Single nucleotide polymorphisms within the 8q24 region of the genome were examined as risk factors for prostate cancer. Several variants were found to be associated with prostate cancer risk, and these varied among races.
Keywords: Prostatic neoplasms, Continental population groups, Single nucleotide polymorphisms, Meta-analysis, Risk assessment
Abstract
Recent studies implicate single nucleotide polymorphisms (SNPs) within the 8q24 region as a risk factor for prostate cancer (PCa). New developments suggest that 8q24 encodes regulators of the nearby MYC gene, a known oncogene. In order to better understand the implications of SNPs in this region, we performed meta-analyses, stratified by race, of seven SNPs and one microsatellite marker previously identified as risk loci on the 8q24 region of the genome. In addition, we reviewed the literature examining the possible associations between these polymorphisms and clinicopathological features of PCa. The results of the meta-analyses indicate that rs6983267, rs1447295, rs6983561, rs7837688, rs16901979, and DG8S737 are significantly associated with a higher risk for PCa for at least one race, whereas the variants rs13254738 and rs7000448 are not. The degree of association and frequency of the causative allele varied among men of different races. Though several studies have demonstrated an association between certain 8q24 SNPs and clinicopathological features of the disease, review of this topic revealed conflicting results.
Introduction
Prostate cancer (PCa) is the most common noncutaneous cancer among men in the U.S., with an estimated 240,890 new cases diagnosed and 33,720 deaths in 2010 [1]. The risk for prostate cancer varies considerably among men of different racial backgrounds. The rate per 100,000 men, is 146.3 in whites, 231.9 in African Americans, 82.3 in Asian Americans and Pacific Islanders, 82.7 in American Indians and Native Alaskans, and 131.1 in Hispanics [1]. In recent years, research has placed a considerable emphasis on the role of genetics in the development, progression, and treatment of this disease. The 8q24 region was first shown to confer a PCa risk in a genomewide linkage scan of 871 Icelandic men [2]. Since then, >30 studies have examined the relationship between 8q24 and PCa risk. Importantly, some of these investigations have compared this association in a variety of races including African Americans (AAs), American whites (AWs), European whites (EWs), Asians (As), and Hispanics (Hs). These studies suggest that single nucleotide polymorphism (SNP) genotype frequencies on 8q24 may vary by race and thus 8q24 may at least partially account for racial differences in PCa risk.
The mechanism by which 8q24 influences the course of PCa is poorly understood. The 8q24 region has been described as a “gene desert;” the 600-kbp gene-poor region appears to have little or no transcriptional activity. Despite this, evidence suggests that 8q24 may play an active role in PCa. First, 8q24 is a highly conserved genomic region. In addition, POU5F1P1 has been identified on 8q24. Originally thought to be a pseudogene, it is now believed that POU5F1P1 can encode a functional protein that contributes to carcinogenesis through its role as a weak transcriptional activator [3]. Additionally, recent studies have shown that 8q24 encodes enhancers of the nearby oncogene MYC [4–7]. Wasserman et al. [5] and Sotelo et al. [4] both identified an enhancer of MYC that contained rs6983267, a SNP located in region 3 of 8q24. Ahmadiyeh et al. [7] provided additional support, finding that all three regions of 8q24 independently interact with both MYC and with one another.
These new findings demonstrate a renewed importance in exploring the SNPs on 8q24 because of their potential predictive value and ability to guide the development of therapies. Though many studies have assessed these SNPs in men with PCa, the results have been inconsistent. In addition, the two previously published meta-analyses of 8q24 SNPs only included a small subset of the relevant studies [8, 9], assessed only a small number of SNPs [8], or were not stratified by race [8]. Consequently, we conducted a systematic review of the relationship between PCa and 8q24 SNPs as well as a meta-analysis stratified by race in order to identify those SNPs that are associated with PCa risk and thus warrant further investigation.
Methods
We obtained data from all available studies in MEDLINE that reported 8q24 genotype frequencies in patients with PCa and in healthy controls. Only those studies that stratified their analysis by race were included in the present study. We grouped studies by SNP and stratified by race (AA, AW, EW, A, and H) within each SNP. The only exception was that Australian Caucasians were grouped with EWs. Our final analysis only included those SNPs whose 8q24 allele frequencies were reported in at least three different studies. In some studies, multiple cohorts of a single race were examined. In these instances, each cohort was assessed separately in the meta-analysis, which is represented in Figure 1.
