Abstract
CTX-M β-lactamases are considered a paradigm in the evolution of a resistance mechanism. Incorporation of different chromosomal blaCTX-M related genes from different species of Kluyvera has derived in different CTX-M clusters. In silico analyses have shown that this event has occurred at least nine times; in CTX-M-1 cluster (3), CTX-M-2 and CTX-M-9 clusters (2 each), and CTX-M-8 and CTX-M-25 clusters (1 each). This has been mainly produced by the participation of genetic mobilization units such as insertion sequences (ISEcp1 or ISCR1) and the later incorporation in hierarchical structures associated with multifaceted genetic structures including complex class 1 integrons and transposons. The capture of these blaCTX-M genes from the environment by highly mobilizable structures could have been a random event. Moreover, after incorporation within these structures, β-lactam selective force such as that exerted by cefotaxime and ceftazidime has fueled mutational events underscoring diversification of different clusters. Nevertheless, more variants of CTX-M enzymes, including those not inhibited by β-lactamase inhibitors such as clavulanic acid (IR-CTX-M variants), only obtained under in in vitro experiments, are still waiting to emerge in the clinical setting. Penetration and the later global spread of CTX-M producing organisms have been produced with the participation of the so-called “epidemic resistance plasmids” often carried in multi-drug resistant and virulent high-risk clones. All these facts but also the incorporation and co-selection of emerging resistance determinants within CTX-M producing bacteria, such as those encoding carbapenemases, depict the currently complex pandemic scenario of multi-drug resistant isolates.
Keywords: blaCTX-M genes, Kluyvera spp., gene mobilization, ISEcp1, ISCR1, plasmid, bacterial clones, antibiotic selective force
Introduction
Antibiotic resistance is a topic in continuous review and the several studies published every year with new data can modify or redefine our perspective about this problem. β-lactam resistance and particularly overviews about β-lactamases, enzymes able to hydrolyze β-lactam antibiotics, have undoubtedly been the most frequently studied topics in this field. β-lactams, the antibiotics most widely used all over the world, have also given rise to a continuous increase of resistant isolates due to their permanent selective force driving diversification of the resistance mechanisms.
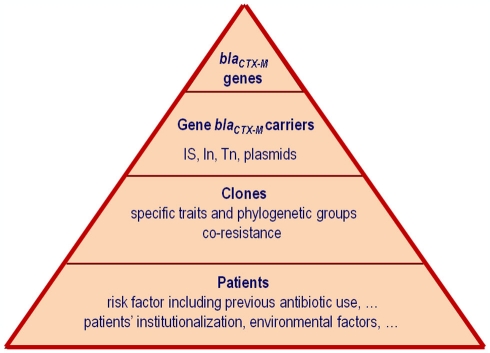
β-Lactamases have been studied from many different aspects, including classification, hydrolytic profiles and kinetic properties, plasticity, frequency, and dissemination. Moreover they have been used as an example of the accelerated evolution following the Darwinian paradigm, where the strong antibiotic selective pressure favors the survival of the most adapted. According to a recent review from Davies and Davies (2010), a dramatic increase in the number of β-lactamases has been described since the 1980s of the last century but this increase is due almost exclusively to class A and D β-lactamases (Bush and Jacoby, 2010). Among the class A β-lactamases, the extended-spectrum β-lactamases (ESBLs) which are able to hydrolyze expanded-spectrum cephalosporins (such as cefotaxime, ceftriaxone, ceftazidime, or cefepime) and monobactams (aztreonam) represent a public health concern (Coque et al., 2008a; Pitout and Laupland, 2008). ESBLs of class A mainly include TEM, SHV, CTX-M, VEB, and GES enzymes. Among them, the highest number of variants described in the last years corresponds to the CTX-M family (123 variants until 2011, last accession December 5, 2011)1. This explosive dissemination of CTX-Ms around the world has been referred as the “CTX-M pandemic” due to their increasing description worldwide (Cantón and Coque, 2006). Although different updates about CTX-M β-lactamases have been published (Bonnet, 2004; Livermore et al., 2007; Rossolini et al., 2008; Hawkey and Jones, 2009; Naseer and Sundsfjord, 2011), new data about dispersion and clonality of CTX-M producing isolates, molecular epidemiology, protein plasticity, evolution and origin of the blaCTX-M genes, influence of antibiotic use, and patients risk factors, justify this new review. Figure 1 illustrates different factors fueling the emergence, maintenance, and spread of the CTX-M ESBLs. We will focus our attention on the epidemiology, origin and evolution of blaCTX-M genes, and influence of surrounding genetic structures participating in the maintenance and spread of these genes and hence organisms producing CTX-M enzymes.
Figure 1.
Factors fueling the emergence, maintenance, and spread of the CTX-M extended-spectrum β-lactamases (ESBLs).
Classification of β-Lactamases and CTX-M Enzymes
β-Lactamases are by far the most important resistant mechanisms in Gram-negative bacilli. With the popularization of molecular techniques, an increasing number of these enzymes have been characterized differing in amino acid sequences and hydrolytic activity against β-lactam antibiotics (Bush, 2010). They are mainly classified using the Ambler scheme (Ambler, 1980), which grades to the so-called molecular classes (A through D) and also currently functional groups which use the characteristic of the enzymes, including their hydrolytic substrate profile, and response to inhibitors (Bush and Jacoby, 2010). Classically, genetic location of the corresponding β-lactamase genes (bla) in plasmids or chromosome was also used to differentiate these enzymes. Nevertheless, this genetic characteristic is no longer used, as chromosomal bla genes can be mobilized and integrated into plasmids or transposons, but a reverse situation of initially describes plasmid-mediated β-lactamases into the chromosome is also increasingly found (Toleman et al., 2006; Coelho et al., 2010). In addition protein regulation characteristics (i.e., constitutive or inducible expression) are also noted when referring to different β-lactamases groups (Livermore, 1995) but this trait depends on the surrounding genes. In Gram-negative organisms, inducible expression of β-lactamases is commonly found in chromosomal β-lactamases whereas plasmid-mediated enzymes are generally constitutively expressed. Enhancement expression of their hydrolytic activity is often regulated by promoters present in upstream genes.
Within the β-lactamases, the ESBLs have been worthy of the attention of the scientific community and clinicians over the last three decades since their first description in 1983 (Knothe et al., 1983). In general, ESBLs are located in plasmids and are characterized for their ability to hydrolyze oxy-imino-cephalosporins (third and fourth generation cephalosporins) and monobactams but not cephamycins such as cefoxitin and carbapenems including imipenem, ertapenem, meropenem, or doripenem (Philippon et al., 1989; Jacoby and Muñoz-Price, 2005; Bush and Fisher, 2011). In addition, they are generally susceptible to β-lactamase inhibitors such as clavulanic acid, sulbactam, and tazobactam. Mainly, they are included in the 2be Bush-Jacoby and Medeiros functional group belonging to the molecular class A β-lactamases from the Ambler’s classification (Ambler, 1980; Bush et al., 1995; Bush and Jacoby, 2010). Enzymes showing an “ESBL phenotype” have been mainly described in TEM-, SHV-, CTX-M, GES, and VEB families into class A β-lactamases, and OXA-ESBLs into class D β-lactamases (Paterson and Bonomo, 2005).
The TEM-, SHV-, and OXA-type ESBL enzymes derive from point mutations from the classical plasmid-mediated enzymes TEM-1, TEM-2, SHV-1, and OXA-10 which are not of greater importance in hydrolyzing extended-spectrum cephalosporins (Paterson and Bonomo, 2005; Bush and Jacoby, 2010). The TEM- and SHV-ESBLs were predominant in the ESBL landscape over the 1980s and 1990s in the past century, mainly associated with outbreaks in hospitals involving Klebsiella pneumoniae and to a lesser extent in Escherichia coli and other Enterobacteriaceae whereas the CTX-M were less prevalent (Bradford, 2001; Paterson and Bonomo, 2005). Although first discovered in 1989 (Bauernfeind et al., 1990, 1992), the CTX-M enzymes did not reach prominence over the other ESBL enzymes until the first decade of the 2000s when accelerated evolution and extraordinary dispersion of these enzymes were observed (Cantón, 2008). They were confined not only to the hospital setting but also to the community, with E. coli being the most important pathogen producing these enzymes (Cantón and Coque, 2006; Coque et al., 2008a).
Unlike other ESBLs, CTX-M family constitutes a complex and non-homogeneous group of enzymes. The first view and alignment of the amino acid sequence of the different CTX-M variants allowed to classify these enzymes in five clusters but recent publications report at least two more additional clusters (Bonnet, 2004; Rossolini et al., 2008; see also below). Phylogenetic analyses suggest that CTX-Ms were not originated by mutations from previous plasmid meditated enzymes but through mobilization of chromosomal bla genes from Kluyvera spp. when they were incorporate into mobile genetic elements (Cantón, 2008). These original mobilized blaCTX-Ms genes affected cefotaxime to a higher degree than ceftazidime. Nevertheless and from an evolutionary point of view, CTX-Ms as other ESBLs later diverged by punctual mutations probably as consequence of antibiotic selective pressure once Kluyvera spp. blaCTX-M genes were mobilized and were incorporated into mobile genetic elements. This also gave the CTX-M enzymes the opportunity to enhance the hydrolytic activity against ceftazidime and a new variants were described (Bonnet, 2004; Poirel et al., 2008).
Epidemiology of CTX-M β-Lactamase-Producing Bacteria
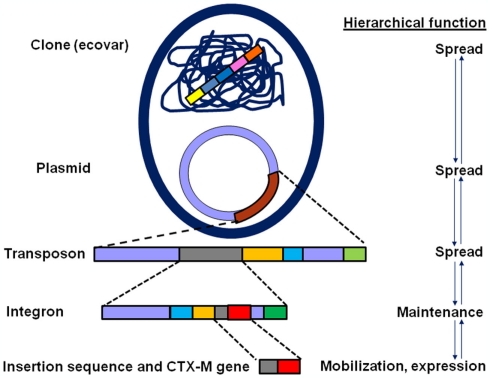
Studies over the last 10 years have revealed that unlike some exceptions, the CTX-M enzymes have nearly displaced other ESBLs enzymes in Enterobacteriaceae, including TEM and SHV ESBL variants (Cantón, 2008; Coque et al., 2008a; Angel Díaz et al., 2009; Hawkey and Jones, 2009; Bush, 2010; Rodriguez-Villalobos et al., 2011). This displacement might have occurred not only as a consequence of the extraordinary dissemination of the corresponding blaCTX-M genes in highly mobilizable genetic platforms, including plasmids and transposons, but also because of these platforms within successful clones (Cantón and Coque, 2006; Rogers et al., 2011; Woodford et al., 2011; Figure 2). Another reason for this increase is the co-resistance phenomenon in CTX-M producing organisms, particularly to aminoglycosides and fluoroquinolones, which might facilitate co-selection processes (Morosini et al., 2006; Cantón and Ruiz-Garbajosa, 2011).
Figure 2.
Hierarchical complexity of blaCTX-M genes within genetic structures and bacterial clones participating in the mobilization, spread, and maintenance of these genes (see the text for further explanation).
Apart from this general overview, within the CTX-M enzymes, the CTX-M-15, and CTX-M-14 are by far the most important ones, virtually invading all human and animal compartments as well as the environment all over the world (Cantón et al., 2008; Hawkey and Jones, 2009; Dolejska et al., 2011; Hiroi et al., 2012). Nevertheless, temporal emergence and penetration of these enzymes in different epidemiological scenarios might also explain the current epidemiology of CTX-M enzymes. Antibiotic consumption and dissimilar risk factors in different geographic areas and groups of patients and particularities of different compartments might have also contributed to the current CTX-M scenario (Carattoli, 2008; Rodríguez-Baño and Navarro, 2008; Rodríguez-Baño and Pascual, 2008; Oteo et al., 2010a; Naseer and Sundsfjord, 2011). In this sense three different periods of CTX-M can be differentiated. The first one includes emergence of different CTX-M β-lactamases in different and distant geographic areas and might have occurred until the mid-1990s of the last century. The second was characterized by the emergence of the most widespread CTX-M enzymes, including CTX-M-3, CTX-M-9, CTX-M-14, and CTX-M-15 enzymes and might have occurred from 1994 to 2000. Some of these enzymes, but not exclusively, are variants of previously described CTX-M β-lactamases. Finally, the third period through 2000 is characterized by the universal dispersion and globalization of these β-lactamases. The second and third periods will be presented together bellow.
The emergence of CTX-M β-lactamases: Different CTX-M enzymes in different countries
The first recognition of the emergence of CTX-M β-lactamases occurred nearly simultaneously in Europe and South America at the beginning of 1989 (Bauernfeind et al., 1990, 1992). The first publication recognizing an ESBL from the CTX-M group was that reporting a cefotaxime-resistant but ceftazidime susceptible E. coli isolate recovered from the ear of a 4-month-old child suffering from otitis media in Munich (Germany; Bauernfeind et al., 1990). The enzyme responsible for this particular ESBL phenotype not affecting ceftazidime was named as CTX-M-1 in reference to its preferential hydrolytic activity against cefotaxime (CTX as its acronym, -M from Munich). This enzyme inaugurated a group of enzymes that during the 1990s were also grouped as cefotaximases due to their particular phenotypic and hydrolytic profile (Bonnet, 2004). In South America, the first isolates with a “cefotaximase pattern” were detected in Salmonella typhimurium recovered in hospitalized patients suffering from meningitis, septicemia, or enteritis (Bauernfeind et al., 1992). The enzyme responsible for this phenotype had a different isoelectric point (pI 7.9) than the one (pI 8.9) described in Germany and was named as CTX-M-2. Later sequencing of blaCTX-M-2 gene in 1996 confirmed a different amino acid sequence of these two enzymes but a shared homology of 84% (Bauernfeind et al., 1996).
