Abstract
Hepatitis B virus (HBV) infection is a leading risk factor for hepatocellular carcinoma (HCC). HBV integration into the host genome has been reported, but its scale, impact and contribution to HCC development is not clear. Here, we sequenced the tumor and nontumor genomes (>80× coverage) and transcriptomes of four HCC patients and identified 255 HBV integration sites. Increased sequencing to 240× coverage revealed a proportionally higher number of integration sites. Clonal expansion of HBV-integrated hepatocytes was found specifically in tumor samples. We observe a diverse collection of genomic perturbations near viral integration sites, including direct gene disruption, viral promoter-driven human transcription, viral-human transcript fusion, and DNA copy number alteration. Thus, we report the most comprehensive characterization of HBV integration in hepatocellular carcinoma patients. Such widespread random viral integration will likely increase carcinogenic opportunities in HBV-infected individuals.
More than 350 million people are infected by hepatitis B virus (HBV) worldwide (Lavanchy 2004). HBV is a leading risk factor for hepatocellular carcinoma (HCC), with over eighty percent of HCC cases occurring in the regions where HBV is endemic (Michielsen and Ho 2011). Approximately 30%–50% of the estimated 320,000 annual HBV-related deaths are due to hepatocellular carcinoma (Farazi and DePinho 2006). Despite clear evidence supporting the involvement of HBV in HCC (Farazi and DePinho 2006; Chemin and Zoulim 2009; Bouchard and Navas-Martin 2011), the underlying nature of viral-host interaction remains elusive (Block et al. 2003). HBV integration into the host genome has been reported both in tumors (Gozuacik et al. 2001; Murakami et al. 2005; Saigo et al. 2008) and in nontumor liver tissue from HBV-infected individuals (Mason et al. 2010), although such integration is not essential for HBV replication. The relative extent, mutation model, and the functional impact of HBV integration in host genomes is not clear due to the lack of an unbiased approach to identify and quantify genome-wide HBV integration sites. Recent advances in sequencing technologies (Meyerson et al. 2010) provide an opportunity to investigate the global extent, mutation model, and functional impact of viral integration in the host genome. Recently, a primary hepatitis C virus-infected HCC patient has been subjected to whole-genome sequencing, and many somatic mutations were reported (Totoki et al. 2011). However, as an RNA virus, HCV never integrates into the host genome during its life cycle; therefore, liver cancer with HCV infection is not an optimal model to study viral-human genomic interactions. To that end, sequencing the genome and transcriptome of an HBV-positive HCC patient provides a great opportunity to reveal the functional impact of viral integration on the host genome.
Results
Detection of HBV integration based on whole-genome sequencing
We performed whole-genome deep sequencing (>80× coverage) and transcriptome sequencing on primary HCC tumors and matched adjacent non-neoplastic liver tissue from four patients (Supplemental Table S1; Supplemental Fig. S1; Supplemental Material Sections 1–3). Three of these patients are HBV-positive, and one is HBV-negative. For comparison, we also sequenced blood samples from one HBV-positive and one HBV-negative patient. Deep coverage sequencing enabled detection of rare integration events, quantification of the abundance of each event, and investigation of the genomic impact of HBV integration on the human genome. Besides, transcriptome sequencing (RNA-seq) enabled us to evaluate the transcriptional impact of the integration events.
To detect HBV sequences in our samples, we aligned all short reads from whole-genome sequencing against a comprehensive list (n = 73) of HBV reference genomes (genotype A-I and a strain of Woolly Monkey HBV) (Supplemental Table 2; Zöllner et al. 2006; Mulyanto et al. 2011). HBV sequences were not detected in samples from the HBV-negative individual but were clearly present in both tumor and nontumor liver samples from the HBV-positive patients (Fig. 1A,B). We found that these patients were infected by three different strains of HBV (B, C, and D genotypes) (Supplemental Material Section 4), based on the clear majority representation, in each patient, of only one out of the constellation of HBV variants.
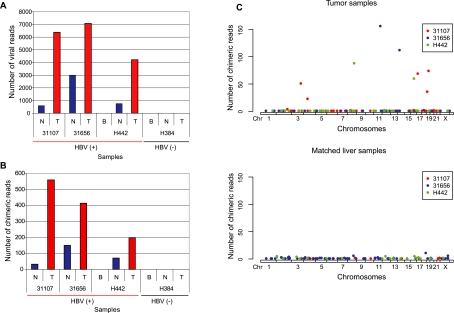
Figure 1.
Tumor-specific clonal expansion of virus-integrated hepatocytes in HCC. For each sequenced human genome, viral integration is quantified as the total number of paired-end reads where at least one arm maps to the HBV genome (A), the number of human-viral chimeric reads (B), and the number of chimeric reads as a function of the genomic location of HBV integration sites in the human genome (C). In contrast to the matched nontumor samples, tumor samples carry a few loci with a substantially larger number of chimeric reads. (T) Hepatocellular carcinoma tumor samples. (N) Matched nontumor liver samples. (B) Blood samples. The legend in panel C indicates internal identifiers for the three HBV positive patients in this study.