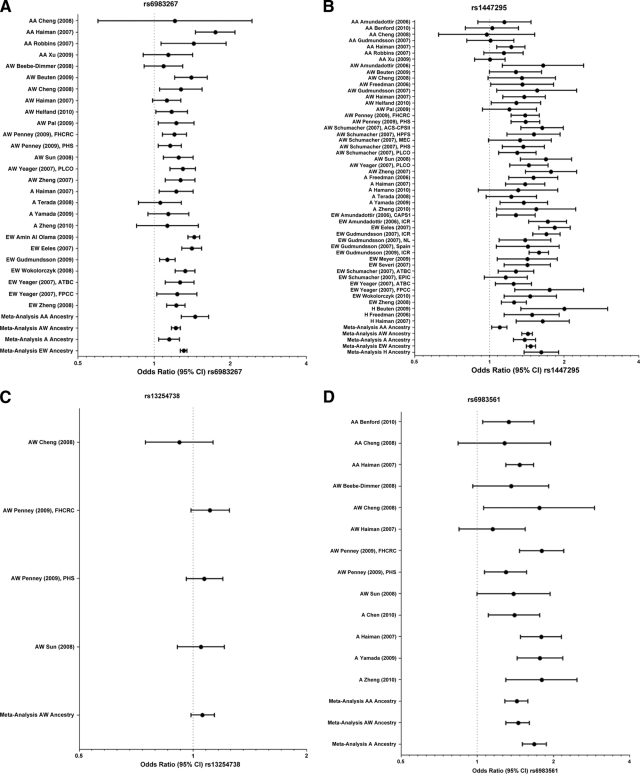
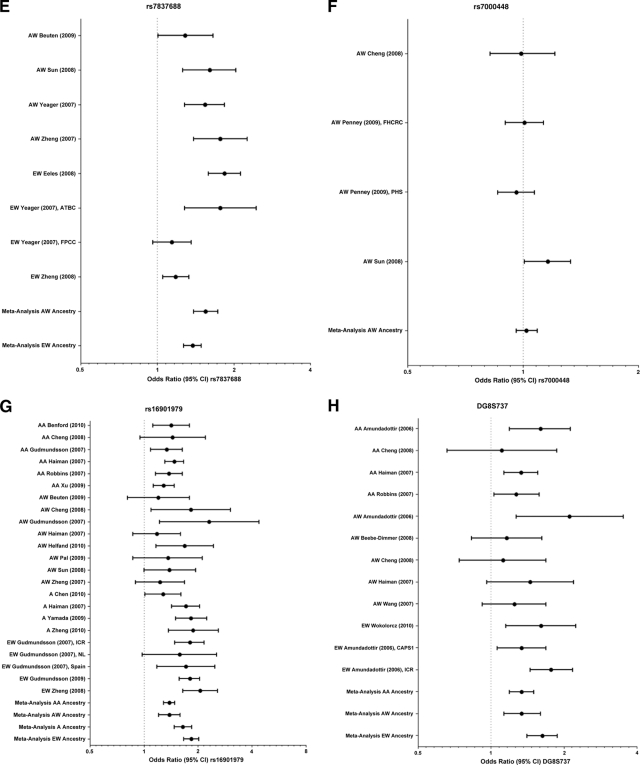
Figure 1.
Forest plots of odds ratios (95% confidence interval [CI]) of individual studies and a meta-analysis for the variant allele at rs6983267 (A), rs1447295 (B), rs13254738 (C), rs6983561 (D), rs78737688 (E), rs7000448 (F), rs16901979 (G), and DG8S737 (H).
Abbreviations: A, Asian; AA, African American; ACS-CPSII, American Cancer Society Cancer Prevention Study II; ATBC, Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study; AW, American white; CAPS1, Cancer of Prostate in Sweden 1; EPIC, European Prospective Investigation into Cancer and Nutrition Cohort; EW, European white; FHCRC, Fred Hutchinson Cancer Research Center King County; FPCC, French Prostate Case-Control Study; H, Hispanic; HPFS, Health Professionals Follow-up Study; ICR, Icelandic Cancer Registry; MEC, Multiethnic Cohort; NL, Netherlands; PHS, Physicians' Health Study; PLCO, Prostate, Lung, Colon, Ovarian Trial.
