Abstract
Over the course of ontogenesis, the human brain and human cognitive abilities develop in parallel, resulting in a phenotype strikingly distinct from that of other primates. Here, we used microarrays and RNA-sequencing to examine human-specific gene expression changes taking place during postnatal brain development in the prefrontal cortex and cerebellum of humans, chimpanzees, and rhesus macaques. We show that the most prominent human-specific expression change affects genes associated with synaptic functions and represents an extreme shift in the timing of synaptic development in the prefrontal cortex, but not the cerebellum. Consequently, peak expression of synaptic genes in the prefrontal cortex is shifted from <1 yr in chimpanzees and macaques to 5 yr in humans. This result was supported by protein expression profiles of synaptic density markers and by direct observation of synaptic density by electron microscopy. Mechanistically, the human-specific change in timing of synaptic development involves the MEF2A-mediated activity-dependent regulatory pathway. Evolutionarily, this change may have taken place after the split of the human and the Neanderthal lineages.
Cognitive differences between humans and chimpanzees are striking (Povinelli and Preuss 1995). However, humans and chimpanzees differ by only about 1.2% in their primary DNA sequences, representing the divergence of the two species over the last 4–6 million yr (Chen and Li 2001; Ebersberger et al. 2002). The major question is, therefore, how has human cognition evolved within such a short evolutionary time?
Simple alterations in developmental processes, such as shifts in the timing of events, can result in dramatically novel phenotypes (Gould 1977; McNamara 1997). Cognitive abilities are no exception, and variations in neurodevelopmental timing can significantly affect cognitive function (Johnson 2001; Langer 2006). It was therefore hypothesized that the remodeling of brain development, and specifically changes in gene expression levels during human neurodevelopment, could underlie the evolution of human-specific cognitive abilities (King and Wilson 1975; Gould 1977). Indeed, it is possible that cognitive features characteristic of human postnatal development might be mirrored by human-specific changes in anatomical and functional brain development. These changes, in turn, would be both regulated by, as well as reflected by, human-specific gene expression changes over the course of human brain development and maturation.
In support of this hypothesis, it was recently shown that some, but not all, gene expression changes taking place during human postnatal prefrontal cortex (PFC) development showed timing differences relative to chimpanzees and macaques (Somel et al. 2009). Although no specific mechanism associated with cognitive development was identified in that study, it reported that genes showing human-specific delays tended to be neuron-associated. This finding might be linked to earlier observations from stereological studies showing that synaptic density in the PFC peaks around 4–5 yr of age in humans but within the first months in macaques (Rakic et al. 1986; Huttenlocher and Dabholkar 1997). Whether or not delayed frontal cortex synaptogenesis is human-specific or is shared with chimpanzees remained unknown.
Human-specific timing changes in brain development could potentially be related to novel patterns of cognitive maturation in humans. Human cognition and the human brain develop in synchrony during postnatal ontogenesis. For example, the infant human brain grows at a pace unmatched by the chimpanzee during the first few years of life (Leigh 2004). This is also the period when assimilation of cultural knowledge from conspecifics is an essential part of cognitive development in human infants. Differences between humans and nonhuman primates in their ability to assimilate such knowledge are already apparent by 2.5 yr of age (Tomasello 2008). Another notable fact is that human children, in contrast to offspring of other primates, remain dependent on others after weaning, between 3 and 7 yr of age. This ontogenic period, during which the infant brain remains highly plastic, has been hypothesized to be unique to humans (Bogin 1997).
Here, to gain a deeper understanding of the evolution of human cognition, we studied the function and regulation of human-specific expression changes during brain development by using multiple large-scale gene expression data sets. By using both microarray and sequencing platforms, we compared postnatal brain ontogenesis in humans, chimpanzees, and rhesus macaques in two functionally and evolutionary distinct brain regions: the dorsolateral PFC and lateral cerebellar cortex (CBC).
Results
Gene expression profiles specific to human brain development
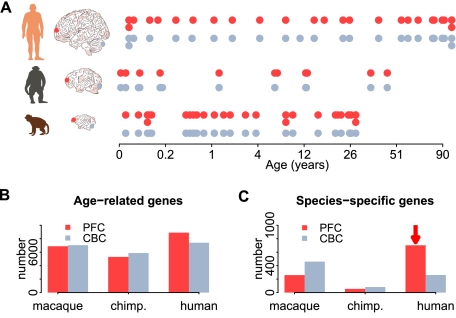
We analyzed mRNA expression levels measured in the PFC and the CBC of humans, chimpanzees, and rhesus macaques using Affymetrix Gene 1.0ST microarrays, in 12–26 individuals per species, with both brain regions derived from the same individual in almost all cases (PFC/CBC: 23/22 humans, 12/12 chimpanzees, 26/24 macaques) (Somel et al. 2011). The samples span the bulk of each species' lifespan, with a particular focus on the first years of life (humans, 0–98 yr; chimpanzees, 0–44 yr; rhesus macaques, 0–28 yr) (Fig. 1A; Supplemental Table S1).
Figure 1.
Age-related gene expression change in the PFC and CBC. (A) Age distribution of samples used in this study. Each point represents an individual, with technical replicates shown as a second point below the first. Only one of the two replicates was used in the main analysis. The colors indicate brain regions (red, PFC; gray, CBC). The x-axis represents individual age in fourth root (age1/4) scale. Numbers of age-related genes (B) and genes with species-specific expression profiles (C) identified in the PFC (red) or CBC (gray). The red arrows highlight excess human-specific expression changes in the PFC.
By using microarray probes matching all three genomes perfectly, we reliably detected expression from more than 12,000 genes (PFC, 12,447; CBC, 12,853) (Supplemental Table S2). By using polynomial regression analysis, we identified ∼8000 genes that show significantly changed expression levels with age in at least one brain region of at least one species (PFC, 8613; CBC, 7988; F-test P < 0.01; permutation-based false-discovery rate [FDR]<0.10) (Fig. 1B; Supplemental Table S3). Among these genes, we identified species-specific age-related expression changes using analysis of covariance. For instance, genes were classified as having human-specific expression patterns if their pattern of expression change during ontogenesis differed significantly between humans and chimpanzees, as well as between humans and macaques, but did not differ between chimpanzees and macaques. We ignored expression differences between species that were constant across lifespan (Supplemental Fig. S1A), since constant expression differences predominantly follow neutral divergence patterns and tend to affect multiple tissues (Somel et al. 2011). They are, therefore, less likely to be related to cognitive development. Furthermore, constant expression differences are more likely to be caused by technical and environmental artifacts (Gilad et al. 2006; Khaitovich et al. 2006).
In the CBC, we found 260 genes with human-specific expression profiles and 82 genes with chimpanzee-specific expression profiles. In the PFC, 702 genes had human-specific expression profiles and 55 genes had chimpanzee-specific expression profiles (Fig. 1C). Thus, there is a threefold excess of genes showing human-specific expression changes in the CBC and a 12-fold excess in the PFC. This was not caused by differences in sample numbers, in the age distributions of samples, or in maturation rates among species, as quantitatively similar results were obtained using subsets of 10 individuals per species where age distributions were matched with regard to chronological age or to age normalized to the species' lifespan (Supplemental Fig. S1C,D; Supplemental Table S4). Notably, we have also detected an excess of human-specific developmental changes in the PFC using a different statistical procedure, as reported recently (Somel et al. 2011). Finally, there was no difference in quality of the PFC and CBC data sets, as the total numbers of age-related changes identified in the two brain regions in all three species were comparable (Fig. 1B; Supplemental Fig. S1B).