The total number of viral reads is substantially higher in the HCC tumors than in their matching nontumor liver tissue (Fig. 1A) at similar overall coverage (Supplemental Table S1). Based on total number of viral and human reads, we estimate that, on average, the tumor samples contain at least two copies of the viral genome per diploid human genome. We identified HBV integration sites by searching for human-virus chimeric paired-end reads, where one end mapped to the human genome and the other mapped to the viral genome. We found such chimeric reads in both tumor and nontumor liver tissue but not in blood. The tumor samples again harbor a much higher number of chimeric reads (Fig. 1B) than the nontumor samples. Chimeric reads supporting the same viral integration event were then clustered, yielding 255 unique HBV insertion sites in the three HBV-positive patients (Supplemental Table S3A; Supplemental Material Section 4), 48 of which are supported by multiple chimeric reads (Supplemental Table S3A).
Next, we questioned whether the number of detected viral integration events is dependent on the sequencing depth. We selected patient 31656 for additional sequencing, bringing the total coverage to 234× for the tumor and 243× for the nontumor sample. In total, we detected 142 integration sites in the tumor sample and 136 sites in the nontumor tissue (Supplemental Fig. S2A; Supplemental Table S4). After simulating lower-coverage sequencing from this high-coverage data, we found that the number of unique viral integrations detected was proportional to the sequencing depth for both the normal and tumor samples (Supplemental Fig. S2B; Supplemental Material Section 9). This supports many stochastic viral integration events without clonal expansion, which are likely to be underestimated by previous PCR-based approaches (Bréchot et al. 2000; Gozuacik et al. 2001; Saigo et al. 2008).
Both high-depth coverage (∼80×) and ultra high-depth (∼240×) sequencing reveals a heterogeneous, widespread viral integration landscape in tumor as well as in nontumor liver tissue from HCC patients. However, HCC tumor samples and their adjacent nontumor liver tissues exhibit strikingly distinct patterns of viral insertion (Fig. 1C). Based on high-depth coverage (∼80×) data, we found nontumor tissues contained 107 viral integration sites among the three HBV-positive patients, each with 11 or fewer supporting chimeric reads. This suggests that viral DNA integration occurs commonly and at many sites in nontumor liver tissue with HBV infection, resulting in a heterogeneous collection of insertion-carrying hepatocytes, each representing a small proportion of the population. In contrast, the tumor samples contained 148 insertion sites, with a small subset (nine sites) at much higher frequencies (supported by 23 or more chimeric reads) (Fig. 1C; Supplemental Table S3A), designated as Major Integration Sites (MIS) (Supplemental Fig. S3; Supplemental Material Section 5). The tumor samples also contain large numbers of low-frequency viral insertion sites, likely due to contamination of nontumor tissue or late viral integration in the expanding tumor. Occurrence of MIS in each of the three HBV-positive HCC tumors we examined is most likely the result of clonal expansion of hepatocytes carrying these insertions, suggesting that the events leading to MIS occur fairly early during tumorigenesis. Therefore, any functional genomic impact of viral integration would be restricted to these few clonal sites.
Transcriptional and genomic impact of HBV integration
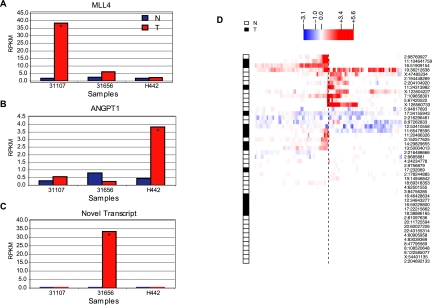
Detailed expression analysis, using RNA sequencing, of the genomic region flanking the most abundant MIS in each patient revealed a distinct transcriptional impact of viral integration. In patient 31107, we observed a MIS within the Mixed-Lineage Leukemia 4 (MLL4) gene, which has been previously reported as a recurrent HBV integration target among HCC patients (Saigo et al. 2008). MLL4, a histone-lysine N-methyltransferase, is a part of the ASC-2 complex implicated in the p53 tumor suppressor pathway (Lee et al. 2009). Other members of the MLL family are frequently mutated in solid tumors (Lee et al. 2008; Natarajan et al. 2010). This viral integration within the MLL4 gene is accompanied by a >20-fold increase in the MLL4 transcript level (Fig. 2A; Supplemental Fig. S4A). Detailed sequence analysis reveals that the inserted HBV sequence contains two partial copies of the HBV genome, driving adjacent increased expression on both strands (Supplemental Fig. S4A). In patient H442, the most abundant viral insertion occurs ∼10 kb upstream of ANGPT1 (Supplemental Table S3A), leading to a greater than eightfold increase in expression compared to that of the matched liver sample (Fig. 2B). Overexpression of ANGPT1 in HCC has been previously reported (Zeng et al. 2008). Interestingly, in patient 31656, the most abundant MIS is in a nongenic region. However, we observed novel transcription precisely next to the viral insertion site that was not observed in the samples without this viral insertion (Fig. 2C; Supplemental Fig. S5).
