Abstract
Developmental genes are regulated by complex, distantly located cis-regulatory modules (CRMs), often forming genomic regulatory blocks (GRBs) that are conserved among vertebrates and among insects. We have investigated GRBs associated with Iroquois homeobox genes in 39 metazoans. Despite 600 million years of independent evolution, Iroquois genes are linked to ankyrin-repeat-containing Sowah genes in nearly all studied bilaterians. We show that Iroquois-specific CRMs populate the Sowah locus, suggesting that regulatory constraints underlie the maintenance of the Iroquois–Sowah syntenic block. Surprisingly, tetrapod Sowah orthologs are intronless and not associated with Iroquois; however, teleost and elephant shark data demonstrate that this is a derived feature, and that many Iroquois–CRMs were ancestrally located within Sowah introns. Retroposition, gene, and genome duplication have allowed selective elimination of Sowah exons from the Iroquois regulatory landscape while keeping associated CRMs, resulting in large associated gene deserts. These results highlight the importance of CRMs in imposing constraints to genome architecture, even across large phylogenetic distances, and of gene duplication-mediated genetic redundancy to disentangle these constraints, increasing genomic plasticity.
The high complexity of transcriptional control in animals is known to have a significant impact on how their genomes are shaped through evolution. In particular, key developmental genes show an exceptionally complex regulation, with very specific and intricate spatio-temporal expression patterns. Numerous cis-regulatory modules (CRMs) create vast regulatory landscapes around these loci (Nelson et al. 2004) that often extend to neighboring genes. This imposes constraints on genomic restructuring, creating “solid” regions, recognizable as conserved regulatory blocks within phyla, with very low rates of rearrangement (Becker and Lenhard 2007; Engstrom et al. 2007; Kikuta et al. 2007). Nevertheless, it is unclear whether long-range cis-regulatory interactions may have had an impact on genome architecture over deeper evolutionary distances (i.e., across different phyla) (Koonin 2009).
Iroquois (Iro/Irx) genes encode highly conserved homeobox transcription factors (TFs) of the TALE class with multiple and fundamental roles in animal development (Cavodeassi et al. 2001). In addition, they provide arguably one of the most paradigmatic examples of the distinct evolutionary fate followed by genomic regions surrounding major developmental regulators. First, Irx genes are flanked by large genomic regions devoid of genes (gene deserts) (Nobrega et al. 2003; Ovcharenko et al. 2005) in all studied bilaterians (Irimia et al. 2008). In vertebrates, as well as in other lineages, Conserved Noncoding Regions (CNRs) (Aparicio et al. 1995; Bejerano et al. 2004; Woolfe et al. 2005; Pennacchio et al. 2006) are highly enriched in these unusually long intergenic distances (Sandelin et al. 2004; de la Calle-Mustienes et al. 2005; Irimia et al. 2008; Tena et al. 2011), and they often act as tissue-specific enhancers (de la Calle-Mustienes et al. 2005; Tena et al. 2011). Second, Irx genes have independently evolved a cluster organization in at least four bilaterian groups (Irimia et al. 2008; Takatori et al. 2008; Kerner et al. 2009). Finally, Irx genes show the most widespread and ancient conserved linkage to a phylogenetically unrelated gene known to date: Iroquois and the ankyrin-repeat-containing Sosondowah (Sowah) genes are tightly associated in the genome in nearly all studied bilaterians, including fast-evolving species such as flies and nematodes, but with the intriguing exception of tetrapods (Supplemental Fig. S1; Irimia et al. 2008; Kerner et al. 2009).
Each of these features might be explained by the presence of strong regulatory constraints, such as shared enhancers or global control regions (de la Calle-Mustienes et al. 2005; Irimia et al. 2008). Consistently, functional studies in vertebrates and flies have shown that some regulatory elements are shared between clustered Irx genes (Gomez-Skarmeta et al. 1996; Tena et al. 2011), providing an explanation for the preservation of the cluster organization in these lineages, although functional data is still missing for other groups. In addition, no studies have yet provided an explanation for the striking linkage of Irx genes to Sowah in invertebrates and their dissociation in tetrapods. The strong conservation of synteny in invertebrates suggests that the Sowah loci could be part of the Irx regulatory landscape. Therefore, a full understanding of Irx regulation and evolution might not be complete without addressing this issue.
Here, we have studied the Irx-Sowah regulatory landscape in different animal groups. First, we show that Sowah genes are widely constrained, showing several deeply conserved intron positions and relative lengths, with high density of CNRs. Second, we demonstrate that noncoding sequences within the Sowah loci from amphioxus and Drosophila can act as transcriptional regulators, and are crucial for the proper expression of Irx genes, at least in Drosophila. Third, we show that despite the general absence of Irx-associated Sowah in vertebrates, several key Irx regulators in this lineage were originally located within a Sowah gene that specifically lost its coding sequence aided by the genetic redundancy produced by an early retroposition event. Finally, we discuss the impact of Sowah linkage on Irx regulation and evolution, showing how the recurrent remodeling of the Sowah loci has shaped the Irx regulatory landscape in different bilaterians.
Results
Irx–Sowah linkage and Sowah gene structure are highly conserved in metazoans
To investigate the extent of conservation of the Irx–Sowah linkage during bilaterian evolution, we identified and characterized Sowah genes in 39 genomes (Supplemental Figs. S1, S2; see Methods). Irx complexes are linked to Sowah genes in nearly all bilaterian lineages, with the exception of tetrapods, tunicates, and the leech Helobdella robusta. Alignment of intron/exon structures of metazoan Sowah genes revealed that several intron positions (ancestral introns 5–9, Supplemental Fig. S3) are conserved across all Irx-linked genes, including nematodes (except for intron 5) and insects, which are known to have dramatically divergent intron/exon structures (Rogozin et al. 2003; Coulombe-Huntington and Majewski 2007). In addition, the lengths of these introns are often exceptionally large, up to 70 times the species average (Supplemental Table S1). A possible explanation for the extraordinary conservation of these intron positions and relative lengths may be the presence of regulatory elements within them, which would prevent intron loss in diverse lineages (Irimia et al. 2011). In addition to a potential regulatory function, we found two groups of highly conserved microexons (one of 12 and four of three nucleotides) within the ankyrin-repeat domain (introns 6 and 7, Supplemental Fig. S3) in species with enough available expression data (including cnidarians, ecdysozoans, lophotrochozoans, and chordates).
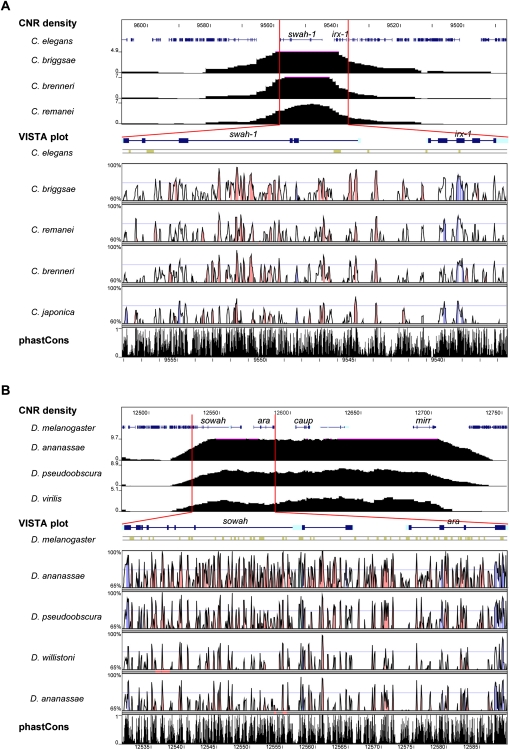
Sowah loci are populated by CNRs that are likely associated with Irx
As potential indicators of regulatory sequences, we analyzed CNRs within Sowah loci in different groups. We performed interspecies comparisons within flies and nematodes, lineages with availability of an appropriate taxon sampling (i.e., within the phylogenetic ranges of CNR detection) (Boffelli et al. 2004). We first determined CNR density in the Irx–Sowah genomic region using Ancora (Engström et al. 2008). Major peaks of CNR density often overlap with developmental genes and can be used to delimit the regulatory landscapes associated with these genes (Engstrom et al. 2007, 2008). In both nematodes and flies, Irx genes were located in regions with very high CNR density. Importantly, the neighboring Sowah genes were fully embedded within these Irx-associated CNR-dense peaks, indicating that Sowah intronic sequences are as enriched in CNRs as sequences immediately flanking Irx (Fig. 1A,B). Indeed, VISTA analyses of the Sowah regions using stringent parameters revealed dozens of CNRs within the long Sowah introns and in the intergenic regions between Irx and Sowah, consistent with previous analyses in nematodes (Vavouri et al. 2007) and with the high number of evolutionarily constrained elements detected by phastCons (Fig. 1A,B; Siepel et al. 2005). These results further emphasize the potential functional importance of Sowah intronic sequences.
Figure 1.