These CTX-M-1 and CTX-M-2 β-lactamases were the first reported CTX-M like enzymes but not chronologically the enzymes belonging to this group. They were preceded by the isolation in 1986 of a cefotaxime-resistant E. coli isolate recovered from the fecal flora of a laboratory dog which was used for pharmacokinetic studies of β-lactam antibiotics in Japan (Matsumoto et al., 1988). The enzyme responsible for this phenotype was named as FEC-1 (Fecal E. coli), which was later recognized to be related with the CTX-M-3 enzyme initially found in Poland (Bonnet, 2004). Moreover, and also at the beginning of 1989 but in France, an E. coli with an identical phenotype was isolated from a Italian patient (Barthélémy et al., 1992; Bernard et al., 1992). The β-lactamase, named as MEN-1 (named after the patient in who it was isolated), was the first available sequence of a CTX-M β-lactamase (Barthélémy et al., 1992). In 1996 sequencing of both the original CTX-M enzymes, CTX-M-1 and CTX-M-2, revealed that MEN-1 was identical to CTX-M-1 (Bauernfeind et al., 1996).
Other CTX-M related β-lactamases that were contemporarily isolated during the 1990s were Toho-1 and Toho-2 enzymes (Toho refers to Toho University School of Medicine Omori Hospital in Tokyo). The former were characterized in a cefotaxime-resistant E. coli isolate recovered in 1993 from a 1-year-old female child who developed cystitis in Japan (Ishii et al., 1995) and the latter also in a cefotaxime-resistant E. coli isolate recovered in Japan in 1995 from the urine of a 69-year-old male who suffered from colon cancer (Ma et al., 1998). Sequence alignment revealed their linkage with CTX-M-2. Both enzymes were later renamed and designated as CTX-M-44 (Toho-1) and CTX-M-45 (Toho-2; last accession December 5, 2011)2
All these earliest descriptions of CTX-M enzymes showed hot areas for their emergence (Central Europe, South America, and Japan) that could reflect not only specific selective driving forces such as antibiotic consumption but also geographic specificity for different enzymes that could be related with their presence in specific clones or abundance of organism from which they derived (see below). Unfortunately, clonality of ancestral CTX-M producing isolates was not performed as well as plasmid characterization. These studies would have been relevant for tracing the emergence and dissemination of the first CTX-M ESBLs. Moreover, epidemiological surveys including molecular characterization at the end of the 1980s from other geographic areas, such as South East Asia that is nowadays relevant for emerging β-lactamases, including carbapenemases (enzymes that are able to hydrolyze carbapenems), were absent. These would also have been important for the better understanding of the current dissemination of epidemiologically important CTX-Ms that were described through 1994, such as CTX-M-3, CTX-M-9, CTX-M-14, and CTX-15. Table 1 includes epidemiological data of the earliest descriptions of different CTX-M ESBLs during the 1980s and 1990s. This data are relevant for understanding current epidemiological scenario.
Table 1.
Initial description of CTX-M enzymes belonging to different CTX-M groups.
| Year | Country | CTX-M | CTX-M group | Comment | Reference |
|---|---|---|---|---|---|
| 1986 | Japan | FEC-1 | CTX-M-1 | Escherichia coli from the fecal flora of laboratory dogs used for pharmacokinetic studies and previously administered cephem antibiotics | Matsumoto et al. (1988) |
| 1989 | Germany | CTX-M-1 | CTX-M-1 | Escherichia coli isolate recovered from the ear of a 4-month child suffering from otitis media in Munich. The β-lactamases involved were firstly named as cefotaximases | Bauernfeind et al. (1990) |
| 1989 | Argentina | CTX-M-2 | CTX-M-2 | Salmonella Thypimurium isolates from hospitalized patients with meningitis, septicemia, or enteritis | Bauernfeind et al. (1992) |
| 1989 | France | MEN-1 | CTX-M-1 | Escherichia coli isolate in France in an Italian patient. Later sequencing reveals identity with CTX-M-1 | Bernard et al. (1992), Barthélémy et al. (1992) |
| 1993 | Japan | Toho-1 | CTX-M-2 | Escherichia coli isolate recovered from a 1-year-old female child who developed cystitis. Toho-1 was later renamed as CTX-M-44 | Ishii et al. (1995) |
| 1996 | Poland | CTX-M-3 | CTX-M-1 | Citrobacter freundii and Escherichia coli isolates recovered in Warsaw in patients with urinary tract infections | Gniadkowski et al. (1998) |
| 1999 | Taiwan | CTX-M-3 | CTX-M-1 | Different Escherichia coli isolates from several sources in hospitalized and non-hospitalized patients | Yan et al. (2000) |
| 1994 | France | CTX-M-9 | CTX-M-9 | A retrospective study showed presence of Escherichia coli with this enzyme in this country before its first recognition in Spain | Saladin et al. (2002) |
| 1996 | Spain | CTX-M-9 | CTX-M-9 | First description of CTX-M-14 in an Escherichia coli isolate from a patients with an urinary tract infection | Sabaté et al. (2000) |
| 1996 | Brazil | CTX-M-9 | CTX-M-9 | Escherichia coli isolate recovered from an hospitalized patient in Rio de Janeiro | Bonnet et al. (2001) |
| 1997 | China | CTX-M-9 | CTX-M-9 | Escherichia coli and Klebsiella pneumoniae recovered as part of an antimicrobial resistance monitoring project | Chanawong et al., 2002 |
| 1996 | Korea | CTX-M-14 | CTX-M-9 | Detected in Shigella sonnei and in blood isolates of Escherichia coli and Klebsiella pneumoniae | Pai et al. (2001) |
| 1997–1998 | China | CTX-M-14 | CTX-M-9 | Escherichia coli, Klebsiella pneumoniae and Enterobacter cloacae recovered as part of an antimicrobial resistance monitoring project | Chanawong et al. (2002) |
| 1998 | Taiwan | CTX-M-14 | CTX-M-9 | Escherichia coli recovered during a multicenter antimicrobial resistance surveillance study | Ma et al. (2002) |
| 1998 | Poland | CTX-M-15 | CTX-M-1 | A retrospective study showed the presence of Serratia marcescens and Escherichia coli before the first description of this enzyme in India. Sequence and plasmid analysis revealed potential evolution from CTX-M-3 | Baraniak et al. (2002) |
| 1999 | India | CTX-M-15 | CTX-M-1 | First description of this enzyme | Karim et al. (2001) |
| 2001 | United Kingdom | CTX-M-15 | CTX-M-1 | Spread in the community of Escherichia coli isolates with CTX-M-1. A major clone was later identified as belonging to the international 25:H4-ST131 clone | Mushtaq et al. (2003), Woodford et al. (2004) |
| 1996–1997 | Brazil | CTX-M-8 | CTX-M-8 | Enterobacter cloacae, Citrobacter amalonaticus, and Enterobacter aerogenes from different sources in hospitalized patients | Bonnet et al. (2000) |
| 2000 | Canada | CTX-M-25 | CTX-M-25 | Escherichia coli isolate recovered from an hospitalized patient | Munday et al. (2004) |
Penetration and globalization of CTX-M enzymes all over the world
During the 1990s, different reports showed the appearance nearly at the same time of the same enzymes in very distant countries, denoting the potential independent emergence of these enzymes but also suggesting a rapid dissemination. During these years, diversification was also noted with the ever increasing description of new CTX-M enzymes from different groups. This has been well illustrated with CTX-M-3, a closely related enzyme with CTX-M-1 differing in four amino acid positions (V77A, D114N, S140A, and N288D). This enzyme was detected in E. coli and K. pneumoniae isolates recovered in 1996–1997 in Poland (Gniadkowski et al., 1998; Palucha et al., 1999), in 1998 in France (De Champs et al., 2000; Doucet-Populaire et al., 2000) and in 1999 in Taiwan (Yan et al., 2000). Unlike genetic determinants harboring blaCTX-M-3 gene which were not sequenced at that time, CTX-M plasmid size and plasmid restriction patterns showed similarity demonstrating the potential diffusion of the same plasmids within isolates obtained from Poland and Taiwan (Yan et al., 2000). Despite this result, importation of this enzyme to Taiwan from Europe or vice versa was not possible to rule out as clonality was not addressed. Nevertheless, in both countries rapid dissemination of this enzyme was illustrated after its emergence (Baraniak et al., 2002; Yu et al., 2002; Ma et al., 2005).
Diversification of CTX-M-1 cluster was exemplified with increasing descriptions of new variants over the 1990s. For instance, CTX-M-10 was mainly described in the Mediterranean area (Spain and France; Oliver et al., 2001; Leavitt et al., 2009) and the CTX-M-15, initially identified in 1999 in enteric isolates recovered from a hospital in New Delhi, India (Karim et al., 2001), but nowadays found all over the world (Hawkey and Jones, 2009; Rogers et al., 2011). The CTX-M-10 differs from CTX-M-3 in two amino acids (A27V and R38Q) whereas CTX-M-15 in only one amino acid position (D240G). Nevertheless, diversification of CTX-M-3, CTX-M-10, and CTX-M-15 should have been produced from a common ancestor (Novais et al., 2010).
A similar example is the CTX-M-9 enzyme, which was initially described in Spain and Brazil in 1996 (Sabaté et al., 2000; Bonnet et al., 2001), followed by China in 1997 (Chanawong et al., 2002). A retrospective study performed in France revealed the presence of this enzyme in strains isolated during 1994 (Saladin et al., 2002). In Spain, this enzyme became one of the most prevalent ESBL but co-existing with CTX-M-14 enzyme (a variant of CTX-M-9 differing in one amino acid, A231V). This enzyme emerged in Korea in 1995 (Pai et al., 2001) and was rapidly disseminated over the world. In China, it was detected in 1997–1998 (Chanawong et al., 2002), in Taiwan in 1998–1999 (Yu et al., 2002), in France in 1999 (Dutour et al., 2002) and in Brazil in 1999 (Bonnet, 2004).
Surveys performed in different countries during that period revealed that once a CTX-M β-lactamase penetrates in a specific geographic area it becomes prevalent with displacement or superimposition over TEM and SHV ESBL variants. This was the case of CTX-M-2 in South America, CTX-M-3 in Poland and CTX-M-14 in Spain, Taiwan and China (Bonnet, 2004; Livermore and Hawkey, 2005). This landscape was also the case for CTX-M-15 in United Kingdom (Paterson and Bonomo, 2005; Cantón and Coque, 2006; Hawkey and Jones, 2009). As previously noted, it was first isolated in India in 1999 (Karim et al., 2001) but also emerged in the United Kingdom in 2001 and probably in Japan early in the 2000s [designated as UOE-1 in GeneBank (accession number AY013478; Mushtaq et al., 2003; Livermore and Hawkey, 2005)]. Moreover, in Poland a retrospective study of a collection of Enterobacteriaceae resistant to expanded-spectrum cephalosporins revealed the presence of the CTX-M-15 in Serratia marcescens and E. coli since 1998 (Baraniak et al., 2002). Interestingly plasmid analysis suggested that this enzyme could have evolved directly from CTX-M-3 rather than being imported from another country and independently in different hospitals.
In the United Kingdom, isolation of CTX-M was rare before 2001 and only sporadic cases of CTX-M-9 were described as well as an important outbreak due to CTX-M-26, a variant of CTX-M-25 with three amino acid substitutions (V80A, Q225R, and G242D), which was previously described in Canada (Munday et al., 2004). The CTX-M-15 rapidly disseminated in the United Kingdom since its first detection. Isolates were particularly associated with different E. coli clones, A to F, the first one representing the majority and later identified as belonging to the international O25:H4-ST131 clone that has been detected all over the world (Lau et al., 2008). Again a dramatic increase of CTX-Ms was demonstrated mainly due to the CTX-M-15 enzymes (Coque et al., 2008b).
In the United States, CTX-Ms were rare during the 1990s but increased from 25% in 2000 to nearly 90% in 2005 (Lewis et al., 2007). Interestingly, CTX-M-15 rapidly penetrated over a minority of CTX-Ms that include CTX-M-8, CTX-M-14, and CTX-M-16 (Lewis et al., 2007; Johnson et al., 2010; Peirano et al., 2010). Within this country and as in other parts of the world, the CTX-M-15 enzyme has been identified in hospitalized and extrahospitalary patients and also in companion animals in which CTX-M-15 was the most prevalent CTX-M enzyme (Castanheira et al., 2008; Urban et al., 2010; Shaheen et al., 2011).
The model of CTX-M-15 dissemination could have been different in countries in which other CTX-M β-lactamases were well established prior to the detection and penetration of this enzyme. This is the case of Spain and South America. In our institution in Spain, CTX-M-10 was first detected in 1990 in K. pneumoniae and in 1991 in E. coli. The first detection of CTX-M-9 was in E. coli in 1996 and in K. pneumoniae in 1999, CTX-M-14 in 2001 in K. pneumoniae and in E. coli and CTX-M-15 in 2001 in E. coli and in 2002 in K. pneumoniae. It is of note that in a national survey in 2000 in Spain involving 40 centers, CTX-M-15 was not found. However, in 2006 in a similar study, CTX-M-15 was found in different regions and was one of the most prevalent enzymes after CTX-M-14 and SHV-12 (Díaz et al., 2010). More recently, in our institution CTX-M-15 represents 34% of total ESBL producing E. coli (Novais et al., unpublished data), 37% of CTX-M-14, and 20% of SHV-12, denoting the increasing prevalence of CTX-M-15 but still co-existing with other CTX-M enzymes. This data illustrates that when CTX-M-15 penetrates in a landscape where other ESBLs are present the increment of this enzyme and displacement of other ESBLs is a slow process. This model can also be the case of South America where CTX-M-2 has been the most prevalent CTX-M over years. Nevertheless, reports of CTX-M-15 producing isolates are increasing in this continent, including Colombia (Valenzuela de Silva et al., 2006; Ruiz et al., 2011), Brazil (Cergole-Novella et al., 2010), Uruguay (Bado et al., 2010), and Peru (Pallecchi et al., 2004, 2007).