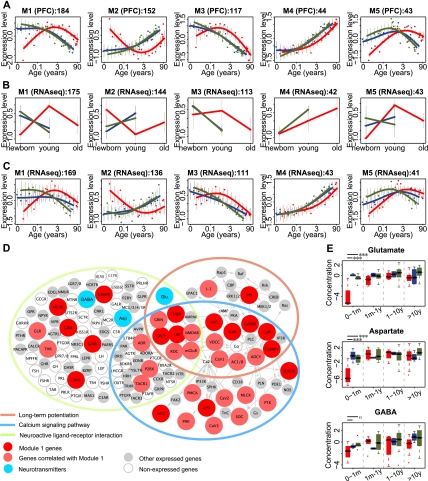
The excess of human-specific developmental changes, compared with chimpanzee-specific ones, observed in the two brain regions is intriguing, as is the fact that the excess is greater in the PFC. Both of these brain regions have been previously implicated in complex and possibly human-specific cognitive functions: the PFC in processes such as social behavior, abstract thinking, and reasoning (Clark et al. 2001; Wood and Grafman 2003; Rilling 2006; Barbey et al. 2009) and the CBC with language and manual abilities (Rilling 2006). Our results, however, suggest that biological and/or functional remodeling of the human PFC since divergence from chimpanzees has been more extensive than that of the CBC. To investigate this further, we checked whether genes with human-specific developmental expression profiles form discrete coexpressed modules that might share functional and regulatory characteristics. By using nonsupervised hierarchical clustering, we indeed found five such modules in the PFC and two in the CBC (Fig. 2A; Supplemental S2A,B; Supplemental Table S5). Importantly, genes showing human-specific patterns had similar probe numbers and probe location distributions across gene regions such as coding sequences (CDS), 5′ UTR, and 3′ UTR as did other genes expressed in the brain (Supplemental Fig. S3; Supplemental Table S6).
Figure 2.
Patterns and functions of genes with human-specific expression in PFC. (A) Five major human-specific gene expression modules found in the PFC. Expression profile of PFC modules measured using RNA-seq data set 1 (B) and data set 2 (C). In A through C, each point represents an individual (red, human; blue, chimpanzee; green, macaque), the lines in A and C show cubic spline curves. (Error bars) SD across genes in a module. (x-axis) Age in log2 scale. Expression levels of all genes were standardized to mean = 0 and SD = 1 before averaging. The titles on top of each panel show module information; numbers in A show number of genes in each module and in B and C show the number of module member genes expressed in RNA-seq. (D) Three KEGG pathways (Kanehisa et al. 2008) significantly enriched in module 1 genes: neuroactive ligand-receptor interaction, calcium signaling, and long-term potentiation. Proteins or protein complexes containing module 1 genes are shown in red, containing module 1–like genes in light red, showing expressed genes in gray, and without expressed genes as empty circles. Genes with Pearson correlation r > 0.5 between their expression change with age, and the module 1 mean expression change with age in all three species was classified as module 1–like genes. (Blue) Three neurotransmitters (glutamate [Glu], aspartate [Asp], and GABA) associated with the pathways and showing human-specific profiles. (Arrows) Interactions based on the KEGG pathway annotation. (E) Concentration profiles of the three human-specific neurotransmitters: glutamate, aspartate, and GABA, in the PFC of the three species (red, human; blue, chimpanzee; green, macaque). To reduce individual variation, individuals with similar ages were combined in four age groups, separated by a gray dashed line. The distribution of neurotransmitters' concentrations from samples within each age group is shown in a boxplot. In the youngest age group (0–1 mo), the neurotransmitters' concentrations in humans were lower than in chimpanzees and macaques. Significance of the difference between human–macaque or human–chimpanzee comparisons is shown by the long or short lines above the youngest age group (one-sided z-test: ***P < 0.001; oP < 0.1; Supplemental Methods).
To test the validity of these results and the reproducibility of the module expression profiles, we measured mRNA expression in the PFC and CBC in human, chimpanzee, and rhesus macaque newborns and adults (data set 1) and in the PFC of 14 humans, 14 chimpanzees, and 15 rhesus macaques (data set 2), using high-throughput RNA sequencing (RNA-seq) (Supplemental Tables S1 and S7). Age-related expression changes measured by microarrays and RNA-seq correlated strongly and positively on a gene-by-gene basis (Supplemental Fig. S4). Furthermore, all human-specific expression patterns found in the PFC and CBC were reproduced in the RNA-seq data sets (Fig. 2B,C; Supplemental Fig. S2C,D).
Timing of human-specific expression patterns
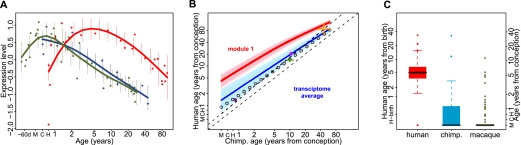
In many of the identified modules, human developmental trajectories differed drastically from the chimpanzee and macaque trajectories. For instance, in modules 1 and 3, gene expression increased after birth in humans, followed by a decrease starting at ∼5 yr of age for the module 1 genes and at ∼1 yr for module 3 genes. In contrast, in both chimpanzees and macaques, gene expression started decreasing soon after birth (Fig. 2A). Do these species differences represent developmental patterns that are uniquely evolved in humans, as has been reported for a number of morphological and life history characteristics (Penin et al. 2002; Bogin 2009)? Or do they represent a shift in timing of the pre-existing patterns? To distinguish between these two possibilities, we extended the macaque expression patterns into the prenatal stage by measuring PFC expression in six fetal samples at ∼70 to ∼30 d before birth and in six newborn macaque samples with ages between 0.5 d and 3 yr (Supplemental Table S1). This revealed that both module 1 and module 3 genes follow the same developmental trajectories in humans and macaques, separated by a temporal delay in humans (Fig. 3A; Supplemental Fig. S5A). The expression profiles of genes in the other PFC modules also follow similar developmental trajectories in humans and macaques, with different degrees of delay in humans.
Figure 3.
Time-shift in expression of module 1 genes among species. (A) Module 1 gene expression including the fetal rhesus macaque samples. Points indicate individuals (red, human; blue, chimpanzee; green, macaque), lines show cubic spline curves. (Error bars) SD across genes in a module. (B) Difference in developmental timing between humans and chimpanzees for PFC module 1 genes (red) and for all age-related genes in the PFC (transcriptome average; blue). The x- and y-axes show human and chimpanzee age in years from estimated conception event. H, C, and M axis marks show the time of birth for human, chimpanzee, and macaque, respectively. The curves show at what age human expression levels correspond to those of chimpanzee, estimated by aligning the chimpanzee and human expression profiles using the dynamic time warping algorithm. The light red and blue areas show variation in the module 1 time-shift estimate and the transcriptome average's time-shift estimate, respectively, obtained by bootstrapping module 1 genes or random assignments of PFC age-related genes to module 1, 1000 times. The other lines and symbols show timing of life history landmarks: Lower and upper black dashed lines show the diagonal line passing through the origin and the maximum lifespan point, respectively. Empty gray circles represent the timing of human brain growth relative to that of chimpanzees, using the dynamic time warping algorithm (data from Leigh 2004). The symbols represent the following life history landmarks: maximum lifespan (orange), female sexual maturity (green), and eruption of first deciduous and last permanent dentition (dark and light purple, respectively) (data from Smith et al. 1994; de Magalhães and Costa 2009). Note that the transcriptome average (dark blue curve) being above the diagonal indicates that humans reach the same expression levels at a later age than chimpanzees. Further, this developmental delay is significantly greater for module 1 (red curve) than for the transcriptome average. (C) The boxplot shows the distribution of ages at which expression of PFC module 1 genes reaches its maximum (red, human; blue, chimpanzee; green, macaque). The left and right y-axes show ages in years from birth for humans and from estimated conception time, respectively. H, C, and M indicate birth age for humans, chimpanzees, and macaques, respectively.