The random effects model described by DerSimonian and Laird (1986) was used for the meta-analyses [10]. In this method, each study in a population is weighted by the inverse sum of the estimated variances of the study itself and of all the individual studies from the population mean. The allelic odds ratio (OR) values and confidence intervals (CIs) were calculated using the Mantel–Haenszel method. This test assumes homogeneity, so we first used the Breslow–Day test to determine whether or not the assumption of homogeneity was valid in each population. If the test for homogeneity was not significant, the population was said to be homogeneous. The results of the Mantel–Haenszel test were deemed significant if the OR 95% CI did not include 1.
In addition to allele frequencies, we recorded any data that assessed the relationship between 8q24 SNPs and the clinicopathological features of PCa, including aggressiveness, prostate-specific antigen (PSA) level at diagnosis, Gleason score, tumor grade, surgical margin, age at onset, tumor stage, and hereditary or familial versus sporadic PCa. Because of a scarcity of studies on this topic, we were not able to perform any meta-analyses.
Associations Between 8q24 SNPs and PCa
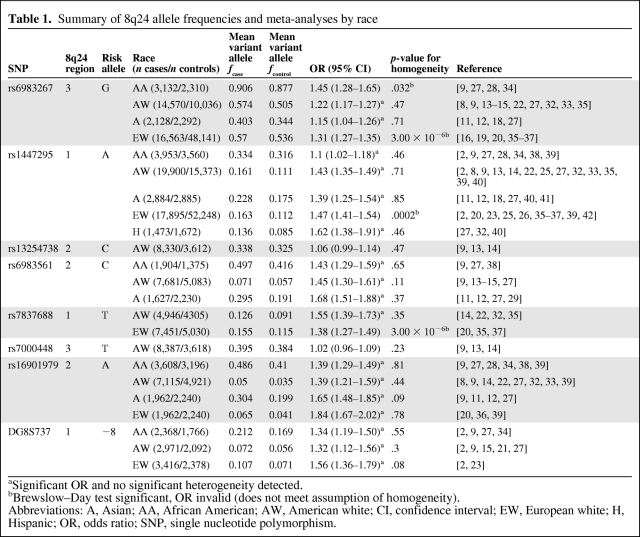
In total, 29 studies examining seven SNPs and one microsatellite marker were included in the analyses. The location on 8q24, variant allele, total number of cases and controls, number of studies assessed, mean allele frequencies, and meta-analysis results are presented in Table 1.
Table 1.
Summary of 8q24 allele frequencies and meta-analyses by race
aSignificant OR and no significant heterogeneity detected.
bBrewslow–Day test significant, OR invalid (does not meet assumption of homogeneity).
Abbreviations: A, Asian; AA, African American; AW, American white; CI, confidence interval; EW, European white; H, Hispanic; OR, odds ratio; SNP, single nucleotide polymorphism.
rs6983267
We identified 20 studies that met the inclusion criteria for this SNP. Two of four AA, nine of 11 AW, one of four A, and all seven EW studies showed a significant association between the rs6983267 risk allele and PCa risk. The results of the meta-analysis indicated that the association between PCa risk and the variant allele at rs6983267 was significant in AWs and As (Fig. 1A, Table 1). The AA and EW groups had significant heterogeneity and therefore valid ORs could not be calculated.
rs1447295
Twenty-six studies reporting genotype frequencies for rs1447295 met the criteria for inclusion. Only one of seven studies of AAs found a significant association between this SNP and PCa risk whereas four of six studies of As, 15 of 18 studies of AWs, 14 of 15 studies of EWs, and all three studies of Hs were significant. Our meta-analysis (Fig. 1B, Table 1) showed a significant association in AW, A, H, and AA populations. We could not assess the EW subgroup because of heterogeneity. The significant OR for the AA subgroup contradicts the fact that only one of seven studies of AAs found that the association was significant. This may be explained by the fact the one significant study had the largest sample size, contributing to 32% of the total number of participants. In addition, though significant, the OR was only moderately greater than one.
rs13254738
Our literature search only identified five studies that reported genotype frequencies for rs13254738. Four of those studies assessed this SNP in AWs and two reported data for As. Yamada et al. [11] and Zheng et al. [12] both found a significant relationship between this SNP and PCa risk in As, but there was an insufficient number of studies to conduct a meta-analysis. No evidence suggests that the rs13254738 marker is associated with PCa risk in AWs [9, 13, 14]. Similarly, the present meta-analysis found no association between the variant allele at this SNP and PCa risk (Fig. 1C, Table 1).
