Abstract
Previously conducted sequence analysis of Arabidopsis thaliana (ecotype Columbia-0) reported an insertion of 270-kb mtDNA into the pericentric region on the short arm of chromosome 2. DNA fiber-based fluorescence in situ hybridization analyses reveal that the mtDNA insert is 618 ± 42 kb, ≈2.3 times greater than that determined by contig assembly and sequencing analysis. Portions of the mitochondrial genome previously believed to be absent were identified within the insert. Sections of the mtDNA are repeated throughout the insert. The cytological data illustrate that DNA contig assembly by using bacterial artificial chromosomes tends to produce a minimal clone path by skipping over duplicated regions, thereby resulting in sequencing errors. We demonstrate that fiber-fluorescence in situ hybridization is a powerful technique to analyze large repetitive regions in the higher eukaryotic genomes and is a valuable complement to ongoing large genome sequencing projects.
Evidence for mitochondrial-to-nuclear DNA transfer events has been found in all eukaryotic genomes studied in detail (1). Mitochondrial-to-nuclear DNA transfer events in plants are believed to often occur as single gene transfers through an RNA intermediate, as many mtDNA-derived nuclear sequences appear in the form of edited versions of the intact mtDNA sequences (2). The majority of mtDNA transfers tend to be only a few hundred base pairs (3). Some events, however, have been reported to exceed 1,000 bp (4, 5).
The recent sequence analysis of Arabidopsis thaliana (ecotype Columbia-0) chromosome 2 revealed the surprising discovery of a mitochondrial-to-nuclear DNA transfer of nearly the entire mitochondrial genome into the pericentric region on the short arm (6). This insertion was verified by PCR amplification across the nuclear-mitochondrial DNA junctions followed by sequencing of the PCR fragments (6). Sequencing of a bacterial artificial chromosome (BAC) contig, which was believed to cover the entire length of the insertion, indicated a mtDNA insertion size of 270 kb, significantly larger than any previously reported mitochondrial-to-nuclear DNA insertions (3, 5). The present study offers a cytological characterization of this mtDNA integration. Fiber-fluorescence in situ hybridization (fiber-FISH) analysis has revealed significant sequence misrepresentations of the genome organization at this locus due to the repetitive nature of the inserted mtDNA. We demonstrate that fiber-FISH analysis is a valuable and complementary tool for sequencing complex and repeated regions in higher eukaryotic genomes.
Materials and Methods
Materials.
For fiber-FISH analysis, we used a 491-kb Arabidopsis BAC contig, which was used for sequencing of chromosome 2 (6). This contig consists of six BAC clones, including four mtDNA-related clones (T5M2, T17H1, T18C6, and T5E7) and two BACs flanking this mtDNA insert (telomeric-end flanker F9A16 and centromeric-end flanker T18A9). Fig. 1A shows the order and overlap of the six clones. The centromere-proximal end of the mtDNA insert (on BAC T5E7) is ≈92 kb away from the 180-bp centromeric repeats. The published sequence for the region related to clone T18A9 was derived from BAC T12J2. However, because T12J2 contains a block of the 180-bp centromeric repeat that hybridizes to other regions of the genome, we used BAC end sequences to select another flanking clone (T18A9) that lacks this repeat. T18A9, replacing T12J2, was used for fiber-FISH analysis. Detailed insert size and sequence information are available at http://www.tigr.org/tdb/ath1/htmls/index.html. A set of overlapping cosmid clones spanning the entire A. thaliana ecotype C24 mitochondrial genome was provided by Axel Brennicke, Universität Ulm, Germany.
Figure 1.