CNRs in nematode and drosophilid Sowah genes. Ancora plots of CNR density (top), VISTA plots (middle), and phastCons tracks (bottom) of the Irx-Sowah region of nematodes (A) and flies (B). (Red bars) Region depicted in Ancora plots zoomed in on VISTA and phastCons tracks. VISTA colored peaks (blue, coding; turquoise, UTR; pink, noncoding) indicate regions of at least 50 bp and ≥90% similarity (≥85% in the case of Caenorhabditis japonica) in nematodes and 60 bp and ≥90% similarity in flies. Only gene symbols corresponding to Sowah (swah-1 in nematodes) and Irx (ara, caup, and mirr in Drosophila) are indicated. Numbers at the left correspond to the percentage of base pairs covered by CNRs in Ancora plots, percentage identity in VISTA analyses, and conservation scores in phastCons.
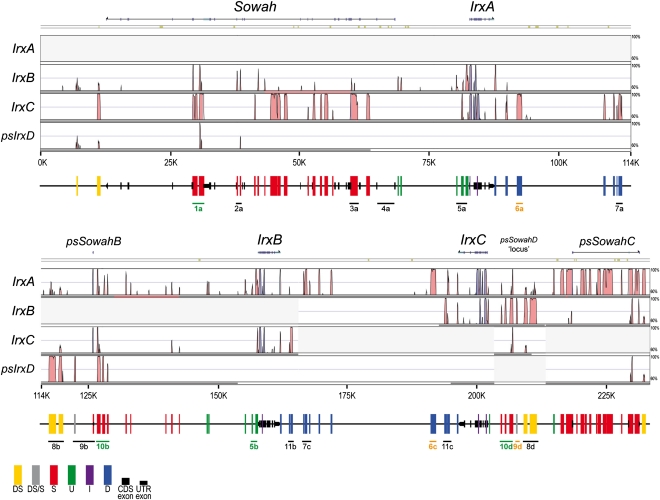
In the case of the cephalochordate amphioxus, there are currently no sequenced genomes within the appropriate phylogenetic range to perform informative interspecies comparisons (Pascual-Anaya et al. 2008), and therefore Ancora and phastCons tools cannot be used in this lineage. However, VISTA analysis between paralogous regions have been successfully applied before in this species (Jiménez-Delgado et al. 2006; Irimia et al. 2008). We had previously identified several highly conserved noncoding sequences by crossed VISTA alignments between the regions surrounding the three Irx genes in the amphioxus cluster (Irimia et al. 2008). Here, we have expanded these analyses to include Sowah (Fig. 2). We divided the amphioxus cluster plus Sowah into four regions. Three contained one of the Irx genes (IrxA, IrxB, or IrxC), their surrounding noncoding sequences and, in the case of IrxA, the entire Sowah locus, which is immediately next to it. We also defined a fourth region corresponding to a putative “IrxD” locus that was lost during amphioxus evolution, as suggested by CNR complements after preliminary analyses (see below and Methods for further details). Crossed VISTA comparisons of these regions revealed many repeated blocks with high-sequence similarity that were not detected in our previous survey (Irimia et al. 2008). Importantly, these novel conserved repeated blocks have one of their copies lying either within Sowah introns or in the intergenic region between Sowah and IrxA (Fig. 2; Supplemental Fig. S4; Supplemental Data S1). Like the previously identified CNRs, most conserved elements are present in two copies, and some in three, indicating differential losses after the gene duplications that gave rise to the cluster (note that potentially functional single-copy sequences cannot be detected with this analysis). In addition, some CNRs were present in four copies, despite the fact that there are only three Irx genes. The implications of these findings are threefold.
Figure 2.
Internal organization of the Irx-Sowah complex in B. floridae. VISTA plot of the alignments between each of the three Irx genes (plus a fourth region corresponding to a putative IrxD locus lost during amphioxus evolution) and their respective surrounding noncoding regions, including Sowah in the case of IrxA. Colored peaks (blue, coding; turquoise, UTR; pink, noncoding) indicate regions of at least 100 bp and ≥70% similarity. High-copy number elements (such as repeats and mobile elements) are masked and their presence is indicated by khaki segments above the VISTA plot. Vertical bars of different colors below the VISTA plot represent the different conserved repeated blocks, indicating their respective location to Sowah and Irx loci (DS, Downstream Sowah; S, within Sowah; U, Upstream Irx; I, Introns of Irx; D, Downstream Irx. DS/S bars indicate elements of uncertain identity [DS or S]). Black rectangles and arrows indicate the exon sequences of Irx, Sowah, or the remains of Sowah duplicates. The blocks tested for transcriptional enhancer activity are indicated and named with a number and letter code. The letter refers to the Irx locus with which they are associated; color indicates whether they are tissue-specific enhancers (green), unspecific enhancers (yellow), or negative elements (black).
First, it was not the ancestral Irx alone, but the pair Irx–Sowah that duplicated in tandem to generate the amphioxus cluster. In fact, remains of three exons from pseudogenized Sowah copies are still present next to IrxB and IrxC, as can be noticed in the VISTA plots (Fig. 2; Supplemental Fig. S4; Supplemental Data S1).
Second, the four-copy CNRs and the respective order and orientation of both genes and CNRs suggest that, in addition to the three Irx genes and their respective Sowah genes and pseudogenes, a fourth duplicate of the tandem Irx–Sowah may have been present in the cluster during amphioxus evolution. Despite the loss of an IrxD gene, this “D” paralogous region is clearly identifiable as a CNR array of sequences that were originally in the intronic and downstream regions of a SowahD locus (Fig. 2; Supplemental Fig. S4). Since CRMs located in gene clusters may target multiple paralogous genes (Tena et al. 2011), the CNRs may have been conserved due to interactions with other Irx genes, despite the loss of their original target gene. These results, together with data from phylogenetic analysis (Irimia et al. 2008) and CNR similarity (Supplemental Fig. S4) indicating that A duplicate is more closely related to C, and that B is more similar to D, provide a more detailed picture of the evolutionary origin of the amphioxus Irx cluster (Supplemental Fig. S5; Supplemental Discussion S1).
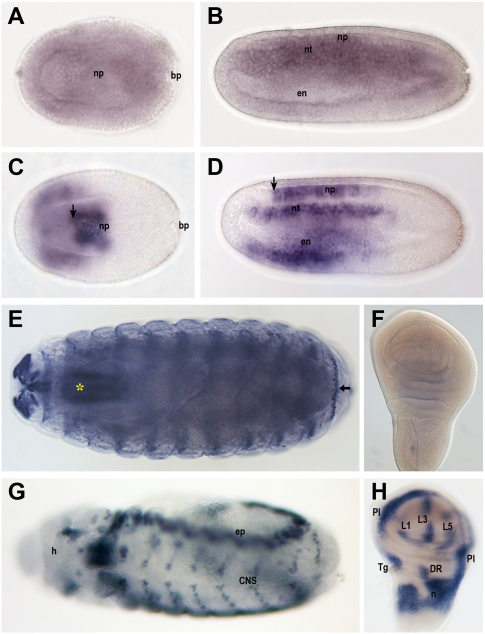
Third, the selective loss of Sowah exons versus their intronic conserved elements suggests that the CNRs in the Irx–Sowah genomic region are most likely associated with Irx, not with Sowah, providing a potential explanation for the conservation of the Iroquois–Sowah syntenic block. Adding to this idea, the expression pattern of amphioxus Sowah differs widely from those of Irx genes (Fig. 3A–D; Kaltenbach et al. 2009; Irimia et al. 2010), suggesting distinct regulations, and thus, that Sowah and Irx genes are not likely to share major transcriptional enhancers. A comparable situation is found in Drosophila, where the expression pattern of Sowah is not similar to that of Irx complex genes, araucan (ara), caupolican (caup), and mirror (mirr) (Gomez-Skarmeta et al. 1996; McNeill et al. 1997), indicating that Irx and Sowah genes are probably not coregulated (Fig. 3E–H; data not shown).
Figure 3.
Comparison of the expression patterns of Sowah and Iroquois genes in amphioxus and fly. In situ hybridization of B. lanceolatum Sowah (A,B) and IrxB (C,D) genes in 15-h early neurulas (A,C) and 21-h neurulas (B,D) in dorsal and lateral views, respectively. Sowah transcripts were detected almost ubiquitously, with stronger expression in the dorsal half of the embryos. In contrast, IrxB showed a very defined and restricted pattern in the endoderm (en), notochord (nt), and neural plate (np). The anterior limit of expression in the neural plate, which is conserved in evolution (Irimia et al. 2010), is indicated by an arrow. The expression of IrxA and IrxC was similar at these stages (data not shown). (bp) Blastopore. In situ hybridization of sowah (E,F) and caup (G,H) in D. melanogaster stage 17 (E) and stage 12 (G) embryos (dorsal views) and third instar larvae wing imaginal discs (F,H, anterior is to the left). (E,F) sowah is expressed in the pharynx (*) and cephalic nervous system of late embryos (arrow in E points to nonspecific staining of the cuticular denticle belts), but is undetectable in the wing imaginal discs. (G,H) During embryonic development, ara and caup display coincident dynamic expression patterns in the epidermis (ep), central nervous system (CNS) and mesoderm, as well as in the head (h). In the wing disc, caup is expressed in the prospective regions of the 1, 3, and 5 longitudinal veins (L1, L3, L5), pleura (Pl); tegula (Tg); dorsal radius (DR); alula and lateral notum (n).
