauer J., Margolis M., Schreiner C., Edgell C. J., Azizkhan J., Lazarowski E., Juliano R. L. (1992) In vitro model of angiogenesis using a human endothelium-derived permanent cell line: contributions of induced gene expression, G-proteins, and integrins. J. Cell. Physiol. 153, 437–449 [DOI] [PubMed] [Google Scholar]
- 16. Grotegut S., von Schweinitz D., Christofori G., Lehembre F. (2006) Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 25, 3534–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCawley L. J., O'Brien P., Hudson L. G. (1997) Overexpression of the epidermal growth factor receptor contributes to enhanced ligand-mediated motility in keratinocyte cell lines. Endocrinology 138, 121–127 [DOI] [PubMed] [Google Scholar]
- 18. Fitzgerald J. S., Busch S., Wengenmayer T., Foerster K., de la Motte T., Poehlmann T. G., Markert U. R. (2005) Signal transduction in trophoblast invasion. Chem. Immunol. Allergy 88, 181–199 [DOI] [PubMed] [Google Scholar]
- 19. Xian C. J., Zhou X. F. (2004) EGF family of growth factors: essential roles and functional redundancy in the nerve system. Front. Biosci. 9, 85–92 [DOI] [PubMed] [Google Scholar]
- 20. Schlegel J., Stumm G., Brandle K., Merdes A., Mechtersheimer G., Hynes N. E., Kiessling M. (1994) Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J. Neurooncol. 22, 201–207 [DOI] [PubMed] [Google Scholar]
- 21. Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., Wei S., Hashimoto A., Yamada A., Yano H., Mazaki Y., Kodama H., Nio Y., Manabe T., Wada H., Kobayashi H., Sabe H. (2008) GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10, 85–92 [DOI] [PubMed] [Google Scholar]
- 22. Avraham R., Sas-Chen A., Manor O., Steinfeld I., Shalgi R., Tarcic G., Bossel N., Zeisel A., Amit I., Zwang Y., Enerly E., Russnes H. G., Biagioni F., Mottolese M., Strano S., Blandino G., Borresen-Dale A. L., Pilpel Y., Yakhini Z., Segal E., Yarden Y. (2010) EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci. Signal. 3, ra43. [DOI] [PubMed] [Google Scholar]
- 23. Tait L., Soule H. D., Russo J. (1990) Ultrastructural and immunocytochemical characterization of an immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6087–6094 [PubMed] [Google Scholar]
- 24. Gilles C., Polette M., Zahm J. M., Tournier J. M., Volders L., Foidart J. M., Birembaut P. (1999) Vimentin contributes to human mammary epithelial cell migration. J. Cell Sci. 112, 4615–4625 [DOI] [PubMed] [Google Scholar]
- 25. Irie H. Y., Pearline R. V., Grueneberg D., Hsia M., Ravichandran P., Kothari N., Natesan S., Brugge J. S. (2005) Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J. Cell Biol. 171, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sheehan K. M., Calvert V. S., Kay E. W., Lu Y., Fishman D., Espina V., Aquino J., Speer R., Araujo R., Mills G. B., Liotta L. A., Petricoin E. F., 3rd, Wulfkuhle J. D. (2005) Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics 4, 346–355 [DOI] [PubMed] [Google Scholar]
- 27. Wells A., Gupta K., Chang P., Swindle S., Glading A., Shiraha H. (1998) Epidermal growth factor receptor-mediated motility in fibroblasts. Microsc. Res. Tech. 43, 395–411 [DOI] [PubMed] [Google Scholar]
- 28. Klemke R. L., Cai S., Giannini A. L., Gallagher P. J., de Lanerolle P., Cheresh D. A. (1997) Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 137, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potempa S., Ridley A. J. (1998) Activation of both MAP kinase and phosphatidylinositide 3-kinase by Ras is required for hepatocyte growth factor/scatter factor-induced adherens junction disassembly. Mol. Biol. Cell 9, 2185–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moelling K., Schad K., Bosse M., Zimmermann S., Schweneker M. (2002) Regulation of Raf-Akt Cross-talk. J. Biol. Chem. 277, 31099–31106 [DOI] [PubMed] [Google Scholar]
- 31. Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 32. Sharrocks A. D. (2001) The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 33. Conacci-Sorrell M., Ngouenet C., Eisenman R. N. (2010) Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kasneci A., Kemeny-Suss N. M., Komarova S. V., Chalifour L. E. (2009) Egr-1 negatively regulates calsequestrin expression and calcium dynamics in ventricular cells. Cardiovasc. Res. 81, 695–702 [DOI] [PubMed] [Google Scholar]
