Abstract
Although retroviruses are relatively promiscuous in choice of integration sites, retrotransposons can display marked integration specificity. In yeast and slime mold, some retrotransposons are associated with tRNA genes (tDNAs). In the Saccharomyces cerevisiae genome, the long terminal repeat retrotransposon Ty3 is found at RNA polymerase III (Pol III) transcription start sites of tDNAs. Ty1, 2, and 4 elements also cluster in the upstream regions of these genes. To determine the extent to which other Pol III–transcribed genes serve as genomic targets for Ty3, a set of 10,000 Ty3 genomic retrotranspositions were mapped using high-throughput DNA sequencing. Integrations occurred at all known tDNAs, two tDNA relics (iYGR033c and ZOD1), and six non-tDNA, Pol III–transcribed types of genes (RDN5, SNR6, SNR52, RPR1, RNA170, and SCR1). Previous work in vitro demonstrated that the Pol III transcription factor (TF) IIIB is important for Ty3 targeting. However, seven loci that bind the TFIIIB loader, TFIIIC, were not targeted, underscoring the unexplained absence of TFIIIB at those sites. Ty3 integrations also occurred in two open reading frames not previously associated with Pol III transcription, suggesting the existence of a small number of additional sites in the yeast genome that interact with Pol III transcription complexes.
The genomic patterns of retrovirus and retrotransposon integration sites provide useful insights into host factors that influence retroelement proliferation as well as the potential impact of these integrations on host cell biology. Despite the relatively low selectivity of retroviruses for integration into complex genomes, high-throughput sequencing has revealed influences of DNA sequence and structure, transcription factors, and epigenetic modifications of DNA and histones on retrovirus integration site preferences (e.g., Berry et al. 2006). Analysis of integration patterns of non-native transposable elements has also provided insights into chromatin structure and mechanisms of integration, for example, the targeting of nucleosome-free chromatin by the Hermes cut-and-paste DNA element expressed in Saccharomyces cerevisiae (Gangadharan et al. 2010). In some contrast to these systems, some retrotransposons in single-celled organisms have evolved efficient targeting mechanisms. These include the global targeting of RNA polymerase II (Pol II) promoters in Schizosaccharomyces pombe (e.g., Guo and Levin 2010), the targeting of heterochromatin in S. cerevisiae (Baller et al. 2011), and the targeting of Pol III promoters in S. cerevisiae and Dictyostelium.
In S. cerevisiae, long terminal repeat (LTR) retrotransposons Ty1-4 and isolated LTR remnants cluster upstream of tRNA genes (tDNAs). Ty1 and Ty3, the most extensively studied, both require active Pol III promoters, but associate in distinct patterns (Sandmeyer et al. 2002; Voytas and Boeke 2002). Ty1 clusters from ∼60 bp to 750 bp upstream of the tDNA transcription start site (TSS). Integration frequency decreases with distance from the target with an 80-bp periodicity (Bachman et al. 2004). Consistent with the interpretation that a particular aspect of nucleosome structure is favored, Ty1 insertion patterns are sensitive to chromatin organization. For example, truncation of the Bdp1 component of TFIIIB or loss of Isw2, a chromatin-remodeling factor that maps to tDNAs, reduces the amplitude of the characteristic periodicity (Bachman et al. 2005; Gelbart et al. 2005). Other long-range factors including the local concentration of tDNAs and isolated LTRs also influence Ty1 integration patterns (Bachman et al. 2004).
Other retrotransposons target the Pol III transcription complex more directly. In Dictyostelium, non-LTR retrotransposons TRE5 and TRE3 cluster ∼44 to 54 bp upstream and 40 to 150 bp downstream of tDNA-coding sequences, respectively (Winckler et al. 2005). Genomic Ty3 and LTR remnants occur within a few base pairs of the tDNA TSS (Sandmeyer et al. 2002). Although genomic copies of Ty3 are associated with tDNAs, studies using plasmid-borne Pol III–transcribed genes indicate that other genes associated with TFIIIB including 5S (RDN5) and U6 (SNR6) genes are also targets.
Two general types of promoters are recognized by Pol III in the yeast genome (Kassavetis and Geiduschek 2006; Dieci et al. 2007). For most of these genes, the promoter elements are short sequences inside the transcribed region, and terminators are tracks of five or more T's on the template strand (Allison and Hall 1985). Type 1 promoters require factors TFIIIA, TFIIIC, and TFIIIB. TFIIIA loads onto internal box C and box A promoter elements and mediates binding of TFIIIC, which loads TFIIIB upstream of the initiation site (Taylor and Segall 1985; Braun et al. 1989). TFIIIB is the initiation factor proper comprised of Brf1, Tbp1, and Bdp1 (Kassavetis et al. 1990). RDN5 genes are the sole representatives of Type 1 promoters. They are present in 100–200 copies alternating with repeats of RDN37, the gene for the 35S rRNA, on chromosome XII (Johnston et al. 1997).
Pol III Type 2 promoters have box A and box B elements downstream from the TSS. TFIIIC binds Type 2 promoters through interactions with box A and box B sequences and, similar to its activity on Type 1 promoters, mediates binding of TFIIIB upstream of the TSS. For a subset of genes, there is an upstream TATA element that enhances binding of TFIIIB (Eschenlauer et al. 1993; Giuliodori et al. 2003). Informatic analysis and genome-wide chromatin immunoprecipitation (ChIP) have identified more than 280 Type 2 genes (Lowe and Eddy 1997; Percudani et al. 1997; Hani and Feldmann 1998; Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). These include 274 tDNAs and one pseudo-tDNA; SCR1, encoding the RNA component of the signal recognition particle (Dieci et al. 2007); SNR52, encoding a C/D box snoRNA guide for rRNA processing; RPR1, encoding the RNA component of the RNase P1-processing complex; SNR6, encoding the U6 spliceosomal RNA; and three genes of unknown function. These are iYGR033C and ZOD1, which encode ancient tDNAs for tRNAArg and tRNAIle, respectively (Guffanti et al. 2006), and RNA170 (Olivas et al. 1997). For the most part, Type 2 genes are occupied by TFIIIB, TFIIIC, and Pol III in logarithmically growing cells (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). These loci vary with respect to expression, upstream TATA elements, separation of box A and box B and position of box B relative to transcribed sequence.
Recent studies have implicated Pol III transcription factors in functions in addition to Pol III transcription. A small but intriguing set of loci in the S. cerevisiae genome binds TFIIIC, but low or undetectable amounts of TFIIIB. These were dubbed Extra Three C (ETC) (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004) and include RNA170/ETC5 and ZOD1, which are weakly transcribed, and iYGR033c, ETC1-4, and ETC6-8, which are not detectably transcribed (Olivas et al. 1997; Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). About 400 condensin binding sites in S. cerevisiae are associated with TFIIIC, and only a subset of these are tDNAs (D'Ambrosio et al. 2008). Roles for Pol III promoter elements in simple eukaryote chromatin structure have been also observed in S. pombe (Noma et al. 2006). In addition to structural roles for ETCs, tDNAs establish heterochromatic boundaries (Donze and Kamakaka 2001), phase nucleosomes (Morse et al. 1992), bind condensins (D'Ambrosio et al. 2008; Haeusler et al. 2008; Gard et al. 2009), repress transcription of neighboring Pol II–transcribed genes (Kinsey and Sandmeyer 1991; Hull et al. 1994), undergo ectopic recombination (Munz et al. 1982), and disrupt progression of downstream replication forks (Deshpande and Newlon 1996).
Ty3 elements target Pol III TSS (Chalker and Sandmeyer 1990, 1992; Sandmeyer et al. 2002). In vitro, recombinant TFIIIB subunits Tbp1 and Brf1 are sufficient to target strand-transfer of Ty3 cDNA from virus-like particles (Kirchner et al. 1995; Yieh et al. 2000). However, TFIIIC is implicated by in vivo experiments in which truncation of Tfc1, a subunit of TFIIIC, restricts orientation of Ty3 insertions (Aye et al. 2001). Nonetheless, genomic Ty3 elements have only been observed in association with tDNAs. The present study was undertaken to saturate de novo Ty3 genomic transposition targets and determine the overlap with genes transcribed by Pol III. The results show that Ty3 integrates with high specificity at all known Pol III–transcribed genes and at additional sites, two of which were confirmed to be dependent on the presence of a consensus box B (Olivas et al. 1997; Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004; Guffanti et al. 2006; Marck et al. 2006). ETC loci did not support Ty3 integration.