At present, we cannot rule out that international travel and immigration might have contributed during the 1990s and the early 2000s to the rapid emergence and dissemination of CTX-M enzymes in different countries and geographic areas. This has recently been demonstrated with carbapenemases and particularly with the NDM-1 metalo-β-lactamase-producing organisms (Kumarasamy et al., 2010). Nevertheless, the presence of CTX-M enzymes in animals and most importantly in uncooked food products that were transported from one country to another suggests potential routes for dissemination (Matsumoto et al., 2007; Warren et al., 2008; Dhanji et al., 2010).
Finally, globalization of CTX-M enzymes is illustrated by their presence not only in food and companion animals but also in wild animals (Bonnedahl et al., 2010; Literak et al., 2010; Gonçalves et al., 2011; Silva et al., 2011). Additionally, they have been isolated in environmental compartments (Chen et al., 2010; Dhanji et al., 2011a).
CTX-M enzymes not only in enterobacteriaceae
As previously stated CTX-M enzymes were initially described in E. coli, K. pneumoniae, and Salmonella spp. but rapidly emerged in other Enterobacteriaceae. This event occurs in bacterial isolates with chromosomal mechanisms such as inducible chromosomal AmpC in Enterobacter spp., S. marcescens, Citrobacter spp., and Morganella morganii able to destroy oxy-imino-cephalosporins (Cantón et al., 2002; Paterson and Bonomo, 2005). This find suggests the facility for transmission of blaCTX-M genes and also that the acquisition of these genes could have lower fitness cost for the bacteria than selection of derepressed AmpC β-lactamase variants which finally also contribute to cephalosporin resistance.
As CTX-M enzymes increased in prevalence in Enterobacteriaceae in the early 2000s, they also increased in non-fermentative Gram-negative bacilli and other non-Enterobacteriaceae species. The first CTX-M enzyme described in Pseudomonas aeruginosa and Stenotrophomonas maltophilia was observed in isolates recovered in 2004 from sputum samples of a cystic fibrosis patient in Amsterdam (al Naiemi et al., 2006). The CTX-M enzyme from these isolates was characterized as CTX-M-1. In the same 2004 year, CTX-M-43 (a variant of CTX-M-44 carrying the D240G change and belonging to the CTX-M-2 group), was also observed in P. aeruginosa and Acinetobacter spp. in isolates recovered from Bolivian hospitals (Celenza et al., 2006). CTX-M-2 was also found in a P. aeruginosa isolate recovered in a patient hospitalized in Brazil in 2005 (Picão et al., 2009). Despite these descriptions, prevalence of CTX-M enzymes in non-fermenter bacilli is still rare. This fact could be due to incompatibility of plasmids carrying these enzymes or might reflect phenotypic difficulties in recognizing non-fermenter isolates expressing CTX-Ms. On the other hand, Aeromonas spp. or Vibrio spp. isolates with CTX-M enzymes have been also recognized (Soler Bistué et al., 2006; Ye et al., 2010; Chowdhury et al., 2011; Gómez-Garcés et al., 2011).
Origin and Mobilization of the blaCTX-M Genes
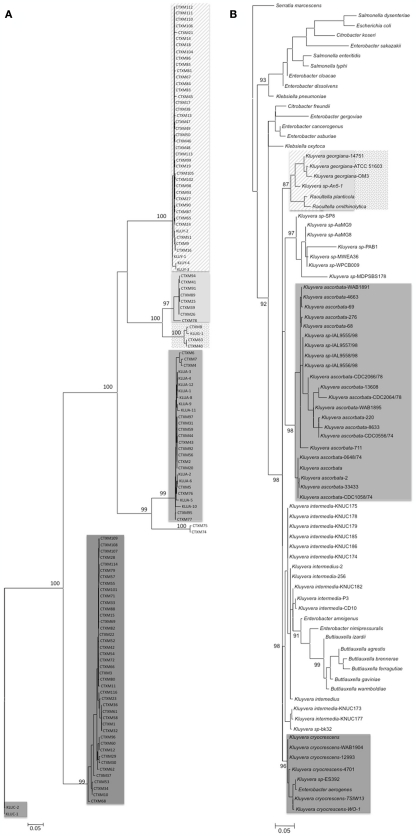
According to phylogenetic trees performed with all CTX-M β-lactamases described, the CTX-M lineage could be differentiated at in least five main clusters (Figure 3). Traditionally, each CTX-M cluster has been related to the chromosomal bla genes present in different Kluyvera species, which are part of the human normal intestinal microbiome, usually at low bacterial counts, and is considered as a saprophytic and an opportunistic pathogen (Farmer et al., 1981). Furthermore, they have been occasionally isolated in humans associated with different infections, mainly affecting the urinary tract and skin and soft tissues (Sarria et al., 2001) and is also present in the environment as free-living organisms in water, soil, sewage, and food products of animal origin (Farmer et al., 1981).
Figure 3.
Maximum likelihood trees of blaCTX-M genes and 16S rDNA of Kluyvera spp. in order to compare their respective phylogenetic topologies. (A) Phylogenetic tree of blaCTX-M genes (n = 109 and 17 chromosomal genes from Kluyvera) was obtained using PhyML_3.0 program (846 nt). The Tamura Nei nucleotide substitution model, used as evolutionary model was selected with the jModeltest program. The robustness of the relevant nodes was estimated with 1000 bootstrap pseudorandom replicates. We considered nodes valid when bootstrap value was >95%. (B) Phylogenetic tree of 16s rDNA of Kluyvera spp. (n = 55) and related species download of www.ncbi.nlm.nih.gov, using PhyML_3.0 (1310 nt). The Hasegawa–Kishino–Yano was the evolutionary model inferred by jModeltest program and invariant site rate of 77.6% (HKY + I + G). We considered nodes valid when bootstrap value was >85%. The phylogenetic trees were represented using MEGA 5.0 program.
Chromosomal bla gene, kluC, present in Kluyvera cryocrescens has been considered the ancestor of the CTX-M-1 cluster (Decousser et al., 2001), kluA gene from Kluyvera ascorbata the origin of the CTX-M-2 cluster (Humeniuk et al., 2002) and three different chromosomal bla genes from Kluyvera georgiana, kluG, kluY, and blaCTX-M-78 the origin of CTX-M-8 (Poirel et al., 2002), CTX-M-9 (Olson et al., 2005), and CTX-M-25 clusters (Rodríguez et al., 2010) respectively. Interestingly, the CTX-M-74 and CTX-M-75, characterized in a survey conducted in Brazil and with single one amino acid change with respect to CTX-M-2 (Minarini et al., 2009), showed a degree of nucleotide divergence that could be considered as a new cluster, reflecting a convergent evolution of these β-lactamases (Stepanova et al., 2008). All these data suggest that the environmental source of CTX-M β-lactamases was the genus Kluyvera. Although the scientific community has unequivocally accepted this point, several questions still arise to guarantee this line of evidence. First, the comparison of the topologies of phylogenetic trees between blaCTX-M and available 16S rDNA sequences of Kluyvera species reveals that the evolutionary relationships among CTX-M clusters is not completely correlate with the phylogenetic distance among the different species of Kluyvera (Figure 3). Whereas all Kluyvera species diversified at the same time from the same ancestral bacteria, this topology is not observed among blaCTX-M clusters, suggesting different evolutionary trajectories. Second, if each chromosomal gene from Kluyvera was the source of a determined CTX-M cluster, why have at least three chromosomal genes from K. georgiana (blakluG, blakluY, and blaCTX-M-78) been proposed as precursor of three different CTX-M clusters? On the contrary, CTX-M-1 cluster has been related to different Kluyvera species (K. ascorbata and K. cryocrescens; Decousser et al., 2001; Rodríguez et al., 2004). These data could also suggest that blaCTX-M genes have been also circulating among different Kluyvera species. Third, upstream of blaklu gene identical genome organization was found in Enterobacter aerogenes, suggesting that Kluyvera could have gained blaklu gene after these species diverged, therefore most of the isolates must carry blaklu gene, including Kluyvera intermedia, where no blaklu gene has been identified. However, despite the presence of the blaklu gene in most Kluyvera isolates, 90% are susceptible to cefotaxime (Sarria et al., 2001). Obviously, blaklu gene could be present but weakly expressed. Unfortunately the presence of blaklu gene in Kluyvera susceptible isolates or their genetic environment has not been searched. These studies could help us to understand previous observations of Kluyvera spp. as the precursor of the CTX-M enzymes and responsible for the current pandemic.
It is well known that chromosomal blaklu genes are weakly expressed in their original context and require the presence of a strong promoter upstream to increase the MIC values and consequently be considered as phenotypically resistant. The insertion sequences (IS) have provided this promoter in clinical cefotaxime-resistant Enterobacteriaceae strains, the most prevalent being ISEcp1 and ISCR1, while IS26 has been only detected in few isolates. Curiously, these IS have not been described in natural isolates of Kluyvera, although the capture of ISEcp1 upstream of chromosomal blaklu/CTX-M genes occurs at high frequency in laboratory conditions. This was demonstrated in an elegant in vitro experiment performed by Lartigue et al. (2006). When ISEcp1 was inserted upstream of blaklu/CTX-M gene, stress situations such as high temperature (40°C) or that exerted by antimicrobials such as ceftazidime or cefotaxime, the mobilization of ISEcp1 was increased, carrying the downstream sequences. It has been speculated that global warming or higher temperature than humans in the intestinal compartment of certain animals and the increasing use of antimicrobials might have also contributed to the extraordinary increase and spread of the blaklu/CTX-M genes (Lartigue et al., 2006; Nordmann et al., 2008). Apart from these speculations, an interesting question is if mobilization of blaklu occurred, how often was it in the nature? Initially and accepting the genus Kluyvera as progenitor of the CTX-M enzymes, we can suspect mobilization of the same number of occasions has CTX-M clusters has been described. Barlow et al. (2008) using phylogenetic trees based on gene distance suggest that this occurred at least eight times (three times each for CTX-M-1 and CTX-M-2 clusters and only one each for CTX-M-9 and CTX-M-8; Barlow et al., 2008). However the description of different IS such as ISEcp1 and ISCR1 related to CTX-M-9 (Valverde et al., 2009), allow us to suspect that more mobilization events may have occurred (see below).
Evolution and Diversification of CTX-M β-Lactamases
Cumulative representation of the new variants described among the most prevalent β-lactamases shows that the highest slope during the last years correspond to the CTX-M and OXA-type β-lactamases (Bush and Fisher, 2011). This might indicate that they have recently suffered the most spectacular diversification process in the β-lactamases world. CTX-M phylogenetic trees suggest an explosive diversification from the original bla genes in a short time when they arrived in the clinical scenario (Novais et al., 2010). The first CTX-Ms identified were able to confer cefotaxime resistance although their capacities to efficiently hydrolyze ceftazidime was very low or null, but nowadays more than 60% of CTX-M variants are able to confer cefotaxime and ceftazidime-resistance simultaneously. Thus, ceftazidime could have been one of the main selective forces contributing to the diversification of CTX-Ms.
An evolution experiment was designed in our laboratory to obtain more efficient CTX-M variants against ceftazidime; the ceftazidime-resistant CTX-M variants carried the changes A77V, P167S, and D240G (Novais et al., 2008). These changes have been described in the nature in several cases including CTX-M-52 and CTX-M-87, which derived from CTX-M-15 and CTX-M-14 respectively (Yin et al., 2009). These results reinforce the idea that ceftazidime has driven the selection of new CTX-M variants. Moreover, we calculated the ratio of the number of non-synonymous substitutions with respect to synonymous substitutions (ϖ = dN/dS) among the blaCTX-M gene variants in order to define positive (accelerate evolution) or purified (slow evolutionary rate) selection processes during the diversification of CTX-Ms. In the deep branches of the CTX-Ms phylogenetic tree, processes of purified selection occurred while positive selection was the mechanism of molecular evolution in the more recent branches. In our experiments, six positions were identified to be under positive selection, including the amino acid positions 77, 167, and 240, which were affected under ceftazidime regimens. Nevertheless, our results also suggest the presence of more evolutionary forces involved in the selection of new and better adapted variants. The construction of mutants carrying combinations of these mutations revealed that the simultaneous presence of cefotaxime and ceftazidime in the environment has modeled the explosive evolution and diversification of the CTX-M enzyme (Novais et al., 2010).
Recombination events also accelerate the evolution. These events have also been described among blaCTX-M genes. As an example, the CTX-M-64 enzyme is the result of a recombination between members of the blaCTX-M-9 and blaCTX-M-1 derived genes, probably blaCTX-M-14 and blaCTX-M-15, as they are the most prevalent variants in the world (Nagano et al., 2009). The recombination requires the simultaneous presence of both blaCTX-Ms in a same habitat. Recently, we described in an Ethiopian 1-year-old boy both CTX-M-14 and CTX-M-15 (Morosini et al., 2010). Moreover, in a national surveillance performed in China, the CTX-M-64 β-lactamase was recovered from three patients co-existing with other CTX-Ms (Sun et al., 2010). As the concomitant presence of different CTX-M β-lactamases in the same bacteria is no longer an unusual event, other recombinant enzymes might emerge in the near future.
blaCTX-M Gene Plasticity and Other Strategies to Increase the Spectrum of Action of CTX-M β-Lactamases
β-Lactamases are highly modeling proteins as few positions are critical to hydrolyze β-lactam compounds. Nowadays, at least 136 polymorphic positions have been found among all CTX-Ms described. Some of these changes play a main role increasing the β-lactam hydrolytic spectrum while others modulate or restore the synergic or antagonist effects caused by other modifications. Furthermore, some mutations are selected depending on the genetic background due to the interactions among the mutations present (epistasis) whereas other changes have an unknown effect on the enzyme and they could be only selected by random drift. For instance, directed mutagenesis studies have revealed new mutations that have not yet been described in nature, which could be selected under ceftazidime challenge such as M135I and A219V (Pérez-Llarena et al., 2011). These results suggest that the adaptive possibilities of CTX-M enzymes have not yet reached their evolutionary stasis.
