Abstract
DNA methylation (DNAm) plays diverse roles in human biology, but this dynamic epigenetic mark remains far from fully characterized. Although earlier studies uncovered loci that undergo age-associated DNAm changes in adults, little is known about such changes during childhood. Despite profound DNAm plasticity during embryogenesis, monozygotic twins show indistinguishable childhood methylation, suggesting that DNAm is highly coordinated throughout early development. Here we examine the methylation of 27,578 CpG dinucleotides in peripheral blood DNA from a cross-sectional study of 398 boys, aged 3–17 yr, and find significant age-associated changes in DNAm at 2078 loci. These findings correspond well with pyrosequencing data and replicate in a second pediatric population (N = 78). Moreover, we report a deficit of age-related loci on the X chromosome, a preference for specific nucleotides immediately surrounding the interrogated CpG dinucleotide, and a primary association with developmental and immune ontological functions. Meta-analysis (N = 1158) with two adult populations reveals that despite a significant overlap of age-associated loci, most methylation changes do not follow a lifelong linear pattern due to a threefold to fourfold higher rate of change in children compared with adults; consequently, the vast majority of changes are more accurately modeled as a function of logarithmic age. We therefore conclude that age-related DNAm changes in peripheral blood occur more rapidly during childhood and are imperfectly accounted for by statistical corrections that are linear in age, further suggesting that future DNAm studies should be matched closely for age.
Epigenetic modifications modulate gene expression via mechanisms that are highly coordinated throughout mammalian development (Reik 2007) and are subject to influence by environmental exposures (Schanen 2006). The best understood epigenetic modification is DNA methylation (DNAm), which in mammals occurs primarily at cytosines located 5′ to guanine in a CpG dinucleotide (Jaenisch and Bird 2003). This modification is less common in CpG-rich areas, known as CpG islands, which are often located in the promoter regions of many genes and involved in transcriptional regulation. Extensive DNAm is associated with chromatin structure, chromosome stability, transcriptional silencing of imprinted genes (genes that are differentially expressed based on their parent of origin), and the maintenance of the transcriptionally inactive X chromosome in females (Robertson 2005).
A growing number of human diseases are linked to disruption of epigenetic information, emphasizing the importance of DNAm. DNAm is a hallmark of human cancer, a link that was first recognized in 1983, when tumor cells were found to be aberrantly methylated relative to their normal counterparts due to a loss of methylation from repetitive regions of the genome (Feinberg and Vogelstein 1983). Aberrant DNAm within imprinting regions results in numerous other diseases, including Prader-Willi, Angelman, and Beckwith-Wiedemann syndromes, as well as transient neonatal diabetes mellitus (Arima et al. 2001). Finally, disorders associated with repeat instability are also linked to aberrant DNAm (Orr and Zoghbi 2007). Several of these syndromes are neurological disorders that arise from allelic expansion, allelic contraction, and/or an increase in local DNAm (Lemmers et al. 2010).
There is also normal variation in DNAm, and a substantial amount of recent evidence points to organismal age as a significant source of this variation (Issa et al. 1994, 1996; Ahuja et al. 1998; Nakagawa et al. 2001; Fraga et al. 2005, 2007; So et al. 2006; Fraga and Esteller 2007; Bjornsson et al. 2008; Christensen et al. 2009). A genome-wide decrease in DNAm with age has been observed in humans (Bollati et al. 2009), whereas some gene-specific CpG dinucleotides located in the promoters of genes become methylated with age (Herman et al. 1994, 1997, 1998; Kane et al. 1997; Ahuja and Issa 2000; Dammann et al. 2000; Esteller et al. 2000a,b,c; Burbee et al. 2001). Notably, all of these studies focused on adults, and it is unclear how much normal age-associated DNAm variation occurs during childhood and what the implications are for disease-based pediatric epigenetic studies. Here we investigate the normal variation of DNA methylation in the peripheral blood of a large pediatric cohort using a highly sensitive genome-wide assay that quantitates methylation level at single CpG dinucleotide resolution and show a more robust age-associated DNAm variation in children than in adults.
Results
Age-associated DNA methylation changes manifest during childhood
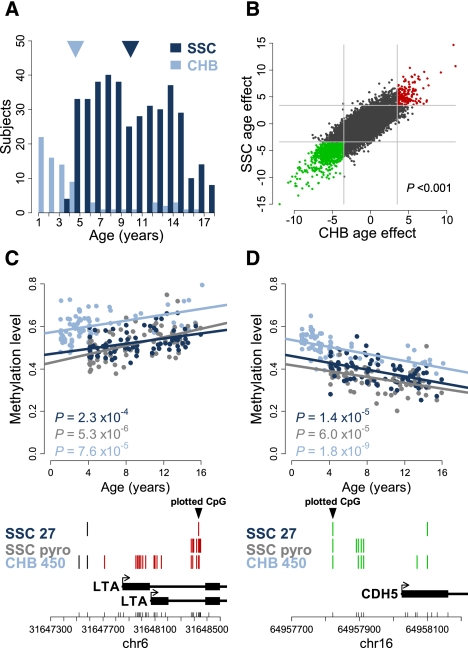
We investigated pediatric age-associated DNAm changes using DNA extracted from the peripheral blood of 398 healthy males from the Simons Simplex Collection (SSC), who are siblings of ASD individuals and do not present a clinical diagnosis of ASD. At the time of collection, these individuals had an age range from 3 to 17 yr (mean 9.9) (Fig. 1A; Supplemental Table 1) and were epityped using Infinium HumanMethylation27 BeadChips, which provide a quantitative measure of DNA methylation denoted as β, calculated as the ratio of methylated to total DNA. This is a highly reproducible (Supplemental Fig. 1A,B) and widely used assay (Bork et al. 2010; Teschendorff et al. 2010; Chen et al. 2011; Koch et al. 2011) that measures β at 27,578 CpG dinucleotides located proximal to the promoters of 14,475 well-annotated genes. To determine if age-associated DNAm is present in these individuals, we analyzed each locus using a linear fixed-effects regression model (see Methods) and identified 2078 pediatric age-associated loci: 1601 age-demethylated and 477 age-methylated loci (FDR < 0.01) (Supplemental Fig. 1C; Supplemental Table 2). To validate our methods statistically, we performed permutation tests and found that the asymptotic P-values were nearly equal to the permutation P-values (Supplemental Fig. 1D; Methods), and that our approach yielded results similar to other published findings (Supplemental Fig. 1E,F; Teschendorff et al. 2010; see Methods: Analysis of Age-Associated CpG Loci).