Conserved sequences from the amphioxus Irx–Sowah locus drive expression consistent with that of Irx in zebrafish stable transgenic lines
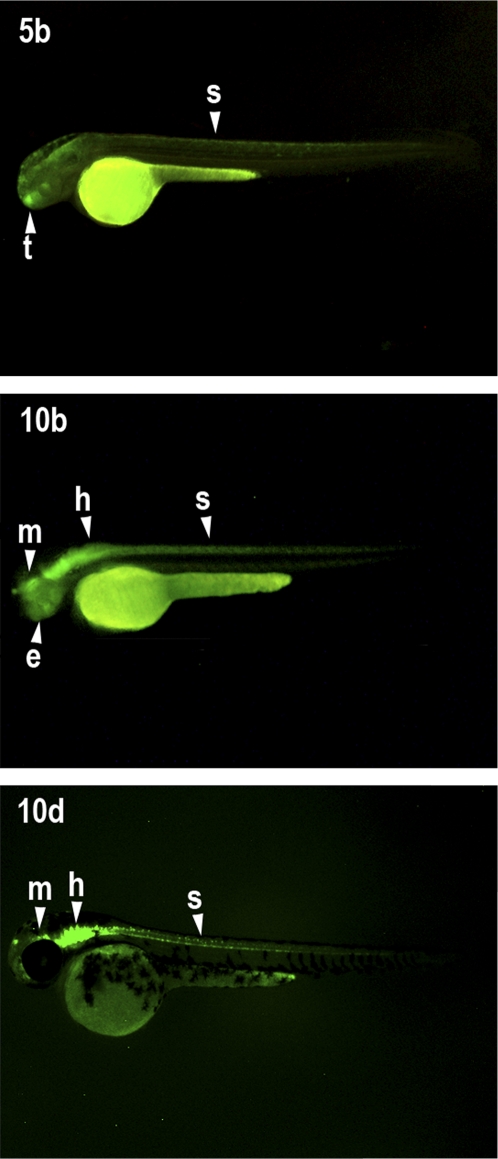
Next, we tested the functionality of the conserved sequences within the amphioxus Irx–Sowah locus. We selected 18 amphioxus CNRs (Supplemental Table S2) from different locations: (1) between two Irx genes, (2) in the intergenic region between Irx and Sowah, and (3) within Sowah introns (Fig. 2). The potential regulatory activity of each of the 18 CNRs was assayed by generating stable transgenic zebrafish lines. To this end, we used the ZED vector (Bessa et al. 2009), which contains the gata2a minimal promoter with a GFP reporter gene, and RFP driven by the cardiac actin promoter as positive control for successful transgenesis. Four of the amphioxus CNRs (1a, 5b, 10b, and 10d [letters indicate the associated Irx loci]) drove reproducible tissue-specific GFP expression in zebrafish embryos in at least three independent founders (Fig. 4; Supplemental Figs. S6, S7). Three other elements (6a, 6c, and 9d) drove GFP expression in a ubiquitous or variable way, suggesting that they may be less-specific enhancers (Supplemental Fig. S6; data not shown; Komisarczuk et al. 2009). No GFP expression could be detected for the other 11 tested elements, for which cardiac and muscle expression of the control RFP was detected in several founders studied for each element (data not shown). These transcriptionally inactive CNRs may not have been recognized by zebrafish TFs or they could be active in developmental stages or tissues not surveyed in this work. Alternatively, they may be involved in negative transcriptional regulation or other regulatory functions that could not be detected in this assay.
Figure 4.
Transcriptional enhancer activity of B. floridae sequences from the Irx-Sowah complex. Lateral views of 48-hpf zebrafish showing GFP expression driven by the 5b, 10b, and 10d CNRs. The 5b-driven expression is detected in the spinal cord and in the telencephalon; 10b and 10d consistently drove expression throughout the CNS (midbrain, hindbrain, and spinal cord) and in the eye. Anterior is to the right. (e) eye; (h) hindbrain; (m) midbrain; (s) spinal cord; (t) telencephalon.
Of the four tissue-specific enhancers, three are located within the introns of Sowah or its pseudogenized copies (two of them, 10b and 10d, are copies of a duplicated element), whereas the other (5b) lies between IrxB and its respective pseudogenized Sowah gene (Fig. 2). The duplicated elements, 10b and 10d, drove similar expression in the central nervous system (CNS) (Fig. 4), in a pattern consistent with Irx genes in both amphioxus (Fig. 3D; Kaltenbach et al. 2009) and zebrafish (Lecaudey et al. 2001; Feijóo et al. 2004). Strikingly, this expression is reminiscent of previously characterized Irx cis-regulatory elements in vertebrates (de la Calle-Mustienes et al. 2005; Visel et al. 2007; Tena et al. 2011), which are also active in wide CNS domains, including anterior regions normally devoid of Irx expression. The CNR 5b drove GFP expression to the spinal cord and the telencephalon (Fig. 4). Again, while the first tissue is a major domain of Irx expression in amphioxus, the second one shows no expression of these genes (Kaltenbach et al. 2009). As it has been proposed in vertebrates (de la Calle-Mustienes et al. 2005), it is likely that other negative cis-regulatory elements contribute to the down-regulation of Irx expression in the telencephalon. Finally, the intronic element 1a was consistently active in a highly restricted domain, the blood islands (Supplemental Fig. S6), a “tissue” with no obvious known counterpart in amphioxus. To date, Irx genes have not been implicated in hematopoiesis in vertebrates (Ferrell et al. 2005), and therefore, it is not clear whether the regulatory activity of element 1a is compatible with either the expression of amphioxus Irx or Sowah genes, or whether this sequence is being read by different regulatory states in vertebrates and amphioxus. This last alternative illustrates the complexity of interpreting trans-species reporter assays at deep evolutionary distances, where the possibility of obtaining reproducible but biologically meaningless results cannot be completely ruled out. Nevertheless, very deep conservation of regulatory states does exist, even across phyla (Royo et al. 2011), and consistent results such as those observed for elements 10b, 10d, and 5b are thus highly suggestive of functional conservation.
Sowah contains regulatory elements essential for proper Iroquois expression in flies
The tissue-specific expression driven by some amphioxus Sowah-derived CNRs in zebrafish embryos, which encompasses Irx expression domains, suggests that these elements may be regulating Irx transcription. To obtain direct evidence that Sowah noncoding sequences were responsible for part of the transcriptional regulation of Irx genes, we then resorted to Drosophila, where the appropriate genetic tools are available. The expression of Drosophila members of the Iro family has been extensively characterized. ara and caup show identical expression patterns, including specific wing and leg imaginal disc regions and several embryonic domains (Figs. 3G,H, 5B,F,H,H',J; Gomez-Skarmeta et al. 1996; Diez del Corral et al. 1999; Letizia et al. 2007; Carrasco-Rando et al. 2011), suggesting that ara and caup share specific CRMs (Gomez-Skarmeta et al. 1996; Letizia et al. 2007). In contrast, mirr expression pattern is slightly different (McNeill et al. 1997; Kehl et al. 1998).
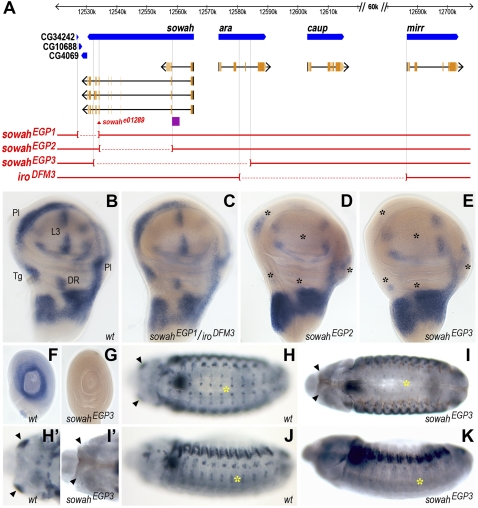
CRMs responsible for ara/caup expression in the prospective notum region of the wing disc are located between caup and mirr (Letizia et al. 2007). On the contrary, no CRMs accounting for the remaining expression domains of the wing disc have been characterized so far. Interestingly, genetic data suggested the presence of L3 vein-specific CRMs upstream of ara (Gomez-Skarmeta et al. 1996), consistent with a putative location within the sowah locus. To find these putative wing CRMs we generated three molecularly defined deletions (sowahEGP1, sowahEGP2, and sowahEGP3) (Fig. 5A) and monitored the expression of ara, caup, and mirr in the wing discs by in situ hybridization (Fig. 5B–E). In every mutant condition examined, ara and caup expressions were undistinguishable from each other, and thus, only caup expression is shown; mirr expression was not affected by any of the deletions (data not shown).
Figure 5.