rs6983561
Genotyping studies for rs6983561 generally indicated that the C allele is associated with a higher risk for PCa. Of those analyzed here, two of three AA, three of six AW, and four of four A studies demonstrated a significant association. Interestingly, some studies that did not find a significant relationship in the overall population did detect a significant association within a subset of patients. For example, Beebe-Dimmer et al. [15] found that this variant was associated with PCa in patients diagnosed before the age of 50 years. Sun et al. [14] showed a significant association for hereditary PCa but not nonhereditary PCa. The meta-analysis indicated a significant association between the risk allele for rs6983561 and PCa risk in AAs, AWs, and As (Fig. 1D, Table 1).
rs7837688
The PCa risk associated with rs7837688 was examined in AW and EW populations. The studies consistently found a significantly higher frequency of the risk allele, T, in cases than in controls for both races. Likewise, most of the studies found a significant association between this SNP and PCa risk. Our meta-analysis confirmed this relationship in AWs (Fig. 1E, Table 1). The EW group did not meet the assumption of homogeneity and therefore the association could not be assessed.
rs7000448
Three studies analyzed rs7000448 in AWs but none found a significant association between this SNP and PCa risk [9, 13, 14]. Consistent with these results, our meta-analysis found no evidence of this relationship (Fig. 1F, Table 1).
rs16901979
This region 2 8q24 SNP was genotyped in men with PCa in 16 studies. The results suggest a relationship between the rs16901979 risk allele and PCa. This pattern is consistent across AA (five of six studies), A (four of four studies), and EW (four of five studies) populations. There appears to be greater variability among studies of AWs because only four of eight studies found the relationship to be significant. Our meta-analysis demonstrated that the association between PCa risk and the variant allele for rs16901979 was significant in all four populations (Fig. 1G, Table 1).
DG8S737
The −8 allele of the DG8S737 satellite marker located on 8q24 has also been analyzed for a possible association with PCa risk. The association was significant in three of four studies of AAs and three of three studies of EWs, but only one of five studies of AWs. However, the results of our meta-analysis indicate that the association is significant for all three races (Fig. 1H, Table 1).
Association Between 8q24 SNPs and Clinical Features of PCa
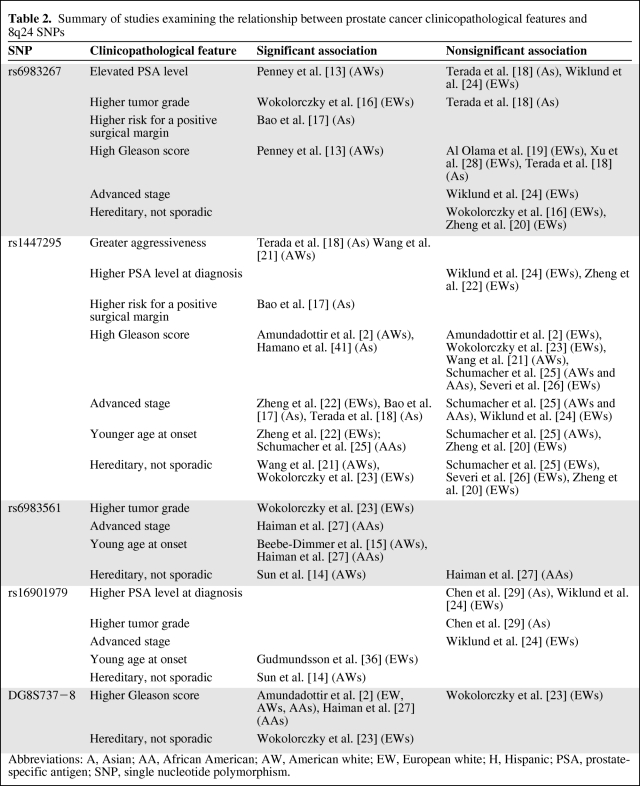
Investigations into the possible links between these polymorphisms and specific clinicopathological features of PCa could reveal whether or not there is an association between these SNPs and a specific subset of PCa cases (i.e., aggressiveness, age at onset, family history, etc.) or suggest possible mechanisms by which 8q24 risk genotypes contribute to PCa risk and disease development. We found 17 papers that assessed the relationship among one or more of the 8q24 SNPs of interest, but there was insufficient data to perform a meta-analysis. The findings are summarized in Table 2.
Table 2.
