Abstract
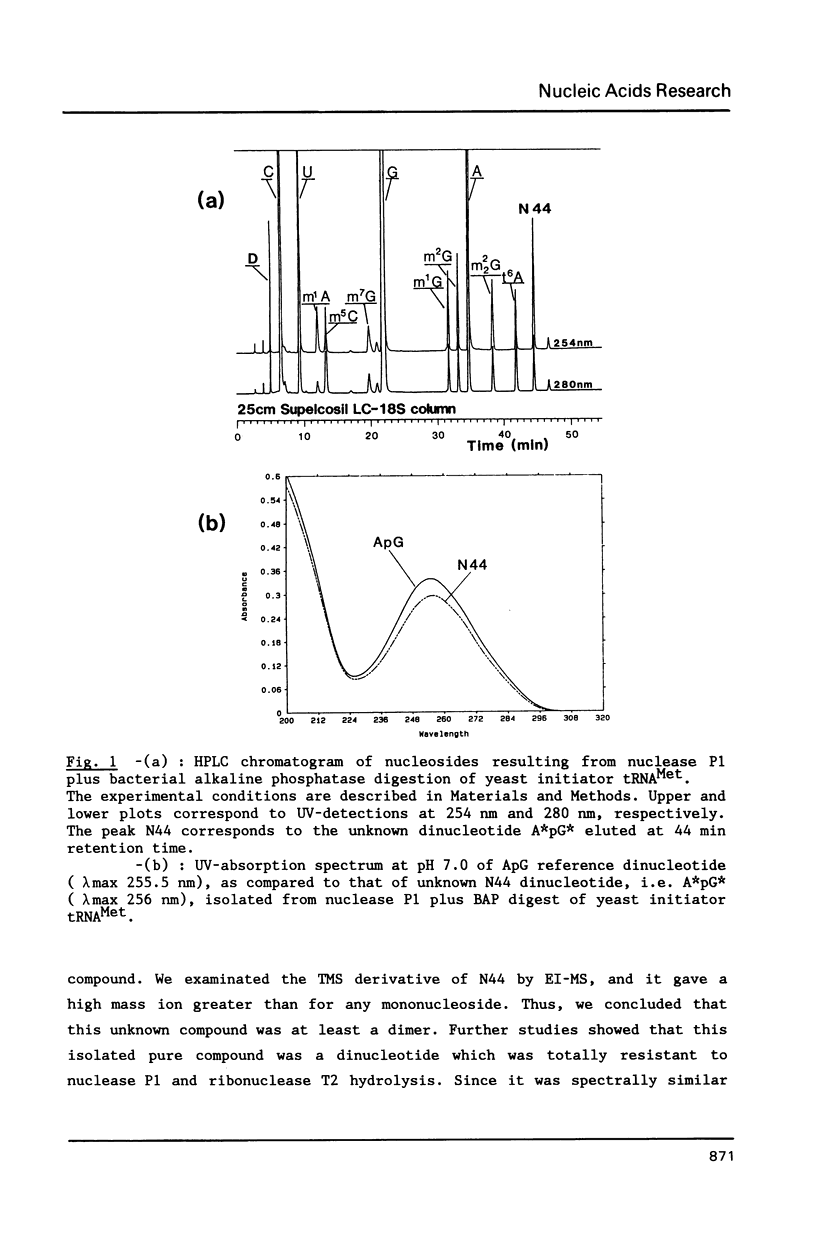
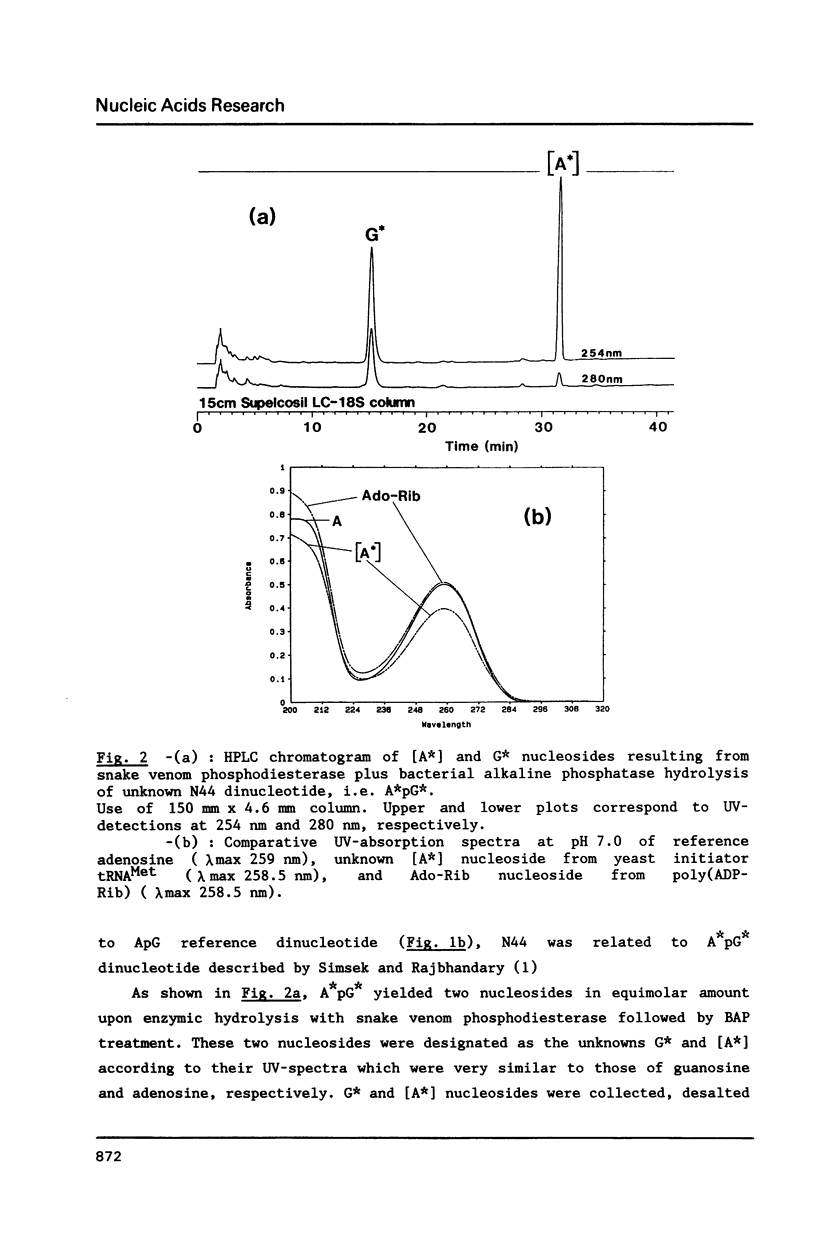
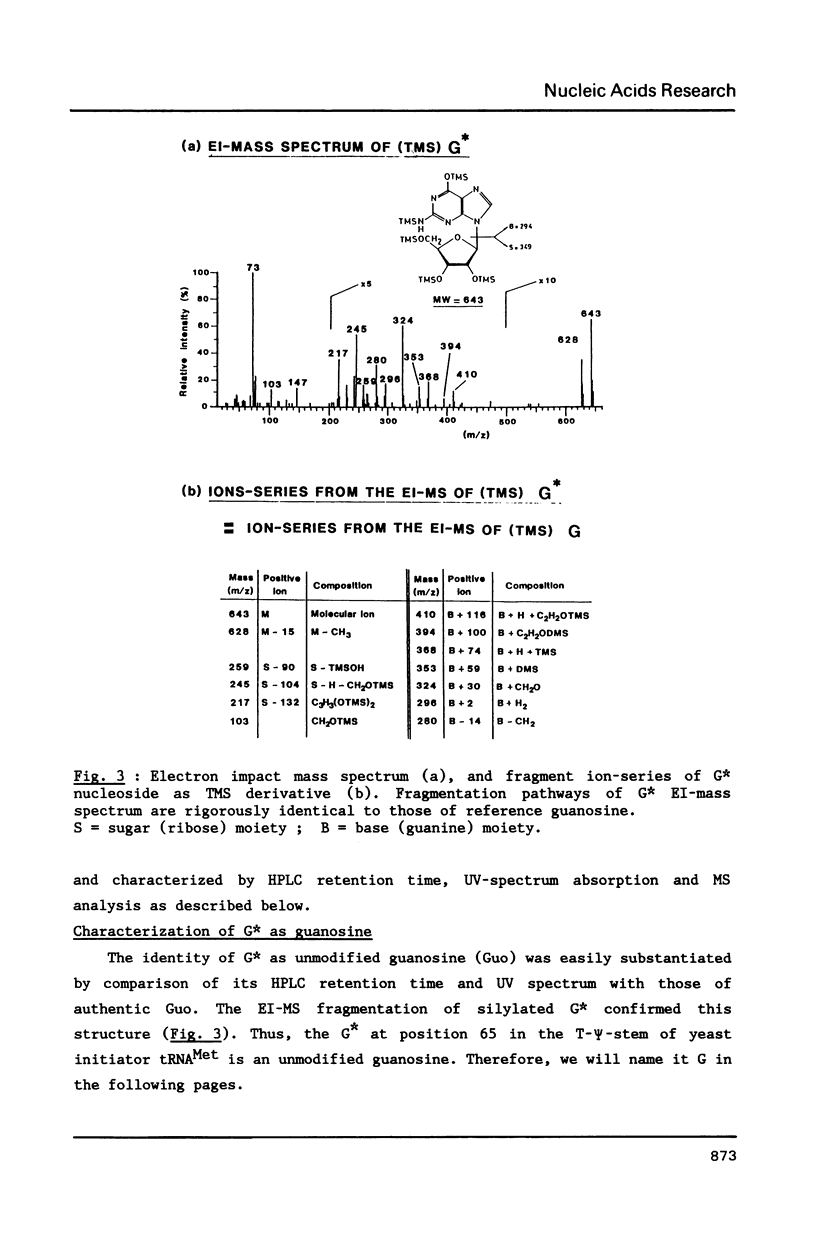
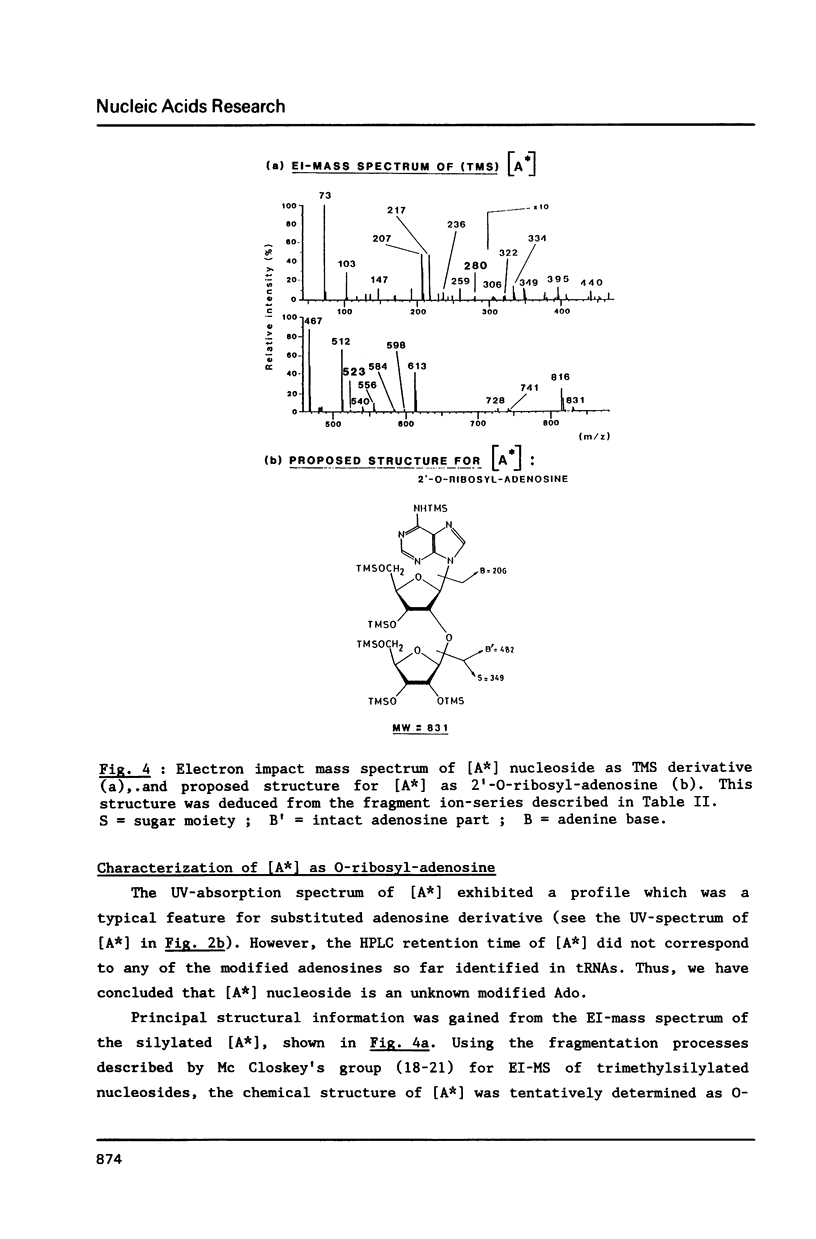
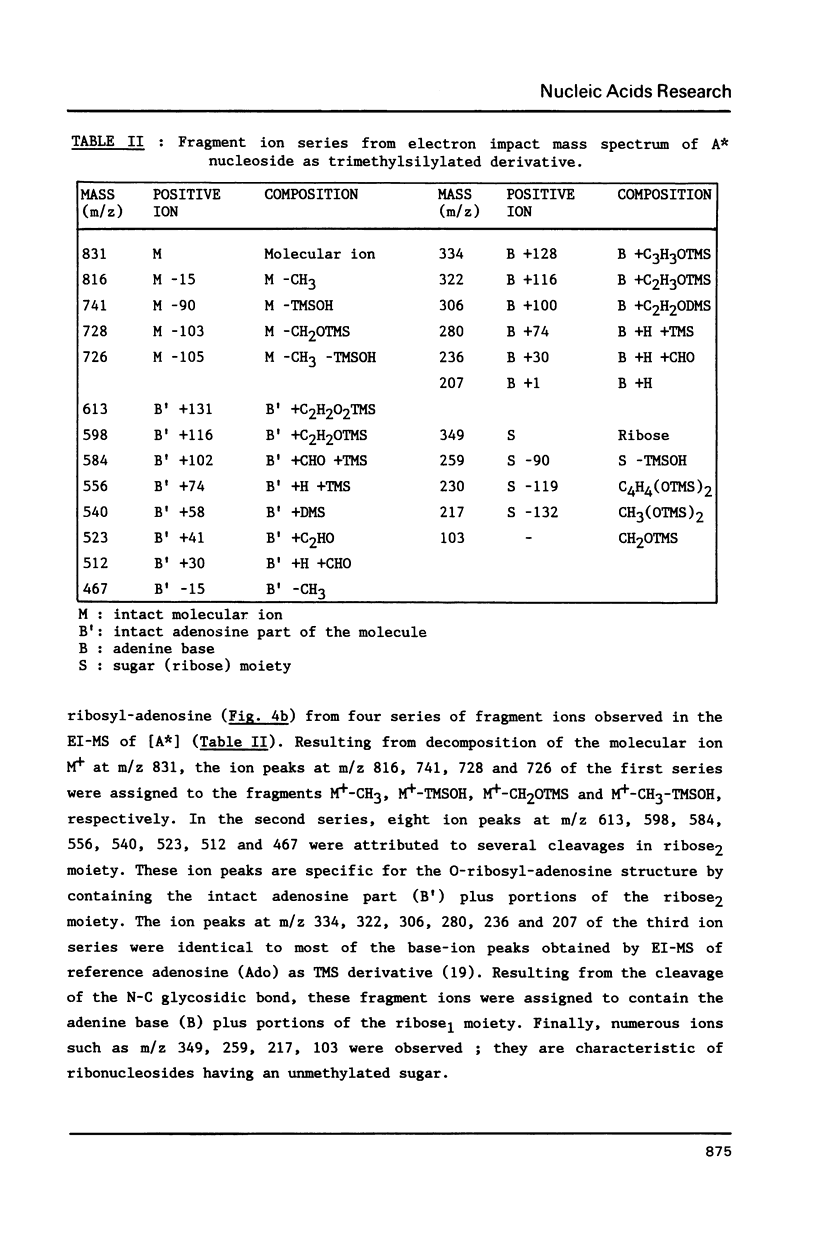
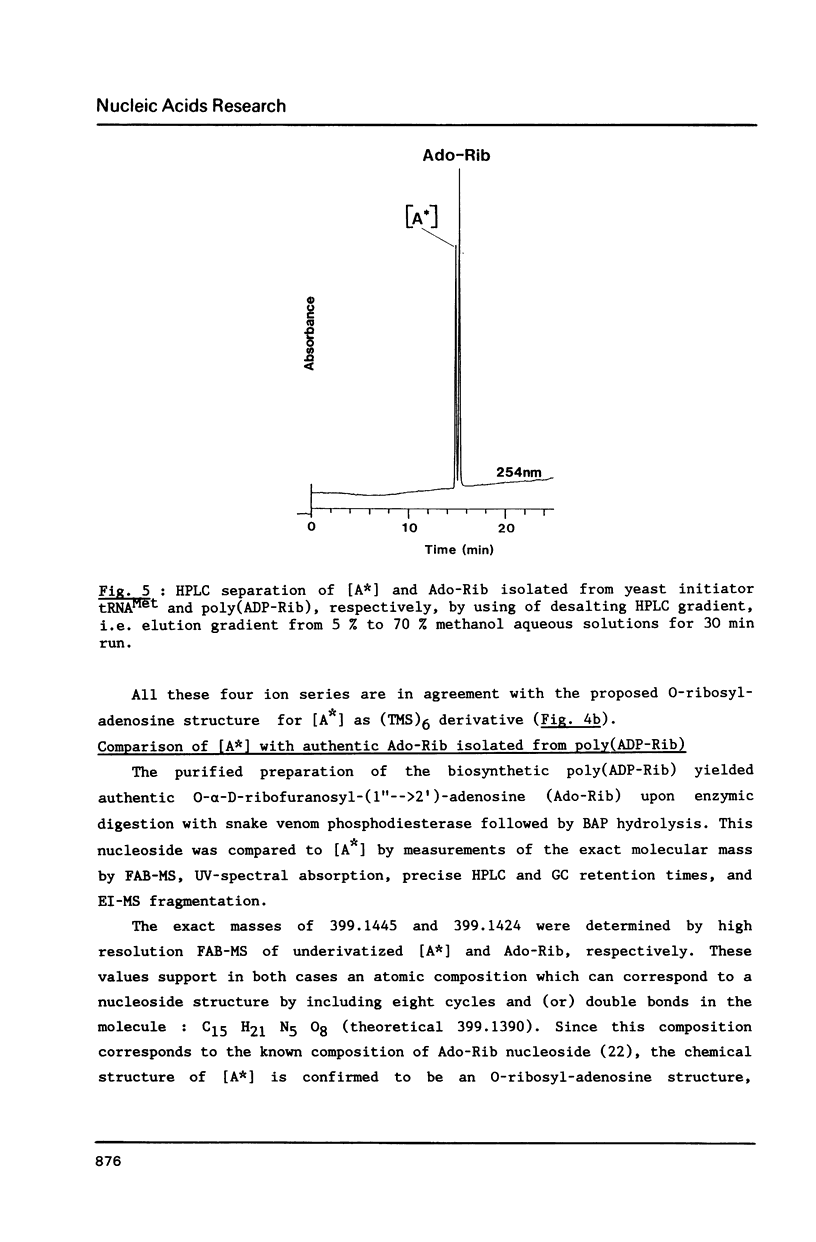
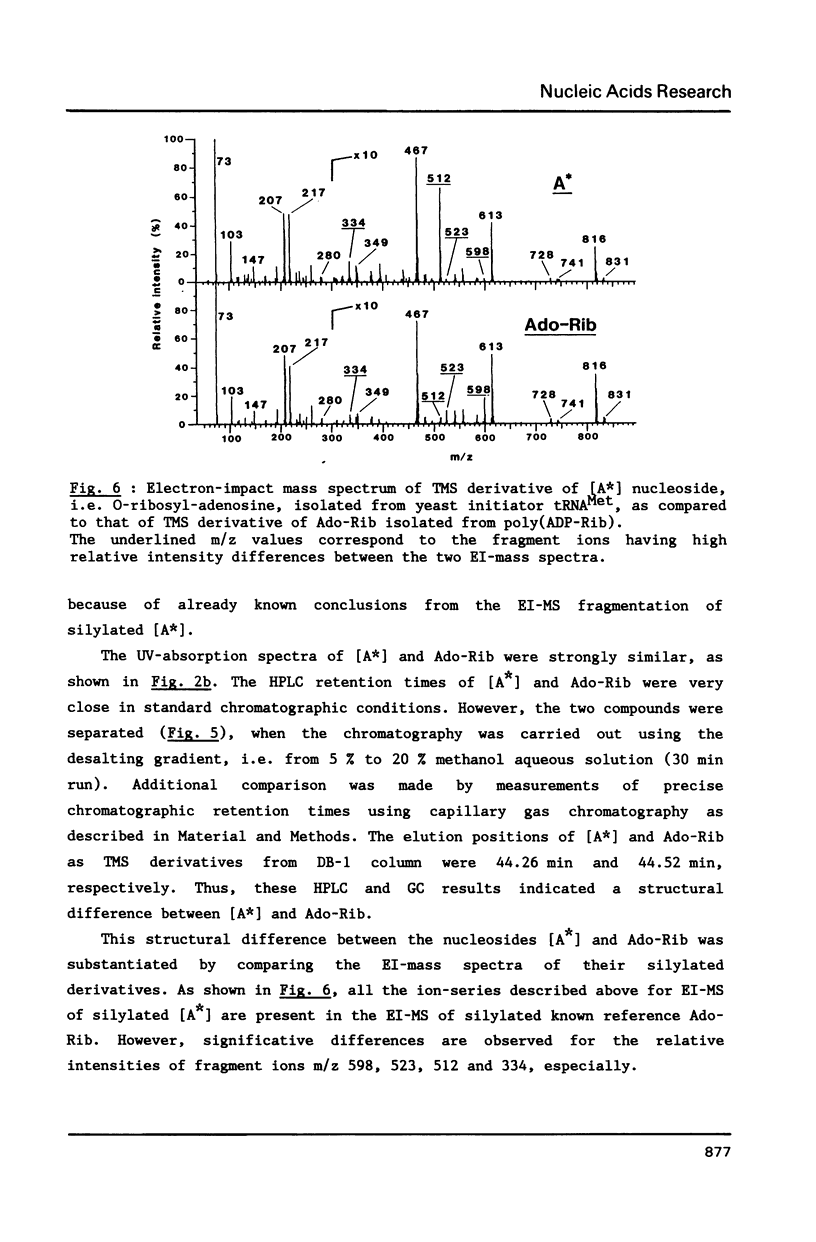
We report in this paper on isolation and characterization of two unknown nucleosides G* and [A*] located in the T-psi-stem of yeast methionine initiator tRNA, using the combined means of HPLC protocols, real time UV-absorption spectrum, and post-run mass spectrometry by electron impact or fast atom bombardment. The G* nucleoside in position 65 was identified as unmodified guanosine. The structure of the unknown [A*] in position 64 was characterized as an isomeric form