Figure 1.
Pediatric age-associated DNA methylation. (A) Age distribution of the 398 Simons Simplex Collection (SCC: dark blue) and 78 Children's Hospital Boston (CHB: light blue) pediatric subjects, with mean ages denoted by arrowheads (dark blue and light blue, respectively). (B) Scatterplot of the age effect as measured by the age-associated t-statistic for the SSC (y-axis) and CHB (x-axis) populations. Each point represents one CpG locus interrogated by both assays and those loci found to be significant (FDR < 0.01) in both populations are red (age-methylated) or green (age-demethylated). (Light gray lines) The significance thresholds in each population; the P-value is the significance of the correlation, as determined by permutation testing. (C,D) Validation of representative age-associated loci in the SSC population, with the CHB data overlaid for comparison. Infinium HumanMethylation27 (SSC Inf27: dark blue) and pyrosequencing (SSC pyro: gray) data are shown for the subset of 75 individuals from the SSC population. In addition, the same loci are shown for the entire CHB population (N = 78), run on the HumanMethylation450 (Inf450: light blue). Lines represent the linear regression of each set of data independently; the P-values are the age-effect significance in the population shown. The y-axis is the methylation level measured by all three assays. Below each plot is a schematic of the interrogated locus and annotated genes in the region. (Black triangle) The CpG locus shown in the above plot; CpG coverage of each assay is denoted by vertical lines, with red denoting a significant (FDR<0.01) age-methylating effect, and green a significant age-demethylating effect; those in black are not significantly associated with age. The chromosome (chr), total CpGs in the region (small vertical black lines), and relative genomic coordinates (NCBI build 36.1) are denoted on the x-axis below the gene schematic.
To corroborate these findings, we analyzed DNA extracted from the peripheral blood of an independent cohort of 78 healthy males collected from Children's Hospital Boston (CHB). At the time of collection, these individuals had an age range from 1 to 16 yr (mean 4.6) (Fig. 1A) and were epityped using Infinium HumanMethylation450 BeadChips that measure β at 485,512 CpG dinucleotides, including 94.2% of those found on the HumanMethylation27 panel. Analysis of age-associated DNA methylation in this group yielded 41,895 age-associated loci (35,997 age-demethylated, 5898 age-methylated, FDR < 0.01) (Supplemental Fig. 1G). Comparison of these results with the loci identified in the SSC group revealed a 62.7% overlap in significant CpG sites (P < 0.001) (Supplemental Fig. 1H), despite differences in age, race, population size, and assay (Fig. 1A; Supplemental Tables 1, 2). Moreover, the individual CpG loci had an overwhelmingly positive correlation in age effects (as measured by age-associated t-statistics) between the two populations (P < 0.001) (Fig. 1B), indicating that the age-associated changes in DNAm are not unique to the SSC group. Finally, pyrosequencing of sodium bisulfite–converted DNA spanning the genomic regions of two representative pediatric age-associated loci, in a subset of the SSC individuals (N = 75), corresponded well with the β values provided by the Infinium assay (Supplemental Fig. 2A,B), again showing significant age-associated DNAm (Fig. 1C,D). In fact, most surrounding CpGs also exhibited age-associated changes in DNAm, suggesting that these changes affect domains of DNA methylation rather than isolated CpGs (Fig. 1C,D, see bottom; Supplemental Fig. 2C–F).
To assess the novelty of these pediatric age-associated DNAm changes, we compared the age-associated loci in the SSC group with previously reported age-associated loci. We first compared age-dependent loci published in two adult populations, each with an equal number of disease and non-disease subjects: one from a type 1 diabetic study (T1D), the other from an ovarian cancer cohort (OC). Age-dependent DNA methylation was found in these adult cohorts independent of disease status, and the results were similar for both genders in the T1D cohort (Teschendorff et al. 2010). The published age-dependent t-statistics from both adult studies showed a stronger correlation with those identified in the SSC group than would be expected by chance alone (P < 0.001) (Supplemental Fig. 3A,B), and the pediatric age-associated loci comprised the majority of those published for either the T1D (53%) or OC (55%) populations (P < 0.001) (Supplemental Fig. 3C; Teschendorff et al. 2010). Similarly, we observed a significant, albeit more modest, overlap of the pediatric age-methylated loci with the age-methylated loci reported in whole blood of another adult study (34/131; P < 0.001) (Rakyan et al. 2010) that had replicated their findings in CD4+ T-cells and CD14+ monocytes. The pediatric age effects corresponding to these adult loci are significantly associated with the published age effects for whole-blood and CD14+ monocytes but not CD4+ T-cells (Supplemental Fig. 3D–F). Finally, the pediatric age-associated loci have a significant overlap with age-related loci identified in a study that compared CD34+ hematopoietic progenitor cells (HPC) from newborn cord blood with the same cells mobilized in peripheral blood of adults (301/542; P < 0.001) (Supplemental Fig. 3G; Bocker et al. 2011). Together, these data indicate that age-associated DNA methylation changes found in adult populations are also present in pediatric populations and suggest that at least some age-related changes are not a result of compositional changes in the cell types found in peripheral blood.