In this section we explore if other mutations or adaptive possibilities could be expected in the CTX-M lineage. Two attractive issues are raised in relation to the capacities of CTX-Ms to confer resistance to non-oxyimino-β-lactam antibiotics, including β-lactam-β-lactamase inhibitor combinations (IR-CTX-M) and/or carbapenems (Girlich et al., 2008; Ripoll et al., 2011), as these variants have not yet been found in nature. In laboratory conditions, IR-CTX-M mutants carrying the S130G, K234R, and S237A mutations have been described (Gazouli et al., 1998; Aumeran et al., 2003; Ripoll et al., 2011). The selection of these mutants conferring resistance to β-lactam plus β-lactamase inhibitors showed an antagonistic pleiotropic effect with a simultaneous loss of the hydrolytic activity against cefotaxime and ceftazidime. Nevertheless, the L169S mutation that was recovered after the S130G was selected, slightly increase the hydrolytic efficiency against cefotaxime and ceftazidime, suggesting that mutations at position 169 could have a restoring effect, similar to the selection of A77V mutation after the emergence of P167S or D240G driving ceftazidime-resistance (Novais et al., 2008; Ripoll et al., 2011). Interestingly, a clinical isolate carrying a novel CTX-M variant (CTX-M-93), which differs from CTX-M-27 in a single L169Q mutation, shows a reduced activity against cefotaxime but increased activity against ceftazidime when compared with its progenitor (Djamdjian et al., 2011). This finding confirms that the mutations in 169-position confers increased hydrolytic activity against ceftazidime and this improvement is higher when the S130G mutation is also present. On the other hand, a recent study using a site-directed mutagenesis strategy found that the V103D and V260L mutations also endow an IR-CTX-M phenotype (Pérez-Llarena et al., 2011), again suggesting that new mutants could be selected in the future.
No mutations in blaCTX-M gene have been involved in resistance to carbapenems. However, clinical carbapenem-resistant strains carrying CTX-M β-lactamases have been isolated. Two types of mutants are related to this carbapenem-resistant phenotype: loss of porins and co-existence with other β-lactamases (Gülmez et al., 2008). The loss of OmpK35 and OmpK36 porins is the most frequent mechanism of carbapenem-resistant associated with CTX-M producing K. pneumoniae (Leavitt et al., 2009) and its functional restoration significantly decreased the MICs of all carbapenems (Doumith et al., 2009). In vivo resistance development to carbapenem has been previously described in two K. pneumoniae strains producing CTX-M-1 due to the lack of OmpK36 expression (Mena et al., 2006). However, the loss of this porin reduced in vitro fitness and attenuated virulence in a murine model (García-Sureda et al., 2011). Moreover, detection of carbapenem-resistant E. coli clinical strains carrying CTX-M due to reduced expression of OmpF and OmpC porins, homologous of OmpK35 and OmpF36, are increasingly recognized (Girlich et al., 2009; Bennett et al., 2010). These results illustrated that porin deficiency is more efficient than potential CTX-M mutation for the increase of carbapenem MICs.
On the other hand, the high worldwide distribution of CTX-M enzymes is giving rise to the co-existence of two or more β-lactamases in the same strain. This possibility is becoming a common bacterial strategy to enhance antibiotic resistance. CTX-Ms have been described associated with VIM-1 (Miró et al., 2010), KPC-2 (Chen et al., 2011), or OXA-48 carbapenemases (Cuzon et al., 2011; Pitart et al., 2011). In fact, the clone belonging to ST395 coproducing CTX-M-15 and OXA-48 has been demonstrated widespread throughout Europe (Potron et al., 2011). An extreme description was the co-existence of KPC-2, VIM-19, CMY-2, and CTX-M-15 in a K. pneumoniae strain isolated in Greece (Pournaras et al., 2010). Moreover, the recently described NDM-1 carbapenemase is normally present in isolates also expressing the CTX-M-15 ESBL and also OXA-type enzymes (Poirel et al., 2011a; Solé et al., 2011). This supports the concept of the genetic capitalism of the multi-drug resistant bacteria where the more resistant bacteria have the higher opportunity to become more resistant (Baquero et al., 2003; Cantón et al., 2003; Baquero, 2004).
Genetic Environments of blaCTX-M Genes Participating in the Mobilization, Spread, and Maintenance of These Genes
A linkage of different blaCTX-M genes with specific surrounding genetic elements, including IS, integrons, and transposons has been described. These genetic backbones are integrated in more complex structures such as different plasmid replicon types as well as specific clones defining a complex hierarchical organization with the possibility of interchanging different modules (Cantón and Coque, 2006; Carattoli, 2011; Woodford et al., 2011). Figure 2 shows hierarchical complexity of blaCTX-M genes within different genetic structures and bacterial clones participating in the mobilization, spread, and maintenance of these genes. As previously described, blaCTX-M genes have been hypothetically mobilized from Kluyvera spp. by IS and to a lesser extent by bacteriophages. The IS also participate in the over-expression of these genes and some of them (i.e., ISCR1) are adjacent to integron structures that are also integrated in transposition units. These supra-structures are often incorporated within conjugative plasmids that might be present in successful clones (or the so-called high-risk clones). Moreover, they can operate as evolutionary units and might individually act as units of selection (Baquero, 2004; Cantón and Ruiz-Garbajosa, 2011).
Some of the genetic elements and plasmids harboring blaCTX-M genes also harbor other resistance genes, including those encoding AmpC β-lactamases (plasmid blaAmpC) and carbapenemases, plasmid-mediated quinolone resistance (PMQR) genes (i.e., qnr genes), or methylases affecting aminoglycosides. All these genes might also give advantage to blaCTX-M for maintenance due to co-selection processes.
Insertion sequences and phage-related sequences involved in the mobilization of blaCTX-M genes
There are a huge number of scientific articles describing new or unique genetic rearrangement associated with blaCTX-M genes but further analysis has revealed that they are only variations of a few different genetic structures (Lartigue et al., 2004; Eckert et al., 2006; Dhanji et al., 2011b; Toleman and Walsh, 2011). The experimental mobilization of blaCTX-Ms genes have been demonstrated by IS located upstream such as ISEcp1 (Lartigue et al., 2006). Moreover, the effect of Pout promoter of these IS increasing the blaCTX-Ms expression suggests that the IS located upstream of these genes have a role in the selection and dissemination of CTX-Ms. Therefore, we have mainly considered in this part of the review the genetic upstream sequences of the blaCTX-M genes. The information of downstream sequences of blaCTX-M genes is less available and less knowledge has been accumulated.
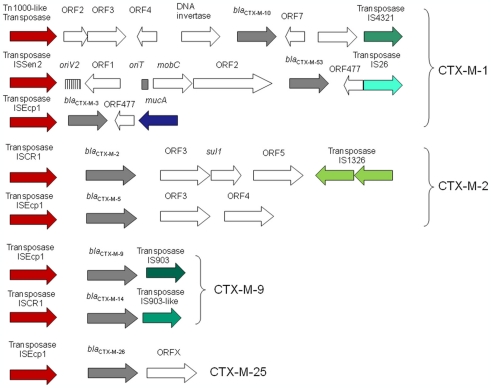
Different IS have been identified upstream of the blaCTX-Ms genes, including ISEcp1, ISCR1, IS10, and IS26. Although other IS elements have also been described upstream, they were the result of subsequent integration events. For instance, IS1 and IS10, and even ISCR1 and IS26, have been described interrupting ISEcp1 (Eckert et al., 2006; Bae et al., 2008). ISEcp1 is the most widely found IS upstream of different blaCTX-M. Although it was first recorded adjacent to the blaCTX-M-15 in 1999, it has been found associated with all clusters of these β-lactamases except CTX-M-8. Moreover, ISCR1 has been related to several members of blaCTX-M-2 and blaCTX-M-9 clusters and IS10 with blaCTX-M-8. The number of new genetic environments upstream of blaCTX-M must be related to the number of mobilization events but unfortunately in many blaCTX-M cases the upstream sequence is unknown (Barlow et al., 2008). Nowadays, we have been able to identify at least nine mobilization events (Figure 4): three in CTX-M-1 cluster (blaCTX-M-10, blaCTX-M-53, and other known blaCTX-M genes included in this cluster), two in CTX-M-2 cluster (blaCTX-M-2 and blaCTX-M-5), two in CTX-M-9 cluster (blaCTX-M-9 and blaCTX-M-14), and one each in CTX-M-8 and CTX-M-25 (blaCTX-M-8 and blaCTX-M-25, respectively).
Figure 4.
Schematic representations of genetic environments surrounding blaCTX-M genes corresponding to suspected cases of mobilization events within different CTX-M groups (CTX-M-1, CTX-M-2, CTX-M-9, and CTX-M-25).
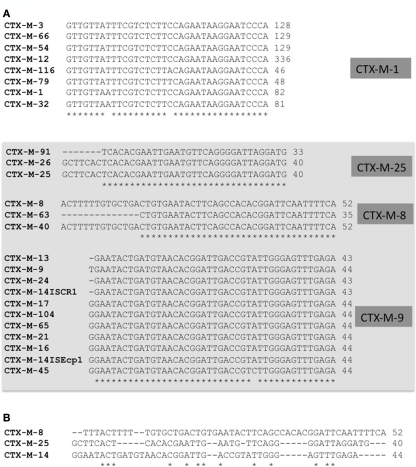
On the other hand, spacer sequences between ISEcp1 and blaCTX-M genes have been well studied. The genetic distance between IS and blaCTX-M gene is related to cephalosporin MIC values (Ma et al., 2011). These distances ranged between 48 and 127 bp in blaCTX-M-1 gene cluster, between 34 and 42 bp in blaCTX-M-9 gene cluster (but more than 300 bp if ISCR1 is upstream of blaCTX-M) and around 40–52 in blaCTX-M-25 and blaCTX-M-8 gene clusters respectively. When the homology among spacer sequences of blaCTX-M-1 gene cluster was analyzed, all but blaCTX-M-10 and blaCTX-M-53 maintained a common region (known as V and W sequences). This suggests that these blaCTX-M genes belonging to CTX-M-1 cluster could derive from a single transposition event. A similar conclusion can be obtained when considering ISEcp1 and blaCTX-M genes belonging to blaCTX-M-9 (Y sequence), blaCTX-M-8 and blaCTX-M-25 (Figure 5). In all these clusters, the ancestral source was K. georgiana. However the lack of homology among spacer sequences of these blaCTX-M gene clusters suggests a different source in each case. These results reveal that non-described Kluyvera species (or another organism source) could be the origin of clusters ascribed to K. georgiana (see Figure 3 about the phylogenetic tree of 16S rDNA of Kluyvera spp. where more species of Kluyvera can be suggested according to bootstrap values).
Figure 5.
Alignment of upstream sequences in different blaCTX-M gene clusters. Different blaCTX-M genes belonging to the same cluster share an identical DNA sequence in upstream region, suggesting the same origin (A). Nevertheless, the loss of alignment among different upstream sequences from different blaCTX-M genes belonging to different clusters (included in the box squared) but theoretically derived from the same ancestral source in Kluyvera georgiana suggesting that these clusters had different sources (B).
Finally, blaCTX-M-10 gene has been associated with phage-related sequences, which underlines bacteriophages involvement in the potential mobilization of this gene (Oliver et al., 2001). Although the occurrence of this event has not been frequently demonstrated, the importance of bacteriophages in the diffusion of bla genes was evaluated in sewage, showing that these sequences may contribute to the spread of some β-lactamases (Muniesa et al., 2004; Colomer-Lluch et al., 2011). Part of these sequences, also known as “phage-like elements,” could have facilitated mobilization of resistance genes into the chromosome (Wozniak and Waldor, 2010; Guglielmini et al., 2011). With the exception of the previous description of the CTX-M-10 enzyme (Oliver et al., 2001), these structures have not been associated with other CTX-Ms despite the fact that these enzymes have also been observed integrated into the chromosome (Coelho et al., 2010; Song et al., 2011). Future in silico analysis might reveal a more frequent presence of these phage-like elements associated with bla genes.
Other integrative and mobilization units
Recently, there has been an increasing interest for the so-called “integrative and conjugative elements” (ICEs) in the mobilization of resistant determinants. They are not homogenous structures and include the “conjugative transposons” or the so-called “integrated conjugative plasmids.” These elements are a diverse group of mobile genetic elements found in both Gram-positive and Gram-negative bacteria. They primarily reside in the chromosome, but retain the ability to excise from and re-integrate into a new chromosome like bacteriophages and to transfer between bacterial cells by conjugation like plasmids. It has been recently stressed that they are closely linked to some of the more powerful resistance mechanisms such as the extended-spectrum-, metallo-, and AmpC type β-lactamases (Toleman and Walsh, 2011).
A characteristic of the ICE elements, as well as complex class 1 integrons and some IS is the inability to consistently recognize one of their own terminal sequences, while they recognize more genetically distant surrogate sequences. This has the effect of mobilizing the DNA sequence found adjacent to their initial location. This process is known as one-ended transposition mechanisms and is exemplified by some IS such as ISCR1 and ISEcp1 as well as Tn3 and Tn21 derivatives, including Tn5090-like transposon (also called Tn402) that in turn harbor the class 1 integrons, ISCR1 and certain blaCTX-M genes. A detailed review of the importance of these elements in the emergence and dispersion of resistance traits, including that exerted by blaCTX-M genes has been recently reviewed (Toleman and Walsh, 2011).