Summary of studies examining the relationship between prostate cancer clinicopathological features and 8q24 SNPs
Abbreviations: A, Asian; AA, African American; AW, American white; EW, European white; H, Hispanic; PSA, prostate-specific antigen; SNP, single nucleotide polymorphism.
rs6983267
Some studies have also analyzed potential associations between rs6983267 and various clinical features of PCa. The findings indicated an association between the risk allele at rs6983267 and PSA level at diagnosis in EWs [13], higher grade tumors in EWs [16], and a higher risk for a positive surgical margin in As [17]. Penney et al. [13] found that AWs heterozygous at rs6983267 had a lower risk for Gleason score 8–10 tumors than men who were homozygous for the risk allele.
Contrary to those findings, Terada et al. [18] found no relationship between the risk allele for this SNP and PSA level or tumor stage in their study of Japanese men. In fact, they found an association between the variant allele and a low Gleason score in PCa patients, compared with controls. Similarly Al Olama et al. [19] failed to detect a relationship between this SNP and Gleason score in EW men.
According to Wokolorczyk et al. [16] and Zheng et al. [20], the risk allele frequency for rs6983267 did not differ significantly between sporadic and familial cases of PCa. However, Sun et al. [14] found that this SNP was associated with nonhereditary PCa but not with hereditary PCa.
rs1447295
The risk allele at rs1447295 may be associated with several PCa clinical features, including greater tumor aggressiveness in As [18] and AWs [21], advanced stage disease in EWs [22] and As [17, 18], and a higher risk for a positive surgical margin [17]. Amundadottir et al. [2] showed that the rs1447295 risk allele tended to be more strongly associated with Gleason score 7–10 tumors than with Gleason score 2–6 tumors in AAs and AWs. This trend does not appear to translate to EW populations [2, 23]. The relationship between PCa risk and this SNP did not appear to be affected by PSA level in EWs [22, 24]. Zheng et al. [22] showed that this allele was associated with a younger age at onset in EWs, but Schumacher et al. [25] and Zheng et al. [20] found no such evidence. The risk allele may be associated with a younger age at onset in AA populations [25].
Schumacher et al. [25], Severi et al. [26], and Zheng et al. [20] failed to find a significant difference in rs1447295 risk allele frequencies between sporadic and familial cases in EWs. However, Wang et al. [21] found that, for AWs, this risk allele was associated with familial PCa but not with sporadic PCa. Likewise, Wokolorczyk et al. [23] found that this risk allele was more strongly associated with familial PCa than with sporadic PCa in EWs.
rs6983561
The A allele at rs6983561 may be associated with clinicopathological features of PCa. For example, Wokolorczyk et al. [16] found that the risk allele was more strongly associated with high-grade tumors than with low-grade tumors in EWs. In addition, the variant allele was moderately more frequent among AA individuals with high-stage PCa than among AA controls [27]. This SNP may also be associated with the risk for younger age at onset in AWs [15] and AAs [27]. Lastly, Sun et al. [14] found that the rs6983561 risk allele was associated with hereditary PCa but not with non-hereditary PCa in AWs, whereas Haiman et al. [27] found that the effect of this SNP was greater in nonhereditary cases among AAs.
rs16901979
According to Wiklund et al. [24], the rs16901979 risk allele is also associated with an elevated PSA level. However, Xu et al. [28] and Chen et al. [29] found conflicting evidence. In addition, this risk allele appears to be associated with lower tumor grade [29]. Finally, according to Sun et al. [14], the association between PCa risk and this variant allele is only significant in hereditary PCa, not nonhereditary PCa.
DG8S737
Some evidence suggests a possible link between this genomic marker and Gleason score. Two studies found that the variant −8 allele frequency was higher among men with high Gleason scores than among those with low Gleason scores in AAs [2, 27], AWs [2], and EWs [2]. However, Wokolorczky et al. [23] did not find the relationship to be significant in EWs. The DG8S727 −8 allele was also more strongly associated with familial PCa than with sporadic PCa.
Conclusions
Many studies have investigated the role of 8q24 variants as genetic indicators of PCa risk and clinicopathological features of PCa. These studies have produced conflicting results, but it is hoped that the present meta-analysis will help elucidate which variants are most critical in specific races.
Interestingly, the frequency of the risk alleles varied considerably across races. For example, the variant allele frequency was highest among AAs for all the SNPs included in the present analysis. For three of the four SNPs, A men had the second highest relative frequency of the variant allele. In general, the frequencies were similar between AWs and EWs and tended to be lower than those of other ethnic populations. As seen in Table 1, in most cases, these differences in allele frequency among races were quite pronounced.
