Abstract
microRNAs (miRNAs) have emerged as regulators of a broad spectrum of neurodevelopmental processes, including brain morphogenesis, neuronal differentiation, and survival. While the role of miRNAs in establishing and maintaining the developing nervous system is widely appreciated, the developmental neurobehavioral role of miRNAs has yet to be defined. Here we show that transient disruption of brain morphogenesis by ethanol exposure results in behavioral hyperactivity in larval zebrafish challenged with changes in lighting conditions. Aberrations in swimming activity persist in juveniles that were developmentally exposed to ethanol. During early neurogenesis, multiple gene expression profiling studies revealed widespread changes in mRNA and miRNA abundance in ethanol-exposed embryos. Consistent with a role for miRNAs in neurobehavioral development, target prediction analyses identified multiple miRNAs misexpressed in the ethanol-exposed cohorts that were also predicted to target inversely expressed transcripts known to influence brain morphogenesis. In vivo knockdown of miR-9/9* or miR-153c persistently phenocopied the effect of ethanol on larval and juvenile swimming behavior. Structural analyses performed on adults showed that repression of miR-153c during development impacts craniofacial skeletal development. Together, these data support an integral role for miRNAs in the establishment of vertebrate neurobehavioral and skeletal systems.—Tal, T. L., Franzosa, J. A., Tilton, S. C., Philbrick, K. A., Iwaniec, U. T., Turner, R. T., Waters, K. M., Tanguay, R. L. MicroRNAs control neurobehavioral development and function in zebrafish.
Keywords: miR-9, miR-153, skeletal development
MicroRNAs (miRNAs) are small, noncoding RNAs that function as regulators of posttranscriptional gene expression. miRNAs control gene expression by binding to complementary sequences in the 3′ untranslated region of target mRNA transcripts, which results in transcript degradation and/or inhibition of translation (reviewed in ref. 1). The developing nervous system is a rich source of miRNAs. A number of miRNA expression profiling studies have identified miRNAs that are enriched in the nervous system and whose expression is tightly controlled, both spatially and temporally, during development (2–4). In animal studies, genomic ablation of dicer, a key ribonuclease involved in miRNA biogenesis, results in the loss of dicer-dependent miRNA synthesis and severe deficits in brain development (5–7). In zebrafish maternal-zygotic dicer mutants, defects in central nervous system (CNS) patterning were partially rescued by exogenous miRNA (miR)-430, underscoring the significance of miRNAs in brain morphogenesis (6). Dicer ablation is also associated with neurodegeneration (8), neuronal cell death (5), survival of differentiating neural crest cells (9), and synaptic plasticity, including deficits in dendritic arborization (10) and axon pathfinding (11). Taken together, these studies emphasize the importance of miRNAs in neural development and disease. While specific miRNAs have been associated with a wide range of neurodevelopmental processes, the neurobehavioral roles of specific miRNAs are less well understood.
The larval zebrafish boasts a rapidly developed nervous system, with the first wave of neurogenesis occurring between 16 and 36 hours postfertilization (hpf; ref. 12). During this window, key developmental events take place, resulting in a fully functional nervous system at the time of hatching. The rapid developmental profile coupled with fertilization external to the mother and genetic tractability render the larval zebrafish an excellent model with which to uncover the role of miRNAs in the establishment and maintenance of the developing neurobehavioral system.
We have shown previously that developmental exposure to ethanol results in characteristic teratogenic effects in embryonic zebrafish (13, 14). In the current study, ethanol exposure was employed to examine the functional role of miRNAs in directing neurobehavioral development. While several studies implicate specific miRNAs in a wide range of neurodevelopmental processes (5, 6, 9, 10), to our knowledge, the functional consequence of miRNA disruption during development has yet to be examined at a behavioral level. However, a single report links a specific miRNA with a functional behavioral phenotype in vivo. Kocerha et al. (15) showed that repression of miR-219 expression in the prefrontal cortex, following acute exposure to an NMDA receptor inhibitor, functions as a compensatory mechanism by which pharmacological locomotor manifestations are abrogated in adult mice. Here we show that transient developmental exposure to ethanol results in persistent behavioral aberrations that are mediated, in part, via misexpression of specific miRNA species during critical windows of nervous system development.
MATERIALS AND METHODS
Fish care and husbandry
Adult tropical 5D strain zebrafish (Danio rerio) were raised according to Institutional Animal Care and Use Committee protocols in the Sinnhuber Aquatic Research Laboratory at Oregon State University. Adults were maintained on a 14/10 h light-dark schedule on a recirculating system in which water was maintained at 28 ± 1°C and pH 7.0 ± 0.2.
Ethanol exposure
Depending on experiment, groups of embryos were exposed to waterborne ethanol (0–300 mM; absolute ethyl alcohol USP, 200 proof; AAPER Alcohol and Chemical, Brookfield, CT, USA; Supplemental Table S1) in buffered embryo medium (16) or embryo medium alone in 20-ml glass vials sealed with Teflon-lined lids to prevent volatilization. Each exposure group was considered a single replicate. Based on previous studies, embryos were exposed to ethanol from 4 to 24 hpf (13) and rinsed, and embryo homogenate was collected or whole embryos were raised until 5, 35, 60, or 100 days postfertilization (dpf), depending on experiment.
Larval locomotion assay
At 5 dpf, automated quantification of larval movement was obtained during alternating periods of light and dark (17, 18). For all experiments, larvae were subjected to locomotion analyses at 12:00 to 3:00 PM in a sound- and temperature-controlled (26°C) behavioral testing room. Movement of 96 individual larvae were simultaneously measured and recorded using an automated video tracking system (Videotrack, quantization mode; Viewpoint Life Sciences, Lyon, France). To distinguish 5-dpf larvae within each individual well from the background, the life-stage-specific size parameter was set at 17, and the number of seconds during which the larvae moved above the movement threshold, set at 7, was analyzed in 60-s bins. The data were processed using a custom PERL script. Data are presented as the total time spent in motion (s) during the initial 5-min dark period. To detect statistical differences between groups, the data were averaged among 16 animals/exposure group, and the experiment was repeated 7 times. Data were analyzed using a repeated-measures analysis of variance (ANOVA) with a Tukey's multiple-comparison post hoc test.