A novel structure named IMU that resides in a plasmid 7-kb IncQ-type plasmid has recently been associated with mobilization of bla genes (Poirel et al., 2009). The IMU structure resembles that of miniature inverted transposable elements (MITEs), which have been identified in eukaryotes such as plants (Chen et al., 2008) and also in bacteria (Buisine et al., 2002; Filée et al., 2007). In Enterobacteriaceae, they form a composite like element consisting of 288-bp elements located in opposite orientation able to mobilize defective class 1 integron structures carrying bla genes. Although they have not yet been associated with mobilization of blaCTX-M genes, they might participate in the future in the mobilization of these genes.
Plasmids and spread of blaCTX-M genes
The importance of plasmids in the spread of blaCTX-M genes is exemplified with certain CTX-M β-lactamases and has been stressed in several publications (Carattoli, 2009). The case of CTX-M-15 enzyme is of particular interest. It has been shown that the blaCTX-M-15 gene is mainly associated with incompatibility group FII plasmids (Coque et al., 2008b). They are a paradigm of narrow host-rage plasmids characterized by a low-copy number in the bacteria. They are mainly found in Enterobacteriaceae and have recently been termed as “epidemic resistance plasmids” due to their propensity to acquire resistance genes and transfer among bacteria (Carattoli, 2011). The FII plasmids are not a homogeneous group and might have evolved by recombinatorial events among different plasmids. It has been shown that within these plasmids different replicon types are present that might facilitate fast evolution and plasmid diversification. Interestingly, initial studies on IncFII plasmids suggest that they were widely distributed within Enterobacteriaceae before antimicrobial use provided that they were well adapted to these organisms (Datta et al., 1980). This fact has undoubtedly facilitated persistence and spread of resistant determinants, including blaCTX-M genes after this acquisition. Moreover, incorporation of genetic determinants mediating non β-lactam resistance mechanisms would have also contributed to the maintenance of these plasmids within the bacteria and hence the blaCTX-M genes under co-selection process (Novais et al., 2007; Woodford et al., 2009).
On the other hand, broad host-range replicon plasmids such as IncN, IncI1, and IncL/M have also been involved in the dissemination and spread of CTX-M enzymes. Some of these plasmids, such as IncN, might have their reservoir in animals as they have been isolated in Salmonella spp. and E. coli from food producing animals (Carattoli, 2011). They often carry qnr and aac(6′)-Ib-cr genes determining low level fluoroquinolone resistance (Shen et al., 2008; Bado et al., 2010). More recently, these plasmids have been able to acquire genes coding the emerging carbapenemase such as blaNDM-1 (Poirel et al., 2011b) and have been identified as epidemic plasmids in countries with a high prevalence of carbapenem resistance (Psichogiou et al., 2008). Other plasmids, such as IncL/M group are also able to carry blaCTX-M genes (i.e., blaCTX-M-3 and blaCTX-M-15) as well as those producing methylase encoding genes involved in aminoglycoside resistance (i.e., armA; Sabtcheva et al., 2008; Naas et al., 2011).
Other plasmid types that have been preferentially associated with blaCTX-M genes are IncK plasmids with blaCTX-M-14 in southern Europe (Valverde et al., 2009; Cottell et al., 2011) and IncHI2 plasmids with blaCTX-M-2 and blaCTX-M-9 in other geographic areas (García-Fernández et al., 2007). Nevertheless, these plasmids have also been responsible for the dispersion of other resistance genes such as IncK and blaCMY genes (Baudry et al., 2009) or IncHI2 and blaVIM and qnr genes (García-Fernández and Carattoli, 2010; Oteo et al., 2010b). These results denote ability of these plasmids for recruitment resistance genes, hence facilitating the increasing prevalence and persistence of the resistance traits of the bacteria. Furthermore, it is expected that persistence over time also gives opportunity to the bacteria to generate gene variation and an increasing the number of blaCTX-M variants in the coming years as has occurred with OXA-type β-lactamases. Some of these variants have been obtained in vitro under antibiotic selective pressure but have not yet been found in clinical isolates (Novais et al., 2008, 2010; Ripoll et al., 2011).
Clonal Dispersion and Co-Selection Processes: Involvement of Multi-Drug Resistant and Virulent High-Risk Clones
One of the most interesting issues in the dispersion of CTX-M enzymes is the participation of specific clones, particularly those from E. coli and K. pneumoniae. Recent approaches based on MLST typing have demonstrated that despite a high diversity among CTX-M producers, a few clones or sequence types (ST) grouped in clonal complexes (CC) have been repeatedly found linked to CTX-M enzymes. This suggests that they are involved in the dissemination of these enzymes and that the adaptive success of some CTX-M enzymes could also depend on specific ST or CC where they are frequently present. These clones have been named as high-risk clones (Woodford et al., 2011). The case of worldwide CTX-M-15 producing E. coli isolates exemplifies this issue as most of these isolates belong to the internationally spread ST131 clone (Coque et al., 2008b; Woodford et al., 2011). ST131 is the predicted founder of a clonal cluster that comprises 13 single locus variants (SLVs) and three double-locus variants (DLVs). It is related to serotype O25:H4 as well as to phylogenetic group B2, which grouped virulent extraintestinal isolates (Coque et al., 2008b; Peirano and Pitout, 2010). In addition, isolates belonging to this clone or CC frequently carry IncFII plasmids suggesting a common co-evolutionary process.
Different population structure studies of E. coli demonstrated that ST131 clone is also not only associated with CTX-M enzymes belonging to group 1 (CTX-M-1, CTX-M-3, and CTX-M-10) but also with others from distinct groups, such as CTX-M-9 or CTX-M-14 (Mora et al., 2010; Cao et al., 2011). This spread is present among human and animal isolates. In addition, very recent studies also showed that ST131 is over represented within non-ESBL producing isolates (Blanco et al., 2011) and that this clone can recruit emerging β-lactamases such as NDM-1 (Peirano et al., 2011). Multiresistance might have facilitated globalization of this clone. Moreover, at individual level the presence of specific virulence traits facilitating host adherence and receptor epithelial binding also contribute to persistence (Peirano and Pitout, 2010).
Other lineages of E. coli that have also contributed to the spread of CTX-M enzymes are ST38, ST393, and ST405 clones and their corresponding single- and DLVs (Coque et al., 2008a; Blanco et al., 2011). These lineages belong to serotype D, which represents also virulent extraintestinal strains (Picard et al., 1999). ST38 and ST393 seem to have been more preferential for the CTX-M-9 and CTX-M-14 enzymes whereas ST405 have been more preferentially with CTX-15 enzyme. Again these clones might also be overrepresented within non-ESBL producing isolates increasing the opportunity to recruit the corresponding blaCTX-M genes. These clones also express other emerging β-lactamases such as OXA-48 and NDM-1 as illustrated with ST38 (Poirel et al., 2011a; Yamamoto et al., 2011).
Many isolates belonging to these E. coli clones are fluoroquinolone-resistant uropathogenic isolates that have their natural environment in the intestinal microbiota. Fluoroquinolones might favor co-selection phenomena that could enrich the intestinal microbiota with these isolates. It has even been speculated that fluoroquinolone resistance either due to plasmid-mediated genes, including qnr variants and acc(6′)-Ib-cr, or to topoisomerase mutations preceded the acquisition of blaCTX-M genes (Jones et al., 2008; Naseer and Sundsfjord, 2011). In addition to this resistant trait, high-risk clones expressing CTX-M enzymes are also more favorable for acquiring other resistance mechanisms, such as those affecting aminoglycosides, trimethoprim–sulfamethoxazole and fosfomycin. Aminoglycoside resistance is related both with aminoglycoside modifying enzymes and methylases (Mushtaq et al., 2011). Fosfomycin resistance involving different resistance mechanisms has been more recently describe in isolates from the community in countries with increasing use of this antibiotic (Oteo et al., 2009). This again illustrates the genetic capitalism of the resistant bacterias (Baquero et al., 2003; Cantón et al., 2003; Baquero, 2004).
Together with these points, these CTX-M associated E. coli clones are highly virulent, easily transmitted among family members in colonized or infected patients (Mihaila et al., 2010) and have been found in wild-life, companion and food animals (Mora et al., 2010; Platell et al., 2011).
In K. pneumoniae more attention has been paid to carbapenemases, mainly KPCs, and the involvement of ST258 clone in their dissemination (Woodford et al., 2011). Nevertheless, different publications have studied the implication of different clones in the dissemination of CTX-M enzymes associated with K. pneumoniae. This is for instance the case of CC11, which has been identified widely spread in Asia associated with different CTX-M enzymes, including CTX-M-14 and CTX-M-15 (Ko et al., 2010; Lee et al., 2011). In Hungary this CC and also ST15 and ST147 were found to be responsible for the country-wide dissemination of ciprofloxacin-resistant CTX-M-15 producing K. pneumoniae (Damjanova et al., 2008). Also in the Copenhagen area of Denmark a K. pneumoniae ST16 clone harboring the blaCTX-M-15 and blaSHV-28 genes was recognized as producing hospital outbreaks. (Nielsen et al., 2011). Some of these clones were able to acquire carbapenemases and other resistant traits due to their particular preferential for specific plasmids, including IncFII, IncN and IncL/M (Andrade et al., 2011). This observation exemplifies once more the genetic capitalism concept for which resistant organisms tend to be more resistant (Baquero et al., 2003; Cantón et al., 2003; Baquero, 2004).
Concluding Remarks
Although in recent years more interest has given to carbapenemases than ESBLs, the CTX-M family still deserve the close attention of researchers and constitute a paradigm of the emergence and spread of a resistant mechanism. Previous descriptions of hierarchical structures that include the blaCTX-M genes delineated the importance of these structures for maintenance and spread of these genes. Comparison of blaCTX-M sequences with sequence databases hypothetically defined that blaCTX-M genes have their ancestors in different Kluyvera spp. Nevertheless, in silico analyses also suggest that blaCTX-M genes might also have circulated among different Kluyvera species and that the incorporation to these organisms might well have occurred after these species diverged. In Enterobacteriaceae, the great adaptive success of blaCTX-M genes has been associated with few surrounding genetic structures, few plasmids and few bacterial clones. Successful associations of these units and co-existence of blaCTX-M genes with other resistance determinants might have contributed to the extraordinary spread of the CTX-M enzymes deserving an uncontrolled pandemic scenario. Future persistence of blaCTX-M genes within bacterial communities will be assured by co-selection processes as other resistance genes, including emerging ones such as carbapenemases, are now often present in CTX-M producing organisms. Persistence will also give new opportunities for evolution.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
José María González-Alba is supported by fellow research contracts from the European Commission funded project PAR, HEALTH-2009-241476). The content and the scientific background of the manuscript were obtained in part from the European Commission funded projects (COBRA, LSHM-CT-2003-503335; TROCAR, HEALTH-F3-2008-223031; and PAR, HEALTH-2009-241476), the Spanish Ministry of Science and Innovation through Instituto de Salud Carlos III (research grant FIS-PI-080624) and the CIBERESP (research network in Epidemiology and Public Health, CB06/02/0053), and the Regional Government of Madrid [DeRemicrobiana Network (CAM.S-SAL-0246-2006)]. We also thank Ana Moreno-Bofarull for substantial administrative support and Mary Harper for helping with the manuscript production.