Humans, chimpanzees, and macaques are known to develop and mature at different rates. For instance, humans reach sexual maturity at 13–14 yr of age, chimpanzees at 8–9 yr, and rhesus macaques at 3.5–4.5 yr (Gavan 1969; Gould 1977; de Magalhães 2006). To test whether these developmental differences explain the observed human-specific expression profiles, we quantified the extent of developmental delay, or time-shift, between humans and the other two species by aligning the species' expression curves using a method based on a modified dynamic time warping algorithm (Yuan et al. 2011) (Supplemental Methods). This algorithm finds the optimal alignment between two time series and estimates both amplitude and significance of time-shift at every time point. By analyzing the expression patterns of all 8613 age-related genes, we found that the transcriptome-wide developmental delay between humans and chimpanzees or macaques closely followed the time-shift trajectories, based either on life history landmarks or on brain growth curves, among the three species (Fig. 3B,C; Supplemental Fig. S5B,C). In contrast, the time-shift in PFC modules 1, 2, and 5, but not in PFC modules 3 and 4, was significantly greater than both the transcriptome-wide delay and the delay expected from life history differences among species (permutation test, P < 0.01). Expression profiles of module 1, 2, and 5 genes did not show an equivalent time-shift in the CBC (Supplemental Fig. S5D). Thus, the human-specific delay in the PFC modules 1, 2, and 5 cannot be explained by the developmental rate differences among species and is not shared between the PFC and CBC.
Regulation of human-specific expression patterns
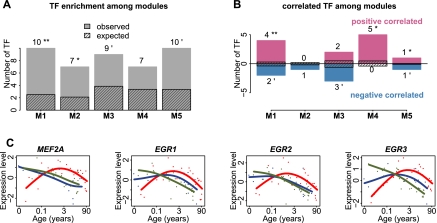
Synchronized expression of genes within each module implies coregulation by shared mechanisms, such as transcription factors (TFs). To test this we estimated enrichment of conserved TF binding sites (TFBSs) in the promoter regions of genes within each module. We observed a significant excess of TFBSs in the PFC modules 1 and 2 (permutation test, P < 0.05) and a marginally significant one in modules 3 and 5 (Fig. 4A; Supplemental Fig. S6A). In addition, the expression profiles of TFs with TFBS enrichment within modules showed a significantly better correlation with their targets in the respective modules (Wilcoxon test, P < 0.05) than expected by chance (permutation test, P = 0.011) (Fig. 4B). Thus, human-specific changes represented by the coexpressed gene modules could be at least partly driven by a limited number of TFs.
Figure 4.
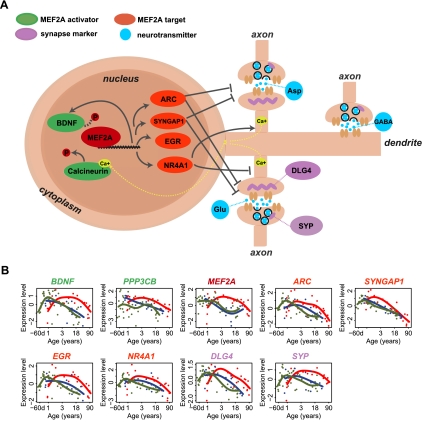
Regulation of human-specific expression patterns by TFs. (A) Number of TFs with TFBSs enriched among genes within each of the five human-specific expression modules in the PFC. The streaked bars represent the mean number of enriched TFs expected by chance, calculated by 1000 random assignments of the PFC age-related genes to the five modules. The numbers and symbols above each bar show numbers of TFs with enriched TFBSs among module genes (one-sided hypergeometric test, P < 0.05), as well as the significance of the number of TFs based on the 1000 permutations (**P < 0.01, *P < 0.05). (B) Number of TFs with TFBSs enriched and correlated with the targets in the same modules. The streaked bars represent the mean number of correlated TFs expected by chance, calculated by 1000 random assignments of the PFC age-related genes to the five modules. The numbers and symbols above each bar show numbers of correlated TFs among module genes (one-sided Wilcoxon test, P < 0.05), as well as the significance of the number of correlated TFs based on the 1000 permutations (**P < 0.01, *P < 0.05). (C) Expression profile of the four TFs showing significant positive correlation with their target gene expression profiles in all species in module 1. Points indicate individuals (red, human; blue, chimpanzee; green, macaque); lines show cubic spline curves. Note that MEF2A is a module 1 member, whereas EGR1-3 failed by a small margin to be classified as human-specific based on our criteria.
Among the five PFC modules, associations between TFs and their predicted target genes were particularly pronounced in module 1. Specifically, four TFs showed both TFBS enrichment and positive correlation with module 1 genes, while less than one would be expected by chance (permutation test, P < 0.05) (Fig. 4B,C). Notably, all four TFs, myocyte enhancer factor-2 (MEF2A) and three early growth response proteins (EGR1, EGR2, and EGR3), are involved in the regulation of neuronal functions, including neuronal survival, synaptogenesis, synaptic transmission, and long-term potentiation (Davis et al. 2003; Flavell et al. 2006; Shalizi et al. 2006; Li et al. 2007), raising the possibility that module 1 genes have roles in synaptic development and neuronal functions.
Module 1 genes are enriched in genes related to neuronal activity
To test whether module 1 genes are enriched in specific biological functions, we compared them to all age-related genes, using Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations (Ashburner et al. 2000; Kanehisa et al. 2008). The result was robust when using all expressed genes as a background. Module 1 genes (N = 184) were significantly enriched in the KEGG pathway “neuroactive ligand-receptor interaction,” “calcium signaling pathway,” and “long-term potentiation,” as well as 21 GO terms, all related to synaptic transmission, signal transduction, ion transport, and cell–cell communication (Bonferroni-corrected one-sided hypergeometric test, P < 0.05) (Supplemental Table S8).
The three KEGG pathways enriched in module 1 genes, “neuroactive ligand-receptor interaction,” “calcium signaling pathway,” and “long-term potentiation,” form a network associated with key cognitive functions, such as memory and learning (Fig. 2D; Bliss and Collingridge 1993; Lynch 2004; Malenka and Bear 2004; Cooke and Bliss 2006). Notably, besides genes, this network includes a number of neurotransmitters. To test whether human-specific changes identified at the mRNA expression level are reflected at the neurotransmitter level, we used a published time-series where metabolite concentrations were measured in the PFC of 50 humans, 12 chimpanzees, and 49 rhesus macaques by gas chromatography and mass spectrometry (Fu et al. 2011). Among six neurotransmitters detected in this experiment, three (glutamate, gamma-aminobutyric acid [GABA] and aspartate) showed human-specific concentration profiles across development. This proportion is significantly greater than expected by chance, given the number of human-specific metabolites (one-sided hypergeometric test, P = 0.019, odd's ratio = 9.2). Importantly, concentration profiles of all three neurotransmitters correlated positively with expression profiles of the module 1 genes in the human PFC (Pearson rmedian > 0.90). Specifically, lower concentrations of glutamate and aspartate were observed in human infants compared with chimpanzees and macaques of the same age (one-sided z-test, P < 0.001), with GABA showing the same trend (Fig. 2E). The positive correlation between concentration profiles of the three neurotransmitters and module 1 expression profiles could not be explained by differences in brain growth trajectories among species (Supplemental Fig. S7; Supplemental Table S9). Furthermore, genes directly associated with the three neurotransmitters were significantly overrepresented in module 1 (one-sided hypergeometric test, P = 0.02, odds ratio = 3.5). The association between gene expression changes and neurotransmitter-level changes both supports the authenticity of human-specific changes observed at the mRNA level and also suggests the influence of these changes on human brain functionality.