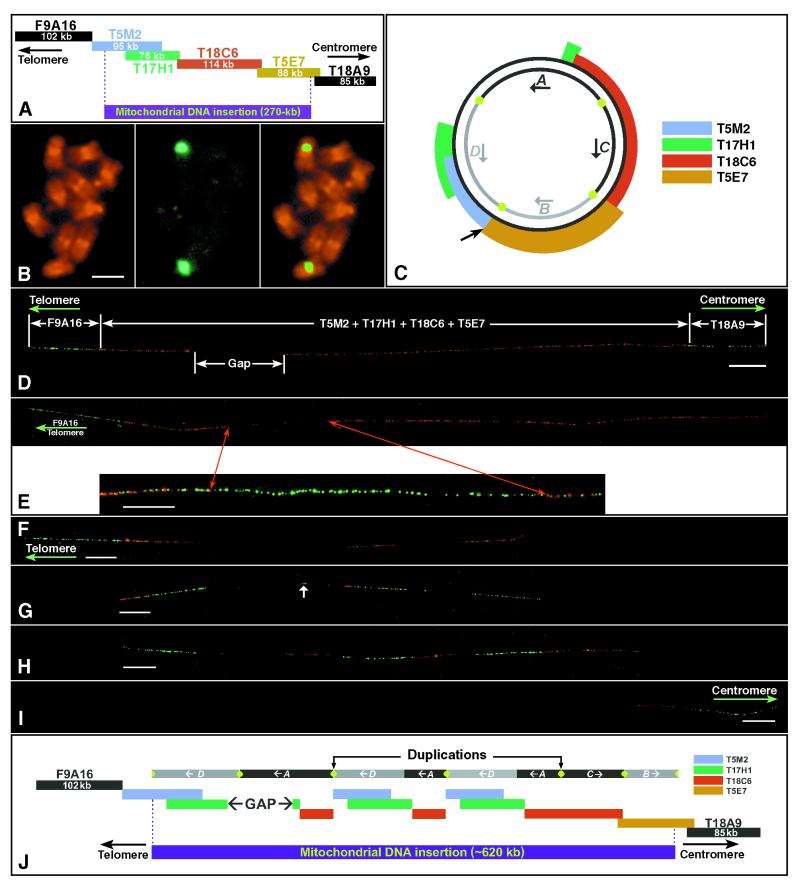
Cytological characterization of the mtDNA-insertion locus on chromosome 2 of A. thaliana. (A) The 491-kb contig of chromosome 2 sequenced by Lin et al. (6) consists of six BAC clones and includes two flanking nuclear BACs and four mtDNA-related BACs (see Results). Kilobase sizes are based on the sequencing data. (B) Hybridization of a mtDNA-related BAC T17H1 to the somatic metaphase chromosomes of A. thaliana (ecotype Columbia-0). (B Left) Propidium iodide-stained somatic metaphase chromosomes. (B Center) FISH signals detected by fluorescein isothiocyanate (green color). (B Right) A merged image of chromosomes and FISH signals. Hybridization signals were detected only in the pericentric regions on one pair of submetacentric chromosomes. (Bar = 2 μm.) (C) Homology of the four mtDNA-related BACs to the published C24 A. thaliana mitochondrial genome (11). Approximately 97 kb of the mtDNA genome is not present in the contig presented in A. The organization of the A, B, C, and D domains was originally proposed by Lin et al. (6) but is incorrectly represented in their figure 7 B and C. The green dots represent the two sets of specific repeats in the mitochondrial genome. The outside arrow points to the possible insertion point (see ref. 6). (D) Hybridization of the six BACs in A to DNA fibers of A. thaliana. The two nuclear flanking BACs are detected by FITC (green) and the four mtDNA-related BACs by rhodamine (red). The mtDNA-related BACs do not hybridize to a gap region. (Bar = 20 μm.) (E Upper) The gap region within the mtDNA insertion is localized to the telomere-proximal end by cohybridization of the telomere-flanking BAC clone F9A16 (green) with the four mtDNA-related BAC clones (red). (E Lower) Cohybridization of four mtDNA-related BACs (in red) with a set of 15 cosmid clones (in green), which comprise the complete mitochondrial genome of A. thaliana. The gap is filled by green signals derived from the cosmids, indicating that regions of the mitochondrial genome not included in the sequenced BAC contig are present within the mtDNA insertion locus on the chromosome. (Bar = 10 μm.) (F) DNA fiber hybridized with telomere-proximal BAC clone F9A16 (green) and mtDNA-derived BAC clone T5M2 (red). T5M2 shows a triplet pattern. (Bar = 20 μm.) (G) DNA fiber hybridized with the first mtDNA BAC T5M2 (red) and the second mtDNA BAC T17H1 (green). These two BACs, which share ≈54-kb sequence, display a similar pattern with the same three units of repetition. A short signal derived from T17H1 (arrow) also was observed between the long telomere-proximal and middle T17H1 signals. (Bar = 20 μm.) (H) DNA fiber hybridized with the second mtDNA BAC T17H1 (green) and the third mtDNA BAC T18C6 (red). Signals derived from T18C6 also appear in triplicate. However, the centromere-proximal unit is longer than the other two. (Bar = 20 μm.) (I) DNA fiber hybridized with the fourth mtDNA BAC T5E7 (red) and centromere-proximal BAC T18A9 (green). The signal derived from T5E7 is not repeated. (Bar = 20 μm.) (J) Proposed structure of mitochondrial insertion locus and its relationship with the four mtDNA domains (the length of each repetitive unit is based on fiber-FISH data and is thus a representation of mean kilobase estimates). As in C, the green dots represent the two sets of specific repeats.