Footnotes
References
- al Naiemi N., Duim B., Bart A. (2006). A CTX-M extended-spectrum β-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J. Med. Microbiol. 55, 1607–1608 10.1099/jmm.0.46704-0 [DOI] [PubMed] [Google Scholar]
- Ambler R. P. (1980). The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289, 321–331 10.1098/rstb.1980.0049 [DOI] [PubMed] [Google Scholar]
- Andrade L. N., Curiao T., Ferreira J. C., Longo J. M., Clímaco E. C., Martinez R., Bellissimo-Rodrigues F., Basile-Filho A., Evaristo M. A., Del Peloso P. F., Ribeiro V. B., Barth A. L., Paula M. C., Baquero F., Cantón R., Darini A. L., Coque T. M. (2011). Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob. Agents Chemother. 55, 3579–3583 10.1128/AAC.01783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel Díaz M., Ramón Hernández J., Martínez-Martínez L., Rodríguez-Baño J., Pascual A., Grupo de Estudio de Infección Hospitalaria (GEIH) (2009). Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Spanish hospitals: 2nd multicenter study (GEIH-BLEE project, 2006). Enferm. Infecc. Microbiol. Clin. 27, 503–510 10.1016/j.eimc.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Aumeran C., Chanal C., Labia R., Sirot D., Sirot J., Bonnet R. (2003). Effects of Ser130Gly and Asp240Lys substitutions in extended-spectrum β-lactamase CTX-M-9. Antimicrob. Agents Chemother. 47, 2958–2961 10.1128/AAC.47.9.2958-2961.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bado I., Cordeiro N. F., Robino L., García-Fulgueiras V., Seija V., Bazet C., Gutkind G., Ayala J. A., Vignoli R. (2010). Detection of class 1 and 2 integrons, extended-spectrum β-lactamases and qnr alleles in enterobacterial isolates from the digestive tract of Intensive Care Unit inpatients. Int. J. Antimicrob. Agents 36, 453–458 10.1016/j.ijantimicag.2010.06.042 [DOI] [PubMed] [Google Scholar]
- Bae I. K., Lee Y. H., Jeong H. J., Hong S. G., Lee S. H., Jeong S. H. (2008). A novel bla(CTX-M-14) gene-harboring complex class 1 integron with an In4-like backbone structure from a clinical isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 62, 340–342 10.1016/j.diagmicrobio.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Baquero F. (2004). From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2, 510–518 10.1038/nrmicro909 [DOI] [PubMed] [Google Scholar]
- Baquero F., Coque T. M., Cantón R. (2003). Antibiotics, complexity, and evolution. ASM News 69, 547–552 [Google Scholar]
- Baraniak A., Fiett J., Sulikowska A., Hryniewicz W., Gniadkowski M. (2002). Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46, 151–159 10.1128/AAC.46.1.151-159.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow M., Reik R. A., Jacobs S. D., Medina M., Meyer M. P., Jr., McGowan J. E., Tenover F. C. (2008). High rate of mobilization for blaCTX-Ms. Emerging Infect. Dis. 14, 423–428 10.3201/eid1403.070405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Bernard H., Tancrède C., Labia R. (1992). Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta. 1122, 15–22 10.1016/0167-4838(92)90121-S [DOI] [PubMed] [Google Scholar]
- Baudry P. J., Mataseje L., Zhanel G. G., Hoban D. J., Mulvey M. R. (2009). Characterization of plasmids encoding CMY-2 AmpC beta-lactamases from Escherichia coli in Canadian intensive care units. Diagn. Microbiol. Infect. Dis. 65, 379–383 10.1016/j.diagmicrobio.2009.08.011 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Casellas J. M., Goldberg M., Holley M., Jungwirth R., Mangold P., Röhnisch T., Schweighart S., Wilhelm R. (1992). A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20, 158–163 10.1007/BF01704610 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Grimm H., Schweighart S. (1990). A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18, 294–298 10.1007/BF01644637 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Stemplinger I., Jungwirth R., Ernst S., Casellas J. M. (1996). Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. W., Mende K., Herrera M. L., Yu X., Lewis J. S., II, Wickes B. L., Jorgensen J. H., Murray C. K. (2010). Mechanisms of carbapenem resistance among a collection of Enterobacteriaceae clinical isolates in a Texas city. Diagn. Microbiol. Infect. Dis. 66, 445–448 10.1016/j.diagmicrobio.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Bernard H., Tancrede C., Livrelli V., Morand A., Barthelemy M., Labia R. (1992). A novel plasmid-mediated extended-spectrum beta-lactamase not derived from TEM- or SHV-type enzymes. J. Antimicrob. Chemother. 29, 590–592 10.1093/jac/29.5.590 [DOI] [PubMed] [Google Scholar]
- Blanco J., Mora A., Mamani R., López C., Blanco M., Dahbi G., Herrera A., Blanco J. E., Alonso M. P., García-Garrote F., Chaves F., Orellana M. Á., Martínez-Martínez L., Calvo J., Prats G., Larrosa M. N., González-López J. J., López-Cerero L., Rodríguez-Baño J., Pascual A. (2011). National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J. Antimicrob. Chemother. 66, 2011–2021 10.1093/jac/dkr235 [DOI] [PubMed] [Google Scholar]
- Bonnedahl J., Drobni P., Johansson A., Hernandez J., Melhus A., Stedt J., Olsen B., Drobni M. (2010). Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum β-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65, 1939–1944 10.1093/jac/dkq222 [DOI] [PubMed] [Google Scholar]
- Bonnet R. (2004). Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14 10.1128/AAC.48.1.1-14.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R., Dutour C., Sampaio J. L., Chanal C., Sirot D., Labia R., De Champs C., Sirot J. (2001). Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45, 2269–2275 10.1128/AAC.45.8.2269-2275.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R., Sampaio J. L., Labia R., De Champs C., Sirot D., Chanal C., Sirot J. (2000). A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44, 1936–1942 10.1128/AAC.44.11.3061-3068.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford P. A. (2001). Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisine N., Tang C. M., Chalmers R. (2002). Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522, 52–58 10.1016/S0014-5793(02)02882-X [DOI] [PubMed] [Google Scholar]
- Bush K. (2010). Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13, 558–564 10.1016/j.mib.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Bush K., Fisher J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A., Medeiros A. A. (1995). A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantón R. (2008). “Epidemiology and evolution of β-lactamases,” in Evolutionary Biology of Bacterial and Fungal Pathogens, eds Baquero F., Nombela C., Casslel G. H., Gutierrez-Fuentes J. A. (Washington: ASM Press; ), 249–270 [Google Scholar]
- Cantón R., Coque T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Cantón R., Coque T. M., Baquero F. (2003). Multi-resistant Gram-negative bacilli: from epidemics to endemics. Curr. Opin. Infect. Dis. 16, 315–325 10.1097/00001432-200308000-00003 [DOI] [PubMed] [Google Scholar]
- Cantón R., Novais A., Valverde A., Machado E., Peixe L., Baquero F., Coque T. M. (2008). Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14, 144–153 10.1111/j.1469-0691.2007.01850.x [DOI] [PubMed] [Google Scholar]
- Cantón R., Oliver A., Coque T. M., Varela Mdel C., Pérez-Díaz J. C., Baquero F. (2002). Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40, 1237–1243 10.1128/JCM.40.4.1237-1243.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantón R., Ruiz-Garbajosa P. (2011). Co-resistance: an opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 11, 477–485 10.1016/j.coph.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Cao X., Cavaco L. M., Lv Y., Li Y., Zheng B., Wang P., Hasman H., Liu Y., Aarestrup F. M. (2011). Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J. Clin. Microbiol. 49, 2496–2501 10.1128/JCM.02503-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2008). Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14, 117–123 10.1111/j.1469-0691.2007.01851.x [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2011). Plasmids in Gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301, 654–658 10.1016/j.ijmm.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Castanheira M., Mendes R. E., Rhomberg P. R., Jones R. N. (2008). Rapid emergence of blaCTX-M among Enterobacteriaceae in U.S. Medical Centers: molecular evaluation from the MYSTIC Program (2007). Microb. Drug Resist. 14, 211–216 10.1089/mdr.2008.0827 [DOI] [PubMed] [Google Scholar]
- Celenza G., Pellegrini C., Caccamo M., Segatore B., Amicosante G., Perilli M. (2006). Spread of bla(CTX-M-type) and bla(PER-2) β-lactamase genes in clinical isolates from Bolivian hospitals. J. Antimicrob. Chemother. 57, 975–978 10.1093/jac/dkl055 [DOI] [PubMed] [Google Scholar]
- Cergole-Novella M. C., Guth B. E., Castanheira M., Carmo M. S., Pignatari A. C. (2010). First description of bla(CTX-M-14)- and bla(CTX-M-15)-producing Escherichia coli isolates in Brazil. Microb. Drug Resist. 16, 177–184 10.1089/mdr.2010.0008 [DOI] [PubMed] [Google Scholar]
- Chanawong A., M’Zali F. H., Heritage J., Xiong J. H., Hawkey P. M. (2002). Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People’s Republic of China. Antimicrob. Agents Chemother. 46, 630–637 10.1128/AAC.46.3.630-637.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shu W., Chang X., Chen J. A., Guo Y., Tan Y. (2010). The profile of antibioticsresistance and integrons of extended-spectrum β-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158, 2459–2464 10.1016/j.envpol.2009.10.042 [DOI] [PubMed] [Google Scholar]
- Chen S., Hu F., Xu X., Liu Y., Wu W., Zhu D., Wang H. (2011). High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum β-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob. Agents Chemother. 55, 2493–2494 10.1128/AAC.00317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou F., Li G., Xu Y. (2008). A recently active miniature inverted-repeat transposable element, Chunjie, inserted into an operon without disturbing the operon structure in Geobacter uraniireducens Rf4. Genetics 179, 2291–2297 10.1534/genetics.108.089995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G., Pazhani G. P., Nair G. B., Ghosh A., Ramamurthy T. (2011). Transferable plasmid-mediated quinolone resistance in association with extended-spectrum β-lactamases and fluoroquinolone-acetylating aminoglycoside-6′-N-acetyltransferase in clinical isolates of Vibrio fluvialis. Int. J. Antimicrob. Agents 38, 169–173 [DOI] [PubMed] [Google Scholar]
- Coelho A., González-López J. J., Miró E., Alonso-Tarrés C., Mirelis B., Larrosa M. N., Bartolomé R. M., Andreu A., Navarro F., Jonson J. R., Prats G. (2010). Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36, 73–78 10.1016/j.ijantimicag.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Colomer-Lluch M., Jofre J., Muniesa M. (2011). Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 6, e17549. 10.1371/journal.pone.0017549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque T. M., Baquero F., Cantón R. (2008a). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13, 19044. [PubMed] [Google Scholar]
- Coque T. M., Novais A., Carattoli A., Poirel L., Pitout J., Peixe L., Baquero F., Cantón R., Nordmann P. (2008b). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerging Infect. Dis. 14, 195–200 10.3201/eid1402.070350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottell J. L., Webber M. A., Coldham N. G., Taylor D. L., Cerdeño-Tárraga A. M., Hauser H., Thomson N. R., Woodward M. J., Piddock L. J. (2011). Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerging Infect. Dis. 17, 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Ouanich J., Gondret R., Naas T., Nordmann P. (2011). Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 55, 2420–2423 10.1128/AAC.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanova I., Tóth A., Pászti J., Hajbel-Vékony G., Jakab M., Berta J., Milch H., Füzi M. (2008). Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005 – the new “MRSAs?” J. Antimicrob. Chemother. 62, 978–985 10.1093/jac/dkn287 [DOI] [PubMed] [Google Scholar]
- Datta N., Dacey S., Hughes V., Knight S., Richards H., Williams G., Casewell M., Shannon K. P. (1980). Distribution of genes for trimethoprim and gentamicin resistance in bacteria and their plasmids in a general hospital. J. Gen. Microbiol. 118, 495–508 [DOI] [PubMed] [Google Scholar]
- Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Champs C., Sirot D., Chanal C., Bonnet R., Sirot J. (2000). A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. The French Study Group. Antimicrob. Agents Chemother. 44, 3177–3179 10.1128/AAC.44.11.3177-3179.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousser J. W., Poirel L., Nordmann P. (2001). Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45, 3595–3598 10.1128/AAC.45.12.3595-3598.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanji H., Murphy N. M., Akhigbe C., Doumith M., Hope R., Livermore D. M., Woodford N. (2011a). Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J. Antimicrob. Chemother. 66, 512–516 10.1093/jac/dkq463 [DOI] [PubMed] [Google Scholar]
- Dhanji H., Patel R., Wall R., Doumith M., Patel B., Hope R., Livermore D. M., Woodford N. (2011b). Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J. Antimicrob. Chemother. 66, 1005–1012 10.1093/jac/dkq463 [DOI] [PubMed] [Google Scholar]
- Dhanji H., Murphy N. M., Doumith M., Durmus S., Lee S. S., Hope R., Woodford N., Livermore D. M. (2010). Cephalosporin resistance mechanisms in Escherichia coli isolated from raw chicken imported into the UK. J. Antimicrob. Chemother. 65, 2534–2537 10.1093/jac/dkq376 [DOI] [PubMed] [Google Scholar]
- Díaz M. A., Hernández-Bello J. R., Rodríguez-Baño J., Martínez-Martínez L., Calvo J., Blanco J., Pascual A., Spanish Group for Nosocomial Infections (GEIH) (2010). Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J. Clin. Microbiol. 48, 2840–2845 10.1128/JCM.02147-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamdjian L., Naas T., Tandé D., Cuzon G., Hanrotel-Saliou C., Nordmann P. (2011). CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity. Antimicrob. Agents Chemother. 55, 1861–1866 10.1128/AAC.01656-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M., Frolkova P., Florek M., Jamborova I., Purgertova M., Kutilova I., Cizek A., Guenther S., Literak I. (2011). CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 66, 2784–2790 10.1093/jac/dkq500 [DOI] [PubMed] [Google Scholar]
- Doucet-Populaire F., Ghnassia J. C., Bonnet R., Sirot J. (2000). First isolation of a CTX-M-3-producing Enterobacter cloacae in France. Antimicrob. Agents Chemother. 44, 3239–3240 10.1128/AAC.44.11.3239-3240.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Ellington M. J., Livermore D. M., Woodford N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63, 659–667 10.1093/jac/dkp029 [DOI] [PubMed] [Google Scholar]