Module 1 overlaps with MEF2A-mediated pathways and reflects a human-specific delay in cortical synaptogenesis
Functional enrichment of module 1 genes in genes associated with synaptic function fits well with the known involvement of its putative regulators, MEF2A, EGR1, EGR2, and EGR3, in synaptogenesis and synaptic transmission regulation. Among the four TFs, MEF2A is an upstream regulator of the other three (Flavell et al. 2008) and might, therefore, be one of the main regulators of module 1 genes. Supporting this notion, expression profiles of predicted MEF2 target genes and MEF2 target genes identified experimentally in rat hippocampal neurons (Flavell et al. 2008) correlated significantly with module 1 expression profiles (Supplemental Fig. S8). Furthermore, experimentally identified MEF2 target genes were overrepresented among module 1 genes (one-sided hypergeometric test, P = 0.043, odd's ratio = 2.7). Notably, among MEF2A targets showing correlated expression with module 1 genes, are ARC, SYNGAP1, and NR4A1 (also known as NUR77), previously shown to inhibit synapse formation and morphogenesis in response to MEF2A and MEF2D activation (Fig. 5A,B; Flavell et al. 2006; Shalizi et al. 2006). Furthermore, expression patterns of two known MEF2A activators, brain-derived neurotrophic factor (BDNF), which is linked to neuronal survival (Liu et al. 2003; Shalizi et al. 2003), as well as PPP3CB (also known as calcineurin A beta), which is involved in calcium influx-dependent dephosphorylation of MEF2A (McKinsey et al. 2002; Shalizi et al. 2006), showed human-specific delays consistent with the MEF2A expression profile and the average expression profile of the module 1 genes (Fig. 5B). Additionally, concentrations of glutamate, the main neurotransmitter activating the calcineurin-mediated signaling pathway, also showed age-dependent changes consistent with the module 1 expression profile (Fig. 2E). Thus, the expression profile of MEF2A during human development is consistent with the developmental profiles of known MEF2A activity-dependent regulators.
Figure 5.
Expression delay in MEF2A-associated pathway in human PFC development. (A) Schematic representation of MEF2A-associated pathway in cortical neurons. MEF2A can be activated by BDNF, which is also a target of MEF2A (Liu et al. 2003; Shalizi et al. 2003; Flavell et al. 2008), or calcium-dependent calcineurin (Flavell et al. 2006). Activity-dependent regulation of MEF2A transcription promotes the expression of target genes, such as ARC (activity-regulated cytoskeletal associated protein) and SYNGAP1 (synaptic localized Ras GAP), to promote elimination of excitatory synapses (Flavell et al. 2006). Activity-dependent regulation of MEF2A can also promote the transcription of target genes, such as NR4A1, to inhibit the postsynaptic differentiation (Shalizi et al. 2006). SYP and DLG4 are the marker genes for presynapse and postsynapse (Masliah et al. 1990; Hunt et al. 1996). (B) Expression profiles of genes shown on panel A during human, chimpanzee, and macaque PFC development. Points represent individuals (red, human; blue, chimpanzee; green, macaque), lines show cubic spline curves. MEF2A, the activators of MEF2A (PPP3CB and BDNF), and the postsynaptic marker gene (DLG4) belong to module 1. Target genes of MEF2A (e.g., EGR), the presynaptic marker (SYP), and three neurotransmitters (glutamate [Glu], aspartate [Asp], and GABA) show human-specific expression delay resembling closely the module 1 profile.
Since MEF2 is known to be involved in activity-dependent regulation of synaptic development and plasticity (Flavell et al. 2006; Shalizi et al. 2006), we hypothesized that module 1 might also be enriched in genes affected by neuronal activation. To test this, we exposed cortical neurons harvested from embryonic day 15 mice to bicuculline (Bic), which leads to calcium influx through NMDA receptors (Hardingham et al. 2002), or to potassium chloride (KCl), which leads to membrane depolarization (Didier et al. 1989). Using Agilent mouse genome oligo microarrays (4x44K), we identified 336 and 947 genes, respectively, that were up-regulated under Bic and KCl stimuli compared with controls (one-sided t-test, P < 0.05, fold-change > 2). In agreement with our prediction, module 1 genes showed significant overlap with genes up-regulated by both types of neuronal activation (one-sided hypergeometric test, P = 0.00008 under Bic stimulus; P = 0.01 under KCl stimulus) (Supplemental Fig. S9; Supplemental Table S10). Among the module 1 genes up-regulated by neuronal activation were several known synaptic genes, including BDNF and two neurotransmitter receptors, GABRG1 and GABRG2 (Supplemental Table S11).
Taken together, these results indicate that module 1 expression profiles reflect a human-specific change in timing of an activity-dependent synaptic development program. Supporting this notion, expression profiles of 724 synapse-related genes annotated by GO and expressed in the PFC, correlated positively and significantly with module 1 expression profiles (Supplemental Fig. S8; Supplemental Table S12). In contrast, profiles of other neuronal genes did not display such a correlation.
A shift in timing of expression of synaptic genes should be reflected by a corresponding shift in timing of synaptogenesis. Previous studies have shown that synaptic density in the human PFC peaks between 3.5 and 10 yr of age (Huttenlocher and Dabholkar 1997; Glantz et al. 2007). In contrast, in macaques, maximal synaptic density in the PFC is reached shortly after birth (Rakic et al. 1986). This suggests that timing of cortical synaptogenesis in human infants is delayed compared with that in macaques (Huttenlocher and Dabholkar 1997). Our expression data agree with these reports. Importantly, our observations indicate that this delay is specific to humans, with both chimpanzees and macaques developing at a faster rate. If extension of synaptic development is indeed specific to human infants, it might be associated with evolution of human-specific cognitive abilities.