FISH and Fiber-FISH.
Ecotype Columbia-0 of A. thaliana was used for chromosome and DNA fiber preparations. Cytological procedures for chromosome and DNA fiber preparation were according to published protocols (7, 8). BAC DNA was isolated by using an alkaline lysis method (9) and labeled with either digoxigenin-11-UTP (Roche Molecular Biochemicals) or biotin-16-UTP (Roche Molecular Biochemicals) by using standard nick translation protocols. Probe preparations and signal detection for FISH and fiber-FISH followed Jackson et al. (10). Each FISH experiment was internally replicated on two different slides. The experiment was replicated if further data collection was necessary. Images were captured digitally by using a SenSys charge-coupled device camera (Photometrics, Tucson, AZ) attached to an Olympus BX60 epifluorescence microscope. Measurements were made on the digital images with iplab spectrum software (Signal Analytics, Vienna, VA). The physical size of the mtDNA insert was calculated relative to the known sizes of the flanking BACs.
Results
The BAC contig believed to cover the inserted mtDNA on chromosome 2 consisted of four clones and included 74% of the reported C24 mitochondrial genome (11), 270 of an expected 367 kb (6). Lin et al. (6) noted that the organization of the inserted mtDNA differed from that of the published mitochondrial genome of A. thaliana ecotype C24 (11) and its predicted alternative forms (12). Lin et al. (6) proposed a novel isoform as the progenitor mitochondrial genome from which the nuclear integration was derived. The nuclear contig and the relationship of each BAC clone to the isoform proposed by Lin et al. (6) are presented in Fig. 1 A and C, respectively.
The four mtDNA-related BAC clones were used as FISH probes to hybridize to the metaphase chromosomes of A. thaliana (Col-0). A single major hybridization site was detected in the pericentric region of a submetacentric chromosome (Fig. 1B). The FISH result showed that the mtDNA insertion on chromosome 2 is the only cytologically detectable mtDNA insertion in the A. thaliana (Col-0) genome.
The order of the four BAC clones within the sequenced contig from telomere-proximal end to centromere-proximal end is T5M2, T17H1, T18C6, and T5E7 (Fig. 1A). T5M2 contains a 95-kb insert, 69 kb of which is 99% identical to the A. thaliana C24 mtDNA. The remaining portion of this BAC consists of unique nuclear DNA sequence that partially overlaps with BAC F9A16, which flanks the insert region at the telomeric end but contains no mtDNA-derived sequences (Fig. 1A). BAC T5E7 is 88 kb, 74 kb of which is derived from the mtDNA whereas the remaining 14 kb consists of unique nuclear DNA sequence that partially overlaps with BAC T18A9. Clone T18A9 flanks the centromeric end of the insert region and contains no mtDNA-derived sequences. BACs T17H1 and T18C6 are located within the interior portion of the insertion and consist entirely of the mitochondrial-derived DNA (Fig. 1A).