of O-ribosyl-adenosine by comparison of its chromatographic, UV-spectral and mass spectrometric properties with those of authentic O-alpha-ribofuranosyl-(1"----2')-adenosine isolated from biosynthetic poly(adenosine diphosphate ribose). Our studies also brought evidence for the presence of a phosphorylmonoester group located on this new modified nucleoside [A*], when isolated by ion exchange chromatography from enzymic hydrolysis of yeast initiator tRNAMet without phosphatase treatment.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dirheimer G., Ebel J. P. Fractionnement des tRNA de levure de bière par distribution en contre-courant. Bull Soc Chim Biol (Paris) 1967;49(12):1679–1687. [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., Davis G. E., Suits R. D., Waalkes T. P., Borek E. Quantitative high-performance liquid chromatography of nucleosides in biological materials. J Chromatogr. 1978 Mar 21;150(2):455–476. doi: 10.1016/s0021-9673(00)88205-9. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., McCune R. A., Gerhardt K. O., Agris P. F. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J Chromatogr. 1982 Jul 9;230(2):297–308. [PubMed] [Google Scholar]
- Gehrke C. W., Patel A. B. Gas-liquid chromatography of nucleosides. Derivatization and chromatography. J Chromatogr. 1976 Aug 4;123(2):335–345. doi: 10.1016/s0021-9673(00)82202-5. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Patel A. B. Gas-liquid chromatography of nucleosides. Effect of silylating reagents and solvents. J Chromatogr. 