Pediatric DNA methylation changes and genomic context
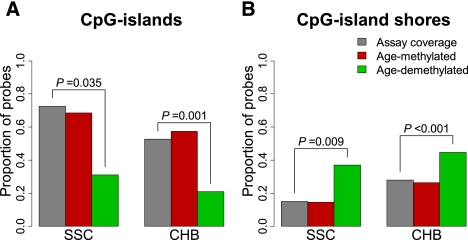
Adult age-associated DNAm changes have been reported to depend on CpG island context; specifically, age-demethylated nucleotides were shown to reside outside of CpG islands and age-methylated nucleotides within CpG islands (Christensen et al. 2009). To test these hypotheses in our pediatric populations, we compared the proportion of age-associated loci residing in CpG islands with the total number of CpG loci included on their respective DNA methylation panels via permutation testing (see Methods). We found fewer pediatric age-demethylated loci in CpG islands than would be expected by chance alone in both SSC (P = 0.035) and CHB populations (P = 0.001) (Fig. 2A). However, we do not observe a higher proportion of age-methylated loci within CpG islands, as was previously reported in adults. Since a substantial amount of DNAm variation has been identified to occur within the 2 kb adjacent to CpG islands or in CpG island “shores” (Irizarry et al. 2009), we examined the proportion of age-demethylated loci on CpG island “shores” and found a significantly larger proportion than expected by chance in both SSC (P = 0.009) and CHB populations (P < 0.001) (Fig. 2B). These data are consistent with those published in the T1D and OC studies where we also found that age-demethylated loci were underrepresented on CpG islands and more prevalent on CpG island shores (Supplemental Fig. 4A,B) (Teschendorff et al. 2010). Thus, the location of pediatric age-demethylation events in the context of CpG islands is consistent with previous reports of adult age-demethylated loci and suggests that DNAm is particularly dynamic in the regions immediately adjacent to CpG islands.
Figure 2.
Proportion of age-associated loci in CpG islands and “shores” in the SSC and CHB cohorts. Barplots of the proportion of age-methylated (red) and age-demethylated (green) loci relative to total assay coverage (gray) found in CpG islands (A) and CpG island “shores” (B). Significant permutation P-values are displayed.
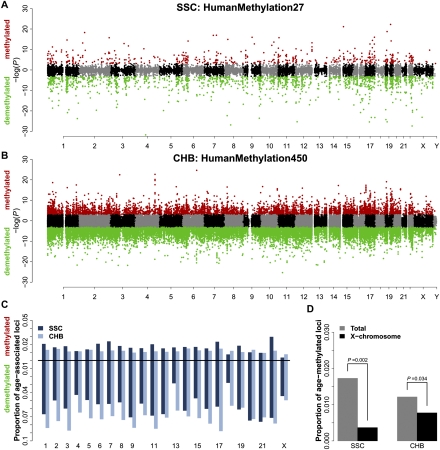
It is well known that specific regions of the genome are differentially methylated based on the biological functions of the genes contained within the region; the most striking example of this is X inactivation in females. To determine if age-associated DNAm is specific to certain regions of the genome, we displayed the genomic locations of the pediatric age-associated loci using a Manhattan plot, separating age-methylating from age-demethylating effects (Fig. 3A,B). These plots indicate that both the SSC and CHB pediatric groups have age-associated loci located throughout the genome, but upon closer examination suggest a deficit of age-associated methylation on the X chromosome.
Figure 3.
Pediatric age-associated loci by chromosome. (A,B) Modified Manhattan plots of age-associated loci in the SSC (A) and CHB (B) populations: loci positively correlated with age are displayed with a positive −log(P-value), and loci negatively correlated with age are displayed with a negative −log(P-value). Loci significantly age-methylated (red) or age-demethylated (green) (FDR < 0.01); otherwise loci are black or gray on alternating chromosomes. (C) Proportion of age-associated loci by chromosome in the SSC (dark blue) and CHB (light blue) populations: the proportion of age-methylated loci is displayed on the positive y-axis and age-demethylated loci on the negative y-axis. The Y chromosome is not shown because only seven probes are available in the SSC data. (D) Proportion of X-chromosome probes (black) that undergo age-methylation events compared with the proportion of all probes (gray) on the available assays for the SSC (left) and CHB (right) data sets. P-values are calculated by permutation testing.
To test the significance of these observations and to determine if other chromosomes contain a disproportionate number of age-associated loci, we compared the proportions of age-associated loci for each chromosome using permutation testing (Fig. 3C; see Methods). This analysis revealed that the X chromosome does indeed have disproportionately fewer age-methylated loci than expected by chance alone in both SSC (P = 0.002) and CHB populations (P = 0.034) (Fig. 3D). The published age-methylated loci from the T1D and OC age studies indicate that these cohorts also have a significant absence of age-associated loci on the X chromosome (P < 0.021, Fisher's exact test) (Supplemental Fig. 5). These data suggest that the X chromosome is not governed by the same age-related DNA methylation kinetics as the rest of the genome, and it appears to be true in both pediatric and adult populations.
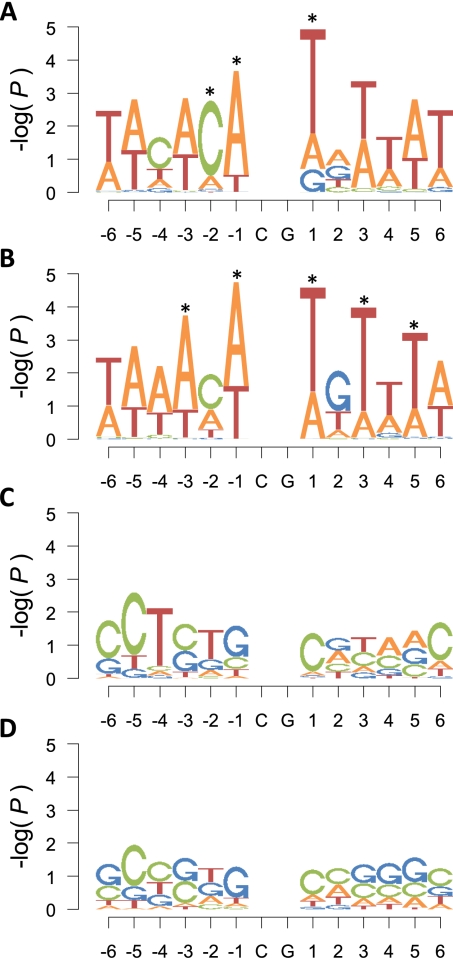
To gain insight into the potential mechanism(s) for age-associated DNAm changes, we analyzed the flanking nucleotides of CpG loci that change with age. Over- or underrepresentation of nucleotides at each position was determined by the proportion of nucleotides surrounding each age-associated locus relative to the nucleotide composition of the assay (Supplemental Fig. 6A,B). Sequences surrounding age-demethylated loci were enriched for adenine (A) and thymidine (T) residues in both pediatric cohorts, which is consistent with our observation that these CpG loci preferentially reside outside of CpG islands (Supplemental Fig. 6C,D). The overrepresentation of specific nucleotides was more significant in sites immediately adjacent to the interrogated CpG loci than in more distal sites, with significant enrichments of C residues at the −2 position (P = 0.004), A residues at the −1 position (P < 0.001), and T residues at the +1 position (P < 0.001). This finding also replicated in the CHB cohort (Fig. 4A,B). Analysis of sequences surrounding the age-methylated loci suggested that cytosines (C) and guanines (G) are overrepresented in regions flanking age-methylated loci (Supplemental Fig. 6E,F). Similar to the age-demethylated loci, the overrepresentation of specific nucleotides was increased immediately adjacent to the interrogated CpG dinucleotide; in fact, both the SSC and CHB data sets indicated a preference for a G at the −1 position and a C at the +1 position (Fig. 4C,D). However, overrepresentation of these individual nucleotides did not reach significance in the SSC cohort (P = 0.10 for G at −1 and P = 0.06 for C at +1) (Fig. 4C). Since it was previously shown that 5′-GCpGC-3′ 4-mers are preferential substrates for the DNA methyltransferase DNMT3A (Handa and Jeltsch 2005), we tested this 4-mer for overrepresentation at age-methylated loci. We found that while 5′-GCpGC-3′ is the most common 4-mer of both the SSC (110/477) and CHB (821/5,898) age-methylated loci, it was not significant in either group (SSC: P = 0.075; CHB: P = 0.137). However, this 4-mer is three to four times more common in age-methylated loci than those demethylated with age.