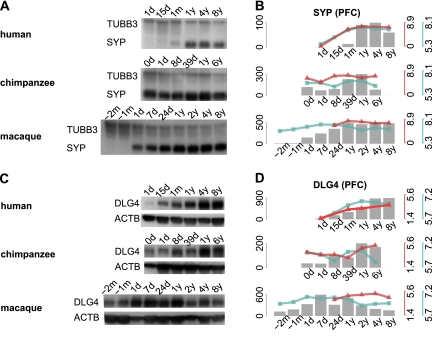
Knowing the timing of cortical synaptogenesis in chimpanzees is critical for evaluating this hypothesis, but has not been reported previously. Thus, to test whether delay in timing of cortical synaptogenesis is specific to humans, we measured synaptic density changes in the three species using two approaches. First, we quantified expression levels of two known synaptic density markers: the presynaptic marker SYP (also known as synaptophysin) and the postsynaptic marker DLG4 (also known as PSD95) (Masliah et al. 1990; Hunt et al. 1996). mRNA expression profiles of both genes coincided with module 1 profiles (Fig. 5B). Notably, in the CBC, mRNA expression profiles of both DLG4 and SYP shown were nearly synchronous in humans and chimpanzees. Furthermore, DLG4 and SYP expression in both species was equally delayed with respect to the macaques (Supplemental Fig. S10). This result indicates that human-specific profiles of these genes in the PFC are not caused by methodological problems, and also confirms that the human-specific delay in synaptogenesis is not observed in the CBC. We further measured protein expression of the two markers in the PFC of the three species by Western blot and by use of label-free proteomics time-series data measured in the PFC of 12 humans, 12 chimpanzees, and 12 rhesus macaques (Fig. 6; Fu et al. 2011). Both the Western blot and proteomic results showed that expression levels of the two synaptic marker proteins peaked at 4–8 yr of age in the human PFC and within the first year of life in the chimpanzee and macaque PFC. These results are in full agreement with previously reported synaptic density profiles measured in human and macaque cortex by electron microscopy (Rakic et al. 1986; Huttenlocher and Dabholkar 1997), as well as with levels of SYP and DLG4 proteins previously measured in human PFC by Western blot (Glantz et al. 2007). More importantly, these results show that both the mRNA and protein expression levels of the two synaptic density markers follow the human-specific profile characteristic to module 1 genes.
Figure 6.
DLG4 and SYP expression during PFC development. (A,C) Western blots conducted, respectively, using SYP- and DLG4-specific antibodies in PFC samples of individuals of different age in the three species. The age of each individual is shown above the lanes (m, months; d, days; y, years). DLG4 and SYP bands were observed at the predicted molecular weight of 95 kDa and 38 kDa. An equal amount of protein was loaded to each lane. ACTB (also known as beta actin) and TUBB3 (also known as Tubulin beta-III) were used as loading controls (bands at the predicted weight of 42 kDa and 55 kDa). (B,D) Protein expression level of SYP and DLG4 in PFC development measured as the integrated optical density (IOD) from the Western blots bands (gray bars) and measured using published mass-spectrometry–based protein time-series (red lines) (Fu et al. 2011), as well as mRNA expression levels measured using microarrays in this study (blue line). The y-axes show the expression values across samples. The axis' colors correspond to the measurements: gray, Western blot; red, mass spectrometry; and blue, microarrays.
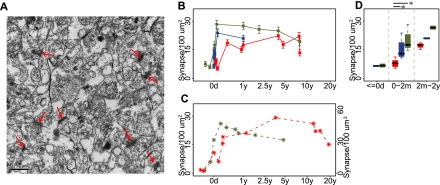
In a second set of experiments, we directly evaluated synaptic density in the PFC of the three species by counting the number of synapses in 12 humans, six chimpanzees, and nine macaques using transmission electron microscopy (Fig. 7A). From a total of 2700 electron micrographs analyzed independently by two investigators (X Jiang and A Oleksiak, unpubl.), 7801–9168 synapses were identified. Based on these data, we observed that synaptic density peaks at ∼4 yr in humans, and shortly after birth in macaques (Fig. 7B, Supplemental Fig. S11A). These observations are in line with previous studies (Fig. 7C; Rakic et al. 1986; Huttenlocher and Dabholkar 1997). In the chimpanzee PFC, due to uneven age distribution and lower histological integrity of the samples, synapse numbers could only be quantified reliably over the first year of life. Even based on this limited sample set, a sharp increase in synapse numbers could be observed in the chimpanzee PFC shortly after birth. This increase mirrored closely the one observed in the macaque PFC and differed significantly from a much smaller increase in synaptic density found in the human PFC (one-sided Wilcoxon test, P < 0.05) (Fig. 7D; Supplemental Fig. S11B). This result further supports our notion of delayed synaptogenesis in the human PFC compared with that of both chimpanzees and macaques.
Figure 7.
Synaptic density changes during human, chimpanzee, and macaque PFC development. (A) Example of synapses viewed by electron microscopy (red arrows), in the PFC of a 32-d-old chimpanzee. (B) Mean synaptic density per 100 μm2 measured in the PFC of humans (red), chimpanzees (blue), and rhesus macaques (green) at different ages. (Error bars) 95% confidence intervals obtained by bootstrapping synaptic density values within samples 1000 times. Independent assessment of synaptic density by another investigator is plotted on Supplemental Figure S11. (C) Mean synaptic density in macaque per 100 μm2 and in human per 100 μm3 measured in previous studies (Rakic et al. 1986; Huttenlocher and Dabholkar 1997). (D) Statistical analysis of synaptic density in three age groups. The distribution of mean synaptic density from samples within each age group is shown in a boxplot. Within the age range of 0–2 mo, PFC synaptic density in humans is significantly lower than in both chimpanzees and macaques (one-sided Wilcoxon test, P = 0.016 in human–chimpanzee comparison and P = 0.018 in human–macaque comparison), while there was no significant synaptic density difference between chimpanzees and macaques (P > 0.1). Sample numbers were not sufficient to estimate statistical significance in the other two age intervals.
Indication of recent positive selection for delayed synaptic development
Taken together, our results indicate that the module 1 expression profiles reflect a human-specific shift in the timing of cortical synaptogenesis, which coincides with a shift in the synaptic development program mediated by MEF2A. The large magnitude of this timing difference, as well as its association with neural development, a process generally known for its evolutionary conservation (Finlay and Darlington 1995; Reichert 2009), suggests an extraordinary change during the last few million years of human evolution.
While gene expression divergence among species might be caused by relaxation of evolutionary constraint, the human-specific shift in timing of module 1 genes is not caused by a lack of conservation in humans: Module 1 genes are highly conserved among mammals, and their promoters contain fewer human single nucleotide polymorphisms (Hinds et al. 2005) than do the promoters of all 8613 age-related genes (P = 0.0002) (Supplemental Fig. S6B–D).
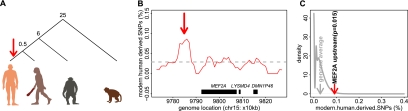
If delayed synaptic development resulted in a beneficial phenotypic trait, such as improved cognitive abilities, the corresponding genomic changes would have been positively selected on the human evolutionary lineage. Signatures of recent positive selection can be identified in the human polymorphism data surrounding the beneficial mutation. Specifically, positive selection that took place within the last 300,000 yr should result in loci with an excess of high-frequency derived SNPs in humans and presence of the ancestral allele in the Neanderthals (Fig. 8A; Green et al. 2010). Among three potential regulators of the synaptic development program, BDNF, PPP3CB, and MEF2A, we identified a significant excess of high-frequency human-derived SNPs in the upstream region of MEF2A (Fig. 8B,C). Assuming that the positive selection signature is associated with the human-specific MEF2A ontogenetic expression profile, this result suggests that the human-specific delay in cortical synaptogenesis had a beneficial effect on human fitness and took place after the separation of the human and the Neanderthal evolutionary lineages.
Figure 8.
Signature of recent positive selection upstream of the MEF2A gene. (A) The phylogenetic relationship of human, Neanderthal, chimpanzee, and rhesus macaque species. (Red arrow) Human lineage. The numbers show approximate divergence time in millions of years (Kumar and Hedges 1998; Pääbo 1999; Chen and Li 2001). (B) Proportion of human-derived SNPs measured using a 50-kb sliding window in the MEF2A gene region (red). SNPs were classified as derived according to the method described by Green et al. (2010). (Gray dashed line) Genome average. (Red arrow) Location, upstream of MEF2A, with significant excess of human-derived SNPs (one-sided Fisher's exact test, P = 0.00006). (C) Distribution of the proportion of human-derived SNPs for all windows across the human genome. (Red arrow) Probability of finding the observed proportion of human-derived SNPs in the location 50–100 kb upstream of MEF2A, estimated from the genome distribution.