Expression of caup in sowahEGP imaginal discs and embryos. (A) Physical map of the Iro-C locus. Genomic DNA is shown as a thick black bar with a 60-kb gap delimited by //. Transcripts are shown as black arrows below the genes (blue). Exons are shown in orange, with protein-coding regions colored darker. Red broken lines within brackets represent deleted regions. The purple box represents a region bound by several transcription factors as determined by ChIP-on-chip assays. (B–G) In situ hybridization with a caup probe of wing (B–E) and leg (F,G) discs of the indicated genotype. caup expression is not affected in the trans-heterozygous combination of sowahEGP1 and iroDFM3, used to rescue the early embryonic lethality of sowahEGP1. In sowahEGP2 (D) and sowahEGP3 (E) discs, caup expression is absent in L3 and DR domains and strongly reduced in Pl and Tg regions (marked with black asterisks). (G) sowahEGP3 leg disc, in which caup wild-type expression in a ring-like pattern (F) is lost. (H–K) Anti-Caup staining of wild-type (H,H',J) and sowahEGP3 (I,I',K) late-stage 13 embryos. Yellow asterisks mark the expression of ara/caup in the nervous system; arrowheads point to the head mandibular segment. The brown signal in I corresponds to a nonspecific staining of the tracheal (respiratory) system. (H,I) ventral, (J,K) lateral views, (H',I') enlarged views of the head.
sowahEGP2 harbors a deletion spanning the central area of sowah, including its longest introns (ancestral introns 5 and 6) (Fig. 5A). In these flies, ara/caup expression patterns were dramatically affected, lacking the expression from the L3 vein, pleura (Pl), tegula (Tg), and dorsal radius (DR) domains of the wing disc (black asterisks in Fig. 5D). This suggests that CRM(s) driving ara/caup expression in these territories are located within this region, most likely within the long introns. Consistently, the sowahf05010 mutation—associated with the insertion of an insulator-containing transposon in the second intron of sowah—shows a similarly altered expression of ara/caup in the wing discs (Supplemental Fig. S8). The presence of this insulator is likely to prevent the interaction of CRM(s) located upstream of the insertion point with the downstream ara and caup promoters. Importantly, sowah transcripts are undetectable in wild-type imaginal discs (Fig. 3F), suggesting that the loss of ara/caup expression is not due to sowah loss of function. This notion is further supported by the fact that sowah exons are unaffected in sowahf05010 flies; however, we cannot completely rule out that the insertion of a transposon within the intronic sequence could have mildly affected sowah activity.
sowahEGP3 has a larger deletion than sowahEGP2. It encompasses the region deleted in sowahEGP2 and spans from the ancestral ninth intron of sowah to the third intron of ara (Fig. 5A). Remarkably, in addition to the domains of ara/caup expression lost in sowahEGP2 and sowahf05010 (black asterisks in Fig. 5E), the sowahEGP3 deficiency also altered the expression of Iro genes in leg discs (Fig. 5G), and in some domains of the embryonic head (arrowheads in Fig. 5I,I') and nervous system (yellow asterisks in Fig. 5I,K). This suggests that CRM(s) responsible for these embryonic expression domains and for the “ring” pattern of the leg discs are located somewhere in the intergenic region between ara and sowah and/or in the first two introns of sowah. Noteworthy, ChIP-on-chip experiments in embryos (http://genome.ucsc.edu/) show that several TFs bind to a region in Drosophila sowah intron 2 (ancestral intron 3, purple box in Fig. 5A), suggesting that this intron could contain the CRM(s) that drive ara/caup expression in the head and nervous system.
In sowahEGP1, which lacks most of the sowah coding region (but not the longest introns) and the three CGs immediately distal to sowah (CG34242, CG10688, and CG4069) (Fig. 5A), and in sowahe01289 flies, which harbor a transposon inserted in the sowah exon 7 (Fig. 5A), ara/caup expression was unmodified (Fig. 5C; data not shown). This further reinforces the idea that the changes in the expression of ara/caup are not related to the absence of sowah function. Thus, our results show conclusively that a substantial part of the complement of CRMs of the Iro family lies within the sowah locus, suggesting that long-range cis-regulatory interactions are the major constraint preventing the disruption of the Iroquois–Sowah genomic block, at least in flies.
Sowah gene complements, intron–exon structures, and synteny in vertebrate genomes
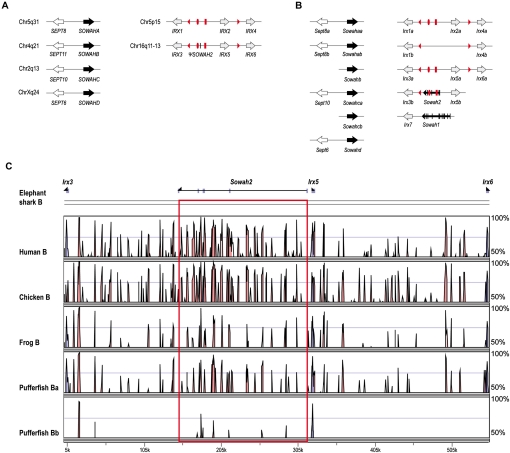
Despite the strong linkage constraint and intron position conservation in other bilaterian lineages, the four Sowah orthologs in the human genome (SOWAHA, SOWAHB, SOWAHC, SOWAHD, also known as ANKRD43, ANKRD56, ANKRD57, and ANKRD58) are not associated with Irx complexes and are intronless. Instead, they are all linked to Septin genes (SEPT8, SEPT11, SEPT10, and SEPT6, respectively), in a head-to-head orientation (Fig. 6A). This suggests that the human Sowah genes originated by a single retroposition event into a new genomic location that occurred before the two ancient rounds of whole-genome duplication (WGD) (Dehal and Boore 2005; Putnam et al. 2008). Therefore, we collectively refer to them as r-Sowah genes hereafter (the letters A–D indicate their retroposition origin, in contrast to the canonical intron-containing sowah1-2 genes in fish, see below). The same pattern was found in all tetrapod species examined (data not shown). However, in the teleost fish fugu and zebrafish, in addition to six Septin-associated intronless copies, two “canonical”, intron-containing Sowah genes are still linked to Irx (Fig. 6B; Supplemental Fig. S9). They are linked to Irx7 (see Supplemental Discussion S2 for the potential implications on the debated Irx7 orthology) (Lecaudey et al. 2001, 2005; Itoh et al. 2002; Dildrop and Rüther 2004; Feijóo et al. 2004) and Irx5b, and we termed them, respectively, Sowah1 (LOC100331692-LOC100148636 in zebrafish) and Sowah2 (previously unannotated and very divergent, since the first four to five and the last two to three ancestral exons could not be detected in any available teleost genome, neither by sequence conservation nor using expression data). Thus, teleosts have partially retained the Irx–Sowah linkage typical of invertebrate animal lineages.
Figure 6.
Genomic organization of Sowah and Irx genes in vertebrates. Schematic representation of the genomic organization of r-Sowah and Sowah (black arrows), Septin (white arrows), and Irx (gray arrows) genes in humans (A) and a generalized teleost (B). Red geometrical shapes represent CNRs within Irx clusters: triangles represent the UltraConserved Regions (UCRs) (de la Calle-Mustienes et al. 2005), and ellipses and rectangles the only two CNRs present within Sowah2 in teleosts. For simplicity, only schematic intron–exon structures are indicated for Sowah1 and Sowah2. (C) VISTA plot of the alignments between IrxB clusters of different vertebrate species, using elephant shark as a reference sequence for the comparison. Colored peaks (blue, coding; turquoise, UTR; pink, noncoding) indicate regions of at least 100 bp and ≥70% similarity. Sowah2-CNRs are demarcated by a red rectangle.
To further explore the early evolution of the Irx–Sowah linkage in vertebrates, we searched the available genomic contigs of a basal jawed vertebrate, the elephant shark (Callorhinchus milii), a species with a slowly evolving genome (Venkatesh et al. 2006; Ravi et al. 2009). We found three intronless r-Sowah genes (Supplemental Fig. S9), but as in the case of teleost fish, we also identified fragments of at least two intron-containing Sowah genes. However, the highly fragmented nature of the available assembly impeded further synteny analyses and gene annotation. We thus screened an elephant shark genomic BAC library and obtained the full sequence of the IrxB cluster, which contains a Sowah2 gene, as in teleosts. The elephant shark IrxB cluster locus contains the three Irx genes (Irx3, Irx5, and Irx6) and their immediate upstream and downstream flanking genes (Fto and Mmp2) (Supplemental Fig. S10), with a total cluster length of 553 kb (similar to zebrafish IrxBa locus and shorter than those of tetrapods, Supplemental Fig. S11). The synteny of Irx and Fto and Mmp2 genes is conserved in the zebrafish IrxBa locus and all sequenced tetrapod IrxB loci (Supplemental Fig. S11). As predicted, we found a Sowah2 ortholog in the intergenic region between Irx3 and Irx5, in the same orientation as teleosts, and corresponding to some of the fragments previously identified in the blast searches. In addition to exons 5–9 of Sowah2 gene, we could also identify the first exon, indicating that the elephant shark Sowah2 gene is more conserved than its teleost counterparts. Nevertheless, as in teleosts, the remaining ancestral exons were either lost or too divergent, raising the question about the functional status of Sowah2 genes. First, Sowah2 genes were transcriptionally active in at least zebrafish and medaka, as evidenced by the presence of Expressed Sequence Tags (five and two ESTs, respectively), and RNAseq data from different zebrafish tissues (see Methods). Second, if still active as protein-coding genes, Sowah2 exonic sequences are expected to reflect the action of selective pressures. Thus, as an indicator of ongoing selection, we estimated the ratio between nonsynonymous (dN) and synonymous (dS) nucleotide substitution rates (dN/dS or ω) for those exons that could be retrieved in at least four different species (exons 5, 7, 8, and 9). In all cases, ω values were very low (ω < 0.12), indicating active purifying selection (Supplemental Fig. S12). These data suggest that, although divergent, Sowah2 is likely a functional protein-coding gene in teleosts and elephant shark.