Figure 4.
Nucleotide composition surrounding age-associated loci. Logo plots of nucleotide composition immediately adjacent to age-demethylated (A,B) and age-methylated (C,D) CpG loci in the SSC (A,C) and CHB (B,D) populations. The height of each letter represents the significance [−log(P-value)] of overrepresentation for each base at the given location. Significance is determined by permutation testing relative to the assay composition.
Pediatric age-associated loci have developmental and immune functions
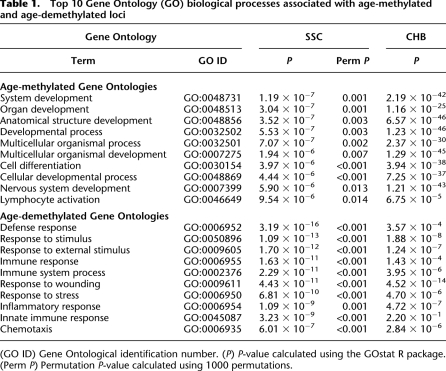
Annotation of the age-associated loci using Gene Ontology revealed several biologically relevant Ontologies that were significantly overrepresented among both SSC age-methylated and age-demethylated loci: specifically developmental processes and immune functions, respectively. These findings were first confirmed using permutation analysis and then independently corroborated in the annotation of the CHB group (Table 1; see Methods). Together, these data are consistent with previous reports identifying age-associated DNAm changes related to the immune system (Richardson 2002; Zhang et al. 2002) and are in accordance with biological processes likely to occur in pediatric populations.
Table 1.
Top 10 Gene Ontology (GO) biological processes associated with age-methylated and age-demethylated loci
Lifelong age-associated DNA methylation trends
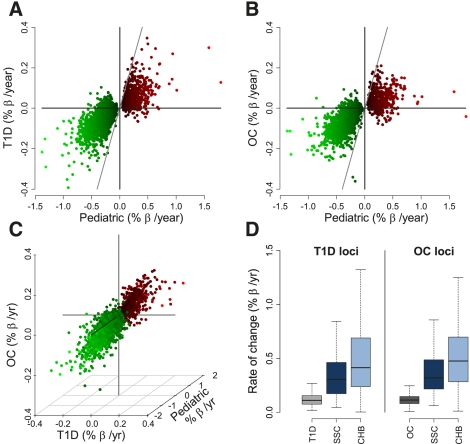
To better understand the dynamics of DNAm throughout life, we analyzed the 2078 pediatric age-associated loci in a meta-analysis of the pediatric SSC (N = 398) and CHB (N = 78) groups as well as the adult T1D (N = 187) and OC (N = 495) cohorts. Our analysis used a linear model with covariates for cohort, batch (BeadChip), gender, and in the T1D and OC cohorts, disease status; additionally, this model included an interaction between cohort and age effect to allow for a different rate of age-associated DNAm in each study (see Methods). This analysis revealed that the rates of age-associated DNAm change are highly correlated between the pediatric and adult cohorts (P < 0.001) (Fig. 5A–C); in fact, although 74.5% of the pediatric age-associated loci were not common to the adult age-associated loci, >88% of them trended in the same direction in both adult cohorts. These findings suggest that the majority of childhood-specific DNAm changes may actually continue into adulthood, but with a reduced rate of change throughout adulthood that does not reach statistical significance. Consistent with this hypothesis, a comparison of the common age-associated loci between the pediatric and the adult T1D and OC cohorts (363 and 325, respectively) revealed that the average rate of age-related DNAm change in the SSC pediatric group is 2.8 and 3.0 times greater than in the adult T1D and OC cohorts, respectively. This rate of change was even more pronounced in the younger CHB group, where it was 4.0 and 4.2 times greater than in the T1D and OC adults, respectively (Fig. 5D). Consequently, we find that 84.7% of the pediatric age-associated loci had significantly smaller rates of DNAm change in adulthood (Fig. 6A–D). These findings led us to hypothesize that age-associated DNAm may have a decaying rate of change with age; thus, we repeated the interaction analysis using the log of age as the explanatory variable. The log of age model overwhelmingly fit the data better, because only 6.5% of the pediatric age-associated loci had significantly different rates of DNAm change from either the T1D or OC cohorts when fit to a log of age model (Fig. 6E–H, FDR < 0.01). Thus, modeling age as a logarithmic variable explains loci with methylation dynamics that cannot be fit by a model linear for age alone, indicating that the rate of age-related DNAm change often decreases throughout life and is therefore imperfectly accounted for by a correction that is only linear for age.
Figure 5.