Results
Ty3 retrotransposition is predominantly mediated by integration rather than homologous recombination
S. cerevisiae LTR retrotransposons reverse-transcribe genomic RNA into a full-length cDNA copy, which transposes into the host genome via Rad52-dependent homologous recombination or integrase (IN)–dependent strand-transfer reactions. The impact of homologous recombination on Ty3 transposition was assessed using S288C-related YMA1322 and its rad52Δ derivative, YMA1356 (Table 1A). YMA1322 and YMA1356 were transformed with a low-copy plasmid marked with URA3 and bearing a galactose-inducible Ty3 modified by the insertion downstream from a Ty3 protein-coding sequence of a copy of the HIS3 gene flanked by unique 60-bp sequence tags (Ty3-ppt HIS3 on plasmid pKN3050) (Table 1B; Supplemental Table S1). Transformants were induced for Ty3 expression by growth in medium containing galactose. Cells that had lost the plasmid but acquired a chromosomal Ty3-ppt HIS3 were selected by growth on medium selecting against the donor plasmid, and for the presence of the HIS3 marker gene. A quantitative version of this assay showed that the frequency of transposition in the wild-type strain was threefold that of the rad52 strain (data not shown). This was similar to the twofold difference between wild-type and rad52 derivative previously reported (Sadeghi et al. 2001). Enrichment for cells that had undergone transposition before production of genomic libraries for sequencing was necessitated by the low percentage of transformants (<1%) that undergo transposition and by the diversity of those events. Approximately 10,000 His+ colonies were collected and pooled for each strain. Ty3 insertion junctions were enriched by an inverse PCR (iPCR) strategy using primers inside Ty3-ppt HIS3 (Fig. 1; Supplemental Table S2; Ochman et al. 1988; Wang et al. 2008; Methods).
Table 1.
Yeast strains and plasmids used in this study
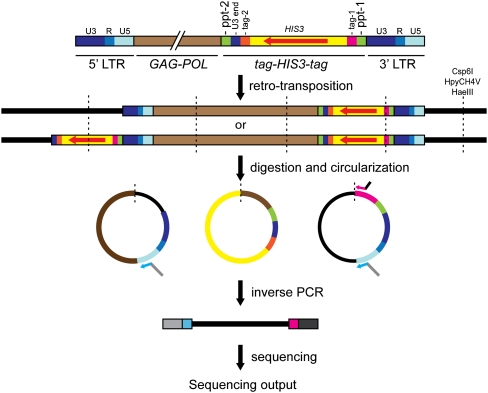
Figure 1.
High-throughput sequencing of de novo Ty3 integration sites. Step 1: Expression of Ty3-ppt-HIS3 containing two unique 60-bp tags for amplification of integration joints. Step 2: Alternative reverse transcription products, dependent on position of ppt. Step 3: Restriction of genomic DNA with one of three restriction enzymes (Csp6I, HpyCH4V, and HaeIII) and circularization. DNA fragments flanking de novo Ty3 integration sites were selectively amplified by inverse PCR (iPCR) using primers that annealed to the Ty3 U5-end LTR and Ty3 tag-1. Primers contained Illumina adaptor sequences and the Ty3 LTR primer contained the Illumina sequencing primer sequence. Step 4: The iPCR products from separate reactions were combined and processed for sequencing using the Illumina GAIIx system. Ty3 transcription begins at the 5′ edge of R and continues through U5 into the internal domain and the 3′ U3 and R regions. Reverse transcription regenerates the complete U3-R-U5 LTR at each end of the cDNA. (Vertical dashed lines) Digestion/ligation sites; (red arrow) HIS3 ORF; (bent arrows) iPCR primers. Arrows indicate primers: (light blue) Ty3 U5-end LTR; (pink) tag-1; (gray) upstream Illumina adaptor with sequencing primer; (black) downstream Illumina adaptor.
Pooled iPCR products were sequenced using Illumina GAIIx technology (Illumina; Tpn-seq). Sequencing primers annealed so that sequence began with the terminal 17 nt of the Ty3 LTR U5 and read into ∼19 nt of flanking genomic DNA (Fig. 2A). Sequences were aligned with the yeast reference sequence from the UCSC Genome Browser (build sacCer2, June 2008) (Rhead et al. 2010) using Bowtie (Langmead et al. 2009). Including data from both experiments, a total of 12.4 × 106 reads were aligned to the Ty3 LTR sequence (Table 2; Supplemental Table S3; NCBI Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31820). Of these, 6.1 × 106 reads aligned to the genome with no mismatches and were analyzed further. A total of 0.37 × 106 reads aligned, but not uniquely (“multiples”), and were separately evaluated. Individual Ty3 insertion sites (“events”) separated by 10 bp or less were clustered, yielding a total of 296 clusters. The number of reads per cluster was defined as the “cluster density” (see example, Fig. 2B), and this value was normalized to total density per experiment. The average density per cluster between the two experiments ranged from ∼100,000 reads per [tT(UGU)P] to six reads per [tV(AAC)L]. Because of the precision of Ty3 targeting, multiple independent transpositions were likely to map to the same insertion site. Therefore, the number of events is likely to underestimate the number of independent insertions (Supplemental Table S3; Chalker and Sandmeyer 1992; Kinsey and Sandmeyer 1995; data not shown).
Figure 2.
Alignment of sequencing reads to yeast genomic sequence. (A) Sequence of Ty3 integration joint. High-throughput sequencing reads began with 17 nt from the U5 end of the Ty3 LTR and continued into genomic sequence. Informative reads aligned uniquely with ∼19 nt of target site DNA in the reference genome. Each unique junction defined an integration event. A sequencing read representing a collection of events at tK(CUU)F is shown. (B) Analysis of target loci. Reads captured by events separated by 10 nt or less were combined into clusters (shaded box). The distribution of reads (hits) among events clustered at tK(CUU)F is shown. (Dashed lines) Ty3 LTR sequences. Depending on the orientation in which Ty3 integrated, sequencing reads aligned with either the Watson or Crick strand of the genomic sequence of yeast reference strain S288C. (Right column) The numbers of reads for the same integration joint (event densities). The most-sequenced junctions in the two strands were offset by the 5-bp stagger characteristic of Ty3 insertion sites (triangles).
Table 2.
Distribution of aligned reads
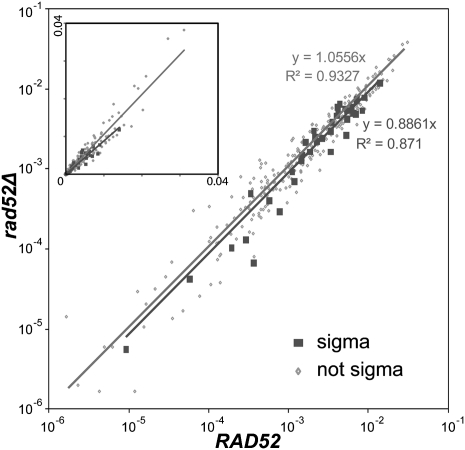
Comparison of normalized cluster densities for RAD52 and rad52Δ strains showed very similar results (Fig. 3). In particular, the plots of Ty3-ppt HIS3 transpositions to loci associated with Ty3 LTRs showed only a slight reduction in slope compared with the plot for loci not associated with Ty3 LTRs (slope = 1.0556; R2 = 0.9327 vs. slope = 0.8861; R2 = 0.871, respectively), indicating that recombining at previously occupied sites is not a major determinant of de novo Ty3 integration.
Figure 3.
Targets with preexisting Ty3 LTRs were used at a slightly greater frequency in the presence of RAD52. Cluster densities determined in RAD52 (x-axis) and rad52Δ (y-axis) strains are shown as the fraction of total cluster densities per experiment in log scale (inset in linear scale). Clusters at targets with preexisting Ty3 LTRs (sigma; solid squares, lower line and description) and clusters at unoccupied sites (not sigma; dots, upper line and description) were plotted.
Ty3 retrotransposition targets the complete Pol III transcriptome
The 296 Ty3 integration clusters were evaluated for proximity to annotated Pol III–transcribed genes (Table 3). This comparison showed that 262 of 274 identified tDNAs were associated with Ty3 integration clusters. In only one case was the cluster closest to the tDNA not within a few bases of a presumptive site of transcription initiation. In that case, tW(CCA)G1, integration mapped near the presumptive initiation site, but also 45 bp downstream from the tRNA-coding sequence.
Table 3.