- 35. Collins S., Lutz M. A., Zarek P. E., Anders R. A., Kersh G. J., Powell J. D. (2008) Opposing regulation of T cell function by Egr-1/NAB2 and Egr-2/Egr-3. Eur. J. Immunol. 38, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Owens D. M., Keyse S. M. (2007) Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 37. Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. (2002) Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844 [DOI] [PubMed] [Google Scholar]
- 38. Weisz L., Zalcenstein A., Stambolsky P., Cohen Y., Goldfinger N., Oren M., Rotter V. (2004) Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 64, 8318–8327 [DOI] [PubMed] [Google Scholar]
- 39. Santos S. D., Verveer P. J., Bastiaens P. I. (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324–330 [DOI] [PubMed] [Google Scholar]
- 40. Zwang Y., Sas-Chen A., Drier Y., Shay T., Avraham R., Lauriola M., Shema E., Lidor-Nili E., Jacob-Hirsch J., Amariglio N., Lu Y., Mills G. B., Rechavi G., Oren M., Domany E., Yarden Y. (2011) Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Mol. Cell 42, 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lawson M. A., Tsutsumi R., Zhang H., Talukdar I., Butler B. K., Santos S. J., Mellon P. L., Webster N. J. (2007) Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop Involving early growth response-1 and Ngfi-A binding protein 1 and 2. Mol. Endocrinol. 21, 1175–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 43. Cohen-Saidon C., Cohen A. A., Sigal A., Liron Y., Alon U. (2009) Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell. 36, 885–893 [DOI] [PubMed] [Google Scholar]
- 44. Tang C., Shi X., Wang W., Zhou D., Tu J., Xie X., Ge Q., Xiao P. F., Sun X., Lu Z. (2010) Global analysis of in vivo EGR1-binding sites in erythroleukemia cell using chromatin immunoprecipitation and massively parallel sequencing. Electrophoresis 31, 2936–2943 [DOI] [PubMed] [Google Scholar]
- 45. Damm S., Koefinger P., Stefan M., Wels C., Mehes G., Richtig E., Kerl H., Otte M., Schaider H. (2010) HGF-promoted motility in primary human melanocytes depends on CD44v6 regulated via NF-κB, Egr-1, and C/EBP-beta. J. Invest. Dermatol. 130, 1893–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cermak V., Kosla J., Plachy J., Trejbalova K., Hejnar J., Dvorak M. (2010) The transcription factor EGR1 regulates metastatic potential of v-src transformed sarcoma cells. Cell. Mol. Life Sci. 67, 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baron V., Adamson E. D., Calogero A., Ragona G., Mercola D. (2006) The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Ther. 13, 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Mestre A. M., Rao S., Hornby J. R., Soe-Htwe T., Khachigian L. M., Hulett M. D. (2005) Early growth response gene 1 (EGR1) regulates heparanase gene transcription in tumor cells. J. Biol. Chem. 280, 35136–35147 [DOI] [PubMed] [Google Scholar]
- 49. Laufs S., Schumacher J., Allgayer H. (2006) Urokinase-receptor (u-PAR): an essential player in multiple games of cancer: a review on its role in tumor progression, invasion, metastasis, proliferation/dormancy, clinical outcome and minimal residual disease. Cell Cycle 5, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 50. Salah Z., Maoz M., Pizov G., Bar-Shavit R. (2007) Transcriptional regulation of human protease-activated receptor 1: a role for the early growth response-1 protein in prostate cancer. Cancer Res. 67, 9835–9843 [DOI] [PubMed] [Google Scholar]
- 51. Kakonen S. M., Selander K. S., Chirgwin J. M., Yin J. J., Burns S., Rankin W. A., Grubbs B. G., Dallas M., Cui Y., Guise T. A. (2002) Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 277, 24571–24578 [DOI] [PubMed] [Google Scholar]
- 52. Sauer L., Gitenay D., Vo C., Baron V. T. (2010) Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene 29, 2628–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scharnhorst V., Menke A. L., Attema J., Haneveld J. K., Riteco N., van Steenbrugge G. J., van der Eb A. J., Jochemsen A. G. (2000) EGR-1 enhances tumor growth and modulates the effect of the Wilms' tumor 1 gene products on tumorigenicity. Oncogene 19, 791–800 [DOI] [PubMed] [Google Scholar]
- 54. Abdulkadir S. A., Qu Z., Garabedian E., Song S. K., Peters T. J., Svaren J., Carbone J. M., Naughton C. K., Catalona W. J., Ackerman J. J., Gordon J. I., Humphrey P. A., Milbrandt J. (2001) Impaired prostate tumorigenesis in Egr1-deficient mice. Nat. Med. 7, 101–107 [DOI] [PubMed] [Google Scholar]
- 55. Oda K., Matsuoka Y., Funahashi A., Kitano H. (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1, 0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.