Juvenile locomotion assay
Juvenile testing was performed on 35-dpf animals using the Viewpoint tracking system as described above. On the day of testing, juveniles were transferred to individual wells of 6-well plates that contained inserts to prevent well-to-well visibility. Following plate acclimatization, movement of 24 animals was assessed concurrently (Viewpoint). For 35-dpf juveniles, the life-stage parameter was set at 30. The amount of time spent in movement above the background threshold, set at 45, was measured in 60-s bins. Data are presented as time spent in motion (s) during 10-min light or dark periods. To eliminate the effect of size bias on swimming activity, juvenile movement data was normalized to animal length (Supplemental Fig. S1).
cDNA and miRNA microarray assays
mRNA was isolated using TRIzol Reagent (Life Technologies, Carlsbad, CA, USA). Each replicate was collected at 24 hpf and derived from pools of 75 embryos that were exposed to 100–300 mM ethanol or embryo medium control from 4 to 24 hpf (n=4). Sample cleanup was performed using MinElute columns (Qiagen, Valencia, CA, USA). cDNA synthesis, labeling, and hybridization were performed by the University of Idaho Initiative for Bioinformatics and Evolutionary Studies core facility (Moscow, ID, USA) according to NimbleGen 5.0 microarray protocol, with the exception of the first-strand synthesis step, in which 50 μM oligo (dT) per reaction and 400 ng of random hexamers per microliter of reaction were used (Roche NimbleGen, Madison, WI, USA). Labeled cDNA samples were hybridized to a zebrafish 12×135K Nimblegen array and NimbleScan software was used for image analysis and quality control. Raw intensity data were quantile normalized by RMA summarization (19), and outliers were identified by correlation analysis based on Pearson correlation coefficient. A single replicate was removed from the 100 mM treatment group and resulting data were subjected to both pairwise analysis of variance (20) with Tukey's post hoc test and 5% false discovery rate calculation (21).
For the miRNA expression study, RNA was isolated using a miRNeasy kit (Qiagen) from pooled tissue (75 embryos) harvested at 12–48 hpf (n=2). The microarray assay was performed by LC Sciences (Houston, TX, USA) as per the manufacturer's specifications (22). Labeled cDNA samples were hybridized to miRZebrafish arrays (LC Sciences; probe content reflects miRBase release 12.0), and images were collected using a laser scanner (GenePix 4000B; Molecular Devices, Sunnyvale, CA, USA) and digitized using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD, USA). Data were background-subtracted, normalized by LOWESS (locally weighted regression; ref. 19), and analyzed for differential expression by t test (P<0.05). All array data are accessible through the U.S. National Center for Biotechnology Information Gene Expression Omnibus (23), series accession number GSE30497 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30497).
Real-time quantitative PCR
Relative gene expression in whole-embryo homogenates was quantified using real-time quantitative RT-PCR (qRT-PCR). Total RNA was isolated (Qiagen) and reverse-transcribed to generate cDNA using a SSIII First Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA). Primer sequences for BCL2 binding component 3 (Bbc3), dual specificity phosphatase 4 (DUSP4), glutathione peroxidase 3 (GPX3), neurogenic differentiation 6 (neuroD6), ornithine decarboxylase 1 (odc1), solute carrier family 6 (neurotransmitter transporter, GABA) member 1 (SLC6A1), solute carrier family 16 (monocarboxylic acid transporters) member 9 (SLC16A9), and carrier family 17 (sodium-dependent inorganic phosphate cotransporter) member 6 (SLC17A6; Eurofins MWG Operon, Huntsville, AL, USA) are listed in Supplemental Table S2. Quantitative fluorogenic amplification of cDNA was performed using the ABI StepOne Plus (Applied Biosystems, Carlsbad, CA, USA), primer sets of interest, and Power SYBER Green PCR Master Mix (Applied Biosystems), and ROX internal reference dye. For miRNA quantification, The miRCURY LNA microRNA PCR System (Exiqon, Woburn, MA, USA) including the universal RT microRNA PCR; polyadenylation and cDNA synthesis kit; SYBR Green master mix; and microRNA primer sets Homo sapiens (hsa)-miR-9 (PN 204513), hsa-204 (PN 204507), or hsa-miR-9* (PN 204620); and control primer set [U6 small nucleolar RNA (snoRNA), PN 203907] were used to quantify miRNA expression. For Danio rerio (dre)-miR-153c, the reverse primer for hsa-miR-153 (PN 204338) was coupled with a custom forward primer named dre-miR-153b/c FwdP (PN 206999).
Bioinformatics
Unsupervised bidirectional hierarchical clustering of microarray data was performed using a euclidean distance metric and centroid linkage clustering to group treatments and gene expression patterns by similarity. Principal component analysis (PCA) was performed on conditions using nontransformed normalized intensity values. PCA was performed on conditions using euclidean distance as a measure of dissimilarity for nontransformed normalized intensity values. For PCA, a covariance matrix was created by measurements between factors, and eigenvectors were determined for the main condition profiles. Eigenvalues, which are representative of the level of explained variance as a percentage of total variance, are reported for the top two components. The clustering algorithms, heat-map visualizations, and centroid calculations were performed in Multi-Experiment Viewer (24) or GeneSpring 11 software (Agilent Technologies, Santa Clara, CA, USA) based on log2 expression ratio values. To query array data, orthologous human, mouse, and rat gene identifiers were imported into Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, USA). Enrichment was calculated in the functional analysis tool using the right-tailed Fisher's exact test, where values of P < 0.05 indicate a statistically significant, nonrandom association. Target prediction algorithms MicroCosm (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl), Target Scan (http://targetscan.org/) and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html) were employed to identify putative targets of candidate miRNAs. To reduce the probability of false positives, targets predicted by <2 programs were excluded from downstream analyses. Putative target lists were merged with empirically derived inversely expressed transcripts.
miRNA knockdown
miR-9, miR-153c, miR-204, and control 3′ fluorescein-tagged antisense oligonucleotide morpholinos (MOs; Gene Tools, Philomath, OR, USA) were microinjected into single-cell stage embryos (Supplemental Table S3). Injected animals were screened at 24 hpf for MO incorporation, and screened morphants were subjected to behavior analysis at 120 hpf or 35 dpf or micro-computed tomography (μCT) analysis at 60 or 100 dpf. Approximately 2 nl of 0.2 mM MO was microinjected into each embryo.