Finally, we characterized the expression of Sowah genes in zebrafish. As in invertebrate lineages, zebrafish sowah1 and r-sowah genes showed ubiquitous and weak expression that was, in some cases, indistinguishable from the in situ hybridization background (data not shown), although expression of all of the genes was detectable by qRT–PCR (Supplemental Fig. S13). Thus, zebrafish sowah expression also differ extensively from the complex expression patterns of zebrafish Irx genes (Feijóo et al. 2004; Lecaudey et al. 2005), suggesting that, regardless of their genomic location, vertebrate Sowah genes are not coregulated with Irx.
Elephant shark and teleost Irx-linked Sowah gene CNRs include cis-regulators of Irx in tetrapods
The identification of exons of Sowah2 gene in elephant shark and teleosts suggests that Sowah2 coding sequences in tetrapods have undergone a process of pseudogenization/loss. VISTA analysis with elephant shark as a reference allowed us to confirm this hypothesis: Highly divergent, pseudogenized exonic remnants are detectable in the chicken, anole lizard, and several mammalian genomes, including human (blue circles in Supplemental Fig. S12). The presence of Sowah2 “pseudoexons” across deeply diverged tetrapod lineages was surprising, since nonfunctional pseudogenic sequences are expected to decay relatively rapidly, and could be suggestive of (ancestral) recruitment of these exons into a cis-regulatory role as CNRs (Dong et al. 2010; Eichenlaub and Ettwiller 2011). However, the degree of degeneration of these pseudogenized remnants is very variable across lineages, with a patchy distribution of pseudoexon presence. For example, three exons (7, 8, and 9) are still clearly identifiable in the chicken genome, and the only evidence of pseudogenization is the lack of splice sites, indicating a very recent inactivation. In contrast, no traces are detectable in the amphibian Xenopus tropicalis, and only one exon is present in lizard (exon 9) and placental mammals (exon 7), in both cases containing frameshift and splice-site mutations. Marsupials present an intermediate situation, with two identifiable exons (7 and 9) in the genome of Monodelphis domestica. We then checked for the presence of epigenetic marks indicative of regulatory enhancer activity in species with available data. Deposition of histone marks was not consistent with enhancer activity in any species, including human cell lines and mouse embryos, as well as sowah2 protein-coding exons in zebrafish embryos (Supplemental Fig. S12; data not shown). In support of this idea, we found no evidence for evolutionary constraint when comparing pseudoexon sequences across closely related species (e.g., within primates, Supplemental Fig. S12). Taken together, these results suggest that the presence of recognizable orthologous pseudogenic exons in deeply diverged tetrapod lineages is probably due to independent and recent Sowah2 inactivation events, and not due to functional conservation. Nevertheless, we cannot rule out that Sowah2 remnants could have cis-regulatory roles in some lineages or developmental stages.
On the contrary, the scenario for noncoding elements within Sowah was completely different: The majority of the CNRs from the IrxB cluster were ancestrally located within the regions orthologous to elephant shark Sowah2 introns (Fig. 6C). The elephant shark Sowah2 locus extends over 167 kb, accounting for >50% of the “gene desert” that separates Irx3 and Irx5 in vertebrates (de la Calle-Mustienes et al. 2005), and almost one-third of the total cluster length (553 kb). This Sowah2 region contains 58% (47 out of 81) of all IrxB cluster CNRs (between elephant shark and human, Supplemental Table S3), and 71% of those between Irx3 and Irx5. In addition, four Sowah2 CNRs are present in both IrxA and IrxB paralogous clusters (McEwen et al. 2006), suggesting that Sowah CNRs predate the WGD event that generated the two vertebrate Irx clusters, and that a Sowah gene was also originally present in the IrxA complex.
Interestingly, CNRs from both IrxA and IrxB clusters have been extensively studied and functionally characterized (de la Calle-Mustienes et al. 2005; Tena et al. 2011), including some of the elements that are now recognized as originally located within Sowah loci. Nine of these Sowah–CNRs (three of them duplicated elements present in both clusters) (McEwen et al. 2006) show transcriptional enhancer activity when assayed in both Xenopus and zebrafish (de la Calle-Mustienes et al. 2005; Tena et al. 2011). These nine CNRs drove consistent tissue-specific expression in subdomains of the endogenous Irx expression patterns (Fig. 7; de la Calle-Mustienes et al. 2005; Tena et al. 2011), and 3C experiments showed that at least one of these elements specifically interacts with the promoters of both Irx1 and Irx2 in the IrxA cluster (element 3240) (Tena et al. 2011). These results demonstrate that cis-regulatory elements that were originally located within a Sowah intron are unequivocally Irx transcriptional regulators in vertebrates and have been maintained despite the loss of the associated Sowah coding regions.
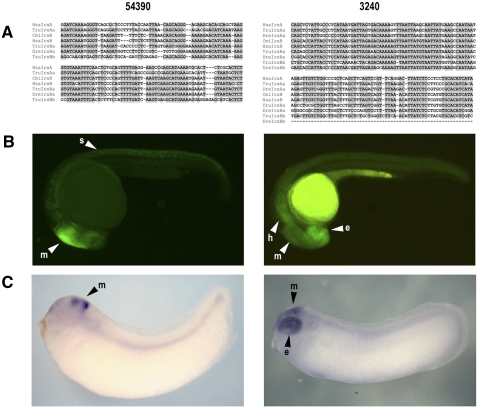
Figure 7.
Sowah intronic CNRs. Two of the Sowah2 pre-WGD CNRs that function as tissue-specific enhancers in reporter assays (represented by red ovals and rectangles in Fig. 6). (Left) CNR 54390 (Sowah2 intron 7); (right) CNR 3240 (Sowah2 intron 8). (A) Sequence alignment of the 54390 and 3240 Sowah-CNRs in different Irx complexes of several species. Shadowed nucleotides correspond to >60% sequence conservation. (B,C) GFP expression driven by the elements 54390 and 3240. Paralogous sequences of the element 54390 in complexes IrxA and IrxB drove similar expression patterns in Xenopus embryos (C) and zebrafish (B) transgenic lines. In the case of the CNR 3240, only that present in the IrxA complex was found to be positive in transgenesis studies. (e) Eye; (m) midbrain; (h) hindbrain; (s) spinal cord.
Discussion
CRMs controlling developmental genes extend along enormous distances, often populating unrelated neighboring transcriptional units (bystander genes) (Engstrom et al. 2007; Kikuta et al. 2007). These intricate and intermingled genomic domains containing a developmental gene and its CRM array encompassing one or several bystander genes are typically known as genomic regulatory blocks (GRBs) (Becker and Lenhard 2007; Kikuta et al. 2007) and create evolutionarily “solid” regions of conserved microsynteny. The presence of conserved GRBs had been previously described only within vertebrates or within insects (Engstrom et al. 2007; Kikuta et al. 2007), but none of them was shown to be conserved between the two lineages. Here, we describe the most ancient and widely conserved GRB, present since the origin of bilaterians. We show that regulatory elements present in the introns of the bystander gene Sowah are likely to regulate Irx in flies, amphioxus, and vertebrates, providing a general explanation for the maintenance of this GRB over 600 MY of independent evolution in several bilaterian lineages. Although we could not find any detectable sequence similarity between the Sowah noncoding regions and CRMs of different phyla (an extremely rare phenomenon) (Royo et al. 2011), most validated Sowah enhancers were located in a common set of conserved and exceptionally long introns (introns 5–9), suggesting the possibility of an ancestral regulatory system for this GRB in all bilaterians. In addition, these results show that GRBs, as a particular type of functional genomic organization, originated in a common ancestor of bilaterians, and that the evolution of complex cis-regulatory systems could have been determining genome remodeling processes since the origin of metazoans. Although the Irx–Sowah syntenic block constitutes the first example, this may well be the tip of the iceberg, and global approaches may reveal a wealth of ancient GRBs.