Fiber-FISH analysis of the mtDNA insertion region was conducted by using these six BACs as probes. The four mtDNA-related BACs were detected with rhodamine (red color) and the two flanking BACs (F9A16 and T18A9) were detected by using FITC (green color) (Fig. 1D). The physical size of the mtDNA insert was calculated relative to the known sizes of the flanking BACs, thus accounting for the variable stretching degree of each fiber. Ten independently calibrated fiber measurements of the entire insertion region revealed an estimated 618 ± 42 kb of the mtDNA at the insertion locus (Fig. 1D), roughly 2.3 times greater than that predicted by contig assembly and sequence analysis. Interestingly, a nonhybridizing region (the gap in Fig. 1D) of ≈100 kb was found, indicating the presence of sequences other than those in the BACs. Using the four mtDNA-related BACs together with flanking BAC F9A16 as probes, we established that the nonhybridizing region is located toward the telomeric end of the insertion (Fig. 1E). We hypothesized that this region might consist of the remaining mtDNA sequence not included in the BAC contig sequenced by Lin et al. (6). To test this hypothesis, fiber-FISH was conducted by using a set of cosmid clones that comprise the entire mitochondrial genome of A. thaliana (12). Fig. 1E shows that the cosmids completely filled the gap. Fiber-FISH analysis using only four cosmids, which are known to contain most of the sequence not included in the original chromosome 2 insertion locus (cosmids 39E9, 30E9, A78, and 7G1), filled in nearly 70% of the gap (data not shown). These results indicate that the regions of the mitochondrial genome not included in the sequenced BAC contig are indeed present within the mtDNA insertion locus.
To better understand the structure of the mtDNA insertion, the location of each BAC sequence was individually analyzed by fiber-FISH. Sequences within BACs T5M2, T17H1, and T18C6 were found to occur more than once within the insertion locus, whereas BAC T5E7 displayed no such repetition (Fig. 1 F–I). Fig. 1F shows the hybridization of BAC T5M2 in red along with the telomeric flanking BAC F9A16 in green. The hybridizations of the T5M2 sequence revealed homologous sequence at three noncontiguous intervals. Fiber measurements of the two centromere-proximal copies of T5M2 estimate that each is ≈69 kb (data not shown), indicating that the entire mtDNA-related sequence of T5M2 is likely included in these two copies. BAC T5M2 is then displayed in red along with BAC T17H1 in green for comparison (Fig. 1G). These two BACs, which share ≈54 kb of sequence, showed the same three units of repetition. In addition, a small portion of T17H1 is independently located between the first and second large repeated regions (Fig. 1G). A side-by-side comparison of BACs T17H1 and T18C6 (Fig. 1H) shows that sequences in BAC T18C6 also appear in triplicate, although the centromere-proximal unit appears to be larger than the other two. The nonrepetitive nature of T5E7 is shown in Fig. 1I.
Discussion
A Model of the mtDNA Insertion.
The A. thaliana mitochondrial genome from the C24 ecotype consists of four regions of unique sequence separated by two pairs of specific repeats of ≈4.4 kb and 6.6 kb, respectively (12). For the purpose of this discussion, we have called the four regions A, B, C, and D (Fig. 1C), as in Lin et al. (6). Recombination across these specific repeats can result in five circular forms of the mitochondrial genome: A-B-C-D = 367 kb; A-C′-B-D′ = 367 kb; A-B-D′-C′ = 367 kb; A-D = ≈240 kb; C-B = ≈120 kb with the symbol ′ representing inverted sequence orientation relative to the published C24 mitochondrial genome (11, 12). These published forms are based on the C24 ecotype. Small variations in mitochondrial genome sequence and structure exist among ecotypes (11, 13). However, for this discussion we assume that the major structural features (two sets of specific repeats separating the four regions) are conserved across the Columbia-0 and the sequenced C24 ecotypes. The BAC contig in the published sequence of chromosome 2 contains these sequences in the order D′-A′-C-B with parts of D′ and A′ postulated to be absent from the insert (6). The relationship between the BACs used in this study and the D′-A′-C-B arrangement is shown in Fig. 1C.