Discussion
The emergence of the fascinatingly complex human cognitive phenotype within a time period restricted to several million years, or even less, has been one of the major conundrums of evolutionary biology (Carroll 2003). Our study provides an insight into potential evolutionary and functional mechanisms underlying this evolutionary innovation.
Our results, based on microarray data and RNA-seq, confirm that developmental expression patterns in the PFC and cerebellum have undergone excessive developmental remodeling since the separation of the human and the chimpanzee evolutionary lineages. This remodeling is particularly pronounced in the PFC: There are 12 times more developmental gene expression changes in humans than in chimpanzees. Here, we focused on one group of 184 coexpressed genes (module 1) representing the most prominent pattern of the human-specific changes observed in PFC. The expression pattern of module 1 differs strikingly between humans and the other two species, with expression peaking at 5 yr of age in the humans and shortly after birth in the chimpanzees and macaques.
Functionally, module 1 genes are involved in pathways associated with synaptic transmission. Although the number of module 1 genes is relatively small, it reflects a general trend: The average expression pattern of all 724 genes associated with synapses and synaptic functions resembles closely the expression of module 1 genes (Supplemental Fig. S8A). This human-specific delay in synaptic development is supported by our analysis of the mRNA and protein levels of the pre- and postsynaptic density markers, SYP and DLG4, as well as by direct quantification of synaptic density in the human, chimpanzee, and rhesus macaque PFC with electron microscopy.
The majority of the human-specific expression changes found in the PFC have similar developmental trajectories in the three species but exhibit a temporal delay in humans. Focusing on module 1 specifically, gene expression increase in human infants during the first 5 yr of postnatal development is mirrored by a similar increase in macaques during the last months of fetal development. Importantly, we find that delay in the peak expression of module 1 genes in the PFC extends far beyond the one expected from life history differences among the three species and reflects previously reported differences in timing of cortical synaptogenesis between humans and macaques (Rakic et al. 1986; Huttenlocher and Dabholkar 1997). Most importantly, our results demonstrate that this delay is not observed in chimpanzees. Thus, extension of human cortical synaptogenesis took place after the separation of the human and the chimpanzee lineages and may therefore be associated with the emergence of human-specific cognitive traits.
The coordinated delay in expression of module 1 genes in the human PFC could be linked to a similar delay in the expression of known regulators of activity-dependent synaptic development programs, such as MEF2A (Lyons et al. 1995; Flavell et al. 2006; Shalizi et al. 2006). MEF2A plays multiple roles in neuronal development, including neuronal survival (Mao et al. 1999; Shalizi et al. 2003), dendritic differentiation (Shalizi et al. 2006), synaptic density of hippocampal neurons (Flavell et al. 2006; Barbosa et al. 2008), spine density in nucleus accumbens (Pulipparacharuvil et al. 2008), and both synapse weakening/elimination and synaptic strengthening (Flavell et al. 2008). In our data, expression profiles that correlated with MEF2A include predicted reported MEF2A target genes associated with both synaptic weakening (e.g., ARC, SYNGAP1, and NR4A1) and synaptic strengthening (e.g., BDNF, LGI1). This may imply that in human cortical ontogenesis, the timing of synaptogenesis and/or subsequent synaptic differentiation and pruning differs from that in other primates. Although MEF2A activity depends on phosphorylation (Shalizi et al. 2006), its activity during synaptic development may also be transcriptionally regulated. Supporting this possibility, we have shown that the MEF2A expression profile is tightly correlated with its downstream targets (see above). Furthermore, the MEF2A promoter region contains an MEF2A binding site, suggesting autoregulation. Further work focusing on phosphorylation and the dynamics of the cellular localization of MEF2A will be necessary to understand its exact roles in the regulation of primate cortex development.
While developmental profile differences between humans and other primates are particularly prominent in the first years of life, they persist throughout adulthood. Specifically, during adulthood, the expression trajectories of both module 1 genes, as well as all synapse-related genes, decline in parallel across the three species. Intriguingly, however, the average expression levels of these genes are consistently higher in humans than in chimpanzees or macaques during adulthood (Supplemental Fig. S2C). Similarly, thrombospondin 4 (THBS4), previously shown to be associated with synaptic functions and synapse formation (Caceres et al. 2003, 2007), also shows significantly higher expression in the human PFC compared with PFCs of the other two species throughout adulthood. This implies that humans might sustain higher levels of synaptogenesis or synaptic activity throughout adult life.
What could be the functional role of such a timing shift? Humans are born in an altricial state and undergo both rapid and extended brain growth compared with that of macaques and chimpanzees (Leigh 2004). A related view is that human ontogeny has acquired a novel phase, between 3 and 7 yr of age, when human infants are not being breast-fed but are still highly dependent on adults (Bogin 1997). It has been proposed that this ontogenic extension/inclusion allows a longer period of interaction between human infants and their social environment and forms the main basis of human intelligence (Gould 1977; Johnson 2001; Langer 2006). Notably, 5–7 yr of age also appears to correspond to a shift in cognitive maturity, such as increased self-regulation (Bogin 1997). Our data suggest that the timing of synaptic development in the human PFC has undergone a parallel extension. This extension might be connected with the roles of the PFC in high-level processes such as abstract thinking and social behavior (Clark et al. 2001; Wood and Grafman 2003; Rilling 2006; Barbey et al. 2009). Further, it is appealing to speculate that shift in timing of cortical synaptogenesis in human children may have resulted in major cognitive differences between human and chimpanzee adults, as well as contributed to maintenance of healthy cognitive activity over the entire duration of human lifespan. If true, this could be an example of a mechanistic connection between two major life history changes in humans: the prolonged growth period and greater longevity (Hawkes et al. 1998; Kaplan et al. 2000).
Studies of early massive brain damage and microcephaly have demonstrated that brain size alone cannot explain the full spectrum of cognitive differences between humans and apes (Mochida and Walsh 2001). Our results suggest that delay in cortical synaptogenesis, extending the period of synapse formation to over 5 yr in humans from a few months in chimpanzees and macaques, could be a potential mechanism contributing to the emergence of human-specific cognitive skills. Notably, we also find that this delay could have happened after the separation of the human and the Neanderthal lineages. While Neanderthal brains were on average 100 mL larger than brains of anatomically modern humans (Stringer and Gamble 1993), our results raise the possibility that cortical synaptic development in Neanderthals may have been faster. Such a notion is concordant with reports that dental maturation and cranial development in Neanderthals was more similar to apes than to modern humans (Smith et al. 2010). Additional work to identify the exact genetic basis of the human-specific developmental delay with subsequent comparison to the genomes of Neanderthals (Green et al. 2010) and Denisovans (Reich et al. 2010) is needed to verify this notion.