Figure 2.
Transcriptional effect of HBV integration on the human genome. Local transcriptional effect of HBV integration on the human genome is shown, for the most abundant integration site for each patient (A–C) and for all integration sites (D), based on RNA-seq data. (A) MLL4 is highly overexpressed in the tumor sample with the HBV integration event (31107). (B) Substantial overexpression of ANGPT1 in the tumor from patient H442 with HBV integration upstream (∼10 kb) of ANGPT1. (C) A novel human-viral fusion transcript in the tumor sample with HBV integration in a nongenic region (patient: 31656). Asterisks indicate samples with viral integration. (D) Transcriptional effect at human-viral junctions defined by DNA-seq. The human-viral junctions supported by multiple DNA-seq chimeric reads (n = 48) are represented as a dotted line at the center. We then used the RNA-seq data to infer the transcriptional changes on each side of these junctions. The color in the heatmap represents the fold-change for each interval, measured as the difference in the generalized log of the RPKM of the altered genome (i.e., the genome containing the viral insertion) versus the unaltered genome. Samples carrying the insertion are indicated as either N (Nontumor) or T (Tumor). The rows were grouped by hierarchical clustering.
Consistent with the MIS described above, adjacent transcriptional activation appears to be a common feature of viral integration. We examined the relative RNA-seq read abundance between paired samples with and without viral insertion, regardless of tumor and nontumor status. In half of the 48 viral integration sites investigated, there is obvious local transcriptional activation. This effect is usually directional (Fig. 2D; Supplemental Material Section 7), suggesting that some of the observed transcriptional activity changes can be attributed to the “run-through” of viral transcripts into human sequences. Indeed, paired-end RNA-seq data reveal the strong presence of chimeric transcripts between the viral HBx gene and the human MLL4 gene in patient 31107 and with the nongenic region in patient 31656.
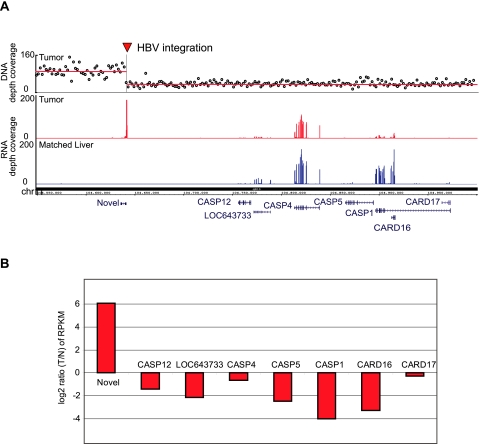
We found that besides direct expression alteration, viral insertion can also lead to genomic instability and introduce copy number changes, indirectly affecting gene expression. As detailed above, the most abundant MIS in the tumor sample of patient 31656 shows local increased human transcript level of an unannotated genomic locus in the immediate vicinity (Figs. 2C, 3A,B). DNA copy number analysis in this region revealed that this viral integration colocalized precisely with the junction of a large DNA copy number loss (chr11q22.3) (Fig. 3A). Interestingly, this deletion leads to the heterozygous loss of a cluster of caspases (CASP12, CASP4, CASP5, CASP1) and caspase recruitment domain family genes (CARD16 and CARD17), proteases that play a central role in the execution phase of cell apoptosis. RNA-seq read coverage shows that a string of these genes are down-regulated in the tumor sample with this viral insertion (Fig. 3B). We reason that a random viral insertion at this site led to genomic instability, resulting in the loss of a large adjacent genomic region, likely due to nonallelic recombination between two copies of integrated viral sequences.
Figure 3.
Genomic instability at the viral integration site near the caspase locus. (A) DNA-seq coverage and transcription around a chr11q22 HBV integration site in patient 31656. (Top panel) Normalized, GC-corrected DNA-seq coverage values in 50-kb windows, with the horizontal red line representing the resulting copy-number segments. There is a copy-number breakpoint right at the HBV integration site, with a copy-number loss of the chromosomal region 3′ from the integration site. Red and blue bar plots show transcription (RNA-seq read coverage) in this locus in the tumor and matched normal, respectively. (B) Genes closest to the integration site within this deletion were significantly down-regulated, including CASP12, CASP4, CASP5, CASP1, CARD16, and CARD17.