De novo Ty3 integration sites identified in HTS
The 13 tDNAs not represented among Ty3 integration clusters were examined individually (summarized in Table 3). We considered four explanations for their absence: (1) Short regions of conserved sequence immediately upstream of the tRNA-coding sequence resulted in alignment of clusters to multiple genomic loci, and exclusion from the uniquely aligned data set. (2) Sequence differences between YMA1322/YMA1356 and the reference S288C strain within the sequenced region led to imperfect alignment and exclusion from further analysis. (3) The tDNA was missing from the YMA1322/YMA1356 strain background. (4) A subset of tDNAs was not targeted because members do not bind factors required by Ty3 or reside in a chromosomal region that excludes access. The data set containing non-unique sequences (“multiples”) (Supplemental Table S3B) was inspected for matches to Pol III–transcribed genes. It contained an insertion junction aligning with two tL(CAA) genes on chromosomes VII and XIII. Thus, either or both of these absent tDNAs were targets of Ty3 integration. The multiples set also included junctions inside four identical tR(UCU) genes. There are four tandem tR-tD genes in the S. cerevisiae genome. The tR(UCU) sequence lies immediately upstream of tD(GUC) on chromosomes II and IV, and two sites on X. Thus, both the upstream member of the pair, tR(UCU), and the downstream member, tD(GUC), were targeted by Ty3. In these cases, it was not possible to assign insertion joints to specific loci. However, given the broad use of other tDNAs and the similar target sequence context, we think it likely that the insertions were distributed among the loci that failed to map uniquely.
We next considered that tDNAs not found as Ty3 targets in the unique or multiple sequence reads [tM(CAU)J3, tP(UGG)O1, tY(GUA)J2, tD(GUC)N, tD(GUC)L2, tK(CUU)C, and tM(CAU)C] (Supplemental Table S3B) occurred in sequences of the host strain that diverged from the sequence of the S288C reference strain. To directly query the representation of these sequences in the host genome used in our experiments, the respective regions from the YMA1322 genome were assayed by PCR using primers flanking the reference sequence. In the first four cases, amplification was successful, and the amplicons were sequenced. This analysis revealed mismatches between the sequence from YMA1322 and the reference S. cerevisiae genome S288C for tM(CAU)J3, tP(UGG)O1, and tY(GUA)J2. Our reference genome sequence was modified to contain these variant sequences and queries of the modified reference sequence identified clusters at each of these loci. The pseudogene tD(GUC)N was also amplified from YMA1322, and this amplicon was identical in sequence to the S288C reference sequence. For the remaining three loci [tK(CUU)C, tM(CAU)C, and tD(GUC)L2], a fragment of the predicted size could not be amplified from the YMA1322 genomic DNA template. Inspection of the reference genome showed that tK(CUU)C and tM(CAU)C were flanked by direct repeats of Ty1 LTRs so that recombination could have deleted both tDNAs, thus accounting for failure to recover those loci by sequencing or by PCR. Sequence analysis of BY4741 indicated that these genes, consistent with previous observations (Harismendy et al. 2003), were also not present in that S288C-related strain (T. Najdi, pers. comm., data not shown). The tD(GUC)L2 locus could not be amplified. However, the S288C reference genome has a single sigma LTR upstream of this gene. Thus, this gene appears to be absent in YMA1322 but shows evidence of being a Ty3 integration target in S288C.
Pol III–transcribed loci that are not tDNAs were also targeted by Ty3 integration (Fig. 4; Table 3). RDN5 is present in the highest copy number. The RDN5-1 copies are repeated within the ∼100–200-copy RDN37 repeat, and RDN5-2 is located in the last repeat and could not be distinguished (Johnston et al. 1997). Four additional orphan copies designated RDN5-3 to -6 flank the rDNA repeat. These orphan genes are expressed (Piper et al. 1984). However, the sequence context diverges 2 bp upstream of the native TSS, the terminal 4 bp differ, and the poly(T) termination tract is shortened. Ty3 insertion clusters were identified in the multiples data set representing RDN5-1 and 2, but not the orphan variant RDN5-3 to 6 genes (Supplemental Table S3B).
Figure 4.
Ty3 integration targets known Pol III–transcribed genes. Ty3 integration sites (filled triangles); Pol III promoter elements, TBP binding site (TATA), box A (unfilled), box B (dark gray), and box C (light gray). TSS are shown only for genes that are transcribed as precursors (bent arrow). TSS is not available for iYGR033C.
All six non-tDNA, Type 2 genes were associated with Ty3 insertions. Similar to tDNAs, SCR1 (Dieci et al. 2002; Guffanti et al. 2006) contains promoter box A and box B elements within the mature RNA-coding sequence. RPR1 and SNR52 (Ishiguro and Kassavetis 2003; Guffanti et al. 2006) contain box A and box B sequences downstream from the TSS and within the transcribed region, but upstream of the mature coding sequence. SNR6 has an upstream TATA element and an internal box A, but terminates upstream of box B (Eschenlauer et al. 1993). Ty3 clusters overlapped the TSS of these diverse Pol III–transcribed genes (Table 3; Supplemental Table S3A).
In addition to Pol III–transcribed genes of known function, genome-wide ChIP experiments using epitope-tagged subunits of TFIIIC, TFIIIB, and Pol III identified sites in the genome that bind amounts of TFIIIC comparable to that bound by tDNAs, but low or undetectable amounts of TFIIIB or Pol III subunits (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). These include ZOD1 and RNA170, which are weakly transcribed, and iYGR033C and ETC1-4 and ETC6-8, for which transcription has not been detected (Olivas et al. 1997; Guffanti et al. 2006). Ty3 integrated close to the TSS of ZOD1 and iYGR033C. Transcription of RNA170 initiates atypically close to the proposed box A (7–15 nt upstream as opposed to ∼20 nt upstream) and terminates downstream from box B, but upstream of a poly(T) tract (Olivas et al. 1997). Surprisingly, Ty3 integrated within the Pol III–transcribed RNA170 sequence between the predicted box A and box B promoter elements. ETC sites (ETC1-4, 6-8) (Olivas et al. 1997; Moqtaderi and Struhl 2004; Guffanti et al. 2006) did not act as Ty3 targets. Thus, ETC sites are neither occupied by significant amounts of TFIIIB and Pol III nor targeted by Ty3.
Our analysis used a HIS3-tagged Ty3 and involved growth of transformants that had undergone transposition on selective medium. While this introduced bias against integrations that affect viability or growth rate, integrations upstream of tS(CGA)C and SNR6, the two essential Pol III–transcribed targets of which we are aware, were recovered. The additional possibility of failure to recover integrations into essential non–Pol III targets cannot be formally excluded.
Correlation of genomic features with Ty3 target activity
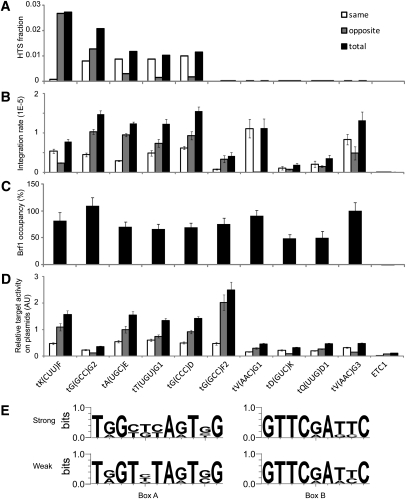
To estimate the range in targeting activity among tDNAs, targets were ranked according to cluster density, and a subset was reevaluated using a quantitative (q)PCR assay for transposition. Five targets from the top [tK(CUU)F, tG(GCC)G2, tA(UGC)E, tT(UGU)G1, and tG(CCC)D] and bottom [tG(GCC)F2, tV(AAC)G1, tD(GUC)K, tQ(UUG)D1, tV(AAC)G3)] deciles (Fig. 5A; Supplemental Table S3A) were tested. The ETC1 locus was used as a negative control. Naive and Ty3 insertion–bearing versions of the loci were cloned and used as standards. Cells were transformed with pKN3097 bearing an unmarked galactose-inducible Ty3-ppt and induced in triplicate for Ty3 expression by growth in SGal-Ura for 24 h. DNA was extracted and subjected to qPCR using primers specific to sequences flanking the 10 genomic target sites and to either the U3 or the U5 end of the Ty3 element. The assay was standardized using a dilution series of the respective cloned Ty3-associated targets, and outcomes were expressed as integrants normalized to an ACT1 control (Fig. 5B). Comparison of the overall pattern of target usage showed that seven targets were recovered with insertions represented at 8 × 10−6 to 16 × 10−6. The remaining three were represented at 2 × 10−6 to 4 × 10−6. The seven targets with activity in the upper range included all five of the targets in the top decile and two [tV(AAC)G1 and tV(AAC)G3] from the bottom decile of Tpn-seq reads. ETC1 was not used. Despite a relative consistency in that top targets in Tpn-seq were more highly used in the qPCR assay, the difference between the different classes of targets was much reduced using the quantitative assay. This result indicated that the sequencing reads provided a sensitive record of positions of integration rather than a quantitative measure of target utilization.