- Dutour C., Bonnet R., Marchandin H., Boyer M., Chanal C., Sirot D., Sirot J. (2002). CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46, 534–537 10.1128/AAC.46.2.534-537.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C., Gautier V., Arlet G. (2006). DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57, 14–23 10.1093/jac/dki398 [DOI] [PubMed] [Google Scholar]
- Farmer J. J., III, Fanning G. R., Huntley-Carter G. P., Holmes B., Hickman F. W., Richard D., Brenner D. J. (1981). Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp nov and Kluyvera cryocrescens sp nov in clinical specimens. J. Clin. Microbiol. 13, 919–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J., Siguier P., Chandler M. (2007). Insertion sequence diversity in Archaea. Microbiol. Mol. Biol. Rev. 71, 121–157 10.1128/MMBR.00031-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández A., Carattoli A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161 10.1093/jac/dkq101 [DOI] [PubMed] [Google Scholar]
- García-Fernández A., Cloeckaert A., Bertini A., Praud K., Doublet B., Weill F. X., Carattoli A. (2007). Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51, 4177–4180 10.1128/AAC.00603-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sureda L., Doménech-Sánchez A., Barbier M., Juan C., Gascó J., Albertí S. (2011). OmpK26, a novel porin associated with carbapenem resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55, 4742–4747 10.1128/AAC.00309-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazouli M., Tzelepi E., Sidorenko S. V., Tzouvelekis L. S. (1998). Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42, 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2008). Do CTX-M β-lactamases hydrolyse ertapenem? J. Antimicrob. Chemother. 62, 1155–1156 10.1093/jac/dkn317 [DOI] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2009). CTX-M expression and selection of ertapenem resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 53, 832–834 10.1128/AAC.01007-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gniadkowski M., Schneider I., Palucha A., Jungwirth R., Mikiewicz B., Bauernfeind A. (1998). Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42, 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Garcés J. L., Saéz D., Almagro M., Fernández-Romero S., Merino F., Campos J., Oteo J. (2011). Osteomyelitis associated to CTX-M-15-producing Aeromonas hydrophila: first description in the literature. Diagn. Microbiol. Infect. Dis. 70, 420–422 10.1016/j.diagmicrobio.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Gonçalves A., Igrejas G., Radhouani H., Estepa V., Alcaide E., Zorrilla I., Serra R., Torres C., Poeta P. (2011). Detection of extended-spectrum β-lactamase-producing Escherichia coli isolates in faecal samples of Iberian Lynx. Lett. Appl. Microbiol. [Epub ahead of print]. 10.1111/j.1472-765X.2011.03173.x [DOI] [PubMed] [Google Scholar]
- Guglielmini J., Quintais L., Garcillán-Barcia M. P., de la Cruz F., Rocha E. P. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7, e1002222. 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gülmez D., Woodford N., Palepou M. F., Mushtaq S., Metan G., Yakupogullari Y., Kocagoz S., Uzun O., Hascelik G., Livermore D. M. (2008). Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31, 523–526 10.1016/j.ijantimicag.2008.01.017 [DOI] [PubMed] [Google Scholar]
- Hawkey P. M., Jones A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64, i3–i10 10.1093/jac/dkp256 [DOI] [PubMed] [Google Scholar]
- Hiroi M., Yamazaki F., Harada T., Takahashi N., Iida N., Noda Y., Yagi M., Nishio T., Kanda T., Kawamori F., Sugiyama K., Masuda T., Hara-Kudo Y., Ohashi N. (2012). Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 74, 189–195 10.1292/jvms.11-0372 [DOI] [PubMed] [Google Scholar]
- Humeniuk C., Arlet G., Gautier V., Grimont P., Labia R., Philippon A. (2002). β-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049 10.1128/AAC.46.9.3045-3049.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Ohno A., Taguchi H., Imajo S., Ishiguro M., Matsuzawa H. (1995). Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Muñoz-Price L. S. (2005). The new β-lactamases. N. Engl. J. Med. 352, 380–391 10.1056/NEJMra041359 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Johnston B., Clabots C., Kuskowski M. A., Castanheira M. (2010). Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51, 286–294 10.1086/656690 [DOI] [PubMed] [Google Scholar]
- Jones G. L., Warren R. E., Skidmore S. J., Davies V. A., Gibreel T., Upton M. (2008). Prevalence and distribution of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli lacking extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 62, 1245–1251 10.1093/jac/dkn406 [DOI] [PubMed] [Google Scholar]
- Karim A., Poirel L., Nagarajan S., Nordmann P. (2001). Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201, 237–241 10.1016/S0378-1097(01)00276-2 [DOI] [PubMed] [Google Scholar]
- Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. (1983). Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11, 315–317 10.1007/BF01641098 [DOI] [PubMed] [Google Scholar]
- Ko K. S., Lee J. Y., Baek J. Y., Suh J. Y., Lee M. Y., Choi J. Y., Yeom J. S., Kim Y. S., Jung S. I., Shin S. Y., Heo S. T., Kwon K. T., Son J. S., Kim S. W., Chang H. H., Ki H. K., Chung D. R., Peck K. R., Song J. H. (2010). Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 59, 822–828 10.1099/jmm.0.018119-0 [DOI] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., Chaudhary U., Doumith M., Giske C. G., Irfan S., Krishnan P., Kumar A. V., Maharjan S., Mushtaq S., Noorie T., Paterson D. L., Pearson A., Perry C., Pike R., Rao B., Ray U., Sarma J. B., Sharma M., Sheridan E., Thirunarayan M. A., Turton J., Upadhyay S., Warner M., Welfare W., Livermore D. M., Woodford N. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue M. F., Poirel L., Aubert D., Nordmann P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50, 1282–1286 10.1128/AAC.50.4.1282-1286.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue M. F., Poirel L., Nordmann P. (2004). Diversity of genetic environment of bla(CTX-M) genes. FEMS Microbiol. Lett. 234, 201–207 10.1111/j.1574-6968.2004.tb09534.x [DOI] [PubMed] [Google Scholar]
- Lau S. H., Kaufmann M. E., Livermore D. M., Woodford N., Willshaw G. A., Cheasty T., Stamper K., Reddy S., Cheesbrough J., Bolton F. J., Fox A. J., Upton M. (2008). UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J. Antimicrob. Chemother. 62, 1241–1244 10.1093/jac/dkn380 [DOI] [PubMed] [Google Scholar]
- Leavitt A., Chmelnitsky I., Colodner R., Ofek I., Carmeli Y., Navon-Venezia S. (2009). Ertapenem resistance among extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 47, 969–974 10.1128/JCM.00651-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Y., Ko K. S., Kang C. I., Chung D. R., Peck K. R., Song J. H. (2011). High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int. J. Antimicrob. Agents 38, 160–163 10.1016/j.ijantimicag.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Lewis J. S., II, Herrera M., Wickes B., Patterson J. E., Jorgensen J. H. (2007). First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob. Agents Chemother. 51, 4015–4021 10.1128/AAC.01449-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I., Dolejska M., Radimersky T., Klimes J., Friedman M., Aarestrup F. M., Hasman H., Cizek A. (2010). Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum β-lactamases in wild boars. J. Appl. Microbiol. 108, 1702–1711 10.1111/j.1365-2672.2009.04572.x [DOI] [PubMed] [Google Scholar]
- Livermore D. M. (1995). β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8, 557–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Cantón R., Gniadkowski M., Nordmann P., Rossolini G. M., Arlet G., Ayala J., Coque T. M., Kern-Zdanowicz I., Luzzaro F., Poirel L., Woodford N. (2007). CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165–174 10.1093/jac/dkl483 [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Hawkey P. M. (2005). CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56, 451–454 10.1093/jac/dki291 [DOI] [PubMed] [Google Scholar]
- Ma L., Chang F. Y., Fung C. P., Chen T. L., Lin J. C., Lu P. L., Huang L. Y., Chang J. C., Siu L. K. (2005). Variety of TEM-, SHV-, and CTX-M-type β-lactamases present in recent clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae from Taiwan. Microb. Drug Resist. 11, 31–39 10.1089/mdr.2005.11.31 [DOI] [PubMed] [Google Scholar]
- Ma L., Ishii Y., Chang F. Y., Yamaguchi K., Ho M., Siu L. K. (2002). CTX-M-14, a plasmid-mediated CTX-M type extended-spectrum β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 46, 1985–1988 10.1128/AAC.46.9.3101-3103.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Ishii Y., Ishiguro M., Matsuzawa H., Yamaguchi K. (1998). Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42, 1181–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Siu L. K., Lu P. L. (2011). Effect of spacer sequences between blaCTX-M and ISEcp1 on blaCTX-M expression. J. Med. Microbiol. 60, 1787–1792 10.1099/jmm.0.033910-0 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Ikeda F., Kamimura T., Yolota Y., Mine Y. (1988). Novel plasmid-mediated β-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32, 1243–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Kitazume H., Yamada M., Ishiguro Y., Muto T., Izumiya H., Watanabe H. (2007). CTX-M-14 type β-lactamase producing Salmonella enterica serovar enteritidis isolated from imported chicken meat. Jpn. J. Infect. Dis. 60, 236–238 [PubMed] [Google Scholar]
- Mena A., Plasencia V., García L., Hidalgo O., Ayestarán J. I., Alberti S., Borrell N., Pérez J. L., Oliver A. (2006). Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44, 2831–2837 10.1128/JCM.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaila L., Wyplosz B., Clermont O., Garry L., Hipeaux M. C., Vittecoq D., Dussaix E., Denamur E., Branger C. (2010). Probable intrafamily transmission of a highly virulent CTXM-3-producing Escherichia coli belonging to the emerging phylogenetic subgroup D2 O102-ST405 clone. J. Antimicrob. Chemother. 6, 1537–1539 10.1093/jac/dkq155 [DOI] [PubMed] [Google Scholar]
- Minarini L. A., Poirel L., Trevisani N. A., Darini A. L., Nordmann P. (2009). Predominance of CTX-M-type extended-spectrum β-lactamase genes among enterobacterial isolates from outpatients in Brazil. Diagn. Microbiol. Infect. Dis. 65, 202–206 10.1016/j.diagmicrobio.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Miró E., Segura C., Navarro F., Sorlí L., Coll P., Horcajada J. P., Alvarez-Lerma F., Salvadó M. (2010). Spread of plasmids containing the bla(VIM-1) and bla(CTX-M) genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J. Antimicrob. Chemother. 65, 661–665 10.1093/jac/dkp504 [DOI] [PubMed] [Google Scholar]
- Mora A., Herrera A., Mamani R., López C., Alonso M. P., Blanco J. E., Blanco M., Dahbi G., García-Garrote F., Pita J. M., Coira A., Bernárdez M. I., Blanco J. (2010). Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl. Environ. Microbiol. 76, 6991–6997 10.1128/AEM.01112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosini M. I., García-Castillo M., Coque T. M., Valverde A., Novais A., Loza E., Baquero F., Cantón R. (2006). Antibiotic coresistance in extended-spectrum-β-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob. Agents Chemother. 50, 2695–2699 10.1128/AAC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosini M. I., Valverde A., García-Castillo M., Nordmann P., Cantón R. (2010). Persistent isolation of Salmonella concord harbouring CTX-M-15, SHV-12 and QnrA1 in an asymptomatic adopted Ethiopian child in Spain also colonized with CTX-M-14- and QnrB-producing Enterobacteriaceae. J. Antimicrob. Chemother. 65, 1545–1546 10.1093/jac/dkq168 [DOI] [PubMed] [Google Scholar]
- Munday C. J., Boyd D. A., Brenwald N., Miller M., Andrews J. M., Wise R., Mulvey M. R., Hawkey P. M. (2004). Molecular and kinetic comparison of the novel extended-spectrum β-lactamases CTX-M-25 and CTX-M-26. Antimicrob. Agents Chemother. 48, 4829–4834 10.1128/AAC.48.12.4829-4834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M., García A., Miró E., Mirelis B., Prats G., Jofre J., Navarro F. (2004). Bacteriophages and diffusion of beta-lactamase genes. Emerging Infect. Dis. 10, 1134–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq S., Irfan S., Sarma J. B., Doumith M., Pike R., Pitout J., Livermore D. M., Woodford N. (2011). Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66, 2002–2005 10.1093/jac/dkr226 [DOI] [PubMed] [Google Scholar]
- Mushtaq S., Woodford N., Potz N., Livermore D. M. (2003). Detection of CTX-M-15 extended-spectrum β-lactamase in the United Kingdom. J. Antimicrob. Chemother. 52, 528–529 10.1093/jac/dkg353 [DOI] [PubMed] [Google Scholar]
- Naas T., Bentchouala C., Cuzon G., Yaou S., Lezzar A., Smati F., Nordmann P. (2011). Outbreak of Salmonella enterica serotype Infantis producing ArmA 16S RNA methylase and CTX-M-15 extended-spectrum β-lactamase in a neonatology ward in Constantine, Algeria. Int. J. Antimicrob. Agents 38, 135–139 10.1016/j.ijantimicag.2011.04.012 [DOI] [PubMed] [Google Scholar]
- Nagano Y., Nagano N., Wachino J., Ishikawa K., Arakawa Y. (2009). Novel chimeric β-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 β-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob. Agents Chemother. 53, 69–74 10.1128/AAC.00227-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer U., Sundsfjord A. (2011). The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17, 83–97 10.1089/mdr.2010.0132 [DOI] [PubMed] [Google Scholar]
- Nielsen J. B., Skov M. N., Jørgensen R. L., Heltberg O., Hansen D. S., Schønning K. (2011). Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur. J. Clin. Microbiol. Infect. Dis. 30, 773–778 10.1007/s10096-011-1153-x [DOI] [PubMed] [Google Scholar]
- Nordmann P., Lartigue M. F., Poirel L. (2008). β-lactam induction of ISEcp1B-mediated mobilization of the naturally occurring bla(CTX-M) β-lactamase gene of Kluyvera ascorbata. FEMS Microbiol. Lett. 288, 247–249 10.1111/j.1574-6968.2008.01359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A., Cantón R., Coque T. M., Moya A., Baquero F., Galán J. C. (2008). Mutational events in cefotaximase extended-spectrum β-lactamases of the CTX-M-1 cluster involved in ceftazidime resistance. Antimicrob. Agents Chemother. 52, 2377–2382 10.1128/AAC.00999-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A., Cantón R., Moreira R., Peixe L., Baquero F., Coque T. M. (2007). Emergence and dissemination of nterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51, 796–799 10.1128/AAC.01070-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A., Comas I., Baquero F., Cantón R., Coque T. M., Moya A., Gonzalez-Candelas F., Galan J. C. (2010). Evolutionary trajectories of β-lactamase CTX-M-1 cluster enzymes: predicting antibiotic resistance. PLoS Pathog. 6, e1000735. 10.1371/journal.ppat.1000735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Pérez-Díaz J. C., Coque T. M., Baquero F., Cantón R. (2001). Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45, 616–620 10.1128/AAC.45.2.616-620.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A. B., Silverman M., Boyd D. A., McGeer A., Willey B. M., Pong-Porter V., Daneman N., Mulvey M. R. (2005). Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49, 2112–2115 10.1128/AAC.49.5.2112-2115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J., Orden B., Bautista V., Cuevas O., Arroyo M., Martínez-Ruiz R., Pérez-Vázquez M., Alcaraz M., García-Cobos S., Campos J. (2009). CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J. Antimicrob. Chemother. 64, 712–717 10.1093/jac/dkp211 [DOI] [PubMed] [Google Scholar]