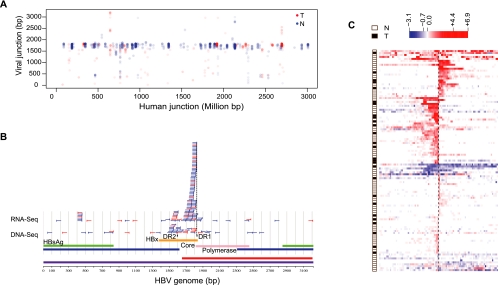
HBV integration at the DR1 site favors fusion transcripts
Viral-human fusion transcripts are prevalent, based on the number of viral-human chimeric RNA-seq reads. Many such events were detected in both tumor and nontumor tissues (Fig. 4A). Based on fusion transcripts supported by two or more chimeric reads, we found that the viral arms of chimeric reads map preferentially to a region between 1500–2000 base pairs on the viral genome (Fig. 4A). Specifically, the fusion junctions obtained from RNA-seq reads predominantly map near the direct repeat 1 (DR1) region located toward the end of the HBx gene (Fig. 4B). In contrast, the majority of viral integration junctions obtained from DNA-seq are close to either DR1 or DR2. We also observe a drop in viral transcription downstream from the DR1 region (Supplemental Fig. S6; Supplemental Material Section 3). The DR1 and DR2 regions have been previously found to be involved in multiple insertion events (Dejean et al. 1984; Mason et al. 2010). We again examined the transcriptional change between paired samples with and without viral-human transcript chimeras and confirmed the trend of unidirectional human transcriptional activation at the sites of RNA fusion (Fig. 4C). In contrast to the strong preference for the viral DR1 site, the human sequences in these chimeric RNA-seq reads map to many distinct locations in the human genome (Fig. 4A). No bias in terms of genomic location or local sequence preference for HBV integration was observed in the human genome, suggesting a stochastic viral DNA integration model (Supplemental Fig. S7; Supplemental Material Section 6).
Figure 4.
Viral-human fusion transcripts are common in both HCC and nontumor samples. (A) The genomic coordinate on the HBV genome of each chimeric RNA-seq read is plotted against its genomic coordinate on the human genome (linearized after concatenating all chromosomes). Only locations supported by two or more chimeric reads are shown. (B) Viral junctions determined from clusters of two or more chimeric reads are shown as vertical bars, with part of the remaining integrated viral sequence (50 bp) indicated as horizontal lines. Reads from both RNA- and DNA-seq are shown. A large majority of the RNA-seq junctions are in close proximity (10 bp) to the DR1 (Direct repeat 1) region on the HBV genome. (Blue) Clusters from nontumor liver samples; (red) clusters from tumor samples. (C) A global view of the transcriptional consequence of viral integration on the flanking human genome. The data were organized in the same manner as in Figure 2D, except that the human junctions in this panel are based on RNA-seq data instead of DNA-seq data. Most of the sites show strong unidirectional transcriptional up-regulation, starting at the integration site, while relatively fewer sites correlate with nondirectional transcriptional down-regulation or up-regulation.
It is worth noting that the preferred DR1 site (1824–1834 bp) of viral-human fusion transcripts is located near the 3′ end of the HBx gene (1374–1838 bp), just before its stop codon. Therefore, many fusion transcripts may extend the open reading frame of the HBx gene. We examined a number of individual cases by local de novo assembly of chimeric RNA-seq reads (Supplemental Material Section 8) and identified 76 precise fusion junctions (Supplemental Table S5). Although the biological consequence of most of these potentially elongated proteins is not clear, it is intriguing that the viral insertion within the MLL4 gene leads to the formation of an in-frame fusion between HBx and truncated MLL4 (Supplemental Fig. S4). Although the overall MLL4 transcription output is much higher in the affected genome (Fig. 2A), the resulting fusion transcript lacks the AT-hook DNA-binding domain of MLL4 (Supplemental Fig. S4B). We speculate that this overexpressed fusion product acts as a dominant negative allele, perhaps replacing the normal MLL4 protein in the ASC-2 tumor suppressor complex without conferring its normal DNA binding activity.