μCT analysis of mineralized tissue
μCT was used for nondestructive 3-dimensional evaluation of mineralized tissue. Fish heads were scanned using a Scanco μCT40 scanner (Scanco Medical AG, Brüttisellen, Switzerland) at a voxel size of 12 × 12 × 12 μm. Head and otolith mineralized tissue volumes were evaluated at a threshold of 215 and 650 (scale 0–1000), respectively.
Statistical analysis
Data are presented as means ± se. Unless indicated otherwise, 2-tailed paired Student's t test or 1-way ANOVA with a Dunnet's or Tukey's multiple-comparison post hoc test was used to evaluate differences between control and treated groups; values of P < 0.05 were considered statistically significant. Results were tabulated with Prism 5.01 (GraphPad, La Jolla, CA, USA).
RESULTS
Transient developmental exposure to ethanol results in persistent neurobehavioral effects in larval and juvenile zebrafish
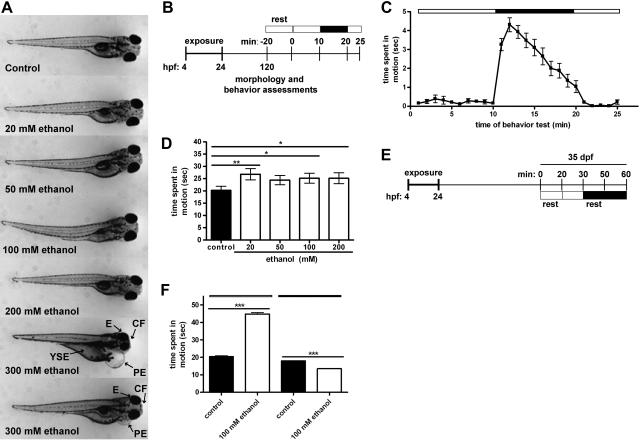
The effects of transient developmental exposure to ethanol are shown in Fig. 1. At 120 hpf, exposure to 20–200 mM ethanol had no visibly discernable effect on larval morphology (Fig. 1A). In contrast, larvae transiently developmentally exposed to 300 mM ethanol presented with a number of malformations, including craniofacial and eye defects, shortened trunk, and yolk sac and pericardial edemas. At 120 hpf, developmentally exposed larvae (0–200 mM ethanol) were subjected to an automated light–dark locomotion test (Fig. 1B). Larval locomotion was more pronounced during periods of dark relative to periods of light (Fig. 1C). Transient developmental exposure to 20, 100, or 200 mM ethanol resulted in behavioral hyperactivity (Fig. 1D).
Figure 1.
Developmental exposure to ethanol results in sustained neurobehavioral effects in larval and juvenile zebrafish. Embryos were exposed to 0–300 mM ethanol from 4 to 24 hpf, and locomotion was assessed at 120 hpf and 35 dpf. A) Representative images at 120 hpf; pericardial edema (PE), yolk sac edema (YSE), and eye (E) and craniofacial (CF) malformations are noted. B) Larval locomotion test, consisting of alternating periods of light (open bar) and dark (solid bar). C) Movement data from a single experiment consisting of 16 larvae; data are represented as time spent in motion (s). D) Total movement during the initial 5 min of the dark period was measured in control and ethanol-exposed groups (n=7). E) Juvenile behavior test. F) Juvenile movement during 10-min dark or light periods, represented as time spent in motion (s) normalized to individual weight-to-length ratios. (n=12). *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether ethanol-induced behavioral abnormalities persist, a subset of 100 mM exposed and clutch-matched embryo medium-exposed embryos were raised until 35 dpf, and subjected to a juvenile light–dark locomotion test (Fig. 1E and Supplemental Fig. S1). Movement data was normalized to individual animal lengths to control for differences in size. Transient developmental exposure to ethanol resulted in general behavioral aberrations, with pronounced hyperactivity under conditions of light and hypoactivity in the absence of light (Fig. 1F). Taken together, these data suggest that, in the absence of physically discernable malformations, transient developmental exposure to ethanol results in persistent locomotor abnormalities in response to changes in lighting conditions.
Exposure to ethanol induces widespread changes in developmental gene expression
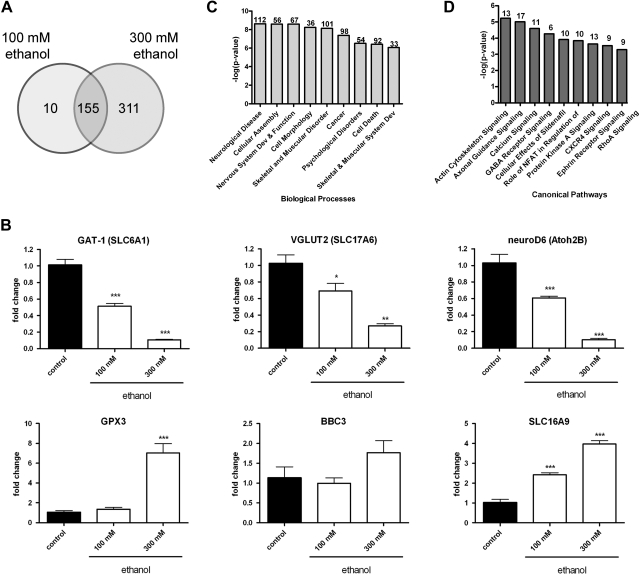
In order to obtain a general assessment of the effects of ethanol on developmental gene expression, we next generated gene expression profiles by microarray analysis of pools of embryos developmentally exposed ethanol. Of the 165 transcripts, 155 differentially expressed in embryos exposed to 100 mM ethanol were similarly misexpressed in the 300 mM ethanol group (Fig. 2A). Validation of microarray results was performed by qRT-PCR to measure levels known to be pivotal to neurobehavioral development or associated with ethanol toxicity: neuroD6, VGLUT2, atonal homolog 2B (Atoh2B), bbc3, GPX3, and SLC16A9 (Fig. 2B).