1977 Jan 11;130:103–114. doi: 10.1016/s0021-9673(00)89786-1. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., Zumwalt R. W., McCune R. A., Kuo K. C. Quantitative high-performance liquid chromatography analysis of modified nucleosides in physiological fluids, tRNA, and DNA. Recent Results Cancer Res. 1983;84:344–359. doi: 10.1007/978-3-642-81947-6_26. [DOI] [PubMed] [Google Scholar]
- HALL R. H. A GENERAL PROCEDURE FOR THE ISOLATION OF "MINOR" NUCLEOSIDES FROM RIBONUCLEIC ACID HYDROLYSATES. Biochemistry. 1965 Apr;4:661–670. doi: 10.1021/bi00880a008. [DOI] [PubMed] [Google Scholar]
- Keith G., Dirheimer G. Primary structure of Bombyx mori posterior silkgland tRNAPhe. Biochem Biophys Res Commun. 1980 Jan 15;92(1):109–115. doi: 10.1016/0006-291x(80)91526-0. [DOI] [PubMed] [Google Scholar]
- Keith G., Pixa G., Fix C., Dirheimer G. Primary structure of three tRNAs from brewer's yeast: tRNAPro2, tRNAHis1 and tRNAHis2. Biochimie. 1983 Nov-Dec;65(11-12):661–672. doi: 10.1016/s0300-9084(84)80030-9. [DOI] [PubMed] [Google Scholar]
- Keith G., Roy A., Ebel J. P., Dirheimer G. The primary structure of tRNA trp from brewer's yeast. I. Complete digestion with pancreatic ribonuclease and T 1 ribonuclease. Biochimie. 1972;54(11):1405–1415. doi: 10.1016/s0300-9084(72)80082-8. [DOI] [PubMed] [Google Scholar]
- Miwa M., Ishihara M., Takishima S., Takasuka N., Maeda M., Yamaizumi Z., Sugimura T., Yokoyama S., Miyazawa T. The branching and linear portions of poly(adenosine diphosphate ribose) have the same alpha(1 leads to 2) ribose-ribose linkage. J Biol Chem. 1981 Mar 25;256(6):2916–2921. [PubMed] [Google Scholar]