Mutation spectrum of HCC revealed by whole genome sequencing
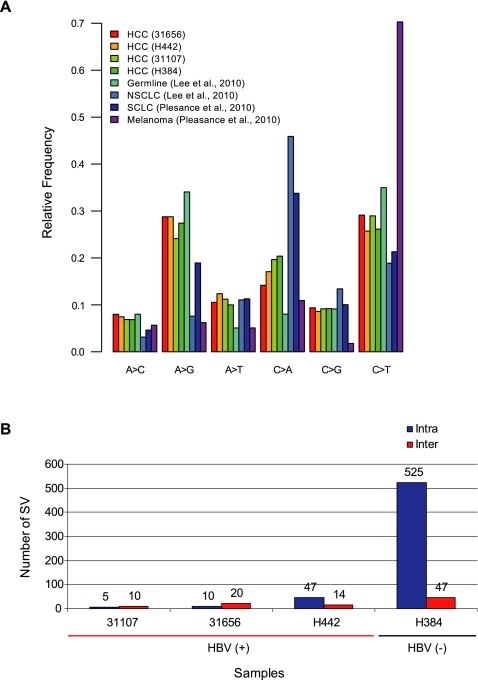
In addition to identifying viral integrations, whole genome sequencing also provides us the opportunity to identify other somatic alterations that may not be directly related to viral integration in these tumor genomes and to compare the somatic changes between HBV-infected and noninfected HCC patients. To obtain a collection of high-confidence somatic mutations, we systematically identified somatic single-base mutations and attempted to validate (Supplemental Fig. S8) a large number (1319) of these mutations (Supplemental Material Section 10). The three HBV-positive tumors had 3180–5862 somatic point substitutions, while the HBV negative patient had 6362 such substitutions. The number of nonsynonymous mutations in these four HCC samples ranges from 22 to 54 (Supplemental Table S6). The only gene mutated in all four tumors is TP53 (Supplemental Fig. S9), with all four patients carrying predicted protein-altering mutations in TP53 (Supplemental Table S7). The nucleotide substitution pattern (designated as mutation signature) that we observed in HCC (Fig. 5A) is distinct from that associated with tobacco smoking (Lee et al. 2010; Pleasance et al. 2010b) or UV damage (Pleasance et al. 2010a; Fig. 5A). The most prevalent substitutions are A→G and C→T transition events, a pattern similar to the one recently reported in a hepatitis C virus-infected HCC patient (Totoki et al. 2011). The mutation signature in HCC patients was the same, irrespective of HBV infection status.
Figure 5.
Mutation signature and structural variations in HCC patients. (A) High confidence, somatic single-base substitutions were classified into all six categories of base substitutions. The fraction of mutations belonging to each category is shown for the four HCC patients, and compared to signatures previously found in NSCLC (Lee et al. 2010), SCLC (Pleasance et al. 2010b), melanoma (Pleasance et al. 2010a), and germline variations. (B) Number of predicted structural variations detected in both HBV positive and negative HCC patients. (Intra) Intrachromosomal SVs. (Inter) Interchromosomal SVs.
We computationally predicted (Fig. 5B) a large number of structural variations and then experimentally validated a subset of these structural variations (Supplemental Table S8; Supplemental Material Section 11) in the four HCC patients. The number of intrachromosomal somatic structural variations in the uninfected individual was at least 10-fold higher than the HBV-infected individuals. Similarly, the uninfected individual carried at least a threefold higher number of interchromosomal structural variations compared to the infected individuals (Fig. 5B). Noteworthy experimentally confirmed structural variations included a fusion between the AXIN1 and LUC7L genes in patient H442, resulting in a truncated AXIN1 and up-regulation of LUC7L (Supplemental Fig. S10). Point mutations and/or epigenetic silencing of AXIN1, a Wnt antagonist, were reported in various types of human solid tumors (Baeza et al. 2003; Segditsas and Tomlinson 2006; Zucman-Rossi et al. 2007). In this case, the truncated AXIN1 presumably disrupts the normal function of the APC-depend destruction complex and consequently activates the Wnt signaling pathway. It is not clear whether the LUC7L gene, the fusion partner, plays any functional role in liver cancer development except that it was reported as a survival predictor of breast cancer (Crawford et al. 2008). DNA copy number variation (CNV) and allele-imbalance (AIB/LOH) (Supplemental Material Section 12) indicated a copy number loss of TP53 (chr17p13 region) in all four patients and amplification of CCND1 (chr11q13 region) in the HBV-negative patient H384 (Supplemental Fig. S11).
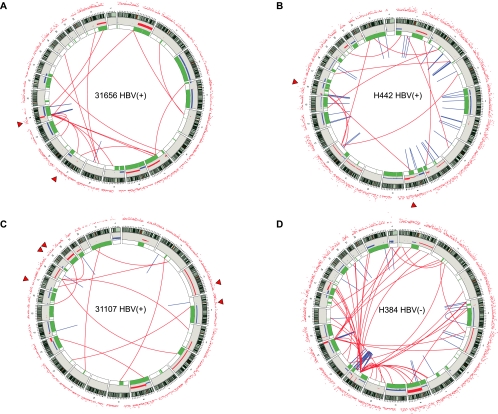
Interestingly, in HBV-infected patients, the MIS sites tend to coincide with boundaries of copy number alterations (Fig. 6A–C; Supplemental Table S9), suggesting an underlying mechanistic connection between HBV integration and genomic instability. The association of MIS with copy number boundaries is statistically significant (P < 10−5) (Supplemental Material Section 13). We note that the single HBV-negative patient we sequenced has a higher rate of mutation and a larger number of structural variations (Supplemental Table S6; Fig. 5B). Incidentally, genes involved in telomere maintenance (PARP1, BLM, and MLH3) were mutated in this patient but not in the HBV-positive patients (Supplemental Fig. S9). In addition, the same patient showed a pattern of structural catastrophe typical of chromothripsis (chromosome 11) (Fig. 6D; Stephens et al. 2011), while the HBV-infected patients did not show such an event. We believe the higher mutation rate that we observed in the HBV-negative patient is likely due to mutations of telomere maintenance genes rather than the HBV status.