Figure 2.
Global misregulation of gene expression in zebrafish embryos developmentally exposed to ethanol. cDNA microarray analysis was performed on samples obtained at 24 hpf from pools of embryos exposed to 0, 100, or 300 mM ethanol from 4 to 24 hpf (n=4). A) Venn diagram depicting the overall number of statistically significant differentially expressed genes relative to stage-matched controls (P<0.05, 5% FDR). B) GAT-1, VGLUT2, nueroD6, GPX3, BBC3, and SLC16A9 mRNA levels were measured by qRT-PCR at 24 hpf in pools of embryos exposed to 0–300 mM ethanol. Data were normalized to ODC1 levels; values reflect fold change relative to unexposed controls (n=5). *P < 0.05, **P < 0.01, ***P < 0.001. C, D) Significant biological processes (C) and canonical pathways (D) enriched in the 100 mM ethanol vs. control array dataset (P<0.05, 5% FDR).
Functional enrichment statistics and network analyses were performed to identify the most significant biological processes affected by 100 mM ethanol treatment (Fig. 2C). Using IPA, 112 of 165 transcripts differentially expressed in embryos exposed to 100 mM ethanol are associated with neurological disease. Of relevance to the locomotion phenotypes observed in ethanol-exposed cohorts, transcripts broadly associated with both the nervous and musculoskeletal systems were also significantly misexpressed relative to controls. When examining the top 10 canonical signaling pathways perturbed in the 100 mM ethanol-exposed cohort, the majority of pathways were critical to nervous system development and function, including actin and cytoskeleton, axonal guidance, calcium, GABA receptor, protein kinase A, ephrin, and RhoA signaling pathways (Fig. 2D). These data point to a subset of genes that may be involved in the complex neurobehavioral manifestations observed in larvae and juveniles transiently exposed to ethanol during development.
Misregulation of miRNAs observed in ethanol-exposed embryos
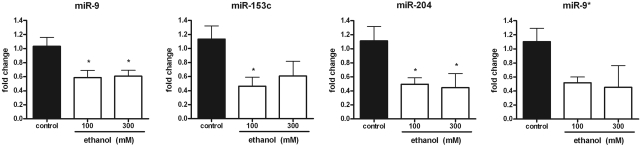
We performed miRNA expression profiling studies to detect developmental differences in miRNA expression in embryos exposed to ethanol from 4–24 hpf. A total of 35 miRNA transcripts were significantly differentially expressed relative to controls with the greatest number of misexpressed miRNAs detected at 24 hpf (Supplemental Fig. S3A). A subset of these miRNAs was selected for further examination based on their previously reported enriched expression patterns in the CNS (2, 25) and sequence homology with mouse and human homologs (Table 1). miR-9 and miR-9* are encoded on the 5p and 3p arms of the same hairpin precursor molecule. As expected, repression of miR-9, miR-153c, and miR-204 was observed by qRT-PCR in ethanol-exposed animals (Fig. 3). Together with the mRNA profiling study, these data suggest that transient developmental exposure to ethanol results in targeted misexpression of genes that encode both miRNAs and, possibly, their target transcripts.
Table 1.
Misexpressed miRNAs display conserved CNS expression and seed sequence homology
| miRNA | log2 | P | Mature miRNA sequence |
|---|---|---|---|
| dre-miR-9 | −3.7 | 0.024 | |
| Human | ucuuugguuaucuagcuguauga | ||
| Mouse | ucuuugguuaucuagcuguauga | ||
| Zebrafish | ucuuugguuaucuagcuguauga | ||
| dre-miR-153c | −2.8 | 0.048 | |
| Human | uugcauagucacaaaagugauc | ||
| Mouse | uugcauagucacaaaagugauc | ||
| Zebrafish | uugcauagucacaaaaaugauc | ||
| dre-miR-204 | −2.7 | 0.016 | |
| Human | uucccuuugucauccuaugccu | ||
| Mouse | uucccuuugucauccuaugccu | ||
| Zebrafish | uucccuuugucauccuaugccu | ||
| dre-miR-9* | −2.6 | 0.057 | |
| Human | auaaagcuagauaaccgaaagu | ||
| Mouse | auaaagcuagauaaccgaaagu | ||
| Zebrafish | uaaagcuagauaaccgaaagu |
For all miRNAs listed, CNS expression is conserved in humans, zebrafish, and mice; seed sequences are underscored. Sequences derived from miRBase (http://www.mirbase.org).
Figure 3.
Exposure to ethanol affects developmental miRNA expression. miR-9, miR-153c, miR-204, and miR-9* transcript levels were assessed by qRT-PCR in pools of embryos exposed to 0, 100, or 300 mM ethanol from 4 to 24 hpf. Data were normalized to U6 snRNA levels; values reflect fold change relative to unexposed controls (n=6–9). *P < 0.05.
Misexpressed transcripts are putatively targeted by inversely expressed miRNAs
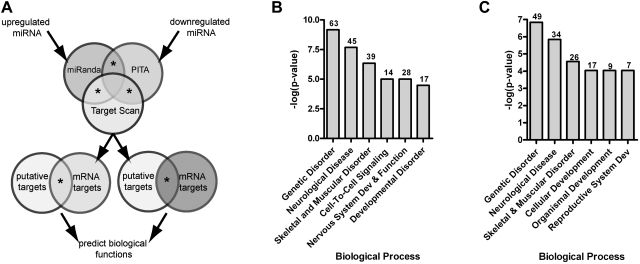
To identify changes in gene expression that may be subject to miRNA regulation, we employed a bioinformatics approach (Fig. 4A). As shown in Fig. 4B, biological processes enriched in misexpressed transcripts predicted to be regulated by inversely expressed miRNA include genetic disorder, neurological disease, skeletal and muscular disorder, cell-to-cell signaling, nervous system development and function, and developmental disorder. When down-regulated or up-regulated transcripts putatively targeted by inversely expressed miRNA were examined in isolation, the top biological processes were similar but not identical (Fig. 4C and Supplemental Fig. S4).