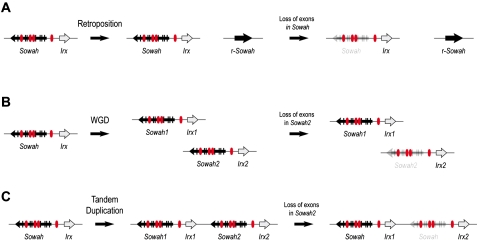
Intriguingly, despite this striking syntenic conservation, a few lineages evolved ways to disentangle the regulatory constraints and break Irx and Sowah apart. We found three different evolutionary paths, all of them involving the creation of genetic redundancy, resulting in the recurrent specific loss of the coding region of the bystander gene, while keeping the intervening CRMs (Fig. 8) (a loss process that could be a general mechanism for the formation of gene deserts, whose origins remain largely mysterious). First, in early vertebrates, a Sowah retroposition event facilitated the disruption of the Irx–Sowah linkage by allowing the specific loss of the parental, Irx-linked coding sequence, and maintenance of the functional noncoding elements. This is, to our knowledge, the first example in which a retroposition event can be related to the genomic restructuring and loss of its parental locus and to the dismantlement of functionally constrained genomic linkages. Second, in the teleost lineage, the extra Irx clusters provide an example of the most classical and commonly reported case of disruption of synteny and GRBs after WGDs (Kikuta et al. 2007). Finally, the amphioxus case illustrates a third mechanism by which the coding regions of bystander genes containing CRMs can be lost: tandem duplication. Interestingly, the only other lineages in which Sowah is not linked to Irx genes—the tunicates and the leech Helobdella robusta—have multiple and genomically disperse Irx duplicates (Supplemental Fig. S1), suggesting that similar processes of gene duplication helped to disrupt the GRBs in these species.
Figure 8.
Evolutionary scenarios for the convergent loss of Sowah coding exons near Irx genes in chordates. (A) A retroposition event to other parts of the genome (indicated as a solid black arrow, r-Sowah) allows the original, Irx-linked Sowah to lose the coding sequences (black bars), while retaining the functional noncoding regions (red). A similar event occurred at the base of the vertebrates. (B) A polyploidization (a WGD) creates redundancy of Sowah genes. Therefore, some Sowah genes can lose their coding sequences. This event was observed in teleosts, after the third round of WGD. (C) Gene redundancy is acquired by tandem duplication of Irx and Sowah, as reported for amphioxus. Subsequently, one of the Sowah copies loses its coding sequences, whereas the functional noncoding regions are maintained.
These processes illustrate the high degree of mutual interdependence between the extremely complex transcriptional regulation of development and genome architecture through bilaterian evolution.
Methods
Search for Sowah genes in metazoan genomes
In species where Sowah genes were not previously described or available gene predictions were fragmentary or poorly annotated, we built new manually curated predictions as described before (D'Aniello et al. 2008). We used the following databases: v1.0 of the genomes of Trichoplax adhaerens, Nematostella vectensis, Branchiostoma floridae, Lottia gigantea, Capitella teleta, and H. robusta, v1.0 and v2.0 of Ciona intestinalis, v4.1 of Xenopus tropicalis, and v4.0 of Takifugu rubripes from JGI (http://genome.jgi-psf.org/euk_home.html), Homo sapiens Build37.1, Gallus gallus Build2.1, Danio rerio Zv8 and Zv7, Strongylocentrotus purpuratus Build2.1, Anopheles gambiae AgamP3.3, Apis mellifera Amel4.5, Nasonia vitripennis Build1.1, and Tribolium castaneum Build2.1 from NCBI (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), Saccoglossus kowalevskii 2008-Dec-09 scaffolds from HGSC Baylor College of Medicine (http://blast.hgsc.bcm.tmc.edu/blast.hgsc?organism=20), Gasterosteus aculeatus 1.0, Oryzias latipes 1.0, Anolis carolinensis 2.0, Ornithorhynchus anatinus 1.2, Monodelphis domestica 2.2, Loxodonta africana 3.0, Canis familiaris 2.0, Mus musculus 37, and Pan troglodytes 2.1 from UCSC (http://genome.ucsc.edu/), Drosophila melanogaster, D. ananassae, D. pseudoobscura, D. willistoni, D. virilis from FlyBase (http://flybase.org), Caenorhabditis elegans, C. briggsae, C. brenneri, C. remanei, and C. japonica from WormBase (http://www.wormbase.org), and Oikopleura dioica v3 from Genoscope (http://www.genoscope.cns.fr). C. milii contigs were searched at the NCBI genomic BLAST webpage for unfinished eukaryotic genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=eukaryotes). Manually curated Sowah mRNA sequences from selected metazoan species are included as Supplemental Data S2. All other final gene annotations, including the correspondent genomic sequences and alternative splice variants are available upon request. In the study of the syntenic and regulatory interactions between Sowah and Irx loci, we applied a strict definition of GRB, requiring the presence of a bystander gene (Engstrom et al. 2007; Kikuta et al. 2007).
Phylogenetic analyses
We aligned Sowah protein sequences from multiple species using MAFFT (Katoh et al. 2002, 2005) as implemented in Jalview 2.4 (Waterhouse et al. 2009), and manually curated the alignments (available upon request) using information on intron positions (Irimia and Roy 2008). We performed two different phylogenetic analyses. First, to establish orthology of all studied Sowah genes, we used an alignment containing only the highly conserved ankyrin-repeat domain (Supplemental Fig. S2B). As there is no published information on closely related genes that could be used as outgroups, we performed BLASTP searches against the “Non-redundant protein sequences” database at NCBI. We selected BLAST hits with the highest score after applying two filters: (1) The gene is not one of our previously identified putative Sowah members, (2) the gene must have the same number of ankyrin-repeat domains as Sowah (four repeats, two highly conserved central domains, and two more divergent flanking ones spanning ancestral exons 5–9, Supplemental Fig. S3). Through this procedure we selected Ilk genes as outgroups. Orthology was further supported by the presence of a highly conserved domain in the N terminus of Sowah proteins that could not be identified in any other protein family (see the three first ancestral exons in Supplemental Fig. S3). This domain was clearly identifiable in all surveyed Sowah sequences except in C. elegans, teleost Sowah2, and vertebrate SowahD, whose orthology was otherwise well supported by the Bayesian trees and synteny data.
Second, to allow confident assignment of r-Sowah genes as in-groups within chordate Sowah genes, we increased the number of informative alignment positions, adding the aforementioned N-terminal domain and excluding the Ilk outgroups, the genes without the conserved N-terminal domain and divergent sequences from fast-evolving species (Supplemental Fig. S2B). We generated gene trees with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) using two independent runs (each with four chains). Model selection using ProtTest (Drummond and Strimmer 2001; Guindon and Gascuel 2003; Abascal et al. 2005), convergence determination, burn-in, and consensus tree calculations were done as previously described (D'Aniello et al. 2008).
Analysis of CNRs in Sowah–Irx loci
For amphioxus, we downloaded the B. floridae Irx cluster from JGI (http://genome.jgipsf.org/Brafl1/Brafl1.home.html). Two different haplotypes are normally present in the v1.0 assembly (Putnam et al. 2008). In the case of the Irx–Sowah cluster, one is located in scaffold-90, whereas the other is scrambled into several scaffolds: 632, 2884, 229, 975, and 90 (downstream from the other haplotype). We generated a consensus sequence of both haplotypes using scaffold-90 as default. We used the other scaffolds to remove gaps, correct assembly errors, and remove polymorphic repetitive elements (Supplemental Data S1). Preliminary blast searches allowed us to identify the presence (or traces of) of four tandem Irx–Sowah loci. Accordingly, we divided the nucleotide sequence of B. floridae cluster (i.e., from the end of the gene model preceding Sowah [Rpgrip] to the start of the gene model immediately after IrxC, the CAVIII gene) in four regions, each containing one of the Irx genes and surrounding noncoding sequences, plus a fourth region that contained the segment corresponding to the ancient IrxD–SowahD locus, that has been partially lost. In the case of IrxA, we also included the entire Sowah locus. With these four sequences, we performed crossed VISTA analysis using default parameters (Frazer et al. 2004). It should be noted that regulatory blocks that are not repeated cannot be detected with this analysis. We also searched repeated sequences using blast-2-sequence (bl2seq) software at http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi. Evidence for loss of IrxD was thoroughly verified using data from the assembly of both haplotypes to discard possible artifacts due to gaps or assembly errors.
For nematodes and flies, genome-wide VISTA analyses have not been done before. Therefore, we chose appropriate parameters for these species based on the length and percentage identity of CNRs previously identified in these lineages. In nematodes we used C. elegans as a reference sequence, using a window size of 50 bp and 90% minimum identity (85% in C. japonica), based on previously published data on worm CNRs (Vavouri et al. 2007) with slight modifications to take into account the inclusion of more divergent species. For flies, following previously reported CNR definitions (100% identity over 50 bp) (Glazov et al. 2005), we used a less-stringent identity criterion to account for the inclusion of D. virilis (90% identity) and a larger minimum window size (60 bp) following works reporting longer CNRs in the vicinity of developmental genes such as Hox genes (Negre et al. 2005). In vertebrates, VISTA analyses were performed using default parameters (≥70% identity over 100 bp), except for the detection of Sowah2 pseudogenic sequences (see below). In the case of teleosts IrxBb clusters, we complemented the VISTA analysis with bl2seq and ClustalW.