Figure 6.
Summary of somatic genomic alterations in HCC patients. Various types of somatic alterations in the four HCC patient genomes using circos plots (Krzywinski et al. 2009) (A–D). High confidence somatic structural variations (SVs) are shown as lines, with red lines representing interchromosomal SVs and blue lines indicating intrachromosomal SVs. (Green bars) Regions of loss of heterozygosity and allelic imbalance. Somatic copy number alterations are shown as bar plots with copy number gain shown in red and copy number loss in blue (the scale ranges from −2 to 4). Each surrounding red dot represents the number of high-confidence somatic SNVs within a 1 million base pair window. (Triangles) Major HBV integration sites. Patient's identifier and HBV status are shown at the center of each circular view.
Discussion
HCC is a consequence of multiple complex mutation processes (Farazi and DePinho 2006). To the best of our knowledge, this study provides the first comprehensive analysis of multiple dimensions of genomic alterations in HBV-infected and uninfected HCC patients, including viral integration, single nucleotide changes, and large genomic alterations (Fig. 6). While conventional PCR-based methods can be used to detect the presence of viral integration, only a small subset of insertions can be detected (Saigo et al. 2008), or only insertions close to targeted human (Murakami et al. 2005) or viral sequences (Mason et al. 2010) can be found. Whole genome and transcriptome sequencing provides an unbiased and sensitive method for comprehensively identifying viral insertion events and quantifying their frequencies, thus providing the first opportunity to interrogate the global extent of viral impact on the human genome and transcriptome. We found that HBV integration occurs frequently in both tumor and nontumor hepatocytes, but they show distinct patterns of integration. Clonal expansion of MIS-carrying hepatocytes was found specifically in the tumor samples but not in the matched liver samples. However, a heterogeneous background population of cells harboring low-frequency viral integrations was detected both in the tumor and matched liver samples. This finding is consistent with a random integration model followed by a positive selection of MIS-carrying hepatocytes during hepatocarcinogenesis, resulting in more virus-integrated hepatocytes (clonally expanded subpopulation) in the tumor samples when compared to their matched nontumor counterparts. We argue that the impact of HBV integration is multifaceted. Given the observed stochastic nature of viral integration, it appears that HBV integration effectively surveys the human genome, exerting insertional mutation pressure, and thus may expand the oncogenic opportunities for patients infected by HBV. In these samples, the most dominant HBV integration sites occur: within the MLL4 gene, a frequently observed target for HBV insertion among HCC patients (Saigo et al. 2008); near the ANGPT1 gene, a key player in angiogenesis; and next to the cluster of caspase genes on chromosome 11, causing a copy number loss at this locus. Recurrence of integration sites in the MLL4 gene argues for a causative role of HBV integration in HCC. Other recurrent integration sites such as ones within the h-TERT gene, PDGFRB, and MAPK1 have also been reported (Paterlini-Bréchot et al. 2003; Murakami et al. 2005). We also examined the gene expression profile of ANGPT1 in an independent large collection of tumor samples across 35 different types of tissues. Significant overexpression of ANGPT1 was found, specifically in the liver cancer samples, which argues ANGPT1 might play an important functional role during tumorigenesis in a tissue-specific manner (Supplemental Fig. S12). In order to check if the deletion at the chromosome 11 caspase locus is an isolated event, we examined copy number alteration in two independent liver cancer data sets (GSE34957 and GSE9829) and found ∼10% of liver cancer samples in these two data sets show copy number loss at the caspase locus (Supplemental Fig. S13).
The whole genome sequencing data show that viral integration frequently occurs near the DR1 or DR2 sites (Fig. 4B). A hot spot of integration on the viral genome might suggest a sequence-specific integration mechanism. However, on the human side, we did not observe any specific sequence features near the fusion breakpoints (Supplemental Fig. S7). Since DRs are 5′ ends of the minus and plus DNA strands of the linearized HBV genome, it is reasonable to argue that the frequent use of the DRs as integration sites is likely due to the preferred use of the free ends of replication intermediates of HBV. Although fusions can occur close to both DR1 and DR2, the viral-human fusion transcripts were strongly biased to regions near DR1 (Fig. 4B). Previous studies (Guo et al. 1993; Raney and McLachlan 1997; Yu and Mertz 2001) reported several important cis-elements near the DR1 regions, such as the enhancer II, the preC promoter, and the hormone response element. Members of the nuclear receptors, a superfamily of transcription factors, can bind to the DR1 hormone response element and regulate the transcription and replication of HBV. A base-pair–level view of fusion transcripts breakpoints (Supplemental Fig. S14) shows that the majority of fusion breakpoints mapped close to DR1, resulting in intact DR1 cis-elements. In contrast to integration using DR2 as a breakpoint, a fusion at DR1 region will also juxtapose the DR1 cis-elements close to the flanking human genomic sequences. We, therefore, speculate that intact DR1 cis-elements and a short physical distance between DR1 elements and the human fusion partner are two critical factors for a successful viral-human transcript formation. In addition, the linearization of the viral genome after integration also explains the significant down-regulation of Polymerase (Supplemental Fig. S6, right panel), a gene otherwise located downstream from the DR1 region in a circular viral genome. In the linearized viral genome, the Polymerase gene would be at the 5′ end, disconnected from the cis-elements upstream of the DR1 region, which would now be at the 3′ end of the linearized genome.