Figure 4.
Misexpressed miRNAs predicted to target inversely expressed transcripts. A) Putative miRNA target lists were merged with empirically derived inversely expressed transcripts. B) Biological processes enriched in misexpressed mRNA transcripts predicted to be regulated by inversely expressed miRNA (P<0.05). Numbers of genes associated with each process are indicated above bars. C) Processes associated with down-regulated mRNA transcripts predicted to be regulated by inversely expressed miRNA (P<0.05).
miR-9/9* and miR-153c control neurobehavioral development in zebrafish
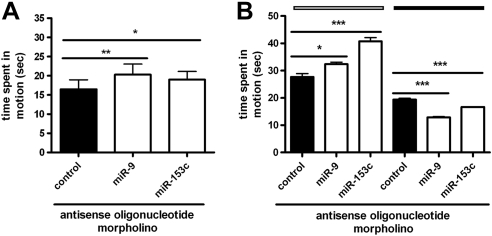
We next examined whether misexpressed miRNAs influence locomotion in larval and juvenile zebrafish. Larvae injected with anti-miR-9/9* or anti-miR-153c MOs were hyperactive during periods of dark, relative to control MO-injected populations, phenocopying the effects of ethanol on larval swimming activity (Fig. 5A). In support of these findings, miR-9/9* and miR-153c morphants (animals injected with morpholinos), raised to the juvenile stage, recapitulated abnormal swimming patterns observed in ethanol-exposed animals (hyperactivity under light conditions and hypoactivity in the dark; Fig. 5B). These data suggest that the effects of ethanol exposure on locomotion may be mediated, in part, via repression of miR-9/9* and miR-153c levels during early neurogenesis.
Figure 5.
miR-9/9* and miR-153c regulate neurobehavioral development in zebrafish. miR-9/9* and miR-153c MOs were injected into single-cell-stage embryos. A) At 120 hpf, miR-9/9*, miR-153c, and control morphants were subjected to the larval light-dark locomotion test (n=7). B) A subset of miR-9/9*, miR-153c, and control morphants, raised until 35 dpf, was subjected to the juvenile locomotion assay; time spent in motion (s) during 10-min dark or light periods was normalized to individual weight-to-length ratios (n=8). *P < 0.05 and ***P < 0.001.
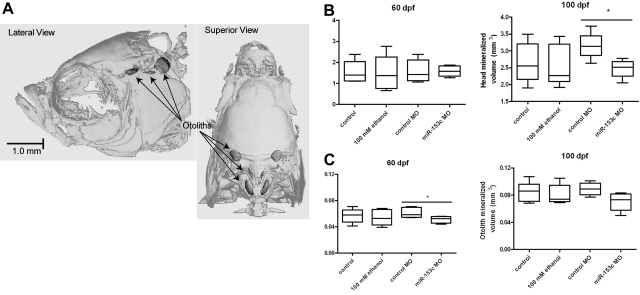
miR-153c promotes skeletal development in zebrafish
To examine the role of miR-153c on skeletal development, we performed μCT analysis on adult zebrafish (60 and 100 dpf) that experienced developmental miRNA knockdown of miR-153c. Clutch-matched adults injected with a control MO or exposed to embryo medium or 100 mM ethanol were included for comparison. Relative to unexposed controls, significant differences in mineralized head or otolith tissue volumes (Fig. 6A) were not detected at either 60 or 100 dpf in ethanol-exposed animals (Fig. 6B, C). In contrast, a significant reduction in head mineralized tissue volume, relative to control morphants, was observed in miR-153c morphant fish at 100 dpf (Fig. 6B). Reduced otolith volume was additionally observed in miR-153c morphants at 60 dpf (Fig. 6C).
Figure 6.
miR-153c influences skeletal development in zebrafish. Embryos injected with miR-153c or control MOs were raised until adulthood. A subset of clutch-matched embryos was exposed to embryo medium control or 100 mM ethanol from 4 to 24 hpf for comparison. At 60 or 100 dpf, adult females were analyzed by μCT to assess skeletal structure of the head. A) Representative μCT depicting head and otolith mineralized tissue volume. B, C) Head (B) and otolith (C) mineralized volumes represent 4–9 females. *P < 0.05.
DISCUSSION
The mechanisms by which miRNAs facilitate or regulate neurobehavioral development and function are not well understood. miRNAs are critical to a variety of neurodevelopmental processes (6, 10, 11, 26, 27). Therefore, we hypothesized that miRNAs are similarly required to establish a functional neurobehavioral system during development. In the present study, we used ethanol as a tool to disrupt normal miRNA-dependent signaling during development to identify for the first time a number of miRNAs that control vertebrate neurobehavioral development.
Genetically encoded sensory-motor circuitry is hard-wired into embryonic development, resulting in a simple set of behaviors that enables the organism to locate food and detect and avoid predators (28). In vertebrates, locomotion is controlled by acute light exposure in addition to endogenous circadian rhythms (28, 29). Darkness also serves as a robust stimulus for vertebrate photoreceptors (30). In larvae, alternating the two lighting conditions results in a unique and reproducible swimming activity pattern recently fit to a successive induction model that describes the lulls and bursts of movement as quantifiable and predictable reflexes to changes in lighting condition (31).
We employed automated locomotion tests to individually quantify swimming activity in larval and juvenile zebrafish stimulated with changing lighting conditions. Resulting changes in locomotion were therefore used as a readout of neurobehavioral function, capturing key developmental events spanning the junction between the nervous and musculoskeletal systems. Thus, a measure of locomotor output can be used to rapidly identify whether an exogenous compound (18, 32), gene (33), or miRNA modifies or controls the development of either of these closely interacting systems.