DNA methylation changes in pediatric and adult populations. Scatterplot showing the rates of DNAm change in the SSC pediatric population (x-axis) as compared with the adult T1D (A) and OC (B) populations. Each point represents one of the 2078 CpG loci found significant in the SSC pediatric cohort; points are colored by the rate of DNAm change in the SSC population (larger-to-smaller shown in light to dark colors; green: age-demethylated, red: age-methylated). The diagonal line represents the one-to-one line. (C) Three-dimensional scatterplot showing correlation of the average age-associated rate of DNA methylation in the SSC (z-axis), T1D (x-axis), and OC (y-axis) populations. Units and colors are as in A and B. (D) Boxplot of rates of DNA methylation change for the 363 loci found in common between the SSC and T1D studies (left) and the 325 loci found in common between the SSC and OC studies. The rates of DNAm change for these loci are also shown for the CHB population.
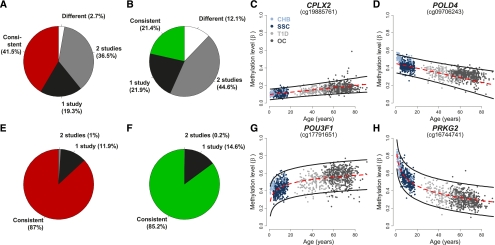
Figure 6.
Meta-analysis of pediatric and adult populations: modeling rates of DNAm change via an interaction model. (A,B) Pie charts displaying the proportion of SSC age-methylated (A) and age-demethylated (B) loci that have significantly different rates of DNAm change from the rates found in the CHB, T1D, and OC populations (different: white); in two of these studies (gray); and in one of these studies (black). Loci with similar rates of DNAm change across all four populations are shown in red (A: age-methylated), and green (B: age-demethylated). (C,D) Representative age-methylated (C) and age-demethylated (D) loci that have a similar rate of DNAm change in all populations. (Red dashed line) The linear regression on the SSC data only; (black lines) the 95% predication interval based on this regression. All individuals are denoted by dots and their colors are described in the inset of C. Each plot is titled by the name of the gene associated with the CpG interrogated by the Illumina platform. (E,F) Pie charts similar to those in A and B but modeling the log of age. (G,H) Representative age-methylated (G) and age-demethylated (H) loci that follow a log of age (but not linear) trend in all populations.
Discussion
Our results offer several important insights into the normal variation of DNAm over the human life span. Here we report 2078 loci that exhibit age-associated DNAm differences in DNA from the peripheral blood of 398 boys aged 3–17 yr, collected from 13 North American sites, making them unlikely to reflect any locale-specific environmental influences. These results replicate in an independent pediatric group who are unrelated to disease individuals, are significantly younger, and are primarily composed of different races/ethnicities than the initial pediatric group, indicating that the association between age and DNA methylation in children is generalizable (Fig. 1B). Furthermore, validation using pyrosequencing identifies a significant age effect at every CpG interrogated in the region, suggesting that age-related changes in DNA methylation occur in domains of CpGs, although further analysis is needed to confirm this finding (Fig. 1C,D). This is the first report, to our knowledge, of age-related DNAm changes from a sizable pediatric population. The pediatric age-associated loci identified here overlap significantly with previously published adult age-associated loci found in multiple studies, indicating that consistent age-related DNAm changes can be seen throughout human life.
The annotations and genomic characterization of these age-associated loci indicate systematic methylation events. We find that a higher proportion of age-demethylated loci are located outside of CpG islands (Fig. 2A) and, specifically, are more likely to reside within the 2 kb immediately adjacent to CpG islands (Fig. 2B), which is consistent with earlier studies (Christensen et al. 2009). Analysis of chromosomes with a disproportionate number of age-associated DNAm probes revealed a significant deficit of age-methylated loci on the X chromosome (Fig. 3). Although the biological significance of this will require further study, it is possible that males, being hemizygous for X-linked loci, are less tolerant of DNAm variation on the X chromosome.
Sequence motif analysis finds a significant overrepresentation of nucleotides surrounding the age-demethylated CpG dinucleotides, suggesting that systematic demethylation events are governed by the immediate nucleotide landscape (Fig. 4A). However, their primary location outside of CpG islands and the AT-rich nucleotide composition is not consistent with the recently identified TET family of proteins involved in 5-hydroxymethylation and subsequent active DNA demethylation by deamination and base excision repair (Tahiliani et al. 2009; Cortellino et al. 2011; Wu et al. 2011), suggesting these age-related demethylation events are governed by an alternative active demethylation pathway or passive DNA demethylation. In contrast, analysis of age-methylated loci did not reveal a significant pattern among the surrounding nucleotides; however, the most common motif independently identified in both pediatric groups, 5′-GCpGC-3′ (Fig. 4B), is a preferential substrate of DNMT3A (Handa and Jeltsch 2005), a de novo DNA methyltransferase that facilitates transcription of neurogenic genes (Wu et al. 2010), which would be consistent with the Gene Ontology results of these loci (e.g., see term “nervous system development” in Table 1).
Gene Ontology annotation of the SSC age-methylated and age-demethylated loci revealed a remarkable prevalence of functions in the developmental and immune systems, which were initially validated by permutation testing (see Methods) and then in an independent set of individuals by annotation of the age-associated loci from the CHB group (Table 1). These findings correspond well with biological events that are likely occurring during childhood, suggesting that DNAm may be a key modulator of dynamic changes in these developmental processes early in life.
Our meta-analysis of age-associated DNAm changes revealed that a sizable proportion (1537 of 2078 or 74.0%) of age-associated loci in our pediatric group does not follow the same linear trends throughout life compared with both the T1D and OC cohorts (Fig. 6A,B). In contrast, only 209 of 2078, or 14.3%, of age-associated changes in DNAm failed to follow logarithmic lifelong trends, suggesting that age-associated variation in DNAm is best modeled by a function of logarithmic age (Fig. 6E,F). This is a direct result of the threefold to fourfold greater rate of DNAm change in children versus adults (Fig. 5D). The noticeably different rate of change between the pediatric populations is likely the result of the younger age of the CHB population; this finding is consistent with previous reports that showed dramatic DNA methylation change rates during the perinatal period of human brain development (Siegmund et al. 2007). Together, these data provide evidence that the dynamics of DNAm variation are much more pronounced throughout childhood than previously appreciated, allowing for the identification of more age-associated loci than previously recognized by adult studies.