In summary, it is evident that viral integration affects the human genome via insertional mutagenesis, viral promoter-driven transcriptional up-regulation, and induction of genomic instability. Despite the diversity of insertion sites and their varied effects, it is conceivable that virus-mediated mutagenesis functions, in conjunction with other genomic alterations such as TP53 mutation, drive hepatocarcinogenesis. Our observations support a model wherein the frequent assault of the human genome by widespread viral integration significantly widens “oncogenic opportunities” in patients with chronic HBV infection.
Methods
Sample preparation
All specimens were obtained from patients with appropriate consent. Tissue samples were examined by pathologists. All tumor samples contained >80% of tumor content. The HBV infection status of patient samples was confirmed by a polymerase chain reaction (PCR) assay. DNA and RNA were extracted by using a standard DNA/RNA extraction kit (Supplemental Material Section 1).
Whole genome and transcriptome sequencing
Whole genome DNA paired-end sequencing was performed by “unchained combinatorial probe anchor ligation sequencing” as described previously (Drmanac et al. 2010). The average coverage was >80×. The single nucleotide variation (SNV) calls for each sample with respect to the human reference genome (NCBI Build37) were made as described previously (Drmanac et al. 2010). Detection of structural variation (SV), copy number variation, and loss of heterozygosity (LOH) are described in Supplemental Material Sections 11 and 12.
Transcriptome sequencing of both tumor and matched liver samples was performed on the Illumina HiSeq Platform using a standard paired-end protocol. On average, 25–35 million 75-bp reads were obtained per sample. The reads were mapped to both the human genome and a collection of HBV reference genomes by GSNAP (Wu and Nacu 2010). The number of reads mapped to the exons of each RefSeq gene was calculated, and the corresponding RPKM (reads mapping to the genome per kilobase of transcript per million reads obtained from sequencing) value was derived (Supplemental Material Section 3).
Statistical analysis of differentially expressed genes was performed using the DEseq package from Bioconductor (Anders and Huber 2010; Supplemental Material Section 14).
HBV integration detection
We first selected a HBV reference genome from a collection (n = 73) of HBV reference sequences by finding the best match. We then utilized the paired-end nature of reads to search for human-virus chimeric reads, an indication of HBV integration in the human genome. Adjacent chimeric reads were clustered to obtain nonredundant integration events (Supplemental Material Section 4).
Experimental validation
Validation of candidate single nucleotide variants was performedusing a Sequenom MassARRAY platform (Supplemental Material Section 10). Validation of HBV insertions and a subset of structural variations was performed by PCR, followed by Sanger sequencing (Supplemental Material Sections 4 and 11).
Data access
Sequencing data can be accessed at dbGAP (accession ID phs000384.v1.p1). Copy number data for HCC patients have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE34957.
Acknowledgments
We thank Eric Brown for valuable discussions; Peter Dijkgraaf, Julie Rae, Carlo Santos, and May Wittke for sample handling; Robert Soriano for generation of microarray data; and Florian Gnad, Kiran K. Mukhyala, Colin Watanabe, Jim Fitzgerald, Meg Green, and Albion Baucom for computational assistance.
Authors' contributions: Z.J.: study design, project coordination, overall data analysis, and preparation of the manuscript; S.J.: overall data analysis and preparation of the manuscript; J.L.: transcriptome sequencing data analysis and preparation of the manuscript; P.M.H.: CNV and LOH data analysis and preparation of the manuscript; W.L.: viral integration breakpoint analysis and preparation of the manuscript; M.I.K., K.P.P., and P.C.: whole genome sequencing and HBV integration site analysis; T.D.W.: Computational pipeline development for short read mapping; Y.G. and Z.M.: PCR validation of structural variations and viral integration sites; J.S. and S.S.: mutation experimental validation; J.D. and S.B.K.: HBV status validation sample preparation; H.M.S. and S.J.: sample handling and histopathological evaluation; S.Y., A.J., and W.Y.: experimentation studies on ANGPT1; D.G.B., R.C.G., and F.J.D.: project coordination and manuscript critiques; Z.Z.: study design, data interpretation, and manuscript preparation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.133926.111.