The effects of acute or chronic ethanol exposure on a range of behavioral endpoints in zebrafish, including locomotion, is the subject of a recent review article (34). Our data showing hyperactivity in larvae transiently developmentally exposed to ethanol is in keeping with previous reports describing hyperactive locomotion in larval zebrafish acutely exposed to 1–2% ethanol for 20 min or 4 h at 6 dpf (18, 35). Also, a recent report shows that developmental exposure to ethanol (10%) for 2-h periods during multiple developmental stages results in reduced swimming in the zebrafish light–dark test (36), but these data include the swimming activity of deformed animals that would be expected to swim less than morphologically intact counterparts. Interestingly, our findings in juveniles suggest that abnormal, yet unique, neurobehavioral responses may persist throughout the life span of animals transiently exposed to ethanol during development. In support of this hypothesis, long-term behavioral changes in response to early life stage exposure to ethanol have been previously shown in a novel shoaling test in adults, in which the distance an animal swam relative to its perceived school of comrade fish was increased (37).
miRNAs are known targets and/or effectors of ethanol toxicity. For example, in immature cortical neurosphere cultures, exposure to ethanol suppresses the expression of miR-21, miR-335, miR-9, and miR-153 (39). In the present study, we similarly observed down-regulation of miR-9 and miR-153c in embryos transiently exposed to ethanol during early brain development. Ethanol-sensitive miRNAs have additionally been linked to neuroadaptation following ethanol exposure (40). The neuronal large-conductance calcium-activated and voltage-gated potassium (BK) channel is a known modifier of ethanol sensitivity (41). BK channels also exhibit tolerance to ethanol, or a decrease in receptor activity as a result of sustained or repeated exposure (40, 41), and deletion of the BK channel gene blocks ethanol-mediated behavioral responses and the development of tolerance (42, 43). Recently, Pietrzykowski et al. (40) showed that miR-9, up-regulated in mature hypothalamic-neurohypophysial system explants on exposure to ethanol, specifically targets a subset of BK channel splice variants that contain miR-9 miRNA recognition elements. The remaining splice variants encode BK isoforms with reduced responsiveness to ethanol, providing a strong mechanistic basis for miR-9-mediated neuroadaptation to ethanol. Taken together, these studies suggest that several miRNAs are sensitive to ethanol exposure and that miR-9 in particular may function as an ethanol-sensitive miRNA switch depending on cell type or maturity.
In our system, we independently identified miR-153c and miR-9 as strong candidates for the development of a functional neurobehavioral system. Relative to miR-9, considerably less is known about the role of miR-153c, a miRNA ubiquitously expressed in the zebrafish CNS during nervous system development (25). With respect to the CNS, a single report has shown that miR-153 posttranscriptionally regulates α-synuclein mRNA levels, a gene whose message and protein accumulation is associated with Parkinson's disease (47). miR-9 is considered to be brain-specific (44), and its expression is well documented in mice and zebrafish (27, 45). Although by no means singular in its distinction, reduction of miR-9 levels has been associated with a number of neurodegenerative disorders, including Huntington's disease (46) and Alzheimer's disease (47), and in animal studies, miR-9 repression has been linked to the development of spinal muscular atrophy (48). Similar to ethanol exposure, we found that putative knockdown of miR-9/9* or miR-153c during development results in larval hyperactivity. Further, movement aberrations persisted in miR-9/9* and miR-153c morphants tested as juveniles. Though the exact mechanism by which miR-9/9* and miR-153c choreograph neurobehavioral development is likely complex and will require further investigation, these data build on previous studies to provide evidence that miRNAs contribute to the establishment of a functional neurobehavioral system during development.
Beyond examining the neurobehavioral relevance of miRNAs, this study may have implications for some aspects of fetal alcohol spectrum disorder (FASD) research. Indeed, on a morphological level, a handful of studies have used the embryonic zebrafish model to uncover mechanisms by which developmental ethanol exposure triggers characteristic FASD-like teratogenic effects, including facial dysmorphia (49–51). In the present study, neurobehavioral endpoints were assessed at ethanol concentrations that fail to induce frank teratogenicty. Clinical and epidemiological studies demonstrate that the majority of children with FASD present with deficits in neurobehavioral development and function, including attention and inhibition problems, but not clinically discernable physical abnormalities (53). Although more work is needed to illuminate any mechanistic underpinnings, our data showing similarly disrupted behavioral profiles in larvae and juvenile animals that were exposed to ethanol during early neurogenesis or experienced putative developmental knockdown of candidate miRNAs is in keeping with the concept that ethanol-sensitive miRNAs may contribute to certain adverse neurobehavioral outcomes associated with developmental exposure to ethanol.
miRNAs have been implicated in osteoprogenitor differentiation in vitro, but few in vivo studies have investigated structural responses to miRNA repression (54–56). To determine whether misregulation of miRNAs modifies skeletal development, we assessed craniofacial and otolith bone volume in adult animals. We detected statistically significant reductions in mineralized tissues in a population of adult animals that experienced putative antisense repression of miR-153c during development. In support of this finding, 101 of 165 transcripts that were expressed differentially in embryos exposed to 100 mM ethanol during development were associated with skeletal and muscular disorders. In addition to reductions in head mineralized skeletal content, we also observed a significant decrease in miR-153c morphant otolith volumes at 60 dpf, suggesting that aberrations in swimming behavior responses result from a mechanism that may involve changes in otolith mass. Otoliths are involved in balance, equilibrium, and linear acceleration and have been shown to be affected by ethanol exposure in zebrafish (13). Taken together, these data provide in vivo evidence that skeletal development is regulated by miR-153c. Interestingly, while chronic ethanol abuse has been associated with decreased bone mass and strength (57), we did not observe any effect on skeletal development following developmental exposure to ethanol. These data suggest that ethanol-induced repression of miR-153c is not sufficiently robust to impede skeletal development in this model system, and more broadly, that ethanol-driven neurobehavioral effects likely involve the concerted actions of multiple, different protein coding and noncoding genes.
miRNAs are critical modulators of brain morphogenesis. Through studies aimed at elucidating the mechanism by which miRNAs promote the establishment and maintenance of a functional neurobehavioral system, we show here that miR-9/9* and miR-153c likely contribute to neurobehavioral development in zebrafish. These findings support an integral role for miRNAs in neurobehavioral and skeletal development and further establish the utility of zebrafish as a model for examining behavioral effects associated with developmental exposure to ethanol.