Based on the fiber-FISH patterns and the known relationships among these BACs and the four domains of the mitochondrial genome, we propose a structural model of this mtDNA insertion in Fig. 1J. The telomere-proximal end of the insertion is consistent with a D′-A′ orientation. This D′-A′ section includes the “gap” that is absent in the published chromosome 2 sequence. The centromere-proximal end is consistent with a perfect C-B domain orientation, in agreement with the published chromosome 2 sequence. BAC T5M2 contains 69 kb of mtDNA. Fiber-FISH measurements estimate that each of the two interior repeats homologous to T5M2 is approximately 69 kb (data not shown). Therefore, we may conclude that these two interior repeats include all of the mtDNA within T5M2. Meanwhile, sequence analysis has shown that the domain A-homologous DNA within BAC T18C6 spans 41 kb. The fiber-FISH measurements showed that the two interior repeats homologous to BAC T18C6 are also ≈41 kb (data not shown). Therefore, our fiber-FISH data suggest that the entire A-domain portion of BAC T18C6 may be present within the two repeats. Taken together, the fiber-FISH results suggest that the duplicated interior of the mtDNA insertion locus may consist of two complete duplications of the D′-A′ region, excluding the ≈97-kb gap region (Fig. 1J).
The specific repeats within the mitochondrial genome are frequently involved in recombination (11, 14). These specific repeats may have undergone break-fusion or other rearrangement events, which eventually gave rise to the duplicated D′-A′ regions. Our data, however, do not provide information as to whether the duplications occurred in the organellar genome before nuclear integration or in the nuclear genome after the integration. Because the mitochondrial genome is highly plastic (12, 15), it is likely that the repetitive interior of the mtDNA insertion originated in the mitochondrial genome before the insertion event or occurred during the course of integration. Lack of recombination at this locus in the pericentric region (16) would then stabilize these tandem duplications.
Errors in Genome Sequencing at Repetitive Chromosomal Regions.
The repetitive nature of the mtDNA insertion caused the construction of an inaccurate but seemingly complete contig by Lin et al. (6). The contig was built by identifying BACs with overlapping fingerprints based on restriction enzyme digestions and by aligning BAC end sequences with previously sequenced BACs. In chromosomal regions containing long-range, large-unit repeats, this method can align BACs “correctly” at several points within the repetitive interval. The process tends to produce a minimal clone path by skipping over duplicated regions and spatially altering the sequencing contig from the true sequence organization. In the work of Lin et al. (6), the region containing the mtDNA insertion was covered by identifying and sequencing BACs that extended inwardly from T5E7 and F9A16 (Fig. 1A). BAC T18C6 is a logically correct extension of T5E7 based on both BAC end match and the proximity of a restriction site (HindIII) used in library construction. Similarly, T5M2 is a logical extension of F9A16 in a centromeric direction. When the ends of BAC T17H1 were found to precisely match parts of T5M2 and T18C6, this region of the genome, including the mtDNA insert, was considered to be closed. However, the fiber-FISH data revealed that ≈69 kb of the sequence found in T5M2 also occurred twice internally in the mtDNA insertion locus. BAC T17H1 was capable of correctly aligning with either of the two ≈69-kb repeats, in addition to BAC T5M2 itself. In this case, the centromere-proximal copy of T17H1 was aligned to the telomere-proximal copy of BAC T5M2. The sequences between the centromere-proximal copy of T17H1 and the telomere-proximal copy of BAC T5M2, including the ≈97-kb gap region, were skipped during contig assembly (Fig. 1J).