References
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza N, Masuoka J, Kleihues P, Ohgaki H 2003. AXIN1 mutations but not deletions in cerebellar medulloblastomas. Oncogene 22: 632–636 [DOI] [PubMed] [Google Scholar]
- Block TM, Mehta AS, Fimmel CJ, Jordan R 2003. Molecular viral oncology of hepatocellular carcinoma. Oncogene 22: 5093–5107 [DOI] [PubMed] [Google Scholar]
- Bouchard MJ, Navas-Martin S 2011. Hepatitis B and C virus hepatocarcinogenesis: Lessons learned and future challenges. Cancer Lett 305: 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P 2000. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol 10: 211–231 [DOI] [PubMed] [Google Scholar]
- Chemin I, Zoulim F 2009. Hepatitis B virus induced hepatocellular carcinoma. Cancer Lett 286: 52–59 [DOI] [PubMed] [Google Scholar]
- Crawford NP, Walker RC, Lukes L, Officewala JS, Williams RW, Hunter KW 2008. The Diasporin Pathway: A tumor progression-related transcriptional network that predicts breast cancer survival. Clin Exp Metastasis 25: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean A, Sonigo P, Wain-Hobson S, Tiollais P 1984. Specific hepatitis B virus integration in hepatocellular carcinoma DNA through a viral 11-base-pair direct repeat. Proc Natl Acad Sci 81: 5350–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81 [DOI] [PubMed] [Google Scholar]
- Farazi PA, DePinho RA 2006. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer 6: 674–687 [DOI] [PubMed] [Google Scholar]
- Gozuacik D, Murakami Y, Saigo K, Chami M, Mugnier C, Lagorce D, Okanoue T, Urashima T, Bréchot C, Paterlini-Bréchot P 2001. Identification of human cancer-related genes by naturally occurring Hepatitis B Virus DNA tagging. Oncogene 20: 6233–6240 [DOI] [PubMed] [Google Scholar]
- Guo W, Chen M, Yen TS, Ou JH 1993. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol 13: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavanchy D 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 11: 97–107 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee J, Lee S-K, Lee JW 2008. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol 22: 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim D-H, Lee S, Yang Q-H, Lee DK, Lee S-K, Roeder RG, Lee JW 2009. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci 106: 8513–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. 2010. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465: 473–477 [DOI] [PubMed] [Google Scholar]
- Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM 2010. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol 84: 8308–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696 [DOI] [PubMed] [Google Scholar]
- Michielsen P, Ho E 2011. Viral hepatitis B and hepatocellular carcinoma. Acta Gastroenterol Belg 74: 4–8 [PubMed] [Google Scholar]
- Mulyanto, Depamede SN, Wahyono A, Jirintai, Nagashima S, Takahashi M, Okamoto H 2011. Analysis of the full-length genomes of novel hepatitis B virus subgenotypes C11 and C12 in Papua, Indonesia. J Med Virol 83: 54–64 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Bréchot C, Paterlini-Bréchot P 2005. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 54: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan TG, Kallakury BV, Sheehan CE, Bartlett MB, Ganesan N, Preet A, Ross JS, Fitzgerald KT 2010. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int 10: 13 doi: 10.1186/1475-2867-10-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini-Bréchot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C, Lagorce D, Bréchot C 2003. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22: 3911–3916 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin M-L, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney AK, McLachlan A 1997. Characterization of the hepatitis B virus major surface antigen promoter hepatocyte nuclear factor 3 binding site. J Gen Virol 78: 3029–3038 [DOI] [PubMed] [Google Scholar]
- Saigo K, Yoshida K, Ikeda R, Sakamoto Y, Murakami Y, Urashima T, Asano T, Kenmochi T, Inoue I 2008. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum Mutat 29: 703–708 [DOI] [PubMed] [Google Scholar]
- Segditsas S, Tomlinson I 2006. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 25: 7531–7537 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, et al. 2011. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet 43: 464–469 [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Mertz JE 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J Virol 75: 11354–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Gouw ASH, van den Heuvel MC, Zwiers PJ, Zondervan PE, Poppema S, Zhang N, Platteel I, de Jong KP, Molema G 2008. The angiogenic makeup of human hepatocellular carcinoma does not favor vascular endothelial growth factor/angiopoietin-driven sprouting neovascularization. Hepatology 48: 1517–1527 [DOI] [PubMed] [Google Scholar]
- Zöllner B, Feucht H-H, Sterneck M, Schäfer H, Rogiers X, Fischer L 2006. Clinical reactivation after liver transplantation with an unusual minor strain of hepatitis B virus in an occult carrier. Liver Transpl 12: 1283–1289 [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Benhamouche S, Godard C, Boyault S, Grimber G, Balabaud C, Cunha AS, Bioulac-Sage P, Perret C 2007. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene 26: 774–780 [DOI] [PubMed] [Google Scholar]