Supplementary Material
Acknowledgments
This work was supported by U.S. National Institute of Environmental Health Sciences (NIEHS) Environmental Health Sciences Core Center grant ES00210, NIEHS training grant T32ES7060, an Oregon State University (OSU) Linus Pauling Institute grant to R.L.T., and Superfund Basic Research Program grant NIEHS P42 ES016465 to R.L.T. and K.M.W. The authors declare no conflicts of interest. The authors are grateful to Katrine Saili and Britton Goodale for their critical review of the manuscript. The authors are grateful to Carrie Barton, Cari Buchner, Brittany McCauslin, and the staff at the OSU Sinnhuber Aquatic Research Laboratory for exemplary fish husbandry and technical expertise. The authors thank Dawn Olsen for her assistance with skeletal analysis, Hao Truong for developing custom PERL software used to process locomotion data, and Gregory Gonnerman for providing body length measurements for the juvenile locomotion studies.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- Atoh2B
- atonal homolog 2B
- Bbc3
- BCL2 binding component 3
- CNS
- central nervous system
- dpf
- days postfertilization
- dre
- Danio rerio
- DUSP4
- dual specificity phosphatase 4
- FASD
- fetal alcohol spectrum disorder
- GPX3
- glutathione peroxidase 3
- hpf
- hours postfertilization
- hsa
- Homo sapiens
- IPA
- Ingenuity Pathway Analysis
- miR
- microRNA
- miRNA
- microRNA
- MO
- antisense oligonucleotide morpholino
- neuroD6
- neurogenic differentiation 6
- odc1
- ornithine decarboxylase 1
- SLC6A1
- solute carrier family 6 (neurotransmitter transporter, GABA) member 1
- SLC16A9
- solute carrier family 16 (monocarboxylic acid transporters) member 9
- SLC17A6
- solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter) member 6
- snoRNA
- small nucleolar RNA
- μCT
- micro-computed tomography.
REFERENCES
- 1. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 2. Kapsimali M., Kloosterman W. P., de Bruijn E., Rosa F., Plasterk R. H., Wilson S. W. (2007) MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 8, R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krichevsky A. M., King K. S., Donahue C. P., Khrapko K., Kosik K. S. (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ling K. H., Brautigan P. J., Hahn C. N., Daish T., Rayner J. R., Cheah P. S., Raison J. M., Piltz S., Mann J. R., Mattiske D. M., Thomas P. Q., Adelson D. L., Scott H. S. (2011) Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genomics 12, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Pietri Tonelli D., Pulvers J. N., Haffner C., Murchison E. P., Hannon G. J., Huttner W. B. (2008) miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P., Schier A. F. (2005) MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838 [DOI] [PubMed] [Google Scholar]
- 7. Huang T., Liu Y., Huang M., Zhao X., Cheng L. (2010) Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J. Mol. Cell Biol. 2, 152–163 [DOI] [PubMed] [Google Scholar]
- 8. Schaefer A., O'Carroll D., Tan C. L., Hillman D., Sugimori M., Llinas R., Greengard P. (2007) Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 204, 1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zehir A., Hua L. L., Maska E. L., Morikawa Y., Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev. Biol. 340, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis T. H., Cuellar T. L., Koch S. M., Barker A. J., Harfe B. D., McManus M. T., Ullian E. M. (2008) Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pinter R., Hindges R. (2010) Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. PLoS One 5, e10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimmel C. B. (1993) Patterning the brain of the zebrafish embryo. Annu. Rev. Neurosci. 16, 707–732 [DOI] [PubMed] [Google Scholar]
- 13. Reimers M. J., Flockton A. R., Tanguay R. L. (2004) Ethanol- and acetaldehyde-mediated developmental toxicity in zebrafish. Neurotoxicol. Teratol. 26, 769–781 [DOI] [PubMed] [Google Scholar]
- 14. Reimers M. J., La Du J. K., Periera C. B., Giovanini J., Tanguay R. L. (2006) Ethanol-dependent toxicity in zebrafish is partially attenuated by antioxidants. Neurotoxicol. Teratol. 28, 497–508 [DOI] [PubMed] [Google Scholar]
- 15. Kocerha J., Faghihi M. A., Lopez-Toledano M. A., Huang J., Ramsey A. J., Caron M. G., Sales N., Willoughby D., Elmen J., Hansen H. F., Orum H., Kauppinen S., Kenny P. J., Wahlestedt C. (2009) MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U. S. A. 106, 3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Westerfield M. (2000) The Zebrafish Book, 4th ed University of Oregon Press, Eugene, OR, USA [Google Scholar]
- 17. Emran F., Rihel J., Dowling J. E. (2008) A behavioral assay to measure responsiveness of zebrafish to changes in light intensities. J. Vis. Exp. 20, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacPhail R. C., Brooks J., Hunter D. L., Padnos B., Irons T. D., Padilla S. (2009) Locomotion in larval zebrafish: Influence of time of day, lighting and ethanol. Neurotoxicology 30, 52–58 [DOI] [PubMed] [Google Scholar]
- 19. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 20. Kerr M. K., Martin M., Churchill G. A. (2000) Analysis of variance for gene expression microarray data. J. Comput. Biol. 7, 819–837 [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y. a. H., Y (1995) Controlling the false discovery rate- a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 22. Gao X., Gulari E., Zhou X. (2004) In situ synthesis of oligonucleotide microarrays. Biopolymers 73, 579–596 [DOI] [PubMed] [Google Scholar]
- 23. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 25. Wienholds E., Plasterk R. H. (2005) MicroRNA function in animal development. FEBS Lett. 579, 5911–5922 [DOI] [PubMed] [Google Scholar]
- 26. Delaloy C., Liu L., Lee J. A., Su H., Shen F., Yang G. Y., Young W. L., Ivey K. N., Gao F. B. (2010) MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leucht C., Stigloher C., Wizenmann A., Klafke R., Folchert A., Bally-Cuif L. (2008) MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648 [DOI] [PubMed] [Google Scholar]
- 28. Burgess H. A., Granato M. (2007) Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 210, 2526–2539 [DOI] [PubMed] [Google Scholar]