These results offer evidence that age-associated DNAm changes occur more rapidly in children than adults. Notably, the results reported here were validated by an independent method and replicated in an independent population, and permutation tests were used for all analyses. Despite these validations, it is possible that the increased rates of age-associated DNAm during childhood are a reflection of a compositional change of the multiple cell types found in peripheral blood, because DNAm is well known to be tissue-specific (Irizarry et al. 2009). However, previous studies have reported a high correlation of age-associated loci identified in multiple tissues and between specific cell types (Rakyan et al. 2010; Teschendorff et al. 2010; Bocker et al. 2011), suggesting that at least some of the age-associated DNAm changes reported here are not a result of changes in cell type composition. Regardless of this possibility, the increased rate of age-associated DNAm changes observed in children makes it especially important to closely match for age in pediatric disease-based studies of DNAm and adjust for age as a covariate. The need for this is evident from published disease-based DNAm studies that lack an age adjustment, which may yield a notable overlap between the reported “disease-associated” DNAm changes and the age-associated loci published here (12 of 37 loci) (Uddin et al. 2010). Today, high-throughput analysis of DNAm data is still in its infancy, and the list of confounding covariates remains incomplete, not unlike early genome-wide association studies, which failed to correct for population stratification. Together, these data provide a more complete understanding of the link between DNAm and age, showing a much stronger relationship in children than in adults, which ultimately will reveal more about the role of DNAm in health and disease.
Methods
DNA methylation profiling
Five hundred nanograms of human genomic DNA was sodium bisulfite–treated for cytosine (C) to thymine (T) conversion using the EZ DNA Methylation-Gold kit (Zymo Research). The converted DNA was purified and prepped for analysis on either the Illumina HumanMethylation27 or HumanMethylation450 BeadChips following the manufacturer's guidelines. Briefly, converted DNA was amplified, fragmented, and hybridized to the HumanMethylation27 or HumanMethylation450 pool of allele-differentiating oligonucleotides. After a series of extension, ligation, and cleanup reactions, the DNA was labeled with a fluorescent dye. The labeled DNA was then scanned using an Illumina BeadArray Reader or iScan. Image analysis and signal determination was performed using the GenomeStudio software, Methylation Module (Illumina).
Pyrosequencing was performed on bisulfite-converted DNA by EpigenDx (http://www.epigendx.com/) using a QIAGEN-Pyrosequencing PSQ MD system following the manufacturer's guidelines. Briefly, the PCR was performed with 0.2 μM primers, one of which was biotinylated for later purification by Streptavidin Sepharose HP (Amersham Biosciences). Sepharose beads containing the PCR product were washed and purified using 0.2 M NaOH and the Pyrosequencing Vacuum Prep Tool (QIAGEN). Ten microliters of the PCR products was sequenced using a PSQ96 HS system (QIAGEN), and methylation status was determined using QCpG software (QIAGEN).
Interpretation and QC of DNA methylation data
CpG DNA methylation (DNAm) data were interpreted using GenomeStudio to quantify methylated (M) and unmethylated (U) signal intensities for genomic DNA. Overall methylation levels (β) were calculated as the ratio of methylated to total signal [i.e., β = M/(M + U)], where β ranges from 0 (unmethylated) to 1 (methylated). Quality control of data resulted in removal of samples with aberrantly low signal intensity (mean <2000) or with fewer than 90% of CpG loci detected, where a given locus was determined not detected if the detection P-value > 0.01 (detection P-value provided by GenomeStudio and calculated relative to background signal). Assay controls were inspected to remove samples with poor bisulfite conversion, staining, extension (single nucleotide extension assay), hybridization, or specificity. Furthermore, outliers identified by hierarchical clustering and/or dissimilarity matrices were removed. In a total of the 458 SSC samples with age, 420 passed QC (398 with age younger than 18 yr); of the CHB samples, 78 of 79 passed QC; and all were below 18 yr of age. Additionally, one control DNA replicate was run on each BeadChip to assess overall assay reproducibility. Methylation profiles of the control DNA correlated well with an average Pearson correlation coefficient (R) of 0.990 between replicates.
Analysis of age-associated CpG loci
To analyze DNAm differences associated with age, we fit a separate regression for each CpG site, where β was modeled as a linear function of age. Linear models were fit using the R function “lm.” Although samples were randomly distributed across BeadChips and experiments with respect to age, BeadChip was also included as a fixed-effect covariate in all analyses to account for potential batch effects. To correct for multiple hypothesis testing, we applied a Benjamini-Hochberg False Discovery Rate (FDR) correction using the R function “p.adjust.” We also conducted this analysis using the “M-value” or log[β/(1 − β)] (Du et al. 2010) and found results that correspond extremely well with those in this study. Analysis of the M-value yields 2111 age-associated CpGs in the SSC group, and 2013 (95.4%) of these overlap with the 2078 CpGs identified using β-values.
Permutation analyses
All permutation analyses were conducted in R using the same linear model as the actual analysis where BeadChip was treated as a fixed-effects covariate, but in each permutation the ages of the subjects were randomly reassigned. In total, 1000 permutations were conducted for both the SSC and CHB groups independently. Permutation P-values for each CpG locus were calculated by assessing the number of times each locus was more significantly associated with age in the 1000 permuted data sets than the actual association (Supplemental Fig. 1D; Supplemental Table 2). Here, strength of association was determined by the absolute value of the age-effect t-statistic from the linear model. The same Monte Carlo strategy was used to compute empirical P-values for correlation of age-effect t-statistics (Fig. 1B; Supplemental Fig. 3A,B,D–F), overlaps of age-affected CpG loci (Supplemental Figs. 1H, 3C,G), CpG island and CpG island shore associations (Fig. 2), chromosome over- and underrepresentation (Fig. 3), nucleotides motifs (Fig. 4), and Gene Ontologies (Table 1). This strategy involved designating as “age-associated” the same number of loci from each permutation by selecting the most positively and negatively age-associated loci, which were chosen regardless of whether those loci were actually significant (FDR < 0.01) in that given permutation. For example, in the SSC group there are 2078 age-associated loci where 1601 are age-demethylated and 477 are age-methylated, so for each permutation the 1601 most age-demethylated and 477 most age-methylated loci are chosen for further analysis. Likewise, for the CHB group, this process was repeated using 41,895 loci for each permutation (35,997 age-demethylated and 5898 age-methylated). These loci are referred to as “permuted age-associated loci” and were chosen to match the numbers observed in the original data. The strength of association was based on the age-effect t-statistic for each locus. These loci were then compared or annotated for the analysis being conducted, and the respective permutation P-value was computed as the proportion of permutations for which the observed association or enrichment was of equal or greater strength than in the original data. For example, the permutation P-value for the overlap between the SSC and CHB pediatric age-associated loci (Supplemental Fig. 1H) was calculated by selecting from the SSC group the top 1601 loci negatively associated with permuted age and the top 477 loci positively associated with permuted age and computing the overlap with the CHB group age-associated list (Supplemental Table 2). In no permutation was the overlap as large as the actual overlap, and thus in this case, the permutation P-value is <0.001. Further details are provided under the analysis specific headings below.