We note that the study presented here is far from being comprehensive: We focused on one major developmental pattern change specific to the human PFC, leaving another four PFC patterns and two CBC patterns unexplored. Most of these patterns are enriched in genes directly associated with neural functions. For instance, human-specific CBC module 2 displays a developmental pattern similar to that of the PFC module 1 and contains genes enriched in the GO category “regulation of synaptic transmission” (Supplemental Table S8). Although these genes do not overlap significantly with genes in the PFC module 1 (one-sided hypergeometric test, P = 0.32), functional similarity between two patterns indicates parallel functional changes in the two brain regions. Thus, the human cerebellum may also have undergone evolutionary changes in the process of synaptogenesis and/or synaptic functionality, but to a more limited extent compared with the PFC. As the CBC and the PFC are functionally interconnected (Rilling 2006), it is plausible that rapid functional evolution in the PFC required corresponding adaptations in other brain regions by means of developmental heterochrony, as well as remodeling of neural connections (Ramnani 2006; Semendeferi et al. 2010). Further work is needed to obtain a comprehensive view of mechanisms and functional consequences of extensive developmental remodeling found in the human brain.
Methods
A complete detailed description of methods is provided in the Supplemental Information.
RNA microarray and sequencing
Samples were obtained from frozen postmortem tissue from healthy individuals. All subjects had suffered sudden deaths. PFC dissections were made from the frontal part of the superior frontal gyrus. All samples had good RNA quality. For microarray experiments, for each sample, total RNA was extracted from 100 mg tissue, and 2 μg of isolated RNA was hybridized to an Affymetrix Human Gene 1.0 ST array. Samples were processed in five batches per brain region. In the pooled sample RNA-seq experiments, we used three human samples (newborn, young, old), two chimpanzee samples (newborn, young), and two macaque samples (newborn, young), prepared by pooling total RNA from five individuals (Supplemental Table S1). Sequencing libraries were prepared according to the Illumina paired-end sample preparation protocol. Each sample was sequenced in a separate lane in the Illumina Genome Analyzer II system, using the 75-bp paired-end sequencing protocol. In the second RNA-seq experiments, sequencing libraries were prepared according to Illumina strand-specific RNA-seq library preparation procedure for humans and Illumina nonstrand specific RNA-seq library preparation procedure for chimpanzees and macaques. Each library was sequenced in a separate lane using the Illumina Genome Analyzer II system to a length of 100 bp. For testing age and species difference in expression levels, we used polynomial regression models and analysis of covariance, following the method described by Faraway (2002).
Estimating time-shift by dynamic time warping
We used a modified dynamic time warping algorithm, DTW-Significance (DTW-S) algorithm, to estimate the time-shifts between the time series expression curves of two species (R package “TimeShift” for DTW-S is available at http://www.picb.ac.cn/Comparative/data.html) (Yuan et al. 2011). A detailed explanation of DTW-S is provided in the Supplemental Information. Briefly, for each gene, we aligned expression-age trajectories from a pair of species. For this, we interpolated cubic spline curves (degrees of freedom, 3) using estimated conception age for each individual. The resulting curves ranged from rhesus macaque birth age to each species' maximum lifespan. We matched points in one time series to the other time series, keeping the age order of individuals, and calculated the distance between the resulting curves. By searching the space of all possible alignments, we chose one with the minimal distance between curves.
Identifying response genes upon neuronal activation
The culture of cortical neurons, RNA isolation, hybridization to microarrays, data preprocessing, and analysis were performed according to the method described by Flavell et al. (2008). Cortical neurons isolated from 15-d-old embryo mouse were cultured in vitro for 12 d. Neurons were then exposed with extracellular Bic or KCl or without stimulus. We measured gene expression profiles of cultured neurons under either stimulus or no stimulus, using Agilent whole-mouse genome oligo microarrays (4x44k). Three biological replicates were performed for each group. To identify the probes that show different expression profiles between stimuli and control, probes with a greater than two times fold-change and a P < 0.05 by t-test were selected.
Western blot analysis
Western blots were performed according to the method described by Glantz et al. (2007). The integrated optical density (IOD) of each band was measured by the Gel-Pro Analyzer.
Electron microscopy
Sample preparation for electron microscopy followed the method described by Huttenlocher and Dabholkar (1997). Briefly, samples <1 mm3 per sample were fixed using 2.5% glutaraldehyde in 0.1 M PBS buffer for >2 h. Samples were then washed in PBS buffer and dehydrated in graded ethanol (50%–90%) and propanol (90%–100%). Next, samples were embedded in 100% samacetone and solidified in an oven. Seventy-nanometer sections were cut and double stained with 3% uranyl acetate and lead citrate and were observed under a JEM-1230 transmission electron microscope and photographed under 20,000 magnification. For each sample, 100 images were taken at randomly chosen positions with good sample quality. Within each image, synapses were identified where a thin continuous presynaptic line plus a thicker, parallel postsynaptic band could be observed according to the method described by Huttenlocher and Dabholkar (1997). Synapses were counted independently by two investigators, blind to the age and species information of individuals.
Data access
The mRNA expression data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE22570/GSE29138/GSE29139. The RNA-seq data have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession numbers SRP005169/SRP009336. Processed RNA-seq data sets and the electron microscopy images are available at http://www.picb.ac.cn/Comparative/data_ms_age_divergence_2010.html.
Acknowledgments
We thank the NICHD Brain and Tissue Bank for Developmental Disorders, the Netherlands Brain Bank, the Chinese Brain Bank Center, and Dr. H.R. Zielke and Dr. J. Dai in particular for providing the human samples; The Yerkes Primate Center, the Biomedical Primate Research Centre, the Anthropological Institute and Museum of the University of Zurich for chimpanzee samples, and Dr. R. Martin and Dr. W. Scheffran in particular for providing the chimpanzee samples; the Suzhou Drug Safety Evaluation and Research Center, and C. Lian, H. Cai, and X. Zheng in particular for providing the macaque samples; the Electron Microscopy Facility of the Institute of Neuroscience, SIBS, CAS, and Q. Song, and Y. Kong in particular for assistance with electron microscopy experiments; G. Xie, P. Mazin, Z. He, E. Lizano, F. Xue, and A. Xu for assistance and S.M. Myles for sharing data; J. Boyd-Kirkup for editing the manuscript; and J.E. Dent, M. Lachmann, and all members of the Comparative Biology Group in Shanghai for helpful discussions and suggestions. We thank the Ministry of Science and Technology of the People's Republic of China (grant no. 2007CB947004 [to P.K.]), the Chinese Academy of Sciences (grant no. KSCX2-YW-R-094 and KSCX2-YWR-251 [to P.K.]), the Shanghai Institutes for Biological Sciences (grant no. 2008KIT104 [to P.K.]), the National Science Foundation of China (grant nos. 31010022 and 31050110128 [to P.K. and M.S.]), 973 program (grant no. 2011CBA00400 [to Z.Q.]), the Hundred Talent program from the Chinese Academy of Sciences (to Z.Q.), the Max Planck-Society, and the Bundesministerum fuer Bildung und Forschung for financial support. M.S. was supported by fellowships from the Chinese Academy of Sciences (2009Y2BS12) and the European Molecular Biology Organization (EMBO ALTF 1475-2010).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.127324.111.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey AK, Krueger F, Grafman J 2009. An evolutionarily adaptive neural architecture for social reasoning. Trends Neurosci 32: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. 2008. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci 105: 9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL 1993. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39 [DOI] [PubMed] [Google Scholar]
- Bogin B 1997. Evolutionary hypotheses for human childhood. Yearb Phys Anthropol 40: 63–89 [Google Scholar]
- Bogin B 2009. Childhood, adolescence, and longevity: a multilevel model of the evolution of reserve capacity in human life history. Am J Hum Biol 21: 567–577 [DOI] [PubMed] [Google Scholar]
- Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C 2003. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci 100: 13030–13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM 2007. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb Cortex 17: 2312–2321 [DOI] [PubMed] [Google Scholar]
- Carroll SB 2003. Genetics and the making of Homo sapiens. Nature 422: 849–857 [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DA, Mitra PP, Wang SSH 2001. Scalable architecture in mammalian brains. Nature 411: 189–193 [DOI] [PubMed] [Google Scholar]
- Cooke SF, Bliss TV 2006. Plasticity in the human central nervous system. Brain 129: 1659–1673 [DOI] [PubMed] [Google Scholar]
- Davis S, Bozon B, Laroche S 2003. How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav Brain Res 142: 17–30 [DOI] [PubMed] [Google Scholar]
- de Magalhães JP. 2006. http://genomics.senescence.info/species AnAge: the Animal Ageing Database, Build 9.