- 29. Aschoff J. (1960) Exogenous and endogenous components in circadian rhythms. Cold Spring Harb. Symp. Quant. Biol. 25, 11–28 [DOI] [PubMed] [Google Scholar]
- 30. Hagins W. A., Penn R. D., Yoshikami S. (1970) Dark current and photocurrent in retinal rods. Biophys. J. 10, 380–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staddon J. E., MacPhail R. C., Padilla S. (2010) The dynamics of successive induction in larval zebrafish. J. Exp. Anal. Behav. 94, 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rihel J., Prober D. A., Arvanites A., Lam K., Zimmerman S., Jang S., Haggarty S. J., Kokel D., Rubin L. L., Peterson R. T., Schier A. F. (2010) Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wolman M., Granato M. (2011) Behavioral genetics in larval zebrafish-learning from the young. [E-pub ahead of print] Dev. Neurobiol. doi: 10.1002/dneu.20872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Echevarria D. J., Toms C. N., Jouandot D. J. (2011) Alcohol-induced behavior change in zebrafish models. Rev. Neurosci. 22, 85–93 [DOI] [PubMed] [Google Scholar]
- 35. Irons T. D., MacPhail R. C., Hunter D. L., Padilla S. (2009) Acute neuroactive drug exposures alter locomotor activity in larval zebrafish. Neurotoxicol. Teratol. 32, 84–90 [DOI] [PubMed] [Google Scholar]
- 36. Ali S., Champagne D. L., Alia A., Richardson M. K. (2011) Large-scale analysis of acute ethanol exposure in zebrafish development: a critical time window and resilience. PLoS One 6, e20037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernandes Y., Gerlai R. (2009) Long-term behavioral changes in response to early developmental exposure to ethanol in zebrafish. Alcohol Clin. Exp. Res. 33, 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blaser R. E., Koid A., Poliner R. M. Context-dependent sensitization to ethanol in zebrafish (Danio rerio). Pharmacol. Biochem. Behav. 95, 278–284 [DOI] [PubMed] [Google Scholar]
- 39. Sathyan P., Golden H. B., Miranda R. C. (2007) Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 27, 8546–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pietrzykowski A. Z., Friesen R. M., Martin G. E., Puig S. I., Nowak C. L., Wynne P. M., Siegelmann H. T., Treistman S. N. (2008) Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Treistman S. N., Martin G. E. (2009) BK channels: mediators and models for alcohol tolerance. Trends Neurosci. 32, 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cowmeadow R. B., Krishnan H. R., Atkinson N. S. (2005) The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin. Exp. Res. 29, 1777–1786 [DOI] [PubMed] [Google Scholar]
- 43. Davies A. G., Pierce-Shimomura J. T., Kim H., VanHoven M. K., Thiele T. R., Bonci A., Bargmann C. I., McIntire S. L. (2003) A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115, 655–666 [DOI] [PubMed] [Google Scholar]
- 44. Gao F. B. (2010) Context-dependent functions of specific microRNAs in neuronal development. Neural. Dev. 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. (2002) Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739 [DOI] [PubMed] [Google Scholar]
- 46. Packer A. N., Xing Y., Harper S. Q., Jones L., Davidson B. L. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 28, 14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cogswell J. P., Ward J., Taylor I. A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C., Prinjha R. K., Richardson J. C., Saunders A. M., Roses A. D., Richards C. A. (2008) Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 14, 27–41 [DOI] [PubMed] [Google Scholar]
- 48. Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P. W., Wills A. M., Wirguin I., Rubin L. L., Misawa H., Tabin C. J., Brown R., Jr., Chen A., Hornstein E. (2010) miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. U. S. A. 107, 13111–13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bilotta J., Barnett J. A., Hancock L., Saszik S. (2004) Ethanol exposure alters zebrafish development: a novel model of fetal alcohol syndrome. Neurotoxicol. Teratol. 26, 737–743 [DOI] [PubMed] [Google Scholar]
- 50. Carvan M. J., 3rd, Loucks E., Weber D. N., Williams F. E. (2004) Ethanol effects on the developing zebrafish: neurobehavior and skeletal morphogenesis. Neurotoxicol. Teratol. 26, 757–768 [DOI] [PubMed] [Google Scholar]
- 51. Marrs J. A., Clendenon S. G., Ratcliffe D. R., Fielding S. M., Liu Q., Bosron W. F. (2010) Zebrafish fetal alcohol syndrome model: effects of ethanol are rescued by retinoic acid supplement. Alcohol 44, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. U.S. Centers for Disease Control and Prevention (2009) Alcohol use among pregnant and nonpregnant women of childbearing age—United States, 1991–2005. MMWR 58, 529–532 [PubMed] [Google Scholar]
- 53. Hoyme H. E., May P. A., Kalberg W. O., Kodituwakku P., Gossage J. P., Trujillo P. M., Buckley D. G., Miller J. H., Aragon A. S., Khaole N., Viljoen D. L., Jones K. L., Robinson L. K. (2005) A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 institute of medicine criteria. Pediatrics 115, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hassan M. Q., Gordon J. A., Beloti M. M., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2010) A network connecting Runx2, SATB2, and the miR-23a∼27a∼24–2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. U. S. A. 107, 19879–19884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Inose H., Ochi H., Kimura A., Fujita K., Xu R., Sato S., Iwasaki M., Sunamura S., Takeuchi Y., Fukumoto S., Saito K., Nakamura T., Siomi H., Ito H., Arai Y., Shinomiya K., Takeda S. (2009) A microRNA regulatory mechanism of osteoblast differentiation. Proc. Natl. Acad. Sci. U. S. A. 106, 20794–20799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Z., Hassan M. Q., Volinia S., van Wijnen A. J., Stein J. L., Croce C. M., Lian J. B., Stein G. S. (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. U. S. A. 105, 13906–13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maurel D. B., Boisseau N., Benhamou C. L., Jaffre C. (2012) Alcohol and bone: review of dose effects and mechanisms. Osteoporos. Int. 23, 1–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.