Ancora data (http://ancora.genereg.net/) were visualized as custom tracks in the UCSC Genome Browser (http://genome.ucsc.edu/). All available Ancora precomputed CNR density tracks (based on different CNR definitions) were checked, yielding equivalent results. For representation in Figure 1, we used the “96% identity over 30 column” density track for nematodes and the “98% identity over 50 columns” for flies.
Conservation tracks by phastCons and PhyloP of Sowah regions were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/).
Sequencing of IrxB locus in elephant shark
We searched the elephant shark C. milli 1.4× assembly (Venkatesh et al. 2007) using TBLASTN with human IRX3 and IRX5 protein sequences. We identified two scaffolds (AAVX01172485.1 and AAVX01359425.1) that contained exons for Irx3 and Irx5 genes, respectively. We cloned different DNA probes for each scaffold using PCR and used them to screen pooled DNA of an elephant shark BAC library (see Ravi et al. 2009). We selected one representative BAC each for Irx3 and Irx5. After sequencing these BACs completely, we identified overlapping BACs by PCR. Altogether, we sequenced six BAC clones (Supplemental Fig. S10) to obtain the complete sequence of elephant shark IrxB locus (GenBank accession number JN228895). BAC sequencing was done using a standard shotgun sequencing technique, with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequences were processed and assembled using phred-phrap and Consed (http://www.phrap.org/phredphrapconsed.html). Elephant shark genes were predicted based on their homology with known genes in other vertebrates and as indicated above.
Functional status of Sowah2 genes
Estimation of ω was performed with the maximum-likelihood (ML) method as implemented in CODEML in PAML v.4.2 (Yang 2007) using the general model and WAG + Γ. Each Sowah2 exon was analyzed independently, since not all of them could be identified in all studied species. Sowah2 pseudogenized exonic sequences in tetrapods were detected with VISTA. Most of them could be retrieved using elephant shark as a reference sequence and low-stringency parameters (60% identity in 60 bp), although in some particular cases we used even a lower stringency (50% in 50 bp for pseudoexon 9 in M. domestica) and a different species as a reference sequence (human in the case of M. musculus pseudoexon 7). These analyses were complemented with bl2seq and ClustalW, and all pseudoexons were aligned and translated to highlight their mutations (data not shown). Selected tetrapod Sowah2 pseudogenic sequences are included in Supplemental Data S3. Expression data for teleost Sowah2 genes were obtained by blasting nucleotide sequences from all teleost species against the teleost EST database at the NCBI blast webpage (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and by mapping available short RNAseq reads from zebrafish head, ovary, and embryos against zebrafish sowah2 mRNA using bowtie (Langmead et al. 2009) with default parameters. Raw ChIP-seq data for H3K4me1 and H3K4me3 in whole zebrafish embryos at 24 hpf was obtained from Aday et al. (2011). Highly enriched regions (peaks) of histone methylation were obtained by the MACS (v.1.3.3) algorithm (Zhang et al. 2008) using standard settings with one modification (mfold = 20). These results were uploaded to the UCSC browser (http://genome.ucsc.edu/) as custom tracks for visualization in Danio rerio Zv8. Sowah2 exon 8 (located in an unmapped contig <2.7 kb long) data were not represented in Supplemental Figure S12 due to the lack of genomic context. Equivalent data from human and mouse were checked in roadmap (http://www.roadmapepigenomics.org/data) and the UCSC Genome Browser, respectively.
Cloning of Sowah genes from amphioxus, D. melanogaster, and zebrafish, and of amphioxus CNRs
We designed primer pairs to span partial coding sequences of D. melanogaster and B. floridae Sowah and zebrafish sowah - r-sowah complement. We screened aliquots of cDNA libraries from different developmental stages of both B. floridae and Branchiostoma lanceolatum by PCR using the B. floridae Sowah primers. D. melanogaster Sowah was amplified by PCR using cDNA from larvae. Zebrafish genomic DNA and cDNA was used as a template to PCR amplify non-Irx linked r-sowah genes and sowah1, respectively. We cloned all genes in TA Cloning pCRII vectors (Invitrogen) and sequenced them using standard M13F and M13R primers. B. lanceloatum Sowah sequence has been submitted to GenBank (accession number JN609218).
For each amphioxus CNR, we designed primers to span the whole conserved sequence, plus a padding of ∼100 nt on each side. We performed PCRs on genomic DNA using iProof High-Fidelity DNA Polymerase (Bio-Rad). We cloned amplicons in pCR8GW/TOPO vector (Invitrogen) according to the manufacturer's instructions. We then transferred sequence-verified clones with the Gateway recombination system (Invitrogen) to the ZED vector (Bessa et al. 2009). We purified the final transgenic constructs using phenolchloroform and normalized at 50 ng/mL in DEPC water prior to microinjection.
Sequences of all primers are provided in Supplemental Table S2.
sowahEGP deficiency generation and fly stocks
The FLP–FRT recombination method and the FRT-bearing piggyBac insertion lines from the Exelixis collection were used to generate w+ sowahEGP deficiencies, which were confirmed by PCR analyses (Parks et al. 2004; Thibault et al. 2004). We combined e03723 and e01289 starting insertions to generate sowahEGP1, e01289, and f05010 for sowahEGP2, and f01127 and e02801b for sowahEGP3. We confirmed the localization of the piggyBac insertion site by PCR in all lines, matching their previously described localization (Thibault et al. 2004). Note that there is an error in FlyBase: f01127 insertion site is reported >16 kb downstream from its actual location. sowahEGP1 is embryonic lethal, while a few LIII escapers are found in sowahEGP2 and sowahEGP3.
iroDFM3 is a deficiency obtained by imprecise excision of the P[LacZ] element of irorF209, which removes ara, caup, and the promoter of mirr (Gomez-Skarmeta et al. 1996). sowahe01289 is a putative null alelle of sowah: due to the transposon insertion at the beginning of the region encoding the highly conserved ankyrin repeat domain (Fig. 5A), this allele could encode a Sowah protein deficient in this domain.
In situ hybridization and inmunohistochemistry in different species
In situ hybridization of whole mounts of Drosophila imaginal discs, amphioxus specimens of the European species B. lanceolatum, and zebrafish embryos with digoxigenin-labeled antisense RNA probes and immunocytochemistry of Drosophila embryos, were performed as previously described (Cubas et al. 1991; Tena et al. 2007; Yu and Holland 2009; Irimia et al. 2010; Carrasco-Rando et al. 2011). The following primary antibodies were used: rat anti-Caup, an antibody that recognizes both Ara and Caup 1:50 (Diez del Corral et al. 1999), and rabbit anti-β-galactosidase (Cappel; 1:5000).
Zebrafish microinjections and husbandry
We injected 50–100 pg of each ZED-CNR vector into one-cell stage embryos together with 50–100 pg of Tol2 mRNA. We observed injected fish at 24 and 48 hpf for GFP expression. As internal injection quality control, we determined muscle RFP expression at 72 hpf. Embryos were selected and raised to sexual maturity. Three or more independent stable transgenic lines were generated for each construct. We cloned PCR fragments in pCR8/TOPO and generated RNA probes using T7 polymerase and standard procedures. Embryos were reared at 28°C in standard E3 medium.
Real time–qPCRs (RT–qPCRs)
We studied expression dynamics of Sowah paralogs by RT–qPCRs. We isolated total RNA from 20 embryos each from 24 and 48 hpf stages, and 30 embryos from 80% of epiboly stage. cDNA was synthesized from total RNA by reverse transcription, and the relative amounts of different gene products were measured by RT–qPCR. We normalized all data using the gene elongation factor 1-alpha. We took the 24-hpf stage sample as reference.
Data access
The sequence data generated for this study have been submitted to GenBank (accession numbers JN228895 and JN609218).
Acknowledgments
We thank Jose L. Ferrán for kindly helping with amphioxus in situ hybridizations and stimulating conversations, Silvia Naranjo, Ana M. Neto, Elisa de la Calle-Mustienes, Ana Ariza, Elisa Rodríguez-Seguel, and all members of the J.-L.G.-S. lab for supporting help and advice, Yolanda Roncero for zebrafish husbandry, Ferdinand Marlétaz for help with ω estimations, Marina Ruiz and Isabel Almudí for technical support, Mar Ruiz-Gómez and Marta Carrasco-Rando for help with Drosophila embryo staining, and Peter W.H. Holland for support and helpful suggestions on the manuscript. M.I., I.M., and J.G.-F. were funded by Grants BFU2005-00252 and BMC2008-03776 and BMC2011-23291 from the Spanish Ministry of Science and Innovation, and J.G.F. by the ICREA Academia Prize. M.I. and I.M. held FPI and FPU grants, respectively. J.-L.G.-S. and J.J.T. were supported by Grants BFU2010-14839, CSD2007-00008 (MEC), and CVI 3488 (Junta de Andalucía). S.C. and E.G.-P. were supported by grants BFU2008-03762/BMC (MICIIN), CDS2007-00008 (MEC), and an institutional grant from Fundación Ramón Areces to the CBMSO. D.T., V.R., and B.V. were supported by the Biomedical Research Council of A*STAR, Singapore.