Figure 5.
Quantitative Ty3 transposition at tDNAs with high and low DNA sequencing reads. (A) Distribution of sequencing reads from five top decile tDNA clusters and five bottom decile tDNA clusters. Yeast strains YMA1322 and YMA1356 transformed with Ty3-HIS3 expression plasmid pKN3050 were induced for Ty3 expression for 24 h. Insertions into tDNA target clusters were ranked by number of reads, and example top and bottom decile genes (shown) were selected. ETC1 was not detected by sequencing and was used as a negative test case. Bars show reads in which Ty3 and target are transcribed in the same (open) and divergent (gray) orientations and the total (black) is shown. (B) YMA1322 transformed with Ty3-ppt expression plasmid pKN3097 was induced for Ty3-ppt expression as described in A, and Ty3 integrations at reporter target loci were measured using qPCR (Methods). Measurements were within the linear range as standardized using plasmid versions of the Ty3 insertions at each locus. Bars represent technical triplicates of biological duplicates. (C) TFIIIB occupancy of reporter Ty3 target loci. Yeast strain YKN1692 expressing TFIIIB subunit Brf1 tagged with 3HA and transformed with empty URA3 vector plasmid YCplac33 was grown under inducing conditions for 24 h, and Brf1 occupancy at reporter loci was measured using ChIP. ChIP values were determined relative to an ACT1 control. The fold of enrichment for tDNA reporter tV(AAC)G3 to ACT1 was 73:1. The data are expressed relative to the fold enrichment at tV(AAC)G3 set at 100%. (D) Ty3 integration frequencies at tDNA reporter loci and flanking sequence. YMA1322 transformed with Ty3-ppt plasmid pKN3097 and vector pXP622 containing reporter target loci and flanking sequence (120 bp upstream and downstream of target gene) was induced to express Ty3 for 36 h, and integrations were quantified by qPCR. Data are expressed as described in B. (E) Based on target activity (D), promoter elements in strong target loci [tK(CUU)F, tA(UGC)E, tT(UGU)G1, tG(CCC)D, and tG(GCC)F2] and weak target loci [tG(GCC)G2, tV(AAC)G1, tD(GUC)K, tQ(UUG)D1, and tV(AAC)G3] were analyzed for consensus using logo sequence analysis software (http://weblogo.berkeley.edu).
Based on in vitro assays, TFIIIB occupancy is the major determinant of Ty3 targeting. ChIP was therefore performed in order to assess the correspondence between TFIIIB occupancy and reporter targeting activity (Fig. 5C). YMA1322 was modified as previously described (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004) to express a 3HA-tagged Brf1 (Brf1-3HA; YKN1692). This strain was induced for Ty3 retrotransposition, and cells were processed for qPCR-ChIP analysis. Eight genes, including the six genes with highest Ty3 targeting by qPCR, showed relatively high Brf1 occupancy; as reported previously (Moqtaderi and Struhl 2004), ETC1 did not show significant TFIIIB association.
To test the contribution of sequences in the immediate vicinity of the reporter tDNAs to targeting, the 10 reporter genes and flanking upstream and downstream 120 bp were subcloned and retested for Ty3 transposition (Fig. 5D). QPCR analysis was performed using primers that annealed within the Ty3 LTR U3 or U5 sequence and within the vector sequence downstream from the target. The signal for plasmid-borne tDNA was adjusted for plasmid copy number and normalized to the signal of the chromosomal tF(GAA) family. This family gives a robust qPCR integration signal after induction of Ty3-ppt (data not shown). The comparison showed that five of the genes, including three used at higher frequency as chromosomal targets, were used at levels threefold to sixfold higher than the other five targets. Only tD(GUC)K and tQ(UUG)D1 were consistently low in Ty3 targeting in chromosomal and plasmid contexts and in Brf1 occupancy. Targeting to the chromosome and plasmid did not correlate (R2 = 0.29), suggesting that local sequence played a greater role in plasmid-based targeting. Box B and box A motifs were generated from high- and low-frequency targets (Crooks et al. 2004; http://weblogo.berkeley.edu). The resulting box B motif conformed to the consensus (Fig. 5E), whereas more active and less active targets showed distinct versions of the consensus box A sequence (TRGYYYAGTNG vs. TRGTNTAGTNG, respectively). If this sample is representative, these findings indicate that chromosomal context and local sequence contribute to tDNA targeting, but targets bind similar amounts of TFIIIB and overall differ by less than eightfold in activity.
Insertions at targets not known to be Pol III–transcribed genes
Previous efforts to identify Pol III–transcribed genes have used computational approaches to identify tDNA structures (Lowe and Eddy 1997), scans for box A and box B sequences and transcripts (Olivas et al. 1997), and ChIP-chip of inter-ORF regions (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). As described above, excluding one tRNA pseudogene, 266 of the 296 clusters identified in our study corresponded to 262 uniquely aligning tDNAs [tK(UUU)O, tN(GUU)P, tS(AGA)J, tY(GUA)M1 have two upstream clusters each]. Seven single Pol III–transcribed non-tDNA loci aligned uniquely with genome sequence. Eighteen other sequence clusters, including one at an unannotated Ty3 LTR at RHB1, were excluded as PCR artifacts generated from preexisting solo Ty3 LTRs. Five clusters identified unique sites in the genome but did not correspond to known Pol III–transcribed genes. Additional PCR using subcloned YJL206C failed to verify it as a reproducible target (Supplemental Table S4). Two were true targets (SCS3 and YIL100W), and two were not cloned [tW(CCA)G1 downstream insertion and YCG1] (Supplemental Table S4).
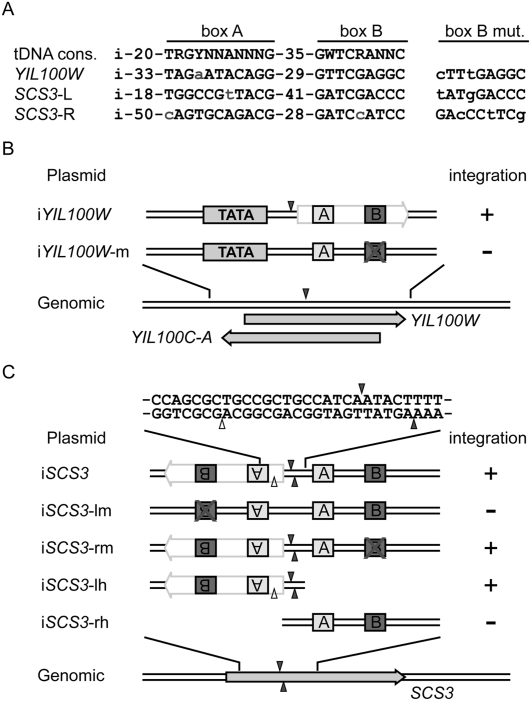
Novel Ty3 clusters neither associated with Pol III transcription nor containing known Ty3 LTRs were verified inside ORFs YIL100 (W and C-A) and SCS3. YIL100W and YIL100C-A designate a bidirectional ∼100-codon ORF classified as dubious (http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=YIL100C; henceforth, YIL100W) (Fig. 6A,B). SCS3 encodes a gene required for inositol prototrophy (Hosaka et al. 1994). Regions of ∼500 bp flanking insertion sites inside YIL100W and SCS3 (iYIL100W and iSCS3, respectively) were cloned. YMA1322 transformants containing Ty3-ppt donor plasmid pKN3097 and each of the target plasmids were assayed for Ty3 transposition. De novo Ty3 insertions were detected using combinations of Ty3- and plasmid-specific primers. Insertions were detected for iYIL100W and iSCS3 in induced, but not uninduced cells. PCR products were cloned and sequenced. Insertions occurred at both cloned targets (Fig. 6B,C). We examined YIL100W and SCS3 sequences for box A and box B promoter elements. In the case of Ty3 insertions upstream of tDNAs, the Ty3 junction is ∼20 bp upstream of box A. Scanning of genomic sequence flanking the iYIL100W cluster revealed a box A–box B pair separated by 29 bp (Fig. 6A). The Ty3 cluster mapped 33 nt upstream of the 5′ end of the box A sequence (Fig. 6A). To test whether, similar to other Ty3 insertion sites, targeting was dependent on active Pol III promoter elements, conserved bases in the box B consensus were mutated to disrupt Pol III promoter activity (Fig. 6A; Marck et al. 2006). YMA1322 transformants containing the Ty3 donor plasmid and wild-type or mutated iYIL100W plasmid were induced for transposition, and samples were assayed by PCR using Ty3- and target plasmid–specific primers. PCR products were generated from insertions upstream of iYIL100W in the wild type but not in the box B mutant clone (Fig. 6B).