- Miwa M., Saikawa N., Yamaizumi Z., Nishimura S., Sugimura T. Structure of poly(adenosine diphosphate ribose): identification of 2'-[1''-ribosyl-2''-(or 3''-)(1'''-ribosyl)]adenosine-5',5'',5'''-tris(phosphate) as a branch linkage. Proc Natl Acad Sci U S A. 1979 Feb;76(2):595–599. doi: 10.1073/pnas.76.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M., Sugimura T. Structure of poly(ADP-ribose). Methods Enzymol. 1984;106:441–450. doi: 10.1016/0076-6879(84)06048-1. [DOI] [PubMed] [Google Scholar]
- Pang H., Smith D. L., Crain P. F., Yamaizumi K., Nishimura S., McCloskey J. A. Identification of nucleosides in hydrolysates of transfer RNA by high-resolution mass spectrometry. Eur J Biochem. 1982 Oct;127(3):459–471. doi: 10.1111/j.1432-1033.1982.tb06894.x. [DOI] [PubMed] [Google Scholar]
- Rogg H., Staehelin M. Nucleotide sequences of rat liver serine-tRNA. 1. Products of digestion with pancreatic ribonuclease. Eur J Biochem. 1971 Jul 29;21(2):235–242. doi: 10.1111/j.1432-1033.1971.tb01461.x. [DOI] [PubMed] [Google Scholar]
- Simsek M., RajBhandary U. L. The primary structure of yeast initiator transfer ribonucleic acid. Biochem Biophys Res Commun. 1972 Oct 17;49(2):508–515. doi: 10.1016/0006-291x(72)90440-8. [DOI] [PubMed] [Google Scholar]
- Sugimura T. Poly(adenosine diphosphate ribose). Prog Nucleic Acid Res Mol Biol. 1973;13:127–151. doi: 10.1016/s0079-6603(08)60102-6. [DOI] [PubMed] [Google Scholar]