Identification of BACs spanning any two interior repetitive units would have revealed the repetitive nature of the inserted mtDNA. For example, identification of a BAC bridging the two ≈69-kb interior T5M2 units would have disclosed the presence of repetitive D domains within the insertion. However, the degree of repetition of this region still cannot be resolved because the clone-by-clone walking approach will infinitely continue aligning repetitive BAC ends. Clone-by-clone approaches, as implemented by Lin et al. (6), have been argued as preferable to whole-genome shotgun approaches, in part, because they presumably avoid sequencing problems arising from distant repeats (17, 18). Our findings, however, indicate that the clone-by-clone approach is still susceptible to errors in the contig assembly of sequences involving large repeats. Recognizing the inherent instability of yeast artificial chromosomes, Venter et al. (19) suggested that the development of BACs capable of accepting inserts up to 350 kb would be critical in addressing multiple issues in contig assembly. In this case, a contig constructed with larger BAC inserts would have been advantageous in mapping the appropriate positions of the large-unit repeats.
The higher eukaryotic genomes are capable of possessing large-scale repeat regions that may generate errors in contig assembly. Large duplications (>150 kb) recently have been reported in the human genome and these duplications exhibit a high degree of sequence similarity (>98%) (20, 21). The large sizes and high sequence similarity makes these duplications particularly problematic for both mapping and sequencing of the human genome (22). Genome sequencing of several diploid species, including yeast (Saccharomyces cerevisiae), worm (Caenorhabditis elegans), and fruit fly (Drosophila melanogaster), indicated that segmental or total genome duplication(s) occurred during the evolution of these species (23–25). Duplicated regions encompass ≈60% of the A. thaliana genome (26). If duplicated segments share significant sequence similarities, similar to the mtDNA insertion locus demonstrated in this study, they can cause difficulties for contig assembly and result in sequencing errors.
Utility of Fiber-FISH in Sequencing of Large Complex Genomes.
Fiber-FISH proved to be an invaluable tool in the analysis of this complex and repetitive locus on A. thaliana chromosome 2. The repetitive nature of the duplicated interior portion of the locus and the degree of repetition of such a region cannot be resolved by BAC end sequence and fingerprints even with a highly rigorous process. There was technically nothing wrong with the tiling path in this region, as both BAC ends and fingerprints matched.
Fiber-FISH has been used in sequencing projects before this study, primarily in estimating gap sizes between assembled contigs (10, 18, 27). Such gaps are hypothesized to result from regions of unclonable DNA and are believed to often be associated with low-copy large repeats (18, 27). We recently found that BAC clones containing tandemly repeated sequences are generally unstable (28). The instability of BAC clones will cause problems in assigning individual clones to specific contigs and results in sequencing gaps in chromosomal regions containing tandem repeats. Fiber-FISH is an effective technique in the physical mapping of repetitive chromosomal regions (7, 10, 29). Large DNA contigs ranging from several hundred kilobases up to 2 Mb can be analyzed by fiber-FISH in a single experiment (7, 10, 30). In general, sequencing through repetitive genomic regions is much easier with the aid of a reliable physical map. Such maps constructed through fiber-FISH and possibly optical mapping analysis (31) will aid attempts to complete complex, repetitive sequence contigs. Fiber-FISH analysis of individual BAC molecules also can contribute to the assembly of sequencing data derived from BAC clones containing complex repeats (32).
Acknowledgments
We are grateful to Dr. Mike Havey for his valuable comments on the manuscript. This work is partially supported by the Hatch Fund 142-E441 (to J.J.). C.D.T. and C.R.B. are supported by funds from the National Science Foundation.