Figure 6.
Ty3 targeting of novel loci requires Pol III promoter element. (A) Box A–box B pairs at iSCS3 and iYIL100W target loci. (Top row) Approximate 20-nt spacing between integration sites (i) and box A and the consensus box A and box B sequence of fungal tDNAs, with an average spacing of 29 bp (Marck et al. 2006). Lowercase indicates divergence from consensus. (Right column, box B mut.) Box B mutations described in B and C. (B) Target activity of novel Ty3 target iYIL100W. YMA1322 transformed with pKN3050 and plasmids containing 500-bp iYIL100W-flanking sequence and a box B mutant derivative was induced to express Ty3 for 36 h. Integration was monitored by PCR (+/−). (Triangles) Insertion sites; (double line) chromosomal DNA; (TATA) upstream TATA-box consensus; (m) mutated box B (mutation sequence listed in A). (C) Target activity of iSCS3. As described in B, 500-bp iSCS3-flanking sequence and variants were tested for targeting. (Filled triangles) Insertion sites determined from sequence analysis; (empty triangles) those recovered from target plasmids; (lm and rm) mutated box B in loci shown; (lh and rh) half truncation as shown.
The Ty3 insertions into SCS3 detected by Tpn-seq were clustered centrally within the ORF. Inspection of the sequence surrounding these insertions showed that they were flanked by inverted box A–box B motifs (Fig. 6A,C). Ty3 insertion into plasmid-borne SCS3 was investigated using an approach similar to that described for YIL100W. De novo Ty3 insertions clustered close to the position identified in Tpn-seq. The two box B motifs in SCS3 were individually mutated (Fig. 6A,C), and the two resulting plasmid-borne targets were retested. Mutations in the rightward box B had no effect. In contrast, mutations in the leftward box B eliminated the majority of Ty3 integration-dependent PCR products. To verify localization of the targeting activity, the left and right halves of SCS3 were subcloned and retested for targeting. As predicted, targeting activity was retained in the clone of the left half of SCS3 but not the right half. This result was consistent with the existence of a Pol III promoter targeting Ty3 integration into the SCS3 ORF (Fig. 6C). These results suggest that there are a limited number of sites in the yeast genome that are not annotated as Pol III transcription units but that may interact with Pol III initiation complexes or Pol III itself.
Discussion
Ty3 accesses the Pol III transcriptome
Before this study, chromosomal Ty3 elements and LTRs had been observed at ∼15% of tDNAs (Chalker and Sandmeyer 1990; Goffeau et al. 1996). This study showed that Ty3 integrates close to the known or predicted TSS known Pol III–transcribed chromosomal loci. The targets include 274 tDNAs, as well as SNR52, SNR6, RPR1, RDN5-1/2, and SCR1. In addition, Ty3 integrated upstream of two loci, ZOD1 and RNA170, predominantly occupied by TFIIIC (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004), at which transcription is enhanced under conditions of nucleosome depletion (Olivas et al. 1997; Guffanti et al. 2006), and one locus, iYGR033C, for which transcription has not been observed (Guffanti et al. 2006).
Because of its distinctive targeting, Ty3 potentially offers an independent marker for sites of Pol III initiation complex assembly. Several arguments support the inference that Ty3 integration and TSS are tightly linked. First, where known for tDNAs, as well as SNR6 and RDN5 (Chalker and Sandmeyer 1992), the gene-proximal strand transfer of Ty3 cDNA coincides with the Pol III TSS and the gene-distal strand transfer is 5 nt upstream. In our study, only 21% of cluster widths exceeded 11 bp, and within these clusters, integrations showed peaks offset by 5 nt on the two strands (e.g., Fig. 2; Supplemental Table S3; GEO http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31820). Second, in vivo mutations in the transcription template that affect TSS selection similarly affect Ty3 integration site selection (Chalker and Sandmeyer 1992, 1993; Kirchner et al. 1995). Third, Brf1 and TBP, which support transcription initiation in vitro on templates with relaxed requirements for promoter opening (Kassavetis et al. 1999), also constitute the minimum Ty3 integration target (Yieh et al. 2000).
Ty3 integrations occurred at a small number of sites not anticipated to be Pol III TSS. These included sites at Pol III–transcribed RNA170, tR(UCU)-tD(GUC), and tW(CCA)G1. It is possible that these insertions represent experimental artifacts, transient atypical interference by nucleosomes, or initiation factor occupancy not predicted by transcript mapping. In the case of RNA170 and tR(UCU)-tD(GUC), loci contain multiple potential matches to box A and box B consensus sequences. The 5′ end of the RNA170 transcript was mapped by primer extension to positions 7 and 15 nt upstream of box A; the minor 3′ end mapped at the poly(T) tract, as is typical, but the major 3′ end mapped upstream of both box B and the poly(T) tract (Olivas et al. 1997). Ty3 insertions mapped between the putative box A and a box B motif implicated by mutagenesis experiments (Olivas et al. 1997). We consider two possible explanations of the seemingly exceptional Ty3 integration site in RNA170. First, alternative positions of nucleosomes in the region of RNA170 affect positioning of TFIIIC and consequently TFIIIB. Consistent with an effect of nucleosome positioning, RNA170 transcripts are observed at low levels but are distinct from some other Pol III promoters in that they change significantly under conditions of nucleosome depletion (Guffanti et al. 2006). Second, alternative box A and box B promoter elements are associated with RNA170. Inspection of the sequence in the vicinity of the Ty3 insertion site shows a candidate box A–box B pair in the opposite orientation to those promoting RNA170. Thirty-nine nucleotides downstream from the Ty3 insertion site is a perfect box A (TGGTATATAAG), 28 nt after which is an imperfect box B (GTTCAgATC; the nucleotide for g is A in consensus). Thus, the Ty3 pre-integration complex could associate with TFIIIB docked via conventional interactions with TFIIIC bound to alternative promoter elements.
A second example of Ty3 integration at a site not associated with initiation is provided by the four tR(UCU)–tD(GUC) genes. A single dimeric pre-tRNA from this locus is observed in vitro (Kjellin-Straby et al. 1984; Engelke et al. 1985). In addition, the tR and tD members of a pair are closely spaced so that simultaneous occupancy of both genes by TFIIIC and TFIIIB would be predicted to be disrupted by steric interference. Surprisingly, integrations occurred associated with both tR(UCU) and tD(GUC) box A motifs. We conclude that it is likely that the downstream tD(GUC) actually binds the initiation factor TFIIIB and is therefore competent to initiate transcription at some frequency.
Ty3 integrations reflect the general activity of Pol III–transcribed genes in yeast
Three ChIP-chip studies in S. cerevisiae (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004) showed that all but approximately a dozen annotated tDNAs bind TFIIIC, TFIIIB, and Pol III (Lowe and Eddy 1997; Olivas et al. 1997; Percudani et al. 1997; Hani and Feldmann 1998). In the Moqtaderi and Struhl (2004) study, quantitative comparison of complexes at nine test tDNA genes using ChIP-qPCR showed robust binding averaging 60% of maximum for the reporter set. Even in the most extreme cases, factors bound with only fivefold to sevenfold differences in efficiency within the test gene set. Similarly, we observed that all Pol III–transcribed candidate genes were used by Ty3, and qPCR sampling of target usage in our study showed an eightfold range in targeting and twofold range in Brf1 occupancy.
Despite significant conservation of the Pol III transcription apparatus between single-celled and metazoan genomes (Huang and Maraia 2001), there is evidence for intriguing differences as well. As noted above, most Pol III genes in yeast are transcriptionally active. This contrasts with the situation in cultured human cells where recent studies showed that only about half of annotated tRNA genes bind TFIIIB, TFIIIC, and Pol III (Canella et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010). Furthermore, in yeast, Pol III–transcribed genes (Hull et al. 1994) are repressive to local Pol II promoters (Kinsey and Sandmeyer 1991; D'Ambrosio et al. 2008; Haeusler et al. 2008), and condensin association, which also occurs at ETCs, is implicated in this repression (D'Ambrosio et al. 2008; Haeusler et al. 2008). In human cells, active Pol III genes and active Pol II promoters are associated to a significant extent (Moqtaderi et al. 2010; Oler et al. 2010), as are ETC and sequences with properties of enhancers (Moqtaderi et al. 2010; Oler et al. 2010).