Age-effect correlations and overlap analyses
The significance of age-effect correlations between groups is permutation-based, where the actual Pearson correlation coefficient (R) of age-effect t-statistics between two cohorts is compared with the permuted correlation coefficients (i.e., the correlation coefficients of permuted age-effect t-statistics for the SSC pediatric group compared with the actual age-effect t-statistics of the CHB, T1D, OC populations or data from Rakyan et al. 2010). Specifically, to test whether the age-effect t-statistics of the SSC pediatric group correlate with the T1D and OC cohorts and more than expected by chance alone (Supplemental Fig. 3A,B), the permuted pediatric age-effect t-statistics and actual age-effect t-statistics published by Teschendorff et al. (2010) for the T1D and OC studies were used to calculate permuted correlation coefficients. In none of the permutations was the permuted correlation coefficient (R) as large as the actual correlation coefficient (Fig. 1C). This same methodology was applied to the published age correlations (ρ) of whole blood, CD4+ T-cells, and CD14+ monocytes from Rakyan et al. (2010).
Significance analysis of the overlap between age-associated loci from our SSC pediatric population and the T1D and OC studies was also permutation-based. Here, the P-values were calculated by first identifying the actual size of the overlap between any two cohorts (Supplemental Fig. 3C) and then using the permuted age-associated loci to calculate the sizes of permuted overlaps. The P-value was determined as the number of times the permuted overlap was greater than the actual overlap, divided by the number of permutations (1000). In no permutation was the overlap as large as the actual overlap (or even close), thus the permutation P-values are <0.001 (Supplemental Fig. 3C). Likewise, the same methodology was applied to the lists published by Rakyan et al. (2010) and Bocker et al. (2011) with the same result.
CpG island analysis
Analysis of age-associated loci relative to CpG islands was based on the NCBI genome (build 36.1) and the Takai-Jones relaxed criteria of CpG islands (Takai and Jones 2002). Specifically, the “seq_cpg_islands.md.gz” file was downloaded from the NCBI website (ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/ARCHIVE/BUILD.36.1/mapview/), and CpG loci interrogated by the Infinium HumanMethylation27 and HumanMethylation450 platforms were mapped relative to CpG islands using a custom R script. This resulted in assignment of CpG island/non-CpG island context consistent with that provided by the Illumina HumanMethylation27 annotation file, and CpG island “shores” were defined as the region within 2 kb of a CpG island. For the HumanMethylation27 panel, 20,006 of 27,578 probes (72.5%) are located in CpG islands, and of the 7572 that are not, 4155 (15.1%) are located in CpG island “shores.” For the HumanMethylation450 panel, 255,455 of the 485,512 CpG loci (52.6%) are located in CpG islands, and of the 230,057 that are not, 135,686 (27.9%) are located in CpG island “shores.” To determine whether age-associated loci were preferentially located in CpG island and CpG island shore contexts, permutation testing was applied based on the number of age-associated loci in CpG islands or CpG island shores for both the SSC and CHB groups. For the SSC group, 327 of the 477 age-methylated loci (69.6%) are in CpG islands, and 69 (14.5%) are in CpG island shores; whereas 497 of 1601 age-demethylated loci (31.0%) are in CpG islands, and 592 (37%) are in CpG island shores. For the CHB group, 3389 of the 5898 age-methylated loci (57.5%) are in CpG islands, and 1560 (26.4%) are in CpG island shores; whereas 7552 of the 35,997 age-demethylated loci (21.0%) are in CpG islands, and 16,118 (44.8%) are located in CpG island shores. These numbers were compared with the locations of the permuted loci for each group, and a permutation P-value was calculated as described in the “Permutation Analyses” section of the Methods (above).
Analysis of genomic location
Loci were mapped to chromosome location using the Illumina annotation files for HumanMethylation27 and HumanMethylation450 panels based on the NCBI Human Genome (build 36.1). Over- and underrepresentation for age-methylated and age-demethylated loci were assessed using both a Fisher's exact test and permutation analysis, which corresponded well (data not shown). Permutation P-values were determined by comparing the proportion of age-associated loci on a chromosome (termed “actual proportion”) with the proportions obtained using the permuted age-associated loci (termed “permuted proportions”). To correct for multiple hypotheses (i.e., 24 chromosomes and both over- and underrepresentation), the actual proportion of each chromosome was compared with the permuted proportions of any chromosome for each permutation. Thus the permutation P-value is the number of times a permuted proportion is more significant than the actual proportion, divided by the number of permutations (1000).
Sequence motif analysis
Sequence data from the Illumina annotation files were used to analyze the 60 bp flanking each CpG dinucleotide. Overrepresentation of nucleotides was based on the proportion of age-associated loci with that nucleotide relative to the nucleotide composition of the assay, termed “relative proportion” (Supplemental Fig. 6). Specifically, this is calculated for each base as the proportion of age-associated loci that are a given nucleotide divided by the proportion of all assay probes that contain that same nucleotide at the given location. Permutation P-values for overrepresented nucleotides were calculated by comparing the actual nucleotide proportion with the permuted nucleotide proportions for age-methylated and age-demethylated loci. Logo plots (Fig. 4) were created by modifying the “seqLogo” R package to display the letter height as the −log(P-value), where P-value is the permutation based as described above (Schneider and Stephens 1990). Permutation P-values of 0 were displayed on graphs as half the minimal increment (i.e., P = 0.0005) (Fig. 4).