- Oteo J., Pérez-Vázquez M., Campos J. (2010a). Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23, 320–326 10.1097/QCO.0b013e3283398dc1 [DOI] [PubMed] [Google Scholar]
- Oteo J., Hernández-Almaraz J. L., Gil-Antón J., Vindel A., Fernández S., Bautista V., Campos J. (2010b). Outbreak of VIM-1-carbapenemase-producing Enterobacter cloacae in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 29, 1144–1146 10.1097/INF.0b013e3181efaa2d [DOI] [PubMed] [Google Scholar]
- Pai H., Choi E. H., Lee H. J., Hong J. Y., Jacoby G. A. (2001). Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39, 3747–3749 10.1128/JCM.39.10.3747-3749.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L., Bartoloni A., Fiorelli C., Mantella A., Di Maggio T., Gamboa H., Gotuzzo E., Kronvall G., Paradisi F., Rossolini G. M. (2007). Rapid dissemination and diversity of CTX-M extended-spectrum β-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrob. Agents Chemother. 51, 2720–2725 10.1128/AAC.01101-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L., Malossi M., Mantella A., Gotuzzo E., Trigoso C., Bartoloni A., Paradisi F., Kronvall G., Rossolini G. M. (2004). Detection of CTX-M-type β-lactamase genes in fecal Escherichia coli isolates from healthy children in Bolivia and Peru. Antimicrob. Agents Chemother. 48, 4556–4561 10.1128/AAC.48.12.4556-4561.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson D. L., Bonomo R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucha A., Mikiewicz B., Hryniewicz W., Gniadkowski M. (1999). Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44, 489–499 10.1093/jac/44.4.489 [DOI] [PubMed] [Google Scholar]
- Peirano G., Costello M., Pitout J. D. (2010). Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int. J. Antimicrob. Agents 36, 19–23 10.1016/S0924-8579(10)70006-6 [DOI] [PubMed] [Google Scholar]
- Peirano G., Pitout J. D. (2010). Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35, 316–321 10.1016/j.ijantimicag.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Peirano G., Schreckenberger P. C., Pitout J. D. (2011). Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob. Agents Chemother. 55, 2986–2988 10.1128/AAC.01763-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Llarena F. J., Kerff F., Abián O., Mallo S., Fernández M. C., Galleni M., Sancho J., Bou G. (2011). Distant and new mutations in CTX-M-1 β-lactamase affect cefotaxime hydrolysis. Antimicrob. Agents Chemother. 55, 4361–4368 10.1128/AAC.00298-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. (1989). Extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 33, 1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picão R. C., Poirel L., Gales A. C., Nordmann P. (2009). Further identification of CTX-M-2 extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 2225–2226 10.1128/AAC.01602-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., Elion J., Denamur E. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67, 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitart C., Solé M., Roca I., Fàbrega A., Vila J., Marco F. (2011). First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob. Agents Chemother. 55, 4398–4401 10.1128/AAC.00329-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Platell J. L., Johnson J. R., Cobbold R. N., Trott D. J. (2011). Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet. Microbiol. 153, 99–108 10.1016/j.vetmic.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Poirel L., Bernabeu S., Fortineau N., Podglajen I., Lawrence C., Nordmann P. (2011a). Emergence of OXA-48-producing Escherichia coli clone ST38 in France. Antimicrob. Agents Chemother. 55, 4937–4938 10.1128/AAC.00413-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Bonnin R. A., Nordmann P. (2011b). Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55, 4224–4229 10.1128/AAC.00413-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Carrër A., Pitout J. D., Nordmann P. (2009). Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53, 2492–2498 10.1128/AAC.00414-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Kämpfer P., Nordmann P. (2002). Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040 10.1128/AAC.46.12.4038-4040.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. (2008). Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 75–81 10.1111/j.1469-0691.2007.01930.x [DOI] [PubMed] [Google Scholar]
- Potron A., Kalpoe J., Poirel L., Nordmann P. (2011). European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin. Microbiol. Infect. 17, E24–E26 10.1111/j.1469-0691.2011.03669.x [DOI] [PubMed] [Google Scholar]
- Pournaras S., Poulou A., Voulgari E., Vrioni G., Kristo I., Tsakris A. (2010). Detection of the new metallo-β-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65, 1604–1607 10.1093/jac/dkq124 [DOI] [PubMed] [Google Scholar]
- Psichogiou M., Tassios P. T., Avlamis A., Stefanou I., Kosmidis C., Platsouka E., Paniara O., Xanthaki A., Toutouza M., Daikos G. L., Tzouvelekis L. S. (2008). Ongoing epidemic of blaVIM-1-positive Klebsiella pneumoniae in Athens, Greece: a prospective survey. J. Antimicrob. Chemother. 61, 59–63 10.1093/jac/dkm443 [DOI] [PubMed] [Google Scholar]
- Ripoll A., Baquero F., Novais A., Rodríguez-Domínguez M. J., Turrientes M. C., Cantón R., Galán J. C. (2011). In vitro selection of variants resistant to β-lactams plus β-lactamase inhibitors in CTX-M β-lactamases: predicting the in vivo scenario? Antimicrob. Agents Chemother. 55, 4530–4536 10.1128/AAC.00178-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez M. M., Power P., Radice M., Vay C., Famiglietti A., Galleni M., Ayala J. A., Gutkind G. (2004). Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 48, 4895–4897 10.1128/AAC.48.12.4895-4897.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez M. M., Power P., Sader H., Galleni M., Gutkind G. (2010). Novel chromosome-encoded CTX-M-78 β-lactamase from a Kluyvera georgiana clinical isolate as a putative origin of CTX-M-25 subgroup. Antimicrob. Agents Chemother. 54, 3070–3071 10.1128/AAC.01615-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Baño J., Navarro M. D. (2008). Extended-spectrum β-lactamases in ambulatory care: a clinical perspective. Clin. Microbiol. Infect. 14, 104–110 10.1111/j.1469-0691.2007.01866.x [DOI] [PubMed] [Google Scholar]
- Rodríguez-Baño J., Pascual A. (2008). Clinical significance of extended-spectrum β-lactamases. Expert Rev. Anti Infect. Ther. 6, 671–683 10.1586/14787210.6.5.671 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalobos H., Bogaerts P., Berhin C., Bauraing C., Deplano A., Montesinos I., de Mendonça R., Jans B., Glupczynski Y. (2011). Trends in production of extended-spectrum β-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J. Antimicrob. Chemother. 66, 37–47 10.1093/jac/dkq388 [DOI] [PubMed] [Google Scholar]
- Rogers B. A., Sidjabat H. E., Paterson D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66, 1–14 10.1093/jac/dkq415 [DOI] [PubMed] [Google Scholar]
- Rossolini G. M., D’Andrea M. M., Mugnaioli C. (2008). The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 33–41 10.1111/j.1469-0691.2007.01867.x [DOI] [PubMed] [Google Scholar]
- Ruiz S. J., Montealegre M. C., Ruiz-Garbajosa P., Correa A., Briceño D. F., Martinez E., Rosso F., Muñoz M., Quinn J. P., Cantón R., Villegas M. V. (2011). First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J. Clin. Microbiol. 49, 1993–1996 10.1128/JCM.00045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaté M., Tarragó R., Navarro F., Miró E., Vergés C., Barbé J., Prats G. (2000). Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44, 1970–1973 10.1128/AAC.44.7.1970-1973.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabtcheva S., Saga T., Kantardjiev T., Ivanova M., Ishii Y., Kaku M. (2008). Nosocomial spread of armA-mediated high-level aminoglycoside resistance in Enterobacteriaceae isolates producing CTX-M-3 beta-lactamase in a cancer hospital in Bulgaria. J. Chemother. 20, 593–599 [DOI] [PubMed] [Google Scholar]
- Saladin M., Cao V. T., Lambert T., Donay J. L., Herrmann J. L., Ould-Hocine Z., Verdet C., Delisle F., Philippon A., Arlet G. (2002). Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209, 161–168 10.1016/S0378-1097(02)00484-6 [DOI] [PubMed] [Google Scholar]
- Sarria J. C., Vidal A. M., Kimbrough R. C., III. (2001). Infections caused by Kluyvera species in humans. Clin. Infect. Dis. 33, 69–74 10.1086/321860 [DOI] [PubMed] [Google Scholar]
- Shaheen B. W., Nayak R., Foley S. L., Kweon O., Deck J., Park M., Rafii F., Boothe D. M. (2011). Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 55, 5666–5575 10.1128/AAC.00656-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Jiang Y., Zhou Z., Zhang J., Yu Y., Li L. (2008). Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 62, 1252–1256 10.1093/jac/dkn397 [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Rodrigues P., Rodrigues T., Gonçalves A., Felgar A. C., Pacheco R., Gonçalves D., Cunha R., Poeta P. (2011). Molecular characterization of vancomycin-resistant enterococci and extended-spectrum β-lactamase-containing Escherichia coli isolates in wild birds from the Azores Archipelago. Avian Pathol. 40, 473–479 10.1080/03079457.2011.599061 [DOI] [PubMed] [Google Scholar]
- Solé M., Pitart C., Roca I., Fàbrega A., Salvador P., Muñoz L., Oliveira I., Gascón J., Marco F., Vila J. (2011). First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55, 4402–4404 10.1128/AAC.00642-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistué A. J., Martín F. A., Petroni A., Faccone D., Galas M., Tolmasky M. E., Zorreguieta A. (2006). Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob. Agents Chemother. 50, 1903–1907 10.1128/AAC.50.5.1903-1907.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Kim J., Bae I. K., Jeong S. H., Seo Y. H., Shin J. H., Jang S. J., Uh Y., Shin J. H., Lee M. K., Lee K. (2011). Chromosome-encoded AmpC and CTX-M extended-spectrum β-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob. Agents Chemother. 55, 1414–1419 10.1128/AAC.00073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova M. N., Pimkin M., Nikulin A. A., Kozyreva V. K., Agapova E. D., Edelstein M. V. (2008). Convergent in vivo and in vitro selection of ceftazidime resistance mutations at position 167 of CTX-M-3 β-lactamase in hypermutable Escherichia coli Straits. Antimicrob. Agents Chemother. 52, 1297–1301 10.1128/AAC.01060-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zeng Z., Chen S., Ma J., He L., Liu Y., Deng Y., Lei T., Zhao J., Liu J. H. (2010). High prevalence of bla(CTX-M) extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 16, 1475–1481 10.1111/j.1469-0691.2010.03127.x [DOI] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Walsh T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70, 296–316 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Walsh T. R. (2011). Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 912–935 10.1111/j.1574-6976.2011.00294.x [DOI] [PubMed] [Google Scholar]
- Urban C., Mariano N., Bradford P. A., Tuckman M., Segal-Maurer S., Wehbeh W., Grenner L., Colon-Urban R., Johnston B., Johnson J. R., Rahal J. J. (2010). Identification of CTX-M β-lactamases in Escherichia coli from hospitalized patients and residents of long-term care facilities. Diagn. Microbiol. Infect. Dis. 66, 402–406 10.1016/j.diagmicrobio.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Valenzuela de Silva E. M., Mantilla Anaya J. R., Reguero Reza M. T., González Mejía E. B., Pulido Manrique I. Y., Darío Llerena I., Velandia D. (2006). Detection of CTX-M-1, CTX-M-15, and CTX-M-2 in clinical isolates of Enterobacteriaceae in Bogota, Colombia. J. Clin. Microbiol. 44, 1919–1920 10.1128/JCM.44.5.1919-1920.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde A., Cantón R., Garcillán-Barcia M. P., Novais A., Galán J. C., Alvarado A., de la Cruz F., Baquero F., Coque T. M. (2009). Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53, 5204–5212 10.1128/AAC.01706-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. E., Ensor V. M., O’Neill P., Butler V., Taylor J., Nye K., Harvey M., Livermore D. M., Woodford N., Hawkey P. M. (2008). Imported chicken meat as a potential source of quinolone-resistant Escherichia coli producing extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 61, 504–508 10.1093/jac/dkm517 [DOI] [PubMed] [Google Scholar]
- Woodford N., Carattoli A., Karisik E., Underwood A., Ellington M. J., Livermore D. M. (2009). Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53, 4472–4482 10.1128/AAC.00688-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Turton J. F., Livermore D. M. (2011). Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755 10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- Woodford N., Ward M. E., Kaufmann M. E., Turton J., Fagan E. J., James D., Johnson A. P., Pike R., Warner M., Cheasty T., Pearson A., Harry S., Leach J. B., Loughrey A., Lowes J. A., Warren R. E., Livermore D. M. (2004). Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54, 735–743 10.1093/jac/dkh424 [DOI] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Takano T., Iwao Y., Hishinuma A. (2011). Emergence of NDM-1-positive capsulated Escherichia coli with high resistance to serum killing in Japan. J. Infect. Chemother. 17, 435–439 10.1007/s10156-011-0232-3 [DOI] [PubMed] [Google Scholar]
- Yan J. J., Ko W. C., Tsai S. H., Wu H. M., Jin Y. T., Wu J. J. (2000). Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38, 4320–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Xu X. H., Li J. B. (2010). Emergence of CTX-M-3, TEM-1 and a new plasmid-mediated MOX-4 AmpC in a multiresistant Aeromonas caviae isolate from a patient with pneumonia. J. Med. Microbiol. 59, 843–847 10.1099/jmm.0.016337-0 [DOI] [PubMed] [Google Scholar]
- Yin J., Cheng J., Sun Z., Ye Y., Gao Y. F., Li J. B., Zhang X. J. (2009). Characterization of two plasmid-encoded cefotaximases found in clinical Escherichia coli isolates: CTX-M-65 and a novel enzyme, CTX-M-87. J. Med. Microbiol. 58, 811–815 10.1099/jmm.0.006007-0 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Winokur P. L., Von Stein D. L., Pfaller M. A., Wang J. H., Jones R. N. (2002). First description of Klebsiella pneumoniae harboring CTX-M β-lactamases (CTX-M-14 and CTX-M-3) in Taiwan. Antimicrob. Agents Chemother. 46, 1098–1100 10.1128/AAC.46.4.1098-1100.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]