Discrimination of ETC loci
Although TFIIIC loading of TFIIIB has been studied extensively in vitro, the process is less well understood in vivo. The observation of Ty3 integration at annotated Pol III–transcribed loci for which transcription factor binding and transcription have barely been detected suggests that it represents a very sensitive probe for bona fide Pol III–transcribed genes. The S. cerevisiae genome has approximately 3700 box A and box B pairs within 100 bp of one another (i.e., similar to SNR6 box A and box B spacing) with a normalized log odds score (NLOD) >0.83 (above the similarity of many tDNAs to consensus) (Xie et al. 2009; data not shown). It was therefore striking that Ty3 integrated virtually exclusively at known Pol III transcription units. What discriminates transcribed loci from ETC and ETC from box A and box B pairs? One possibility is that requirements are compensatory so that the threshold is determined by the synergy of several features. Filtering the 3700 box A/B pairs for those with spacing of 35 bp or less, which is closer to the average distance separating tDNA box A and box B motifs, reduces the number to approximately 1700. With further filtering for a minimum NLOD score of 0.9, the number of pairs drops to about 360, more consistent with the number of Pol III–transcribed genes. One interesting possibility is that a significant subset of these sequences actually are associated with TFIIIC. In examination of just over 400 sites of condensin binding in the yeast genome, D'Ambrosio et al. (2008) found that most contained box B motifs and were associated with at least a low level of TFIIIC (less than found at tDNAs, for example). Interestingly, they also showed that box B alone was sufficient to mediate a low level of TFIIIC association. Moqtaderi et al. (2010) observed an extended box B motif at yeast ETC loci but did not report box A sequences. Inspection of the eight yeast ETC loci shows that few have optimal box A and box B motifs within 35 bp. These observations raise the possibility that box A, which is proximal to the initiation site, also plays a role in the activity of TFIIIC as a TFIIIB loader. Consistent with that, the tDNA pseudogene tD(GUC)N with a single base change in the box A consensus binds only TFIIIC (Harismendy et al. 2003) and is not a Ty3 target. Although we examined only a small number of Ty3 targets by qPCR, weak targets shared a distinct box A motif, potentially supporting a role for the strength and position of box A elements in determining TFIIIB loading by TFIIIC.
Functional significance of association of retrotransposons and Pol III–transcribed genes
Ty3 targeted all genes known or predicted to be transcribed by Pol III. Ty3 clusters were also identified upstream of box A–box B pairs in iYIL100W and iSCS3. Although the transcripts predicted for Pol III transcription from these loci were not detectable by Northern blot analysis (data not shown), based on the overall specificity of Ty3 for Pol III–transcribed genes, we speculate that these loci are occupied at low levels by Pol III initiation complexes. Although iYIL100W is a dubious ORF, iSCS3 is transcribed into poly(A) RNA (Nagalakshmi et al. 2008). These findings raise the possibility that Pol III transcription factor binding sites in Pol II–transcribed regions provide a mechanism for regulatory cross talk. An explicit example of this occurs at ETC6, which was recently implicated in auto-regulation of transcription of the gene encoding a TFIIIC subunit, Tfc6 (Kleinschmidt et al. 2011).
In addition to highlighting potentially novel transcription initiation sites, the comprehensive association we observed between the Ty3 retrotransposon and Pol III–transcribed genes extends the intriguing relationship between retroelements and Pol III–transcribed genes. It was previously proposed that Pol III–transcribed genes could be dispersed by retrotransposition (Weiner et al. 1986). Since that time, LINEs (long interspersed elements) have been demonstrated to mobilize SINEs (short interspersed elements) (Dewannieux et al. 2003). Retrovirus particles, which are analogous to Ty3 virus-like particles, can, depending on the virus, be enriched for 7SL, U6, and 5S Pol III transcripts in addition to primer tRNAs (for review, see Onafuwa-Nuga et al. 2005). The pattern of transcripts hybridizing to Ty3 LTR probes indicates that readthrough from Ty3 into adjacent genomic sequence occurs (Clark et al. 1988). Thus, readthrough transcription from Pol III–specific retrotransposons into downstream genes is a plausible additional mechanism for inclusion of Pol III–transcribed sequences in virus-like particles. If these are reverse-transcribed, they would provide a pool of cDNAs that could promote dissemination or homogenization of Pol III–transcribed gene families.
Summary
In this study, we show that Ty3 provides a highly sensitive and irreversible probe for Pol III transcription initiation complexes. The absence of Ty3 integration into ETC sites underscores the question of what properties distinguish DNA-bound and condensin-associated TFIIIC that loads TFIIIB and allows Ty3 targeting from that which does not. The specificity of Ty3 association with Pol III–transcribed genes make Ty3 a useful tool for future exploration of the association of retrotransposons and Pol III–transcribed genes.
Methods
Bacterial and yeast strains and culture conditions
Yeast strains are described in Table 1A. Strain S288C derivative YPH500 (Johnston and Davis 1984; Sikorski and Hieter 1989) was modified, generating YMA1322 and its rad52Δ derivative YMA1356 (Aye et al. 2001).
Plasmids (Table 1B) were prepared from DH5α [F− ϕ80lacZΔM15 Δ (lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ−] (Invitrogen) transformants.
For ChIP, YMA1322 was modified by epitope tagging Brf1 with three copies of the hemagglutinin epitope (3HA). The 3HA coding sequence was introduced in-frame at the downstream end of BRF1 in YMA1322 (Longtine et al. 1998) to create yeast strain YKN1692. Brf1 was previously tagged using a similar strategy and was shown not to affect growth of the tagged strain (Harismendy et al. 2003; Roberts et al. 2003; Moqtaderi and Struhl 2004). Correct integrants were confirmed by PCR using Brf1-specific primers KN2485 and KN2411 and verified by sequencing with KN2485. Western blot analysis was performed using α-3HA to verify expression of the tagged protein (data not shown).
Yeast and bacterial culture methods were as previously described (Amberg et al. 2005; Ausubel et al. 2007). Synthetic complete medium lacking His, Leu, or Ura (0.67% yeast nitrogen base, 0.2% amino acid drop-out mix) was supplemented to 2% glucose (SD), 2% galactose (SGal), or 1% raffinose, 2% (v/v) glycerol, 2% (v/v) lactic acid (SR) (Burke et al. 2000). Rich medium was 1% yeast extract 2% peptone, 2% dextrose (YPD).
Plasmid constructions
Standard cloning procedures were used throughout (Ausubel et al. 2007). Ty3-ppt-HIS3 was expressed under control of the GAL1-10 UAS from a URA3-marked low-copy plasmid (pKN3050). Ty3 derived from pNB2361 and containing an antisense insertion of HIS3 (Beliakova-Bethell et al. 2009) at the downstream end of POL3 was recloned into YCplac33 (ATCC), and unique 60-bp sequence tags were introduced flanking HIS3. Because Ty3 POL3 extends into the downstream LTR, insertion of the HIS3 gene resulted in an unfavorably long downstream LTR. The LTR sequence was reconstituted downstream from HIS3. This Ty3 expression plasmid was designated pKN3050. Plasmid pKN3050 was cleaved with AflII and SbfI and religated, thereby removing one tag and the HIS3 gene and creating pKN3097 (Table 1A). Positive clones were verified by DNA sequencing. This and other sequence analysis were performed at Genewiz Inc. Details of this construction are provided in the Supplemental Methods.
Testing integrations at specific loci
To test specific loci for target activity, 300–600-bp DNA fragments containing the loci were amplified in PCR reactions. To construct the plasmids used as qPCR standards for measurement of integration at specific chromosomal loci, tDNAs with a segment of flanking Ty3 insertion were amplified and cloned into the pCR2.1 vector (Invitrogen). Details of the constructions are provided in the Supplemental Methods. Oligonucleotide primers are listed in Supplemental Table S2. Product plasmids are listed in Supplemental Table S1.