Gene Ontologies
Gene Ontology (GO) analyses were conducted in R using the “GOstats” package and the lumiHumanAll.db annotation file. Briefly, this package assesses significantly overrepresented Gene Ontologies given a list of significant genes and the list of all genes available on the platform (termed the “gene universe”). GOstats calculates enrichment of Ontologies using the hypergeometric test such that only Ontologies enriched over the gene universe are found significant. Entrez gene IDs were used to identify genes and were provided in the HumanMethylation27 annotation file. The HumanMethylation450 annotation file provided UCSC RefSeq accession numbers, which were mapped to Entrez gene IDs using the NCBI file gene2refseq.gz (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/). Permutation P-values were calculated using the same GOstats package for the permuted age-associated loci.
Meta-analysis of Teschendorff and colleagues' data
Data were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) data sets GSE19711 and GSE20067 for the OC and T1D data sets, respectively. Pediatric, T1D, and the OC data sets were combined and analyzed using a linear fixed-effects model (R/Bioconductor function “lm” of package “stats”) that adjusted for study, BeadChip, gender, and disease status (T1D and OC cohorts only); additionally, a parameter for interaction of age effect and cohort was used to identify those loci that had significantly different rates of age-associated DNAm (Fig. 6A,B,E,F). Rates of DNAm change were calculated using the standardized linear regression for each data set independently (SSC, CHB, T1D and OC) (Fig. 5). Linear models of logarithmic age were fit using a log transform for the age variable and the “lm” function in R. Predictions of the pediatric methylation data were calculated using the “predict.lm” function of the “stats” package also in R (Fig. 6C,D,G,H, see black and red lines).
Data access
We have submitted the data generated from the 398 samples and the 96 controls for this study to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under the Gene Series GSE27097.
Acknowledgments
We thank Julie Mowrey and Brian Lynch for technical assistance and Cheryl Strauss for editorial comments. We also thank Dr. Teschendorff and Dr. Widschwendter for communicating specific samples included in their study; and Dr. Louis M. Kunkel, Dr. Isaac S. Kohane, Dr. Sek Won Kong, and Dr. Yuko Soneoka from Children's Hospital Boston. This work was supported in part by the Emory Biomarker Service Center, as well as a Simons Foundation (SFARI) award and NIH grant MH089606, both to S.T.W. B.G.B. was supported by HHS training grant no. 5T32GM008490-19. We are grateful to all of the families at the participating SFARI Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, and E. Wijsman). We also appreciate the access to phenotypic data on the SFARI Base. Approved researchers can obtain the SSC population data set described in this study by applying at https://base.sfari.org. Disclaimer: The opinions and findings of this report are those of the authors and should not be taken as official positions or findings of the CDC.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/10.1101/gr.125187.111.
References
- Ahuja N, Issa JP 2000. Aging, methylation and cancer. Histol Histopathol 15: 835–842 [DOI] [PubMed] [Google Scholar]
- Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP 1998. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 58: 5489–5494 [PubMed] [Google Scholar]
- Arima T, Drewell RA, Arney KL, Inoue J, Makita Y, Hata A, Oshimura M, Wake N, Surani MA 2001. A conserved imprinting control region at the HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum Mol Genet 10: 1475–1483 [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299: 2877–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F 2011. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117: e182–e189 [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A 2009. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130: 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W 2010. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 9: 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, et al. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Choufani S, Ferreira JC, Grafodatskaya D, Butcher DT, Weksberg R 2011. Sequence overlap between autosomal and sex-linked probes on the Illumina HumanMethylation27 microarray. Genomics 97: 214–222 [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5: e1000602 doi: 10.1371/journal.pgen.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25: 315–319 [DOI] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM 2010. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11: 587 doi: 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Avizienyte E, Corn PG, Lothe RA, Baylin SB, Aaltonen LA, Herman JG 2000a. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene 19: 164–168 [DOI] [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, et al. 2000b. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92: 564–569 [DOI] [PubMed] [Google Scholar]
- Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG 2000c. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res 60: 2368–2371 [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M 2007. Epigenetics and aging: The targets and the marks. Trends Genet 23: 413–418 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Agrelo R, Esteller M 2007. Cross-talk between aging and cancer: the epigenetic language. Ann NY Acad Sci 1100: 60–74 [DOI] [PubMed] [Google Scholar]
- Handa V, Jeltsch A 2005. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol 348: 1103–1112 [DOI] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al. 1994. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci 91: 9700–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, Baylin SB 1997. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res 57: 837–841 [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci 95: 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB 1994. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540 [DOI] [PubMed] [Google Scholar]
- Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB 1996. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci 93: 11757–11762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet (Suppl) 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57: 808–811 [PubMed] [Google Scholar]
- Koch CM, Suschek CV, Lin Q, Bork S, Goergens M, Joussen S, Pallua N, Ho AD, Zenke M, Wagner W 2011. Specific age-associated DNA methylation changes in human dermal fibroblasts. PLoS ONE 6: e16679 doi: 10.1371/journal.pone.0016679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. 2010. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329: 1650–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Nuovo GJ, Zervos EE, Martin EW Jr, Salovaara R, Aaltonen LA, de la Chapelle A 2001. Age-related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res 61: 6991–6995 [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621 [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20: 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Richardson BC 2002. Role of DNA methylation in the regulation of cell function: Autoimmunity, aging and cancer. J Nutr (Suppl) 132: 2401S–2405S [DOI] [PubMed] [Google Scholar]
- Robertson KD 2005. DNA methylation and human disease. Nat Rev Genet 6: 597–610 [DOI] [PubMed] [Google Scholar]
- Schanen NC 2006. Epigenetics of autism spectrum disorders. Hum Mol Genet 15: R138–R150 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S 2007. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2: e895 doi: 10.1371/journal.pone.0000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So K, Tamura G, Honda T, Homma N, Waki T, Togawa N, Nishizuka S, Motoyama T 2006. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci 97: 1155–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci 99: 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S 2010. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci 107: 9470–9475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Deng C, Lu Q, Richardson B 2002. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev 123: 1257–1268 [DOI] [PubMed] [Google Scholar]