- de Magalhães J, Costa J 2009. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol 22: 1770–1774 [DOI] [PubMed] [Google Scholar]
- Didier M, Roux P, Piechaczyk M, Verrier B, Bockaert J, Pin JP 1989. Cerebellar granule cell survival and maturation induced by K+ and NMDA correlate with c-fos proto-oncogene expression. Neurosci Lett 107: 55–62 [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Metzler D, Schwarz C, Pääbo S 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 70: 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraway J. 2002. Practical regression and ANOVA using R. http://cran.rproject.org/doc/contrib/Faraway-PRA.pdf.
- Finlay BL, Darlington RB 1995. Linked regularities in the development and evolution of mammalian brains. Science 268: 1578–1584 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim T, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME 2006. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Giavalisco P, Liu X, Catchpole G, Fu N, Ning ZB, Guo S, Yan Z, Somel M, Pääbo S, et al. 2011. Rapid metabolic evolution in human prefrontal cortex. Proc Natl Acad Sci 108: 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavan JA 1969. A handbook of living primates: morphology, ecology and behaviour of nonhuman primates. J. R. Napier and P. H. Napier. Am Anthropol 71: 357–358 [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA 2006. Natural selection on gene expression. Trends Genet 22: 456–461 [DOI] [PubMed] [Google Scholar]
- Glantz LA, Gilmore JH, Hamer RM, Lieberman JA, Jarskog LF 2007. Synaptophysin and postsynaptic density protein 95 in the human prefrontal cortex from mid-gestation into early adulthood. Neuroscience 149: 582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. 1977. Ontogeny and phylogeny. Harvard University Press, Cambridge, MA. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neanderthal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H 2002. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci 5: 405–414 [DOI] [PubMed] [Google Scholar]
- Hawkes K, O'Connell JF, Jones NG, Alvarez H, Charnov EL 1998. Grandmothering, menopause, and the evolution of human life histories. Proc Natl Acad Sci 95: 1336–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079 [DOI] [PubMed] [Google Scholar]
- Hunt CA, Schenker LJ, Kennedy MB 1996. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J Neurosci 16: 1380–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS 1997. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol 387: 167–178 [DOI] [PubMed] [Google Scholar]
- Johnson MH 2001. Functional brain development in humans. Nat Rev Neurosci 2: 475–483 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H, Hill K, Lancaster J, Hurtado AM 2000. A theory of human life history evolution: diet, intelligence, and longevity. Evol Anthropol 9: 156–185 [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S 2006. Evolution of primate gene expression. Nat Rev Genet 7: 693–702 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB 1998. A molecular timescale for vertebrate evolution. Nature 392: 917–920 [DOI] [PubMed] [Google Scholar]
- Langer J 2006. The heterochronic evolution of primate cognitive development. Biol Theory 1: 41–43 [Google Scholar]
- Leigh S 2004. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol 62: 139–164 [DOI] [PubMed] [Google Scholar]
- Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, Tourtellotte WG 2007. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci 35: 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cavanaugh JE, Wang Y, Sakagami H, Mao Z, Xia Z 2003. ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci 100: 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MA 2004. Long-term potentiation and memory. Physiol Rev 84: 87–136 [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN 1995. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci 15: 5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF 2004. LTP and LTD: an embarrassment of riches. Neuron 44: 5–21 [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME 1999. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286: 785–790 [DOI] [PubMed] [Google Scholar]
- Masliah E, Terry RD, Alford M, DeTeresa R 1990. Quantitative immunohistochemistry of synaptophysin in human neocortex: an alternative method to estimate density of presynaptic terminals in paraffin sections. J Histochem Cytochem 38: 837–844 [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci 27: 40–47 [DOI] [PubMed] [Google Scholar]
- McNamara KJ. 1997. Shapes of time. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Mochida GH, Walsh CA 2001. Molecular genetics of human microcephaly. Curr Opin Neurol 14: 151–156 [DOI] [PubMed] [Google Scholar]
- Pääbo S 1999. Human evolution. Trends Cell Biol 9: M13–M16 [PubMed] [Google Scholar]
- Penin X, Berge C, Baylac M 2002. Ontogenetic study of the skull in modern humans and the common chimpanzees: neotenic hypothesis reconsidered with a tridimensional procrustes analysis. Am J Phys Anthropol 118: 50–62 [DOI] [PubMed] [Google Scholar]
- Povinelli DJ, Preuss TM 1995. Theory of mind: evolutionary history of a cognitive specialization. Trends Neurosci 18: 418–424 [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. 2008. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron 59: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS 1986. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science 232: 232–235 [DOI] [PubMed] [Google Scholar]
- Ramnani N 2006. The primate cortico-cerebellar system: anatomy and function. Nat Rev Neurosci 7: 511–522 [DOI] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H 2009. Evolutionary conservation of mechanisms for neural regionalization, proliferation and interconnection in brain development. Biol Lett 5: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK 2006. Human and nonhuman primate brains: are they allometrically scaled versions of the same design? Evol Anthropol 15: 65–77 [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J 2010. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex 21: 1485–1497 [DOI] [PubMed] [Google Scholar]
- Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y, Bonni A 2003. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci 23: 7326–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A 2006. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017 [DOI] [PubMed] [Google Scholar]
- Smith BH, Crummett TL, Brandt KL 1994. Ages of eruption of primate teeth: a compendium for aging individuals and comparing life histories. Am J Phys Anthropol 37: 177–231 [Google Scholar]
- Smith TM, Tafforeau P, Reid DJ, Pouech J, Lazzari V, Zermeno JP, Guatelli-Steinberg D, Olejniczak AJ, Hoffman A, Radovcic J, et al. 2010. Dental evidence for ontogenetic differences between modern humans and Neanderthals. Proc Natl Acad Sci 107: 20923–20928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. 2009. Transcriptional neoteny in the human brain. Proc Natl Acad Sci 106: 5743–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G et al. 2011. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol 9: e1001214 doi:1001210.1001371/journal.pbio.1001214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Gamble C 1993. In search of the Neanderthals: solving the puzzle of human origins. Thames and Hudson, New York [Google Scholar]
- Tomasello M. 2008. Origins of human communication. MIT Press, Cambridge, MA. [Google Scholar]
- Wood JN, Grafman J 2003. Human prefrontal cortex: processing and representational perspectives. Nat Rev Neurosci 4: 139–147 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chen YP, Ni S, Xu AG, Tang L, Vingron M, Somel M, Khaitovich P 2011. Development and application of a modified dynamic time warping algorithm (DTW-S) to analyses of primate brain expression time series. BMC Bioinformatics 12: 347 doi: 10.1186/1471-2105-12-347 [DOI] [PMC free article] [PubMed] [Google Scholar]