Ty3 transposition
For HTS analysis of Ty3-HIS3 insertion sites, YMA1322 and YMA1356 transformed with the Ty3-HIS3 expression plasmid pKN3050 were grown to log phase in SR−Ura, −His at 24°C (RT). From each culture, 2 × 106 transformants were transferred to each of 10 100-mm plates of SGal−Ura, −His medium or on SD−Ura, −His medium as negative control and maintained for 3 d to allow Ty3 expression. Subsequent steps were performed for both tested conditions. Cultures were replica-plated onto YPD and incubated for 24 h at 30°C. Colonies were replica-plated onto SD−His medium supplemented with 0.1% 5-fluoro-orotic acid (5FOA) (Boeke et al. 1984) to select for cells that had lost the URA3-marked donor plasmid but contained Ty3-HIS3 chromosomal integrants. On average, there were ∼103 integrants per plate for Ty3-induced samples and 20–40 colonies for negative-control samples. Approximately 10,000 colonies each for YMA1322 and YMA1356 cultures were scraped from plates. Harvested cells were inoculated into 10 mL of SD−His and incubated for 24 h at 30°C on a rotating wheel. Genomic DNA was extracted according to standard procedures (Ausubel et al. 2007).
For quantitative comparison of transposition into different chromosomal target loci, yMA1322 transformed with Ty3-ppt donor plasmid pKN3097 was grown to log phase in SR−Ura medium, diluted into 25 mL of SGal−Ura medium to a final concentration of 0.2 OD600, and grown with shaking at 180 rpm, for 24 h at room temperature. As negative control, cultures were diluted into 5 mL of SD−Ura medium to 0.2 OD600 and allowed to grow for 24 h at room temperature. Quantitative comparison of transposition into plasmid-borne targets was performed similarly except that −Ura −Leu medium was used to select for donor and target plasmids. Slower growth necessitated extending the SGal −Ura −Leu incubation from 24 h to 36 h.
Analysis of Ty3 transposition using Illumina GAIIx sequencing (Tpn-seq)
Ty3–host junctions were identified using an inverse (i) PCR strategy (Fig. 1; Ochman et al. 1988; Wang et al. 2008). Samples were processed as follows: (1) gDNA was digested independently with three different 4-bp cutters (Csp6I, HaeIII, and HpyCH4V). (2) Cleaved DNA was circularized in a ligation reaction. (3) DNA containing Ty3 U5 end joint with genomic DNA was amplified by iPCR using primers within the Ty3 U5 and tag1. (4) Samples were submitted to the Genetics Department Sequence Facility, Washington University (St. Louis, MO) for analysis using the Illumina GAIIx. Details are provided in the Supplemental Methods.
Chromatin immunoprecipitation
For quantitative comparison of Brf1 occupancy of target loci, YKN1692 transformed with plasmid YCplac33 was grown under conditions described for Ty3 induction. Samples were processed for ChIP analysis essentially as previously described (Strahl-Bolsinger et al. 1997). Details of the processing are provided in the Supplemental Methods.
Quantitative PCR
Nucleic acid was extracted according to standard procedures (Ausubel et al. 2007). Samples were treated with RNase A (0.2 μg/μL), and DNA was purified using Microspin-30 columns (Bio-Rad). DNA concentrations were measured on a NanoDrop-1000 (Thermal Scientific Inc., NanoDrop products).
QPCR was conducted in an iCycler (Bio-Rad). The total volume of 25-μL reactions contained 1× iQ SYBR Green Super Mix (Bio-Rad), 0.4 μM each primer, and various amounts of DNA template. Serial dilutions of standard plasmids of known concentration were used to construct a standard curve. All reactions were performed in triplicate. Thermal cycling included 40 cycles of extension (30 sec at 95°C, 30 sec at 55°C, 30 sec at 72°C). Fluorescence was captured after each extension step. PCR products were analyzed by melt curves and electrophoresis.
QPCR of DNA samples containing integrations at chromosomal loci were standardized using plasmid templates constructed as described above to contain the U5 or U3 end of the Ty3 LTR juxtaposed to each specific target. QPCR primers are described in Supplemental Table S2. Chromosomal ACT1 copy number determined by qPCR using forward primer XQ3563 and reverse primer XQ3564 was used to normalize the integrations determined in different samples.
For plasmid target experiments, standard curves were used to determine the copy numbers of: (A) Ty3 integrations at the target plasmids using primers XQ2603/XQ3313 for U5 orientation, and using 3135/3582 for U3 orientation; (B) total plasmids using primer pair JY3435/3436; (C) Ty3 integrations at chromosomal tF(GAA) family members using primer pair XQ2603/3621; and (D) total chromosomal DNA, indicated by ACT1 copy number, using primer pair XQ3563/XQ3564. Relative integration was calculated as the ratio on plasmids (A/B) normalized with the integration ratio on chromosome (C/D): (A/B)/(C/D). Duplicate samples were used, and the mean of relative integrations was plotted.
For quantification of Brf1 binding, 2 μL of ChIP DNA sample containing 1–5 ng of DNA was used in a 25-μL qPCR reaction. QPCR was conducted in the linear range for each set of primers determined using a serial dilution of template DNA. Primers are listed in Supplemental Table S2. Occupancy values (in arbitrary units) were calculated as described previously (Moqtaderi and Struhl 2004). The apparent immunoprecipitation (IP) efficiency (i.e., the amount of PCR product in the IP sample divided by the amount of PCR product in the input sample) was divided by the apparent cross-linking efficiency of the ACT1 control DNA segment in each sample. The resulting relative IP efficiency of ACT1, 1.0, was then subtracted from all relative values to yield a background of zero.
Computational analysis
HTS image files were converted to fastq files using Illumina pipeline software. Each sequence began with the last 17 nt of the Ty3 LTR, and HTS reads that did not initiate with this sequence were discarded. For remaining sequences, the LTR barcode was truncated, and the genomic sequence was extracted. Sequence reads with <19 nt of genomic sequence were also excluded from further analysis. Retained reads were mapped to the yeast genome sequence downloaded from the UCSC Genome Browser (build sacCer2 June 2008) (Rhead et al. 2010) using Bowtie (v 0.9.8.1) (Langmead et al. 2009). Only reads with no mismatches were kept for further analysis. Reads that matched multiple genomic loci were tagged for separate analysis.
Mapped sequence reads were converted to insertion sites defined by chromosome number, strand (Watson or Crick), and coordinate. The read density of an insertion site was defined as the number of reads that mapped to that site. Insertion sites with densities less than 5 in either data set were excluded from further analysis. Neighboring insertion sites were linked into clusters by aggregating any two sites that were no more than 10 nt apart, regardless of strandedness. Thus, a cluster is defined by a genomic interval, delimited by two addresses. This procedure yielded a total of 296 clusters using only reads mapping to unique genomic locations. The read density of a cluster is the sum of the read densities of the insertion sites in the cluster. The event density of a cluster is the number of unique insertion sites associated with the cluster. Clusters are described in Supplemental Table S3.
Each of the cluster intervals was used to find nearby genomic features based on publicly available genome-wide annotations. The locations of Pol III–transcribed genes, ORFs, ARSs, and LTRs were downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org/, Dec 2009). Box A and box B sequences associated with Pol III–transcribed genes were from Marck et al. (2006).
Sequence processing and annotation were conducted using a variety of tools. In particular, the MotifMap pipeline (Xie et al. 2009) and the open source Python package Pygr (Alekseyenko and Lee 2007) were used. All data processing and manipulation were done with Python (van Rossum and de Boer 1991), and statistical analyses were computed with R (R Core Development Team 2009).
Feature density for Pol III–transcribed genes, LTRs, and ARS sequences represents the average number of occurrences of a given feature within a sliding window from 7 kb upstream to 7 kb downstream of the cluster center point.
Data access
The original fastq files are available at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE31820.
Acknowledgments
This research was supported in whole or in part by funds from the National Science Foundation Grant 0450159 and National Institutes of Health (NIH), Public Service Grant GM33281 to S.S., and by funds provided by the James S. McDonnell Foundation to M.J. The work of K.D., S.F., P.R., and P.B. was supported by grants NIH LM-07443, NIH LM-010235, NSF EIA- 0321390, NSF IIS-0513376 to P.B., and by the UCI Institute for Genomics and Bioinformatics. The work of R.M. and D.M. was supported by grants 1R01DA025744-01 and 3R01DA025744-02S1. We thank P. Kaiser (UCI) for providing plasmid reagents. We thank the Genome Technology Access Center in the Department of Genetics for sequence analysis. We thank T. Najdi and the UCI Genomics High Throughput Facility for sharing genomic sequence from yeast strain BY4741 (Open Biosystems Inc.). M.J. thanks his family for their unconditional love and support.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131219.111.