These frequency data both complement and contradict variations in the incidence of PCa, suggesting that 8q24 may modify other genetic risk factors within races. For example, AA men have the highest incidence of PCa [30] and a higher frequency of the variant alleles for 8q24 SNPs than men of other ethnicities. In contrast, As have the lowest incidence of PCa but tend to have higher variant allele frequencies than EWs and AWs.
The relative strength of the association between PCa risk and the 8q24 variant alleles also appeared to vary among races and among SNPs within the same race. Although our meta-analysis helps to clarify these associations relative to the null hypothesis (i.e., no effect on risk in individual populations), the overlapping CIs of many of the estimated ORs suggest that comparisons between them should be interpreted with caution. It is worth noting that CIs did not overlap for rs1447295 (lower in AAs than in other races) and rs6983267 (lower in AAs and AWs than in EWs), whereas overlap was observed for all other comparisons. Therefore, future studies should account for the possibility that certain 8q24 SNPs confer greater risk in certain populations.
Despite these differences, the risk alleles at rs6983267, rs1447295, rs6983561, rs16901979, and DG8S737 were consistently associated with PCa risk across races. The only SNPs that did not appear to be related to PCa risk were rs13254738 and rs7000448.
Haplotype analyses indicate that certain SNPs are in linkage disequilibrium; thus, there may be multiple interacting regions on 8q24 that confer a PCa risk [12, 29, 31, 32]. For instance, multiple studies report that rs6983561 and rs16901979 are in strong linkage disequilibrium [19, 29]. Despite these interactions, it is clear that many 8q24 SNPs, including five of the seven SNPs assessed herein, independently predict PCa risk. This is supported by research assessing the cumulative risk associated with 8q24 polymorphisms. These studies suggest that the risk alleles interact additively or even synergistically [11, 13, 14, 17, 20, 22, 32, 33]. For example, one study found that each additional risk allele increased the risk for PCa by 19% [13]. This has important implications because it suggests that 8q24 may contain multiple functional variants.
Many factors aside from race are associated with PCa, including age at onset, family history, tumor aggressiveness, PSA level, Gleason score, tumor stage, and surgical margins. Few studies have examined the relationship between 8q24 SNPs and these clinicopathological features and thus we were not able to include them in the meta-analyses. Of those studies that have been conducted, the results have been generally inconsistent. In subsequent studies of 8q24 polymorphisms in PCa patients, the association between 8q24 SNPs and these prognostic factors must be assessed.
In addition, future studies of 8q24 should look to explain the mechanism by which 8q24 increases the risk for PCa. It will be important to explore the association between these SNPs and gene variants on nearby regions such as MYC and POU5F1P1. A clear association exists between 8q24 and PCa, but the value of this relationship for drug development and prognostics will continue to be limited until future studies can identify the mechanism of action.
Author Contributions
Conception/Design: Sarah M. Troutman
Provision of study material or patients: Sarah M. Troutman
Collection and/or assembly of data: Sarah M. Troutman, Cheryl D. Cropp
Data analysis and interpretation: Tristan M. Sissung, Cheryl D. Cropp, David J. Venzon
Manuscript writing: William D. Figg, Sarah M. Troutman, Tristan M. Sissung, Cheryl D. Cropp, David J. Venzon, Shawn D. Spencer, Bamidele A. Adesunloye, Xuan Huang, Fatima H. Karzai, Douglas K. Price
Final approval of manuscript: William D. Figg
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Panagopoulos I, Möller E, Collin A, et al. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol Rep. 2008;20:1029–1033. [PubMed] [Google Scholar]
- 4.Sotelo J, Esposito D, Duhagon MA, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmadiyeh N, Pomerantz MM, Grisanzio C, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal P, Xi H, Guha S, et al. Common variants in 8q24 are associated with risk for prostate cancer and tumor aggressiveness in men of European ancestry. Prostate. 2009;69:1548–1556. doi: 10.1002/pros.20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: Association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 11.Yamada H, Penney KL, Takahashi H, et al. Replication of prostate cancer risk loci in a Japanese case-control association study. J Natl Cancer Inst. 2009;101:1330–1336. doi: 10.1093/jnci/djp287. [DOI] [PubMed] [Google Scholar]