Abbreviations
- FISH
fluorescence in situ hybridization
- BAC
bacterial artificial chromosome
References
- 1.Thorsness P E, Weber E R. Int Rev Cytol. 1996;165:207–234. doi: 10.1016/s0074-7696(08)62223-8. [DOI] [PubMed] [Google Scholar]
- 2.Schuster W, Brennicke A. EMBO J. 1987;6:2857–2863. doi: 10.1002/j.1460-2075.1987.tb02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard J L, Schmidt G W. J Mol Evol. 1995;41:397–406. doi: 10.1007/BF00160310. [DOI] [PubMed] [Google Scholar]
- 4.Sun C W, Callis J. Plant Cell. 1993;5:97–107. doi: 10.1105/tpc.5.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayliffe M A, Scott N S, Timmis J N. Mol Biol Evol. 1998;15:738–745. doi: 10.1093/oxfordjournals.molbev.a025977. [DOI] [PubMed] [Google Scholar]
- 6.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 7.Fransz P F, Alonso-Blanco C, Liharska T B, Peeters A J M, Zabel P, De Jong J H. Plant J. 1996;9:421–430. doi: 10.1046/j.1365-313x.1996.09030421.x. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, Hulbert S H, Gill B S, Ward D C. Mol Gen Genet. 1996;252:497–502. doi: 10.1007/BF02172395. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatas T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Jackson S A, Wang M L, Goodman H M, Jiang J. Genome. 1998;41:566–572. [PubMed] [Google Scholar]
- 11.Unseld M, Marienfeld J R, Brandt P, Brennicke A. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 12.Klein M, Eckert-Ossenkopp U, Schmiedeberg I, Brandt P, Unseld M, Brennicke A, Schuster W. Plant J. 1994;6:447–455. doi: 10.1046/j.1365-313x.1994.06030447.x. [DOI] [PubMed] [Google Scholar]
- 13.Ulrich H, Lattig K, Brennicke A, Knoop V. Plant Mol Biol. 1997;33:37–45. doi: 10.1023/a:1005720910028. [DOI] [PubMed] [Google Scholar]
- 14.Andre C, Levy A, Walbot V. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- 15.Bendich A J. J Mol Biol. 1996;255:564–588. doi: 10.1006/jmbi.1996.0048. [DOI] [PubMed] [Google Scholar]
- 16.Copenhaver G P, Nickel K, Kuromori T, Benito M I, Kaul S, Lin X Y, Bevan M, Murphy G, Harris B, Parnell L D, et al. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 17.Green P. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- 18.Dunham I, Shimizu N, Roe B A, Chissoe S, Dunham I, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, et al. Nature (London) 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 19.Venter J C, Smith H O, Hood L. Nature (London) 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- 20.Orti R, Potier M C, Maunoury C, Prieur M, Creau N, Delabar J M. Cytogenet Cell Genet. 1998;83:262–265. doi: 10.1159/000015201. [DOI] [PubMed] [Google Scholar]
- 21.Horvath J E, Viggiano L, Loftus B J, Adams M D, Archidiacono N, Rocchi M, Eichler E E. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- 22.Horvath J E, Schwarth S, Eichler E E. Genome Res. 2000;10:839–852. doi: 10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 24.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 25.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 26.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 27.Hattori M, Fujiyama A, Taylor T D, Watanabe H, Yada T, Park H S, Toyoda A, Ishii K, Totoki Y, Choi D K, et al. Nature (London) 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 28.Song, J., Dong, F., Lilly, J. W., Stupar, R. M. & Jiang, J. (2001) Genome, in press. [PubMed]
- 29.Fransz P F, Armstrong S, De Jong J H, Parnell L D, Van Drunen C, Dean C, Zabel P, Bisseling T, Jones G H. Cell. 2000;100:367–376. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- 30.Jackson S A, Cheng Z K, Wang M L, Goodman H M, Jiang J. Genetics. 2000;156:833–838. doi: 10.1093/genetics/156.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aston C, Mishra B, Schwartz D C. Trends Biotechnol. 1999;17:297–302. doi: 10.1016/s0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- 32.Jackson S A, Dong F, Jiang J. Plant J. 1999;17:581–587. doi: 10.1046/j.1365-313x.1999.00398.x. [DOI] [PubMed] [Google Scholar]