References
- Alekseyenko AV, Lee CJ 2007. Nested Containment List (NCList): A new algorithm for accelerating interval query of genome alignment and interval databases. Bioinformatics 23: 1386–1393 [DOI] [PubMed] [Google Scholar]
- Allison DS, Hall BD 1985. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J 4: 2657–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K 2007. Current protocols in molecular biology. Wiley, New York [Google Scholar]
- Aye M, Dildine SL, Claypool JA, Jourdain S, Sandmeyer SB 2001. A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration. Mol Cell Biol 21: 7839–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N, Eby Y, Boeke JD 2004. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res 14: 1232–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev 19: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Baller JA, Gao J, Voytas D 2011. Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5. Proc Natl Acad Sci 108: 20351–20356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N, Terry LJ, Bilanchone V, DaSilva R, Nagashima K, Wente SR, Sandmeyer S 2009. Ty3 nuclear entry is initiated by viruslike particle docking on GLFG nucleoporins. J Virol 83: 11914–11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2: e157 doi: 10.1371/journal.pcbi.0020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-Fluoro-orotic acid resistance. Mol Gen Genet 197: 345–346 [DOI] [PubMed] [Google Scholar]
- Braun BR, Riggs DL, Kassavetis GA, Geiduschek EP 1989. Multiple states of protein–DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci 86: 2530–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Canella D, Praz V, Reina JH, Cousin P, Hernandez N 2010. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res 20: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Sandmeyer SB 1990. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics 126: 837–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Sandmeyer SB 1992. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev 6: 117–128 [DOI] [PubMed] [Google Scholar]
- Chalker DL, Sandmeyer SB 1993. Sites of RNA polymerase III transcription initiation and Ty3 integration at the U6 gene are positioned by the TATA box. Proc Natl Acad Sci 90: 4927–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Bilanchone VW, Haywood LJ, Dildine SL, Sandmeyer SB 1988. A yeast sigma composite element, Ty3, has properties of a retrotransposon. J Biol Chem 263: 1413–1423 [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Dieci G, Giuliodori S, Catellani M, Percudani R, Ottonello S 2002. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J Biol Chem 277: 6903–6914 [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A 2007. The expanding RNA polymerase III transcriptome. Trends Genet 23: 614–622 [DOI] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J 20: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke DR, Gegenheimer P, Abelson J 1985. Nucleolytic processing of a tRNAArg–tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem 260: 1271–1279 [PubMed] [Google Scholar]
- Eschenlauer JB, Kaiser MW, Gerlach VL, Brow DA 1993. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol 13: 3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Salmon K, Shen MWY, Aeling KA, Ito E, Irwin B, Tran UPC, Hatfield GW, Da Silva NA, Sandmeyer S 2011. A vector set for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast 28: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ, Craig NL 2010. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc Natl Acad Sci 107: 21966–21972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol 187: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev 19: 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori S, Percudani R, Braglia P, Ferrari R, Guffanti E, Ottonello S, Dieci G 2003. A composite upstream sequence motif potentiates tRNA gene transcription in yeast. J Mol Biol 333: 1–20 [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. 1996. Life with 6000 genes. Science 274: 563–567 [DOI] [PubMed] [Google Scholar]
- Guffanti E, Percudani R, Harismendy O, Soutourina J, Werner M, Iacovella MG, Negri R, Dieci G 2006. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J Biol Chem 281: 29155–29164 [DOI] [PubMed] [Google Scholar]
- Guo Y, Levin HL 2010. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res 20: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani J, Feldmann H 1998. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res 26: 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O 2003. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J 22: 4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Nikawa J, Kodaki T, Ishizu H, Yamashita S 1994. Cloning and sequence of the SCS3 gene which is required for inositol prototrophy in Saccharomyces cerevisiae. J Biochem 116: 1317–1321 [DOI] [PubMed] [Google Scholar]
- Huang Y, Maraia RJ 2001. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res 29: 2675–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR 1994. tRNA genes as transcriptional repressor elements. Mol Cell Biol 14: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A, Kassavetis GA 2003. A gene-specific effect of an internal deletion in the Bdp1 subunit of the RNA polymerase III transcription initiation factor TFIIIB. FEBS Lett 548: 33–36 [DOI] [PubMed] [Google Scholar]
- Johnston M, Davis RW 1984. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol 4: 1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Hillier L, Riles L, Albermann K, Andre B, Ansorge W, Benes V, Bruckner M, Delius H, Dubois E, et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature (Suppl) 387: 87–90 [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Geiduschek EP 2006. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem Soc Trans 34: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60: 235–245 [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Letts GA, Geiduschek EP 1999. A minimal RNA polymerase III transcription system. EMBO J 18: 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey PT, Sandmeyer SB 1991. Adjacent Pol II and Pol III promoters: Transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res 19: 1317–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey PT, Sandmeyer SB 1995. Ty3 transposes in mating populations of yeast: A novel transposition assay for Ty3. Genetics 139: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Connolly CM, Sandmeyer SB 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Kjellin-Straby K, Engelke DR, Abelson J 1984. Homologous in vitro transcription of linear DNA fragments containing the tRNAArg–tRNAAsp gene pair from Saccharomyces cerevisiae. DNA 3: 167–171 [DOI] [PubMed] [Google Scholar]
- Kleinschmidt RA, LeBlanc KE, Donze D 2011. Autoregulation of an RNA polymerase II promoter by the RNA polymerase III transcription factor III C (TFIIIC) complex. Proc Natl Acad Sci 108: 8385–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H 2006. The RNA polymerase III-dependent family of genes in hemiascomycetes: Comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res 34: 1816–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K 2004. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol 24: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH, Roth SY, Simpson RT 1992. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol 12: 4015–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics 113: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P, Amstutz H, Kohli J, Leupold U 1982. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature 300: 225–231 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas WM, Muhlrad D, Parker R 1997. Analysis of the yeast genome: Identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res 25: 4619–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, King SR, Telesnitsky A 2005. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J Virol 79: 13528–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 268: 322–330 [DOI] [PubMed] [Google Scholar]
- Piper PW, Lockheart A, Patel N 1984. A minor class of 5S rRNA genes in Saccharomyces cerevisiae X2180-1B, one member of which lies adjacent to a Ty transposable element. Nucleic Acids Res 12: 4083–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2009 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci 100: 14695–14700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi N, Rutz ML, Menees TM 2001. Thermal blockage of viruslike particle formation for the yeast retrotransposon Ty3 reveals differences in the cellular stress response. Arch Virol 146: 1919–1934 [DOI] [PubMed] [Google Scholar]
- Sandmeyer SB, Aye M, Menees TM 2002. Ty3: A position-specific gypsylike element in Saccharomyces cerevisiae. In Mobile DNA II (ed. NL Craig et al.). ASM Press, Washington, DC [Google Scholar]
- Sikorski RS, Hieter P 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Segall J 1985. Characterization of factors and DNA sequences required for accurate transcription of the Saccharomyces cerevisiae 5S RNA gene. J Biol Chem 260: 4531–4540 [PubMed] [Google Scholar]
- van Rossum G, de Boer J 1991. Interactively testing remote servers using the python programming language. CWI Quarterly 4: 283–303 [Google Scholar]
- Voytas DF, Boeke JD 2002. Ty1 and Ty5 of Saccharomyces cerevisiae. In Mobile DNA II (ed. NL Craig et al.), pp. 631–662. American Society for Microbiology, Washington, DC [Google Scholar]
- Wang H, Heinz ME, Crosby SD, Johnston M, Mitra RD 2008. ‘Calling Cards’ method for high-throughput identification of targets of yeast DNA-binding proteins. Nat Protoc 3: 1569–1577 [DOI] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A 1986. Nonviral retroposons: Genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem 55: 631–661 [DOI] [PubMed] [Google Scholar]
- Winckler T, Szafranski K, Glockner G 2005. Transfer RNA gene-targeted integration: An adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome. Cytogenet Genome Res 110: 288–298 [DOI] [PubMed] [Google Scholar]
- Xie X, Rigor P, Baldi P 2009. MotifMap: A human genome-wide map of candidate regulatory motif sites. Bioinformatics 25: 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yieh L, Kassavetis G, Geiduschek EP, Sandmeyer SB 2000. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J Biol Chem 275: 29800–29807 [DOI] [PubMed] [Google Scholar]