- 12.Zheng SL, Hsing AW, Sun J, et al. Association of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese men. Prostate. 2010;70:425–432. doi: 10.1002/pros.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penney KL, Salinas CA, Pomerantz M, et al. Evaluation of 8q24 and 17q risk loci and prostate cancer mortality. Clin Cancer Res. 2009;15:3223–3230. doi: 10.1158/1078-0432.CCR-08-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Lange EM, Isaacs SD, et al. Chromosome 8q24 risk variants in hereditary and non-hereditary prostate cancer patients. Prostate. 2008;68:489–497. doi: 10.1002/pros.20695. [DOI] [PubMed] [Google Scholar]
- 15.Beebe-Dimmer JL, Levin AM, Ray AM, et al. Chromosome 8q24 markers: Risk of early-onset and familial prostate cancer. Int J Cancer. 2008;122:2876–2879. doi: 10.1002/ijc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wokolorczyk D, Gliniewicz B, Sikorski A, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 17.Bao BY, Pao JB, Lin VC, et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin Chim Acta. 2010;411:1232–1237. doi: 10.1016/j.cca.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 18.Terada N, Tsuchiya N, Ma Z, et al. Association of genetic polymorphisms at 8q24 with the risk of prostate cancer in a Japanese population. Prostate. 2008;68:1689–1695. doi: 10.1002/pros.20831. [DOI] [PubMed] [Google Scholar]
- 19.Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 20.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 22.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 23.Wokolorczyk D, Gliniewicz B, Stojewski M, et al. The rs1447295 and DG8S737 markers on chromosome 8q24 and cancer risk in the Polish population. Eur J Cancer Prev. 2010;19:167–171. doi: 10.1097/CEJ.0b013e32832945c3. [DOI] [PubMed] [Google Scholar]
- 24.Wiklund F, Zheng SL, Sun J, et al. Association of reported prostate cancer risk alleles with PSA levels among men without a diagnosis of prostate cancer. Prostate. 2009;69:419–427. doi: 10.1002/pros.20908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 26.Severi G, Hayes VM, Padilla EJ, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: Results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 27.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Kibel AS, Hu JJ, et al. Prostate cancer risk associated loci in African Americans. Cancer Epidemiol Biomarkers Prev. 2009;18:2145–2149. doi: 10.1158/1055-9965.EPI-09-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, Huang YC, Yang S, et al. Common variants at 8q24 are associated with prostate cancer risk in Taiwanese men. Prostate. 2010;70:502–507. doi: 10.1002/pros.21084. [DOI] [PubMed] [Google Scholar]
- 30.Howlander N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review. Bethesda, MD: National Cancer Institute; 1975–2008. [accessed August 9, 2011]. Available at http://seer.cancer.gov/csr/1975_2008/, based on November 2010 SEER data submission, posted to the SEER web site, 2011. [Google Scholar]
- 31.Tan YC, Zeigler-Johnson C, Mittal RD, et al. Common 8q24 sequence variations are associated with Asian Indian advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2431–2435. doi: 10.1158/1055-9965.EPI-07-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beuten J, Gelfond JA, Martinez-Fierro ML, et al. Association of chromosome 8q variants with prostate cancer risk in Caucasian and Hispanic men. Carcinogenesis. 2009;30:1372–1379. doi: 10.1093/carcin/bgp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helfand BT, Fought AJ, Loeb S, et al. Genetic prostate cancer risk assessment: Common variants in 9 genomic regions are associated with cumulative risk. J Urol. 2010;184:501–505. doi: 10.1016/j.juro.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins C, Torres JB, Hooker S, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 36.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 38.Benford ML, VanCleave TT, Lavender NA, et al. 8q24 sequence variants in relation to prostate can-cer risk among men of African descent: A case-control study. BMC Cancer. 2010;10:334. doi: 10.1186/1471-2407-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 40.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamano T, Matsui H, Sekine Y, et al. Association of SNP rs1447295 and microsatellite marker DG8S737 with familial prostate cancer and high grade disease. J Urol. 2010;184:738–742. doi: 10.1016/j.juro.2010.03.102. [DOI] [PubMed] [Google Scholar]
- 42.Meyer A, Schr̈mann P, Ghahremani M, et al. Association of chromosomal locus 8q24 and risk of prostate cancer: A hospital-based study of German patients treated with brachytherapy. Urol Oncol. 2009;27:373–376. doi: 10.1016/j.urolonc.2008.04.010. [DOI] [PubMed] [Google Scholar]