Authors' contributions: I.M. and M.I. performed the bioinformatic analyses, the amphioxus analyses, and cloning of CNRs. J.J.T. with the collaboration of I.M. carried out the zebrafish experiments. E.G.-P. performed the Drosophila experiments. D.T. and V.R. cloned and sequenced the elephant shark cluster. D.T., V.R., B.V., I.M., and M.I. analyzed the elephant shark data. All authors participated in the experimental design. I.M. and M.I. wrote the first draft of the manuscript and J.J.T., E.G.-P., B.V., S.C., J.L.G.-S., and J.G.-F. helped with the writing. I.M. and M.I. conceived the work.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.132233.111.
References
- Abascal F, Zardoya R, Posada D 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND 2011. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol 357: 450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S 1995. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci 92: 1684–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Lenhard B 2007. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genomics 278: 487–491 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn 238: 2409–2417 [DOI] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM 2004. Comparative genomics at the vertebrate extremes. Nat Rev Genet 5: 456–465 [DOI] [PubMed] [Google Scholar]
- Carrasco-Rando M, Tutor AS, Prieto-Sánchez S, González-Pérez E, Barrios N, Letizia A, Martín P, Campuzano S, Ruiz-Gómez M 2011. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet 7: e1002186 doi: 10.1371/journal.pgen.1002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL 2001. The Iroquois family of genes: from body building to neural patterning. Development 128: 2847–2855 [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J 2007. Intron loss and gain in Drosophila. Mol Biol Evol 24: 2842–2850 [DOI] [PubMed] [Google Scholar]
- Cubas P, de Celis JF, Campuzano S, Modolell J 1991. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev 5: 996–1008 [DOI] [PubMed] [Google Scholar]
- D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, Bertrand S, Garcia-Fernàndez J 2008. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol 25: 1841–1854 [DOI] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodríguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL 2005. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res 15: 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314 doi: 10.1371/journal.pbio.0030314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral R, Aroca P, Gómez-Skarmeta JL, Cavodeassi F, Modolell J 1999. The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev 13: 1754–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildrop R, Rüther U 2004. Organization of Iroquois genes in fish. Dev Genes Evol 214: 267–276 [DOI] [PubMed] [Google Scholar]
- Dong X, Navratilova P, Fredman D, Drivenes O, Becker TS, Lenhard B 2010. Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons. Nucleic Acids Res 38: 1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Strimmer K 2001. PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics 17: 662–663 [DOI] [PubMed] [Google Scholar]
- Eichenlaub MP, Ettwiller L 2011. De novo genesis of enhancers in vertebrates. PLoS Biol 9: e1001188 doi: 10.1371/journal.pbio.1001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B 2007. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res 17: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P, Fredman D, Lenhard B 2008. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol 9: R34 doi: 10.1186/gb-2008-9-2-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijóo CG, Manzanares M, de la Calle-Mustienes E, Gómez-Skarmeta JL, Allende ML 2004. The Irx gene family in zebrafish: genomic structure, evolution and initial characterization of irx5b. Dev Genes Evol 214: 277–284 [DOI] [PubMed] [Google Scholar]
- Ferrell CM, Dorsam ST, Ohta H, Humphries RK, Derynck MK, Haqq C, Largman C, Lawrence HJ 2005. Activation of stem-cell specific genes by HOXA9 and HOXA10 homeodomain proteins in CD34+ human cord blood cells. Stem Cells 23: 644–655 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS 2005. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res 15: 800–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferré-Marcó D, Modolell J 1996. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85: 95–105 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW 2008. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res 36: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Garcia-Fernandez J 2008. Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol Biol Evol 25: 1521–1525 [DOI] [PubMed] [Google Scholar]
- Irimia M, Piñeiro C, Maeso I, Gómez-Skarmeta JL, Casares F, Garcia-Fernàndez J 2010. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. EvoDevo 1: 7 doi: 10.1186/2041-9139-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Burguera D, Hidalgo-Sánchez M, Puelles L, Roy SW, Garcia-Fernàndez J, Ferrán JL 2011. Contrasting 5′ and 3′ evolutionary histories and frequent evolutionary convergence in meis/hth gene structures. Genome Biol Evol 3: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kudoh T, Dedekian M, Kim CH, Chitnis AB 2002. A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development 129: 2317–2327 [DOI] [PubMed] [Google Scholar]
- Jiménez-Delgado S, Crespo M, Permanyer J, Garcia-Fernàndez J, Manzanares M 2006. Evolutionary genomics of the recently duplicated amphioxus Hairy genes. Int J Biol Sci 2: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach SL, Holland LZ, Holland ND, Koop D 2009. Developmental expression of the three iroquois genes of amphioxus (BfIrxA, BfIrxB, and BfIrxC) with special attention to the gastrula organizer and anteroposterior boundaries in the central nervous system. Gene Expr Patterns 9: 329–334 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl BT, Cho KO, Choi KW 1998. mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development 125: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Kerner P, Ikmi A, Coen D, Vervoort M 2009. Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evol Biol 9: 74 doi: 10.1186/1471-2148-9-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komisarczuk AZ, Kawakami K, Becker TS 2009. Cis-regulation and chromosomal rearrangement of the fgf8 locus after the teleost/tetrapod split. Dev Biol 336: 301–312 [DOI] [PubMed] [Google Scholar]
- Koonin EV 2009. Evolution of genome architecture. Int J Biochem Cell Biol 41: 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaudey V, Thisse C, Thisse B, Schneider-Maunoury S 2001. Sequence and expression pattern of ziro7, a novel, divergent zebrafish iroquois homeobox gene. Mech Dev 109: 383–388 [DOI] [PubMed] [Google Scholar]
- Lecaudey V, Anselme I, Dildrop R, Rüther U, Schneider-Maunoury S 2005. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J Comp Neurol 492: 289–302 [DOI] [PubMed] [Google Scholar]
- Letizia A, Barrio R, Campuzano S 2007. Antagonistic and cooperative actions of the EGFR and Dpp pathways on the iroquois genes regulate Drosophila mesothorax specification and patterning. Development 134: 1337–1346 [DOI] [PubMed] [Google Scholar]
- McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G 2006. Ancient duplicated conserved noncoding elements in vertebrates: A genomic and functional analysis. Genome Res 16: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA 1997. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev 11: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Negre B, Casillas S, Suzanne M, Sánchez-Herrero E, Akam M, Nefedov M, Barbadilla A, de Jong P, Ruiz A 2005. Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Res 15: 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Hersh B, Carroll S 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol 5: R25 doi: 10.1186/gb-2004-5-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413 doi: 10.1126/science.1088328 [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L 2005. Evolution and functional classification of vertebrate gene deserts. Genome Res 15: 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Pascual-Anaya J, D'Aniello S, Garcia-Fernàndez J 2008. Unexpectedly large number of conserved noncoding regions within the ancestral chordate Hox cluster. Dev Genes Evol 218: 591–597 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Ravi V, Lam K, Tay B-H, Tay A, Brenner S, Venkatesh B 2009. Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci 106: 16327–16332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV 2003. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13: 1512–1517 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Royo JL, Maeso I, Irimia M, Gao F, Peter IS, Lopes CS, D'Aniello S, Casares F, Davidson EH, Garcia-Fernàndez J, et al. 2011. Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci 108: 14186–14191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Bailey P, Bruce S, Engstrom P, Klos J, Wasserman W, Ericson J, Lenhard B 2004. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5: 99 doi: 10.1186/1471-2164-5-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N, Butts T, Candiani S, Pestarino M, Ferrier DEK, Saiga H, Holland PW 2008. Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev Genes Evol 218: 579–590 [DOI] [PubMed] [Google Scholar]
- Tena JJ, Neto A, de la Calle-Mustienes E, Bras-Pereira C, Casares F, Gomez-Skarmeta JL 2007. Odd-skipped genes encode repressors that control kidney development. Dev Biol 301: 518–531 [DOI] [PubMed] [Google Scholar]
- Tena JJ, Alonso ME, de la Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, Gómez-Skarmeta JL 2011. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun 2: 310 doi: 10.1038/ncomms1301 [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Vavouri T, Walter K, Gilks WR, Lehner B, Elgar G 2007. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol 8: R15 doi: 10.1186/gb-2007-8-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, et al. 2006. Ancient noncoding elements conserved in the human genome. Science 314: 1892 doi: 10.1126/science.1130708 [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh Y-H, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, et al. 2007. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol 5: e101 doi: 10.1371/journal.pbio.0050101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi: 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yu JK, Holland LZ 2009. Amphioxus whole-mount in situ hybridization. Cold Spring Harbor Protoc doi: 10.1101/ pdb.prot5286 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nusbaum C, Myers R, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]